The catalytic mechanism and substrate specificity of caffeoyl-CoA O-methyltransferase from Sorghum bicolor deduced from crystal structures, site-directed mutagenesis, and kinetic and thermodynamic analyses.
Abstract
Caffeoyl-coenzyme A 3-O-methyltransferase (CCoAOMT) is an S-adenosyl methionine (SAM)-dependent O-methyltransferase responsible for methylation of the meta-hydroxyl group of caffeoyl-coenzyme A (CoA) on the pathway to monolignols, with their ring methoxylation status characteristic of guaiacyl or syringyl units in lignin. In order to better understand the unique class of type 2 O-methyltransferases from monocots, we have characterized CCoAOMT from sorghum (Sorghum bicolor; SbCCoAOMT), including the SAM binary complex crystal structure and steady-state enzyme kinetics. Key amino acid residues were validated with site-directed mutagenesis. Isothermal titration calorimetry data indicated a sequential binding mechanism for SbCCoAOMT, wherein SAM binds prior to caffeoyl-CoA, and the enzyme showed allosteric behavior with respect to it. 5-Hydroxyferuloyl-CoA was not a substrate for SbCCoAOMT. We propose a catalytic mechanism in which lysine-180 acts as a catalytic base and deprotonates the reactive hydroxyl group of caffeoyl-CoA. This deprotonation is facilitated by the coordination of the reactive hydroxyl group by Ca2+ in the active site, lowering the pKa of the 3′-OH group. Collectively, these data give a new perspective on the catalytic mechanism of CCoAOMTs and provide a basis for the functional diversity exhibited by type 2 plant OMTs that contain a unique insertion loop (residues 208–231) conferring affinity for phenylpropanoid-CoA thioesters. The structural model of SbCCoAOMT can serve as the basis for protein engineering approaches to enhance the nutritional, agronomic, and industrially relevant properties of sorghum.
Current approaches for second-generation biofuel production have significant economic and energetic obstacles due to the extensive lignification of the plant cell wall, which renders large portions of cellulose and hemicelluloses inaccessible to hydrolytic enzymes. The presence of lignin sterically blocks the access of cellulases to their substrate, while simultaneously providing a surface onto which the cellulolytic enzymes adsorb irreversibly. As a consequence, energy-intensive thermochemical pretreatments of plant biomass are needed to remove, or at least partially depolymerize, the lignin (Agbor et al., 2011; Hu and Ragauskas, 2012; Leu and Zhu, 2013). Even so, certain pretreatments generate phenolic compounds derived from lignin that act as inhibitors of cellulases (Ximenes et al., 2011). Mature lignins in the plant cell wall exist as polymers formed via radical-initiated oxidative polymerization of monolignols that are synthesized via the phenylpropanoid pathway (Ralph et al., 2004; Vanholme et al., 2010). The three major monolignols are p-coumaryl, coniferyl, and sinapyl alcohols, which, following polymerization, give rise to p-hydroxyphenyl, guaiacyl, and syringyl residues in the lignin polymer (Ralph et al., 2004).
Manipulation of the monolignol biosynthetic pathway has emerged as an attractive option for reducing the intractability of plant lignification in vivo (Zeng et al., 2014; Vermerris and Abril, 2015; Mottiar et al., 2016). Specifically, plants harboring mutations within the genes encoding monolignol pathway enzymes have shown significantly lower levels of lignification, sometimes accompanied by changes in lignin subunit composition, both of which traits have engendered improved biomass conversion to fermentable sugars. Although this approach has been implemented successfully in both dicots (Chen and Dixon, 2007; Studer et al., 2011; Van Acker et al., 2013) and monocots (Saballos et al., 2008; Fu et al., 2011; Jung et al., 2013; Sattler et al., 2014), the impact of altered lignin subunit composition on biomass conversion appears to differ in these two classes of plants, with a high syringyl/guaiacyl (S/G) ratio improving biomass conversion in dicots, whereas in grasses, a low S/G ratio leads to enhanced yields of fermentable sugars.
The S/G ratio in lignin reflects the degree of methoxylation of the phenolic ring in monolignols. The 4-hydroxyl group of all monolignols is generally unmethylated and is involved in radical formation during lignification (Russell et al., 1996). Two general O-methyltransferases are responsible for the methylation of hydroxyl groups introduced at the 3- and 5-positions of the phenolic ring. The first enzyme, caffeoyl-CoA O-methyltransferase (CCoAOMT), has been shown to have higher activity for the 3-hydroxyl group of 3,4-dihydroxy (caffeoyl) precursors than for the 5-hydroxyl group of 5-hydroxyferuloyl precursors (Guo et al., 2001; Parvathi et al., 2001). The second enzyme, caffeic acid O-methyltransferase (COMT), has, despite the name, activity against 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol substrates (Parvathi et al., 2001). We recently characterized the COMT from sorghum (Sorghum bicolor; SbCOMT; Green et al., 2014). These two classes of S-adenosyl methionine (SAM)-dependent O-methyltransferases have been grouped into two major structural categories. Type 1 O-methyltransferases are larger (38–43 kD) and utilize a His-based catalytic dyad to facilitate its methyltransferase activity (Green et al., 2014). In contrast, type 2 O-methyltransferases are smaller (approximately 23–27 kD) and utilize a divalent cation, which has been assumed to be Mg2+ based on work with the homologous mammalian catechol O-methyltransferase enzymes (Noel et al., 2003; Green et al., 2014), for catalysis. The sorghum CCoAOMT (SbCCoAOMT) and SbCOMT have been shown to belong to the type 2 and type 1 O-methyltransferases, respectively (Noel et al., 2003). In the sorghum genome, there are six additional SbCCoAOMT-like genes, although their activities are unknown (Paterson et al., 2009).
The crystal structures of type 2 O-methyltransferases from alfalfa (Medicago sativa) and Mesembryanthemum crystallinum, MsCCoAOMT and McPFOMT, respectively, have been determined (Ferrer et al., 2005; Kopycki et al., 2008). Based on structure-function studies with those two O-methyltransferases, a unique insertion in CCoAOMT among a variety of type 2 plant O-methyltransferases has been proposed as being responsible for CoA-thioester substrate specificity (Kopycki et al., 2008). The N-terminal arm also contributes to substrate specificity, although the mechanism by which both the N-terminal arm and the insertion loop interact with caffeoyl-CoA to assist substrate binding or catalysis is currently unknown (Ferrer et al., 2005; Kopycki et al., 2008).
In tobacco (Nicotiana tabacum), expression of an antisense copy of the CCoAOMT sequence resulted in an 8% to 15% reduction in total lignin content accompanied by a dwarf phenotype as well as altered leaf and flower morphologies (Pinçon et al., 2001). CCoAOMT-down-regulated alfalfa also contained less lignin (8%–44% reduction relative to the wild type) with an increased S/G ratio. The higher S/G ratio was shown to be the result of a reduction in guaiacyl residues. An analysis of cross sections of the stem tissue revealed that the size of the vascular cells had decreased (Nakashima et al., 2008). Furthermore, the biomass conversion efficiency of these alfalfa transgenics following dilute acid pretreatment and enzymatic saccharification was improved (Chen and Dixon, 2007). In Arabidopsis (Arabidopsis thaliana), biomass from two ccoaomt mutants also yielded significantly more Glc compared with the wild-type control after 48 h of enzymatic saccharification, which was likely the combined result of the lower lignin levels and a higher S/G ratio, similar to what was observed in alfalfa. There was no significant difference in Arabidopsis biomass production as a result of these two ccoaomt mutations (Van Acker et al., 2013).
Our focus is on biomass sorghum, an annual C4 grass with typical dry biomass yields of 20 to 25 Mg ha−1 and maximal yields reaching as high as 40 Mg ha−1, which makes this crop attractive for second-generation (lignocellulosic) biofuel production (Rooney et al., 2007; Almodares and Hadi, 2009; Regassa and Wortmann, 2014). Sorghum provides a number of advantages over other bioenergy crops, especially maize (Zea mays) and sugarcane (Saccharum spp.). Sorghum is more water efficient and requires less nitrogen fertilizer for growth, reducing groundwater contamination associated with large-scale crop production and allowing for growth in a wide variety of climates (Geng et al., 1989; Farre and Faci, 2006). Sorghum has a higher sugar yield per land area and shorter growing season than several alternative second-generation bioenergy crops, translating directly to higher yields per season of fermentable sugars for the production of biofuels or chemicals (Propheter et al., 2010; Gill et al., 2014). Moreover, the sorghum genome has been fully sequenced (Paterson et al., 2009), and several brown midrib mutations that favorably affect biomass conversion have been identified (Bout and Vermerris, 2003; Saballos et al., 2009, 2012; Sattler et al., 2009). Additionally, a population of chemically mutagenized sorghum has provided another resource for the identification of mutants with improved bioenergy traits (Xin et al., 2008, 2009; Sattler et al., 2012, 2014; Scully et al., 2016).
Although the results obtained from studies of transgenic alfalfa and mutant Arabidopsis plants do not guarantee similar outcomes in sorghum and other monocot species, it is likely that strategic manipulation of the CCoAOMT gene, guided by comprehensive characterization with a high-resolution crystal structure, an understanding of the reaction mechanism and substrate specificities, and site-directed mutagenesis, will change lignin content and composition. This structure-guided enzyme engineering offers the potential to improve biomass conversion efficiency without affecting plant fitness. Available options for introducing a newly tuned enzyme do not have to rely on transgenic strategies such as antisense/RNA interference, which incur costly regulatory hurdles, especially in sorghum. Alternative options include chemical mutagenesis, where seeds are treated with DNA-modifying chemicals (e.g. ethyl methanesulfonate and diethylstilbestrol). Despite the intrinsic challenge inherent to this approach, it is possible to find the desired mutation if there is a clear idea of a specific residue in the target enzyme (Sattler et al., 2012). The other option is to engineer sorghum in which the target gene is altered with the use of transcription factor-like effector nucleases (Cermak et al., 2011) or the CRISPR/Cas9 system (Gratz et al., 2013; Jiang et al., 2013; Jinek et al., 2013). Here, we report the SbCCoAOMT crystal structure as well as kinetic, site-directed mutagenesis, and biophysical data. We present a reliable model of caffeoyl-CoA binding, identify two residues critical for enzyme function, and propose a reaction mechanism based on these data.
RESULTS
Global Structure and Oligomeric State of SbCCoAOMT
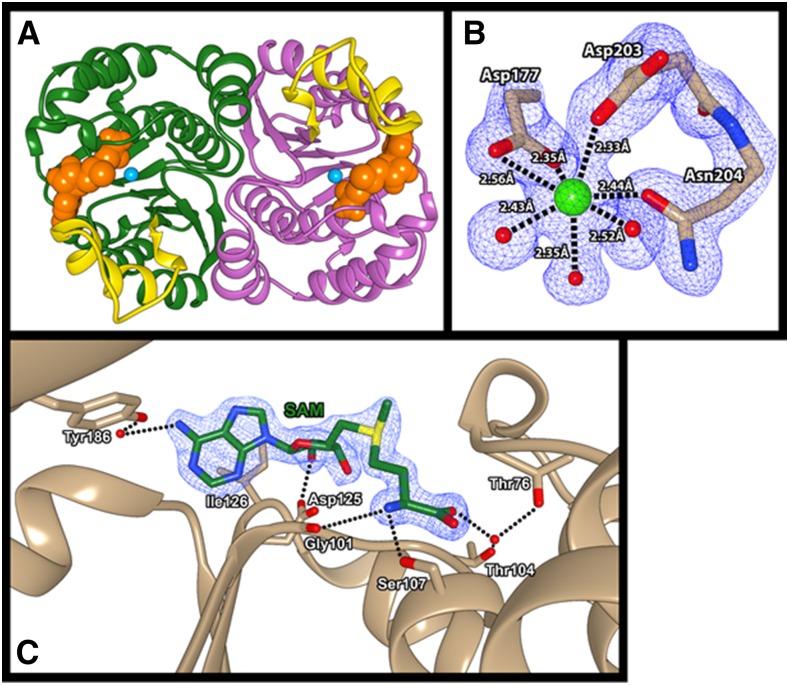
Based on analysis of the crystallographic data, the asymmetric unit of the SbCCoAOMT crystal (PDB ID: 5KVA) was composed of two tightly associated subunits along the pseudo 2-fold axis (Fig. 1A). The structure of each SbCCoAOMT subunit displayed a Rossman-like fold composed of seven β-strands that established a central mixed β-sheet domain core surrounded by 13 α-helices and a series of loop regions that form the exterior surface of the protein, most of which were arranged in an α-β-α pattern. The central β-sheet core was composed of mainly hydrophobic residues. The surrounding α-helices were generally amphipathic and were positioned such that their hydrophobic sides faced toward the β-sheet core and their hydrophilic sides were exposed to the exterior solvent. Specifically, the insertion loop region, which connects the α11 and α12, displayed relatively higher B (atomic displacement)-factors than any other regions, indicating its flexible nature, although the presence of two Pro residues, Pro-216 and Pro-218, seemed to partially stabilize the region.
Figure 1.
A, Global structure of SbCCoAOMT. SbCCoAOMT forms a dimer and adopts a Rossman-like fold to bind SAM. Each subunit of SbCCoAOMT is depicted with ribbon diagrams in green and magenta, and the insertion loop region is highlighted in yellow in both subunits. The space-filling model of SAM and Ca2+ are colored in orange and cyan respectively. Molecular graphics images were produced using the UCSF Chimera package. B, Ca2+ coordination observed in SbCCoAOMT. Asp-177, Asp-203, Asn-204, and three water molecules coordinate the active site Ca2+ of SbCCoAOMT. The purple grid represents an electron density at sigma level 2 of the 2Fo-Fc map. Molecular graphics images were produced using the UCSF Chimera package. C, SAM-binding region of SbCCoAOMT. SAM is coordinated through a combination of hydrogen bonds and hydrophobic interactions. The blue grid represents an electron density at sigma level 2 of the 2Fo-Fc map. Molecular graphics images were produced using the UCSF Chimera package.
The dimer interface of SbCCoAOMT was established by a series of residues located in α1, α5, α13, and β6 through both hydrophobic and hydrophilic interactions. Residues 84 to 92, 199, and 248 to 256 from each subunit established the hydrophobic dimer interface. In addition, a significant amount of hydrophilic interactions flanked these core hydrophobic regions. In detail, the side chains of Glu-244 and Arg-258 on each subunit formed a salt bridge that oriented the Glu-244 carboxylic side chain to form a second salt bridge with the side chain Nζ of Lys-90 from the other subunit. The hydroxyl group of Ser-51 from one subunit appeared to form hydrogen bonds with both the side chain Oε of Gln-247 and the backbone carbonyl oxygen of Asn-234 of the other subunit via an ordered water molecule. There were additional intersubunit hydrogen bonds between the Val-243 backbone carbonyl oxygen and the Arg-55 side chain, the Gln-83 side chain and the backbone nitrogen of Val-243 via a water molecule, and the carbonyl oxygen of Leu-37 and the backbone amide nitrogen of Lys-39. Given these intricate interactions observed in the asymmetric unit of the crystal lattice, it is very likely that SbCCoAOMT exists and functions as a dimer in vivo. In order to confirm our crystallographic observations, the crystal structure of SbCCoAOMT was submitted to PISA (Krissinel and Henrick, 2007), which evaluates the interaction among neighboring monomers in the crystal lattice, thereby allowing for the prediction of biologically relevant oligomeric states. The results of the calculation clearly predicted the presence of a dimer in vivo with a ΔG of formation of −52 kcal mol−1.
Characterization of the Substrate-Binding Pockets
From the beginning stage of phase refinement of the crystal structure of SbCCoAOMT, a high contour-level electron density was observed, which was identified to be a Ca2+ ion through inductively coupled plasma-optical emission spectroscopy (ICP-OES; Supplemental Fig. S1). This Ca2+ ion was located in a solvent-exposed pocket near the N-terminal end of each subunit and was coordinated by the side chain carboxylic moieties of Asp-177 (bidentate manner) and Asp-203 (monodentate manner). Additional coordination by the side chain Oδ of Asn-204 caused a kink that disrupted β5 and thereby formed the structural basis for the insertion loop region (Fig. 1B). Three water molecules also coordinated the positioning of the Ca2+ ion, which overall adopted a heptacoordinate pentagonal bipyramidal geometry. This coordination geometry was significantly different from that of CCoAOMT from alfalfa (MsCCoAOMT; PDB ID: 1SUI; Ferrer et al., 2005). The asymmetric unit of the crystal structure of MsCCoAOMT was composed of two independent dimers, with all four Ca2+ coordination schemes possessing different geometries (Ferrer et al., 2005). The Ca2+ coordination geometry of MsCCoAOMT adopted a highly distorted square pyramidal geometry in three subunits and a hexacoordinated geometry in the fourth subunit. One of the four unique coordination schemes showed a single water molecule as the coordinating ligand, whereas another showed the carboxyl moiety of S-adenosyl homocysteine (SAH) as coordinating the Ca2+ ion. Two of the square planar Ca2+-binding conformations showed coordination by the side chain Nδ atom of Asn-190, which has been shown to be a rare coordination pattern for Ca2+ ions (Katz et al., 1996). The Ca2+ geometry displayed in the MsCCoAOMT structure consistently left one face of the Ca2+ ion uncoordinated, although the significance of this unique coordination geometry is unclear.
We were able to determine the location and conformation of bound SAM by cocrystallization, which yielded clear electron density for the S-methyl moiety of SAM, indicating that the substrate did not convert to SAH during crystallization (Fig. 1C). The SAM-binding region of SbCCoAOMT was established mainly by the residues on β1, β2, α5, and α10. The ribose moiety of the SAM was coordinated at its 2ʹ-OH by the carboxyl side chain of Asp-125. The adenosyl group of SAM was coordinated at its N6 position by the side chain of Tyr-186 through its coordination of a water molecule. This adenosyl group was additionally stabilized through a hydrophobic interaction with the side chain of Ile-126. The amino end of SAM appeared to be coordinated by the backbone carbonyl oxygen of Gly-101 and the side chain Oγ of Ser-107. In addition, the carboxyl moiety of SAM formed a hydrogen bond with a water molecule that was coordinated by the side chains of Thr-76 and Thr-104.
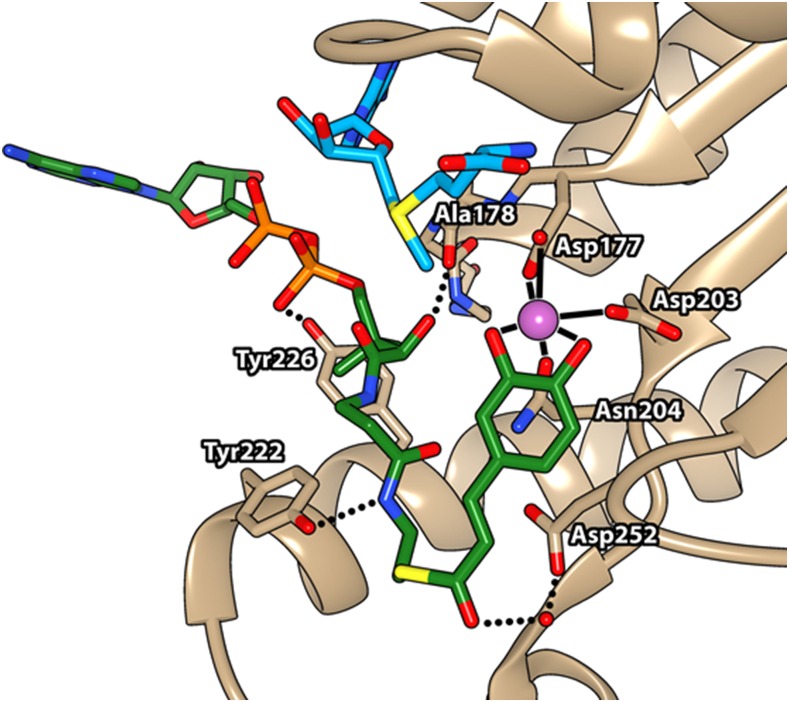
The crystal structures of both the feruloyl-CoA complex (PDB ID: 1SUI) and the sinapoyl-CoA complex (PDB ID: 1SUS) of MsCCoAOMT were further refined with their own x-ray diffraction data to enhance the electron density in order to improve the CoA thioester placement and compare the ligand binding between these two CCoAOMTs. However, further refinement yielded near-zero density coverage of either compound at sigma level 2, with severely broken densities at sigma level 1. Therefore, in order to understand the location and conformation of the CoA thioester substrate in SbCCoAOMT, a molecular docking approach was performed for caffeoyl-CoA using AutoDock Vina (Trott and Olson, 2010). Most of the top conformers were placed between the cleft established between the insertion loop and the SAM-binding pocket, which was extended from the active site Ca2+ (right side in Fig. 2). The best conformer (right side conformation in Fig. 2) displayed a binding enthalpy (ΔH) of −8.8 kcal mol−1, placing its reactive 3-hydroxyl group 2.27 Å from the Ca2+ and 3.87 Å from the SAM methyl group (Fig. 3). This conformation resulted in the displacement of all three water molecules coordinated to the Ca2+, with the caffeoyl-CoA 3,4-dihydroxybenzyl moiety coordinating the Ca2+ in a bidentate manner (Fig. 3). The thioester carbonyl group of CoA was positioned near a water molecule, thus creating a hydrogen bond network with the nearby carboxyl side chain of Asp-252 (Fig. 3). In this conformer, the side chains of Tyr-222 and Tyr-226 and the backbone carbonyl of Ala-78 were within the distance of a hydrogen bond from the amide nitrogen atom, 3ʹ-phosphoryl group, and α-hydroxyl group of the CoA moiety of caffeoyl-CoA, respectively (Fig. 3). The ribosyl group of the CoA moiety did not show any significant interaction with SbCCoAOMT, and the associated adenine ring was facing toward the solvent region (Fig. 3). Overall, this ligand placement in SbCCoAOMT showed that the residues in the insertion loop and SAM-binding pocket were primarily responsible for substrate affinity. This caffeoyl-CoA-binding motif for SbCCoAOMT differed substantially from the reported motifs for feruloyl-CoA and sinapoyl-CoA with MsCCoAOMT (Ferrer et al., 2005). Feruloyl-CoA is a product of SbCCoAOMT, and sinapoyl-CoA is neither a substrate nor a product of SbCCoAOMT.
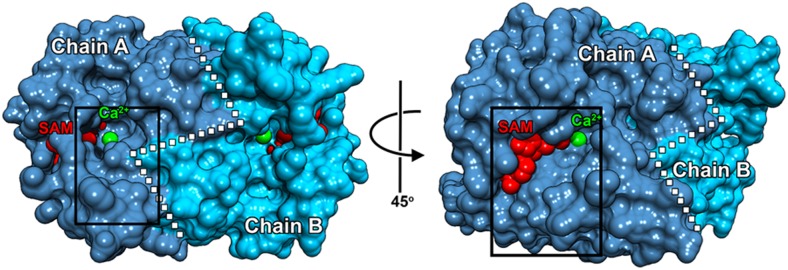
Figure 2.
SbCCoAOMT has two clefts that allow for access to active-site Ca2+. The box on the left image shows a location of one cleft near the dimer interface (white dotted line) of SbCCoAOMT that allows for Ca2+ access. The box on the right image, which is rotated 45° relative to the left image, displays the other cleft that also allows Ca2+ access. SAM and Ca2+ are shown as red and green spheres, respectively. Molecular graphics images were produced using the UCSF Chimera package.
Figure 3.
Binding site of caffeoyl-CoA in SbCCoAOMT. The conformation and location for caffeoyl-CoA binding were established by a molecular docking approach. Caffeoyl-CoA is shown in green and orange, SAM in blue, Ca2+ in magenta, and CCoAOMT in tan. Molecular graphics images were produced using the UCSF Chimera package.
Substrate-Specificity: Isothermal Titration Calorimetry
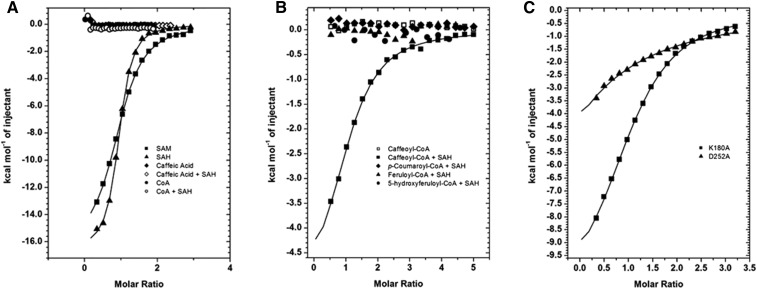
Isothermal titration calorimetry (ITC) was employed in order to understand the binding affinities of several potential substrates for and products of SbCCoAOMT. SAM and SAH were found to bind with dissociation constants (Kd) of 8.6 and 1.9 µm, respectively (Fig. 4A). As shown in Figure 2, there were substantial negative ΔH and entropy (ΔS) effects. On the other hand, neither caffeic acid nor CoA displayed any significant affinity for SbCCoAOMT (Fig. 4A); preequilibrating SbCCoAOMT with a 3:1 molar ratio of SAH prior to analysis did not increase the affinity for either caffeic acid or CoA (Fig. 4A).
Figure 4.
Measurements of binding through ITC experiments. A, SbCCoAOMT displayed significant affinity for SAM and SAH but no affinity for either caffeic acid or CoA. Although SbCCoAOMT was pretreated with a 3:1 molar ratio of SAH, caffeic acid and CoA did not show any affinity to SbCCoAOMT. B, Caffeoyl-CoA showed affinity to SbCCoAOMT only in the presence of SAH, but under the same conditions, feruloyl-CoA, 5-hydroxyferuloyl-CoA, and p-coumaroyl-CoA did not. C, K180A mutant SbCCoAOMT displayed similar affinity to SAM as the wild type, but D252A mutant CCoAOMT has significantly reduced affinity.
Given that neither caffeic acid nor CoA was able to bind to SbCCoAOMT, its substrate-binding mechanism was further elucidated by ITC analysis of its affinity to caffeoyl-CoA. Caffeoyl-CoA was unable bind to SbCCoAOMT, unless the enzyme was preequilibrated with SAH prior to ITC analysis (Fig. 4B). For SbCCoAOMT that had been charged with SAH, the observed Kd of caffeoyl-CoA was 14.8 µm (Table I). In contrast to the thermodynamics of SAM/SAH binding, the ΔH of caffeoyl-CoA association was smaller, but there was a positive ΔS, which may indicate the departure of water molecules, as predicted in our docking models. Our ITC results also showed that feruloyl-CoA, p-coumaroyl-CoA, and 5-hydroxyferuloyl-CoA did not exhibit significant affinity for SbCCoAOMT under the same conditions (Fig. 4B).
Table I. Thermodynamic parameters obtained by ITC.
Enzyme Kinetics and Site-Directed Mutants
Steady-state kinetics were performed by holding SAM concentration constant at 1 mm while caffeoyl-CoA concentration was varied from 1 to 30 µm. For all points in the concentration range used, less than 5% of substrate was converted to product. Fitting the data with a Hill function showed that SbCCoAOMT was positively cooperative, with a Hill coefficient of 2, a Khalf of 4.21 ± 0.134 µm, and a Vmax of 259.8 pkat mg−1 (Fig. 5).
Figure 5.
SbCCoAOMT demonstrates cooperative kinetic activity for caffeoyl-CoA. Plots show kinetic data of wild-type (WT), K180A, and D252A SbCCoAOMT. The two mutant SbCCoAOMTs have no activity toward caffeoyl-CoA, whereas the wild-type curve reflects distinctive cooperative behavior with a Hill coefficient of 2, a Khalf of 4.21 ± 0.134 µm, and a Vmax of 259.8 pkat mg−1, indicated by dotted lines.
According to our SAM complex crystal structure and docking results of SbCCoAOMT, two residues proximal to the active site, Lys-180 and Asp-252, were identified as having a likely role in substrate binding and catalysis. These residues were mutated individually to Ala using site-directed mutagenesis. The mutant proteins were stable and expressed similarly to wild-type SbCCoAOMT. The activities of these mutant proteins were evaluated, and both were shown to be inactive (Fig. 5), confirming the role of both residues in either substrate binding or catalysis.
The enzyme kinetic experiments with 5-hydroxyferuloyl-CoA were performed in order to determine whether the compound also could be a potential methyl acceptor for SbCCoAOMT, producing sinapoyl-CoA as a product. These were performed as activity assays with reaction lengths of 45 s, 2 min, 25 min, and 1 h. Reaction mixtures were analyzed by HPLC to detect the formation of sinapoyl-CoA (Supplemental Fig. S2). The resulting chromatograms did not contain any peak at the retention time corresponding to our sinapoyl-CoA standard. Furthermore, the peak heights of 5-hydroxyferuloyl-CoA did not change between our time points, indicating that SbCCoAOMT has no activity for 5-hydroxyferuloyl-CoA (Supplemental Fig. S2).
Structural Homologs of SbCCoAOMT
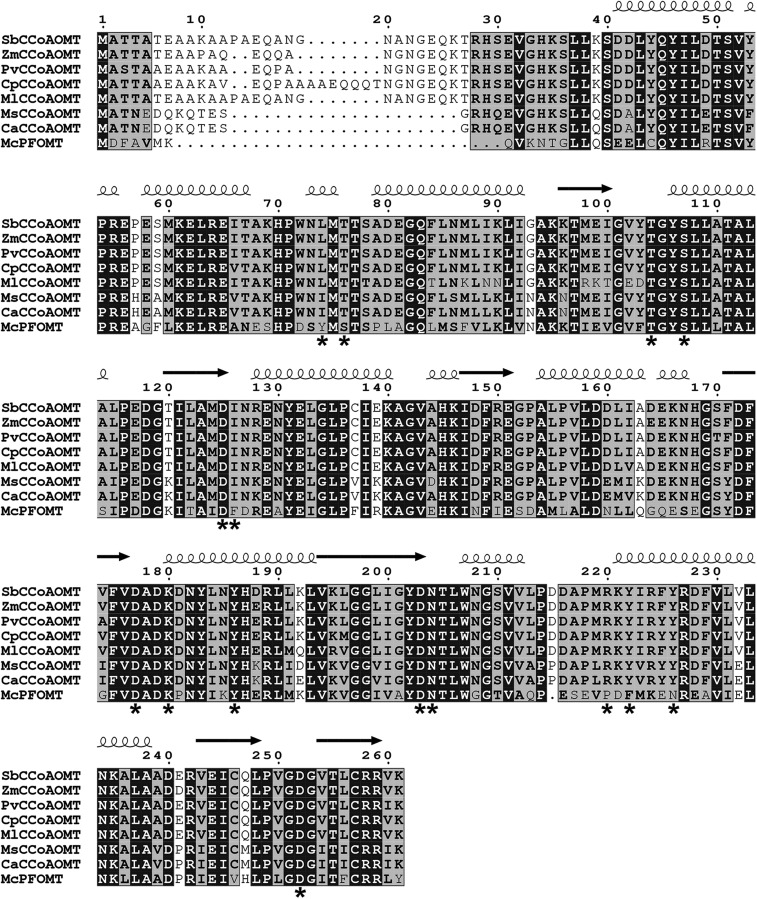
To identify SbCCoAOMT homologs and investigate their binding motifs further, the amino acid sequence of SbCCoAOMT was used to perform a similarity search of the National Center for Biotechnology Information nonredundant sequence database using BLAST. The search identified the CCoAOMTs from maize (query coverage, 100%/identity, 95%), switchgrass (Panicum virgatum; 100%/95%), elephant grass (Cenchrus purpureus; 92%/98%), and silvergrass (Miscanthus lutarioriparius; 93%/93%). When the BLAST search used the sequence of MsCCoAOMT as a query, CCoAOMT from chickpea (Cicer arietinum; 100%/98%) was identified as a top hit. The search also identified the CCoAOMTs from barrelclover (Medicago truncatula; 100%/99%) and peashrub (Caragana korshinskii; 100%/97%) and an uncharacterized protein from birdsfoot trefoil (Lotus japonicus; 100%/95%). Those high-scoring hits obtained from the BLAST search using SbCCoAOMT and MsCCoAOMT were aligned against SbCCoAOMT, MsCCoAOMT, and M. crystallinum phenylpropanoid and flavonoid O-methyltransferase (McPFOMT) and were compared using three-dimensional structural information (Fig. 6). McPFOMT is a plant type 2 O-methyltransferase that exhibits a broad substrate preference that includes both phenylpropanoids and flavonols and also is capable of utilizing caffeoylglucose as a substrate, making it unique among plant type 2 O-methyltransferases (Ibdah et al., 2003; Kopycki et al., 2008). The kinetic profile and structure of McPFOMT have been characterized extensively, making it a useful model to compare with. Aligning these sequences indicated that residues involved in Ca2+ and SAM binding were highly conserved, but the residues contributing to substrate specificity (Leu-74, Tyr-222, and Tyr-226) were not. The alignment also indicated that the sequences of CCoAOMTs from the members of the Poaceae family and the members of the Fabaceae family included were nearly identical outside of their N-terminal regions. In addition, the residues at the above-mentioned nonconserved sites in the alignment were fully conserved with each family (Fig. 6). One of these nonconserved sites is located in the insertion loop region, which was established previously as being responsible for substrate preference in plant type 2 O-methyltransferases (Kopycki et al., 2008).
Figure 6.
Sequence alignment of SbCCoAOMT, MsCCoAOMT, McPFOMT, and related OMTs. Sequences used for the alignment were obtained by BLAST searches using SbCCoAOMT and MsCCoAOMT as query sequences as well as McPFOMT. Above the aligned sequences, α-helices and β-sheets are indicated by coils and arrows, respectively. Fully conserved residues are indicated by black boxes with white letters. Conservative substitutions are shown in black boldface letters, and regions of conservative substitutions are indicated by gray shading. Nonconservative substitutions have no special formatting. Asterisks indicate residues that have been identified as putatively important for catalysis, substrate binding, or substrate specificity.
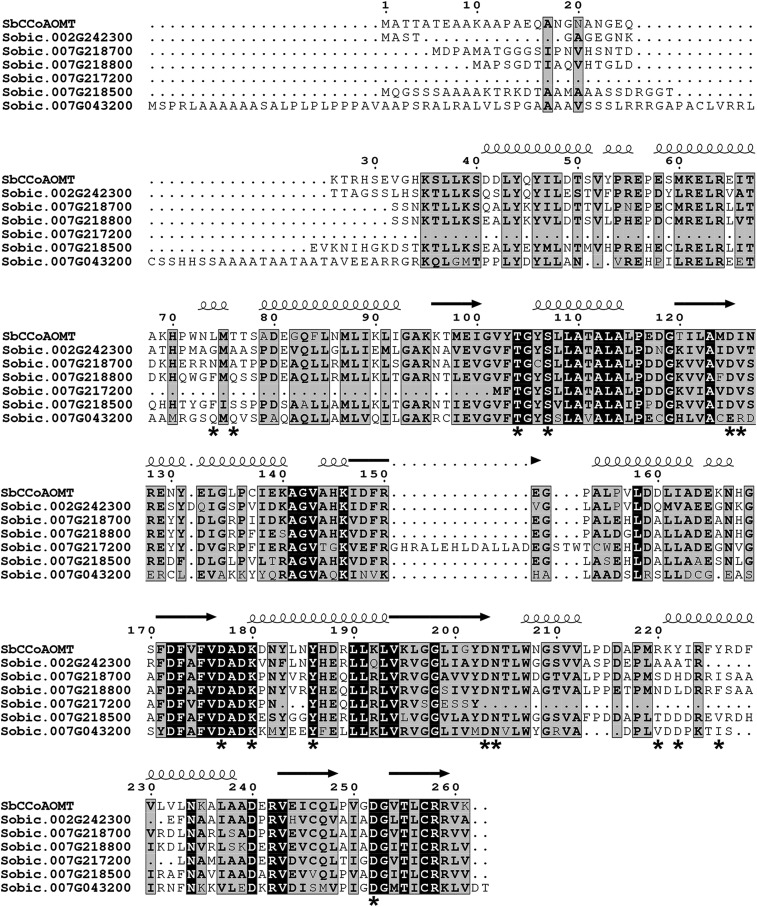
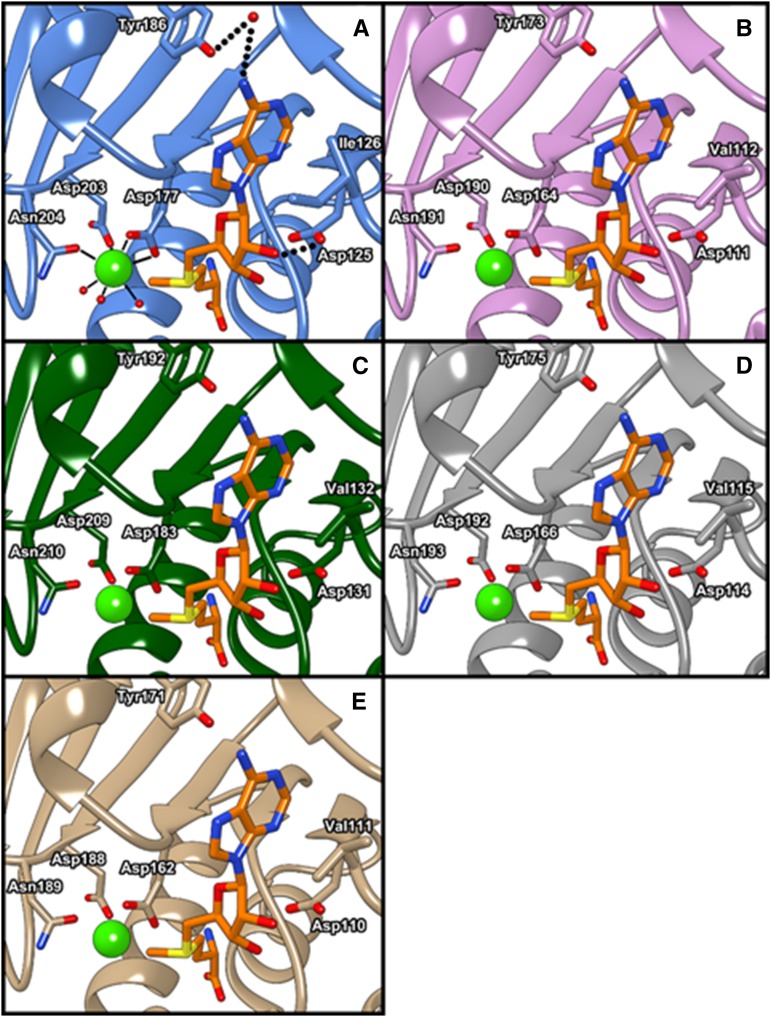
Furthermore, to annotate their catalytic activity, SbCCoAOMT was compared with six proteins encoded by SbCCoAOMT-like genes in the sorghum genome: Sobic.002G242300 (54.2% identity with SbCCoAOMT/70.2% similarity), Sobic.007G043200 (31.4%/43.2%), Sobic.007G217200 (28.4%/37.4%), Sobic.007G218500 (50.9%/68.5%), Sobic.007G218700 (59.1%/69.2%), and Sobic.007G218800 (54%/70.9%; Paterson et al., 2009). Multiple sequence alignment of their amino acids showed that the catalytic Lys, Lys-180, and several residues located at the enzyme active site in SbCCoAOMT were conserved across all SbCCoAOMT-like sequences, with the exception of Sobic.007G217200. Due to its significant deletion relative to the SbCCoAOMT sequence, the amino acid sequence encoded by Sobic.007G217200 lacks Asp-203 and Asn-204, both of which are critical for Ca2+ coordination (Fig. 7). Further distinguishing this protein from the others, the Ile-126 of SbCCoAOMT is substituted as an Arg in Sobic.007G217200, which would prevent substrate access to the SAM-binding pocket. Thus, it is likely that Sobic.007G217200 has no CCoAOMT activity. The amino acid sequences encoded by the remaining five genes were further analyzed by homology modeling using SWISS-MODEL (Schwede et al., 2003). The results showed that Sobic.007G043200 is likely to have severely reduced catalytic activity with caffeoyl-CoA and SAM substrates due to a number of amino acid changes both in the insertion loop and in the SAM-binding site. However, Sobic.002G242300, Sobic.007G218700, Sobic.007G218800, and Sobic.007G218500 appear to have intact SAM-binding pockets, with Thr-77 being substituted for Ser in Sobic.007G218700 and Sobic.007G218800 and for Ala in Sobic.002G242300 and Ile-126 being substituted for Val in all four enzymes. Asp-252, which is involved in coordinating the oxygen atom of the caffeoyl-CoA thioester group, is conserved across these enzymes. Furthermore, the residues involved in Ca2+ binding and their flanking residues were well conserved in Sobic.002G242300, Sobic.007G218700, Sobic.007G218800, and Sobic.007G218500, indicating that they likely maintain a Ca2+ affinity similar to that of SbCCoAOMT (Fig. 8). Given these structural features, Sobic.002G242300, Sobic.007G218700, Sobic.007G218800, and Sobic.007G218500 should be catalytically active enzymes.
Figure 7.
Sequence alignment of SbCCoAOMT and the proteins encoded by six sorghum CCoAOMT-like genes. α-Helices and β-sheets are indicated by coils and arrows, respectively. Fully conserved residues are indicated by black boxes with white letters. Conservative substitutions are shown in black boldface letters, and regions of conservative substitutions are indicated by gray shading. Nonconservative substitutions have no special formatting. Asterisks indicate residues that have been identified as putatively important for catalysis, substrate binding, or substrate specificity.
Figure 8.
Homology-based models of the active sites of four putative sorghum CCOAOMTs: SbCCoAOMT (A), Sobic.002G242300 (B), Sobic.007G218500 (C), Sobic.007G218700 (D), and Sobic.007G218800 (E). Side chains of conserved residues involved in Ca2+ and SAM binding are shown. The active site Ca2+ and SAM are shown as a light green sphere-and-orange stick model. Molecular graphics images were produced using the UCSF Chimera package.
DISCUSSION
Substrate Specificity and Catalytic Reaction Mechanism of SbCCoAOMT
The position and conformation of caffeoyl-CoA established through the SAM binary complex crystal structure and a molecular docking approach indicated that residues in the insertion loop between α11 and α12 and the SAM-binding domains imparted specificity for the CoA moiety observed for SbCCoAOMT, as predicted by structural and kinetic characterization of McPFOMT (Kopycki et al., 2008).
When the SAM-binding modes of SbCCoAOMT and MsCCoAOMT are compared, there are significant differences depending on which subunit of the MsCCoAOMT complex is used for the structural comparison. The MsCCoAOMT structure displays a unique SAM-binding conformation for each of the four subunits in the asymmetric unit (Ferrer et al., 2005). One subunit closely resembled the binding orientation that is present in SbCCoAOMT. However, two other subunits showed the carboxyl moiety of the bound SAM as either involved directly in coordinating the active site Ca2+ or interacting with the carboxyl side chain of Asp-163 in MsCCoAOMT, which corresponds to the Ca2+ coordinating Asp-177 in SbCCoAOMT. The remaining subunit placed the γ-carbon of its bound SAM carboxyl group in close proximity (approximately 2 Å) of the Ca2+ ion at the active site. The effect of these alternate conformations on the activity of MsCCoAOMT and any differences relative to SbCCoAOMT are unclear.
The residues shown to be involved in SAM binding in SbCCoAOMT are generally conserved among all of the enzymes that were surveyed (Fig. 6). The adenyl-coordinating residues Asp-179 and Tyr-186 are fully conserved in the aligned amino acid sequences (Fig. 6). Ile-126, which is likely involved in a hydrophobic interaction with the adenosine ring of SAM, is conserved in all aligned CCoAOMTs, with McPFOMT uniquely possessing a Phe at the same position (Fig. 6). Thr-77, Thr-104, and Ser-107, which provide an affinity for the aminocarboxylic acid moiety of SAM, are fully conserved across all aligned OMTs (Fig. 6). In addition, Asp-125, which coordinates the 2ʹ-OH of the SAM ribose unit, is similarly fully conserved across all compared OMTs (Fig. 6).
Our molecular docking results indicated that the side chains of Tyr-222 and Tyr-226 within the insertion loop contribute to caffeoyl-CoA binding. The side chain of Tyr-222 is oriented in a position that allows it to coordinate the β-mercaptoethanolamine nitrogen of CoA. Tyr-222 is conserved in all aligned CCoAOMT enzymes, but it is substituted by Phe-198 in McPFOMT. The phenolic side chain of Tyr-226 is involved with coordination of the β-5ʹ-phosphoryl group of CoA, which is similarly conserved in the aligned CCoAOMTs but is substituted by Asn-202 in McPFOMT. Thus, those two unique CCoAOMT interactions may be associated with the high catalytic efficiency of SbCCoAOMT and MsCCoAOMT for hydroxycinnamoyl-CoA substrates.
In addition, based on extrapolation of our docking model of SbCCoAOMT, McPFOMT’s observed affinity for caffeoyl-CoA (Kopycki et al., 2008) is likely facilitated by three conserved interactions: (1) between the backbone carbonyl of Ala-155 (SbCCoAOMT Ala-178) and the hydroxyl group of the pantothenic acid moiety in CoA; (2) between the thioester carbonyl group of caffeoyl-CoA and the carboxyl side chain of Asp-228 (SbCCoAOMT Asp-252); and (3) between the bound divalent cation and the 3ʹ-OH and 4ʹ-OH hydroxyl groups of caffeoyl-CoA. These interactions, which are shared among CCoAOMTs and PFOMTs, would orient the reactive oxygen of the substrates, whereas the majority of interactions governing the binding of the CoA moiety are not present in McPFOMT, providing a mechanistic basis for the previously observed weak catalytic activity of this enzyme for caffeoyl-CoA (Kopycki et al., 2008). The amino acid present at the position corresponding to Asp-215 in SbCCoAOMT, which is located in the insertion loop region and shows no apparent direct involvement in caffeoyl-CoA affinity, may have a significant role in substrate specificity and catalytic efficiency. Our sequence alignment indicated that Asp-215 of SbCCoAOMT is substituted by Pro in both MsCCoAOMT and CaCCoAOMT (Fig. 6). The effect of this substitution on CCoAOMT activity is unclear. Significantly, Asp-215 is at the site of a deletion in the insertion loop region of McPFOMT. As our data imply that SbCCoAOMT undergoes significant conformational changes upon binding substrate, it is likely that other CCoAOMTs also undergo similar conformational changes, and this position of Asp-215 might be critical for substrate specificity by controlling insertion loop flexibility, as small structural changes of the insertion loop have been shown to be able to radically alter substrate specificity and catalytic efficiency among type 2 O-methyltransferases (Kopycki et al., 2008).
The monolignol biosynthetic pathway has been revised on multiple occasions based on the identification of previously unanticipated enzyme activities (Humphreys and Chapple, 2002; Vanholme et al., 2013). Even though the current version of the pathway does not include 5-hydroxyferuloyl-CoA as an intermediate, studies of MsCCoAOMT from crude stem materials and recombinant enzyme in Escherichia coli cell lysates have indicated that this enzyme has activity for 5-hydroxyferuloyl-CoA (Inoue et al., 1998; Parvathi et al., 2001). Therefore, we investigated whether 5-hydroxyferuloyl-CoA could be a substrate of SbCCoAOMT, which is of interest for future metabolic engineering efforts, even if this reaction is not part of the monolignol biosynthetic pathway. Superimposition of 5-hydroxyferuloyl-CoA onto our docked caffeoyl-CoA conformer indicates that its 3ʹ-methoxyl moiety has potential steric clashes with the side chain of Glu-81 and the backbone of Gly-251. Accordingly, we observed that SbCCoAOMT has neither binding affinity (Fig. 4B) nor enzymatic activity for 5-hydroxyferuloyl-CoA (Supplemental Fig. S2), which also is consistent with previous reports of in vivo assay in sorghum (Guo et al., 2001). Glu-81 and Gly-251 are conserved across CCoAOMT enzymes, although not across all type 2 O-methyltransferases (Fig. 6). Although those two residues in SbCCoAOMT, which would likely prevent the association of 5-hydroxyferuloyl-CoA, are present in MsCCoAOMT, MsCCoAOMT is apparently able to catalyze 5-hydroxyferuloyl-CoA. It is plausible that the Pro-201 substitution (Fig. 6) in the insertion loop of MsCCoAOMT has a drastic effect on the flexibility of the loop, which could potentially encourage an alternative binding conformation similar to that suggested previously (Ferrer et al., 2005) wherein only the target hydroxyl group of the substrate binds to the catalytic Ca2+, allowing the 5′-methoxyl moiety to point toward the solvent and thereby prevent the steric clash.
Although it could be a crystallographic artifact, there is a unique salt bridge in SbCCoAOMT that is not present in MsCCoAOMT and that is established by the side chain guanidinium moiety of Arg-220 of one subunit and the carboxyl side chain of Glu-117 of another molecule, which orients the Arg-220 side chain to sterically block frontal access of the active site by Ca2+ together with the side chain of Leu-74. Our sequence alignment shows that Arg-220 is conserved in all CCoAOMT enzymes compared (Fig. 6). Leu-74 is also conserved in ZmCCoAOMT, PvCCoAOMT, and CpCCoAOMT but is conservatively substituted to Ile in MsCCoAOMT and CaCCoAOMT. However, in McPFOMT, Leu-74 is substituted to Tyr-51, which has been proposed to be important for flavonoid binding (Kopycki et al., 2008). Given the orientation of the modeled substrate, it is possible that this Tyr residue in McPFOMT acts as a gate to the active site, sterically hindering the binding of longer substrates such as the CoA thioesters, thereby imparting specificity for the shorter flavonoids. It was shown previously that the amino acid present at the Leu-74 position can determine para- or meta-methylation specificity (Wils et al., 2013). A Gly substitution at that position has been shown to encourage para-O-methylation, whereas the Tyr residue at that position in McPFOMT has been shown to encourage meta-O-methylation, likely due to steric clashes that occur between the methyl-acceptor substrate and the Tyr side chain in the binding orientation required for para-O-methylation (Wils et al., 2013). It is tempting to speculate that the combination of the steric presence of the Leu-74 and Arg-220 side chains in SbCCoAOMT causes its observed preference for meta-O-methylation. In addition, it has been established that three CCoAOMTs also have activity for caffeoyl-Glc and flavonoids (Ibdah et al., 2003; Kopycki et al., 2008). Considering the size and shape of the substrate-binding pocket, it is unlikely that SbCCoAOMT has significant activity for either of these substrates, although this needs to be confirmed by enzymatic assays.
Enzyme Reaction Mechanism
The steady-state kinetics (Fig. 5) of wild-type and mutant versions of SbCCoAOMT clearly indicated that Lys-180 and Asp-252 are both essential for catalytic activity. Given that the position of Lys-180 overlaps with the catalytic Lys among the closely related mammal catechol O-methyltransferases, it is likely that Lys-180 performs a similar catalytic role in SbCCoAOMT. Although Lys-180 is distal (approximately 5.5 Å) from the reactive oxygen of the caffeoyl-CoA modeled with our SAM binary complex structure, our kinetic data demonstrated positive cooperativity (Fig. 5), which indicates that significant conformational changes occur upon caffeoyl-CoA binding. Due to the magnitude of this conformational shift, it is likely that the conformation of both the SAM- and caffeoyl-CoA-binding regions of SbCCoAOMT also are altered, moving the side chain Nζ of Lys-180 closer to the reactive oxygen of the caffeoyl-CoA.
Our ITC data (Fig. 4) indicated that SAM binding appears to establish the following caffeoyl-CoA binding, probably by orienting key residues. It is tempting to speculate that SAM binding adjusts the orientation of Tyr-220 and Tyr-222, both of which were located in the flexible insertion loop and were involved in caffeoyl-CoA binding from our docking studies. Moreover, our ITC data indicated that both hydroxyl groups on the phenylpropanoid moiety of caffeoyl-CoA are required for binding (Fig. 4). This is likely to be a mechanism of product dissociation; the weaker interaction of the methoxyl group of feruloyl-CoA with the active site Ca2+, combined with an unfavorable interaction between the newly formed methoxyl group and the nearby catalytic Lys in the active conformation of the enzyme, would discourage feruloyl-CoA binding. Similarly, binding of p-coumaroyl-CoA also would likely displace the three water molecules coordinating the active site Ca2+, leaving the cation in a distorted, energetically unfavorable pentacoordinate geometry. The observed inability of CCoAOMT to bind p-coumaroyl-CoA supports this model (Fig. 4) and implies that two free hydroxyl groups in a catechol-type structure are required for substrate binding, in agreement with our docking results.
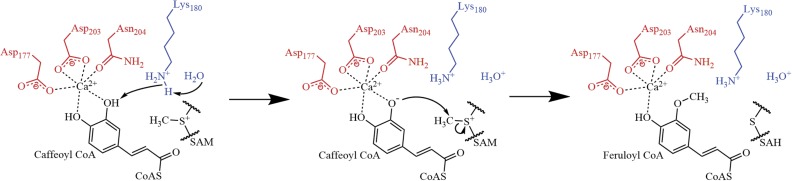
Together, our data indicate that SAM is associated with SbCCoAOMT prior to caffeoyl-CoA, resulting in conformational changes within the dimer and facilitating caffeoyl-CoA binding. In this new conformation induced upon the association of SAM, Lys-180 is positioned to act as a base catalyst and abstract a proton from the 3ʹ-OH of caffeoyl-CoA (Fig. 9), which is in agreement with the mammalian catechol O-methyltransferase reaction mechanism (Vidgren et al., 1994). Although Lys-180 is not at an optimal catalytic distance from the reactive hydroxyl group of caffeoyl-CoA in our modeled structure, our kinetic data indicate that a significant allosteric effect would occur upon substrate binding, which brings the side chain Nζ of Lys-180 within reactive distance of the caffeoyl-CoA 3ʹ-OH. The active site Ca2+ substantially lowers the pKa of the 3ʹ-OH group from caffeoyl-CoA, facilitating its deprotonation by Lys-180. The resulting oxyanion is able to nucleophilically attack the S-methyl group of SAM and thereby form the product, feruloyl-CoA.
Figure 9.
Proposed catalytic reaction mechanism of SbCCoAOMT. Lys-180 deprotonates the 3ʹ-OH of caffeoyl-CoA, allowing it to nucleophilically attack the methyl group of SAM, forming feruloyl-CoA and SAH.
SbCCoAOMT-Like Sequences
Our approaches with multiple sequence alignment and homology modeling indicated that, among the existing six SbCCoAOMT-like genes in the sorghum genome (Paterson et al., 2009), gene products of Sobic.007G217200 and Sobic.007G213200 are unlikely to have any significant CCoAOMT activity. Conversely, Sobic.002G242300, Sobic.007G218700, Sobic.007G218800, and Sobic.007G218500 are expected to have CCoAOMT activity, although these activities may be less relevant if these genes are not expressed in tissues where lignification is occurring (Makita et al., 2015). The enzymes encoded by these four genes have significant amino acid substitutions in their insertion loop, indicating that they may prefer substrates other than caffeoyl-CoA. Sobic.002G242300, in particular, is predicted to have the largest structural change, owing to a significant deletion within the insertion loop that removes key residues for caffeoyl-CoA binding identified from our docking approach. Sobic.007G218700, Sobic.007G218800, and Sobic.007G218500 have nonconservative substitutions at Tyr-222 and Tyr-226, which were identified as putative caffeoyl-CoA-binding residues by our docking approach. Additionally, Glu-81 in SbCCoAOMT is substituted to Ser-87 in Sobic.007G218500. As Glu-81 appeared to sterically hinder the binding of 5-hydroxyferuloyl-CoA in the docking model, the presence of Ser at this position could allow Sobic.007G218500 to accommodate larger substrates. Glu-81 is conserved in Sobic.007G218700, Sobic.007G218800, and Sobic.002G242300. There were no substitutions in the catalytic motifs of four putative O-methyltransferases to prevent or weaken the association of either Ca2+ or SAM. However, it is likely that these four gene products have significantly reduced CCoAOMT activity due to critical nonconservative substitutions and deletions in the insertion loop region that would reduce their affinities for caffeoyl-CoA. It remains to be determined whether these enzymes have other endogenous substrates.
CONCLUSION
The combined results from the experiments aimed at characterizing SbCCoAOMT indicate that this enzyme has catalytic activity only for caffeoyl-CoA. Given that neither 5-hydroxyferuloyl-CoA nor caffeic acid is a substrate for SbCCoAOMT, the primary role of SbCCoAOMT is to methylate caffeoyl-CoA to produce feruloyl-CoA in the monolignol biosynthetic pathway of sorghum, which ultimately leads to the production of both the guaiacyl and syringyl units of lignin. However, SbCCoAOMT may have another endogenous substrate yet to be demonstrated. Transgenic suppression using antisense RNA of CCoAOMT has been shown to both decrease overall lignification and increase the S/G ratio in alfalfa plants, resulting in plant biomass that can be more easily converted to fermentable sugars. Transgenic down-regulation of CCoAOMT in tobacco has shown a strong reduction in lignification accompanied by a dwarf phenotype. A reduction, rather than an elimination, of CCoAOMT activity may offer a way to maximize sugar yield without severely affecting plant growth. Loss-of-function mutations in SbCCoAOMT have not yet been isolated. Although mutations of the residues that are critical for catalysis and substrate affinity, such as K180A and D252A, completely abolished enzymatic activity, the crystal structure of the binary complex of SbCCoAOMT could allow for the identification of certain amino acids as candidates for site-directed mutagenesis in order to engineer sorghum with reduced activity, such as isosteric mutagenesis for residues constituting the substrate-binding pocket, as performed in both SbHCT and SbCOMT (Walker et al., 2013; Green et al., 2014). Given the right balance of enzymatic activity, these plants could exhibit reduced lignification combined with normal plant growth, thus affording biomass with the potential for maximal conversion to fermentable sugars.
MATERIALS AND METHODS
Chemicals
Analytical-grade chemicals were obtained from Sigma-Aldrich, Fisher, and Alfa-Aesar. Crystallization screens were obtained from Hampton Research.
Cloning SbCCoAOMT
The entire coding region of sorghum (Sorghum bicolor) CCoAOMT (Sobic010G052200.1) was amplified by PCR with the following primers (SbCCoAOMT_PciI-F, 5ʹ-CCGACATGTCCACCACGGCGACCGAG-3ʹ; and SbCCoAOMT_EcoRI-R, 5ʹ-TTTGAATTCACTTGACGCGGCGGCA-3ʹ) using Turbo Pfu polymerase (Agilent) and University of Georgia EST clone PH1_9_G09_A002 (GenBank accession no. CF428636) as the template. After generating overhanging ends with the restriction enzymes PciI and EcoRI, the SbCCoAOMT PciI-EcoRI fragment was cloned into vector pET30a (EMD Millipore) that had been linearized with the restriction enzymes NcoI and EcoRI. After heat shock transformation of Escherichia coli DH5α cells, isolated recombinant plasmids were subjected to automated DNA Sanger sequencing at Washington State University to verify the fidelity of the inserted fragment. A verified recombinant plasmid was subsequently introduced in E. coli Rosetta cells (EMD Millipore).
Site-directed mutations were created in the SbCCoAOMT coding region by PCR-based amplification using Phusion High-Fidelity DNA polymerase (New England Biolabs). The amplification was performed using complementary plus- and minus-strand oligonucleotides containing the target mutations and was followed by DpnI (New England Biolabs) digestion to degrade the template prior to the transformation of competent E. coli Rosetta cells (EMD Millipore). Both mutations were confirmed by DNA sequencing (GENEWIZ).
Recombinant Enzyme Expression and Purification
For the expression of recombinant SbCCoAOMT, 200 mL of Luria-Bertani medium containing 100 µg mL−1 kanamycin and 34 µg mL−1 chloramphenicol was inoculated with a freezer stock of E. coli Rosetta cells containing the pET30a-SbCCoAOMT construct and grown overnight at 37°C while shaking. This culture was used to inoculate 3 L of Luria-Bertani medium, which was grown to an OD600 of 0.6 at 37°C with shaking. The cells were then brought to 20°C with shaking, and isopropyl β-thiogalactopyranoside was added to a final concentration of 1 mm. The culture was grown at 20°C while shaking for an additional 18 h. Cells were collected by centrifugation at 6,000 rpm for 12 min at 4°C. The cell pellet was resuspended in 40 mL of lysis buffer (20 mm Tris, pH 8, 150 mm NaCl, and 10 mm imidazole) and was sonicated five times with 15-s pulses (model 450 sonifier; Branson Ultrasonics). The lysate was cleared by centrifugation at 15,000 rpm for 25 min. Cleared supernatant was applied to 15 mL of nickel-nitrilotriacetate agarose (Qiagen), equilibrated with lysis buffer, and placed into a gravity-flow column. The column was washed with 20 column volumes of washing buffer (20 mm Tris, pH 8, 300 mm NaCl, and 20 mm imidazole), and protein was eluted with elution buffer (20 mm Tris, pH 8, 300 mm NaCl, and 250 mm imidazole). Column fractions containing SbCCoAOMT were desalted and concentrated into buffer A (20 mm Tris, pH 8, and 1 mm DTT) using an Amicon 8050 ultrafiltration cell with a 10-kD cutoff membrane (Millipore). Concentrated protein was applied to a Resource-Q column (GE Healthcare) that was preequilibrated with buffer A using a flow rate of 2.5 mL min−1. SbCCoAOMT was eluted from the column with 240 mm NaCl applied in a linear gradient. The fractions containing SbCCoAOMT were desalted and concentrated into buffer B (5 mm sodium phosphate, pH 6.8, 5% glycerol, and 0.05 g dL−1 NaN3) using an Amicon 8050 ultrafiltration cell with a 10-kD cutoff membrane (Millipore). Concentrated protein was applied to a hydroxyapatite column (Bio-Rad) that was preequilibrated with buffer B at a flow rate of 4 mL min−1, and SbCCoAOMT was eluted from the column with approximately 50 mm sodium phosphate applied in a linear gradient. SbCCoAOMT-containing fractions were pooled, and buffer was exchanged into 20 mm MOPS, pH 7.5. K180A and D252A mutant proteins were expressed and purified in an identical manner to the wild-type SbCCoAOMT (Supplemental Fig. S3).
Purification fractions were analyzed by gel electrophoresis using a 12% Tris-Gly-SDS polyacrylamide gel stained with Coomassie Blue dye. All protein concentrations were determined by the Bio-Rad protein assay kit using bovine serum albumin as a standard. The yield of wild-type SbCCoAOMT was 15 mg L−1, and the yields of the K180A and D252A mutants were 11 and 13 mg L−1, respectively.
Crystallization and Structure Determination
SbCCoAOMT was cocrystallized with SAM by hanging-drop vapor diffusion at 4°C. A solution of 0.97 mm SbCCoAOMT and 2.91 mm SAM was mixed with mother liquor (0.2 m calcium acetate hydrate, 0.1 m Tris, pH 8, and 20% PEG 4000) and equilibrated against the reservoir solution. SbCCoAOMT was crystallized at a concentration of 20 mg mL−1 in the space group P21 and had unit cell dimensions a = c = 55.7 Å, b = 91.6 Å, and α = β = γ = 90°. Data were collected to 1.7 Å at the Berkeley Advanced Light Source (beamline 8.2.1) with an exposure time of 1 s and a detector distance of 250 mm. Crystals were transferred to a cryoprotectant solution that contained the reservoir solution mixed with 20 mm MOPS, pH 7.5, in a 1:1 ratio, as well as 12.5% glycerol, prior to data collection at 100 K. Diffraction data were scaled using the program HKL2000 (Otwinowski and Minor, 1997).
Phasing and Refinement
Initial phasing of diffraction data was performed by molecular replacement with the Phaser program in the PHENIX package using Mesembryanthemum crystallinum CCoAOMT as a search model (Kopycki et al., 2008; Terwilliger et al., 2008; Adams et al., 2010). The AutoBuild program in the PHENIX package was used to generate an initial structure after phasing (Terwilliger et al., 2008; Adams et al., 2010). SAM/SAH and Ca2+ were placed manually into the resulting model, and missing amino acid regions were fitted manually using the program Coot and refined using PHENIX (Adams et al., 2010; Emsley et al., 2010). Following refinement of the SAM binary complex, Rwork was 16.3%, Rfree was 19.6%, and root mean square deviations from ideal geometry of the model were 0.007 Å for bonds and 1.173° for angles. The statistics for the diffraction data are listed in Table II. Crystallographic coordinates and structure factors for SbCCoAOMT has been deposited in the Protein Data Bank (PDB ID: 5KVA).
Table II. SbCCoAOMT-SAM complex crystallographic data.
| Parameter | SbCCoAOMT-SAM Complex |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 55.531, 91.562, 55.783 |
| α, β, γ (°) | 90.00, 90.69, 90.00 |
| Resolution (Å) | 47.640–1.827 |
| Rsym | 0.129 |
| I/σI | 7.19 |
| Completeness (%) | 99.02 (91.80) |
| Redundancy | 3.2 |
| Refinement | |
| Resolution | 47.640–1.827 |
| Unique reflections | 48,949 (4,514) |
| Rwork/Rfree | 0.1587/0.1932 |
| No. of atoms | |
| Macromolecules | 3,681 |
| Ion | 2 |
| Ligand | 54 |
| Water molecules | 627 |
| B-factors | |
| Protein | 23.00 |
| Ligand/ion | 19.40 |
| Water | 35.60 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.100 |
| Ramachandran angles | |
| Favored | 98.50 |
| Outliers | 0.22 |
| Clash score | 2.98 |
Synthesis of 5-Hydroxyferulic Acid
In a flame-dried round-bottom flask purged and kept under positive pressure with argon gas, 3,4-dihydroxy-5-methoxybenzaldehyde (168 mg, 1 mmol) was dissolved in pyridine (2 mL). Malonic acid (302 mg, 2.9 mmol) and piperidine (30 μL) were added to the solution, at which point it was heated to 50°C in a sand bath. The mixture was allowed to stir overnight. After removing the volatile components in vacuo, the crude product was taken up in water and acidified to pH approximately 1 with 1 m HCl. The aqueous layer was extracted with EtOAc (3 × 10 mL), and the organics were removed in vacuo to yield a brown solid. After purification by flash column chromatography (10 g of SiO2), 5-hydroxyferulic acid was obtained as a white solid (40 mg, 20% yield): 1H-NMR (acetone-d6) δ 7.54 (d, J = 15.9 Hz, 1H, α), 6.90 (d, J = 1.8 Hz, 1H, 2), 6.83 (d, J = 1.5 Hz, 1H, 6), 6.33 (d, J = 15.9 Hz, 1H, β), 3.89 (s, 3H, OMe); 13C-NMR (acetone-d6) δ 168.19 (γ), 149.16 (3), 146.38 (α), 146.29 (5), 137.39 (4), 126.57 (1), 116.13 (β), 110.44 (6), 104.28 (2), 56.51 (OMe).
Synthesis of CoA Thioesters
Sb4CL was expressed and purified in an identical manner to SbCCoAOMT, with a final concentration of approximately 5 mg mL−1. Caffeoyl-CoA, feruloyl-CoA, 5-hydroxyferuloyl-CoA, sinapoyl-CoA, and p-coumaroyl-CoA were generated by a 10-mL ligation reaction containing 50 mm Na2HPO4 (pH 7.1), 5 mm MgCl2, 2.5 mm ATP, 1 mm HS-CoA, 2 mm p-coumaric, caffeic, ferulic, 5-hydroxy-ferulic, or sinapic acid, and 100 µg of Sb4CL. Reactions were incubated for 18 h at room temperature in the dark and were terminated by boiling for 10 min. Denatured protein was cleared from the reactions by centrifugation at 15,000 rpm for 30 min. Newly synthesized thioesters were separated from the reaction mixture by ethanol precipitation followed by recrystallization using ethanol as a solvent. The identities of CoA thioesters were confirmed with the Sciex 4800 MALDI TOF/TOF Analyzer at the Washington State University Laboratory for Biotechnology and Bioanalysis Unit 1. CoA thioester concentrations were measured using previously determined extinction coefficients (Wengenmayer et al., 1976; Rautengarten et al., 2010).
Molecular Docking
A caffeoyl-CoA model truncated at the β-phosphate of the 3ʹ-phospho-ADP moiety was constructed using GaussView3 (Frisch, 2004) and used as the ligand in the docking search. Files to be used in the AutoDock Vina (Trott and Olson, 2010) calculation were prepared using AutoDock Tools 1.5.6 (Morris et al., 2009). The search space spanned 30 Å in the x dimension, 40 Å in the y dimension, and 26 Å in the z dimension and was oriented to include both potential binding approaches identified from our crystallographic data. The calculation returned nine results, with the top conformer possessing a binding energy of −8.8 kcal mol−1. The truncated docked conformer was extended to the full caffeoyl-CoA structure using GaussView3 (Frisch, 2004), and the undocked ADP moiety was placed manually into the SbCCoAOMT structure.
ITC
Titration was performed using a Micro-ITC (GE Healthcare). For the titrations with SAM and SAH, ligand was prepared at 0.8 mm, and protein was prepared at 0.05 mm in titration buffer; for the caffeic acid and CoA titrations, ligand was prepared at 1 mm, and protein was prepared at 0.02 mm in titration buffer. The caffeic acid and caffeoyl-CoA titrations were repeated at the same ligand and protein concentrations, but both the ligand and protein solutions were incubated on ice for 30 min with 60 µm SAH prior to the titration. The p-coumaroyl-CoA, feruloyl-CoA, and 5-hydroxyferuloyl-CoA titrations were performed by preparing ligand at 1 mm and protein at 0.02 mm in the titration buffer and preequilibrating both solutions with 60 µm SAH prior to the titration. All ligands also were titrated into the titration buffer to account for the heat of dilution.
ICP-OES
ICP-OES was performed using a Perkin-Elmer Optima 3200 RL instrument. The standard curve used for Ca2+ detection consisted of samples with Ca2+ concentrations of 0.1, 0.4, 1, 2, and 10 ppm, as well as a 0-ppm Ca2+ calibration blank, all prepared in 1% HNO3. Protein samples were prepared at 12.8 µm in 30% DMSO and 1% HNO3. Measurements were taken in eight replicates, with intensities averaged over a period of 5 to 20 s.
Standard curve solutions for Mg2+ and Zn2+ were prepared with 0.01, 0.1, 1, 10, and 100 ppm of each respective element as well as 0-ppm calibration blanks dissolved in 1% HNO3. Sample solutions for both Mg2+ and Zn2+ were prepared with 12.8 µm protein in 30% DMSO and 1% HNO3. Measurements were taken in five replicates, with intensities averaged over a period of 2 to 20 s.
Enzymatic Activity Assays of SbCCoAOMT
Kinetic parameters were determined by assay using 60-µL reaction volumes containing 20 mm potassium phosphate, 1 mm SAM, 0.2 mm Ca2+, and concentrations of caffeoyl-CoA varying between 1 and 30 µm. Reactions were initiated by the addition of 1.5 µg of enzyme and incubated at 30°C for 45 s prior to the addition of 24 µL of quenching solution [0.2 µg mL−1 2-methyl-4-(3H)-quinazolinone, 10 mm formic acid, and 50% acetonitrile]. Reaction products were separated by HPLC using a SunFire C8 3.5-µm, 2.1- × 50-mm column running at 0.6 mL min−1 with a gradient of solvent A (5 mm NH4CO2H in deionized water) and solvent B (5 mm NH4CO2H in 95% acetonitrile) varying from 95% A/5% B to 30% A/70% B over a period of 2 min. Products were quantified by mass spectroscopy using an AB SCIEX QTRAP 4000 running in positive ion mode. The analyte, caffeoyl-CoA, and the internal standard, 2-methyl-4(3H)-quinazolinone, were detected using multiple reaction monitoring mode by monitoring the m/z transition from 944.2 to 437.2 and 161 to 120, respectively. Fitting was performed with OriginPro 2015 (OriginLab).
Activity assays of 5-hydroxyferuloyl-CoA were performed using 70-µL reaction volumes containing 20 mm potassium phosphate, 1 mm SAM, 0.2 mm Ca2+, and 10 µm 5-hydroxyferuloyl-CoA. Reactions were initiated by the addition of 1.5 µg of enzyme and incubated at 30°C between 45 s and 1 h prior to termination by the addition of 28 µL of quenching solution (7% trifluoroacetic acid and 50% acetonitrile). Reaction products were separated by HPLC using a Phenomenex Kinetex XB-C18 2.6-µm, 4.6- × 250-mm column. A flow of 1.5 mL min−1 and a gradient of solvent A (0.1% trifluoroacetic acid in deionized water) and solvent B (100% acetonitrile) varying from 5% B to 65% B over a period of 20 min. 5-Hydroxyferuloyl-CoA and sinapoyl-CoA were detected using a Thermo Ultimate 3000 VWD-3400RS monitoring at 352 nm.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Inductively coupled plasma data for SbCCoAOMT calcium content.
Supplemental Figure S2. Chromatograms of 5-hydroxyferuloyl-CoA assays and 100 μm sinapoyl-CoA standard.
Supplemental Figure S3. SDS-PAGE of wild-type SbCCoAOMT and mutants.
Supplementary Material
Glossary
- SAM
S-adenosyl methionine
- ICP-OES
inductively coupled plasma-optical emission spectroscopy
- SAH
S-adenosyl homocysteine
- ITC
isothermal titration calorimetry
- S/G
syringyl/guaiacyl
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB 102114 and CHE 118359 to C.K.), the National Institutes of Health (grant no. 1R01GM11125401 to C.K.) and the M.J. Murdock Charitable Trust (to C.K.); by the Biomass Research and Development Initiative (grant no. 2011–1006–30358 to W.V.); by the U.S. Department of Energy’s Office of Energy Efficiency and Renewable Energy, Bioenergy Technologies Office (grant no. DE–PI0000031 to W.V.); by the Department of Energy Great Lakes Bioenergy Research Center (grant no. DE–FC02–07ER64494 to M.R. and J.R.); and by the U.S. Department of Agriculture (National Institute of Food and Agriculture AFRI grant no. 2011–67009–30026 to S.E.S. and CRIS project grant no. 3042–21220–032–00D).
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbor VB, Cicek N, Sparling R, Berlin A, Levin DB (2011) Biomass pretreatment: fundamentals toward application. Biotechnol Adv 29: 675–685 [DOI] [PubMed] [Google Scholar]
- Almodares A, Hadi MR (2009) Production of bioethanol from sweet sorghum: a review. Afr J Agric Res 4: 772–780 [Google Scholar]
- Bout S, Vermerris W (2003) A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol Genet Genomics 269: 205–214 [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre I, Faci JM (2006) Comparative response of maize (Zea mays L.) and sorghum (Sorghum bicolor L. Moench) to deficit irrigation in a Mediterranean environment. Agric Water Manage 83: 135–143 [Google Scholar]
- Ferrer JL, Zubieta C, Dixon RA, Noel JP (2005) Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol 137: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. (2004) GaussView, Version 3. Guassian, Wallingford, CT [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, et al. (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng S, Hills FJ, Johnson SS, Sah RN (1989) Potential yields and on-farm ethanol-production cost of corn, sweet sorghum, fodderbeet, and sugarbeet. J Agron Crop Sci 162: 21–29 [Google Scholar]
- Gill JR, Burks PS, Staggenborg SA, Odvody GN, Heiniger RW, Macoon B, Moore KJ, Barrett M, Rooney WL (2014) Yield results and stability analysis from the sorghum regional biomass feedstock trial. Bioenerg Res 7: 1026–1034 [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AR, Lewis KM, Barr JT, Jones JP, Lu F, Ralph J, Vermerris W, Sattler SE, Kang C (2014) Determination of the structure and catalytic mechanism of Sorghum bicolor caffeic acid O-methyltransferase and the structural impact of three brown midrib12 mutations. Plant Physiol 165: 1440–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Ragauskas A (2012) Pretreatment and lignocellulosic chemistry. Bioenerg Res 5: 1043–1066 [Google Scholar]
- Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5: 224–229 [DOI] [PubMed] [Google Scholar]
- Ibdah M, Zhang XH, Schmidt J, Vogt T (2003) A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J Biol Chem 278: 43961–43972 [DOI] [PubMed] [Google Scholar]
- Inoue K, Sewalt VJH, Murray GB, Ni W, Stürzer C, Dixon RA (1998) Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Vermerris W, Gallo M, Fedenko JR, Erickson JE, Altpeter F (2013) RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol J 11: 709–716 [DOI] [PubMed] [Google Scholar]
- Katz AK, Glusker JP, Beebe SA, Bock CW (1996) Calcium ion coordination: a comparison with that of beryllium, magnesium, and zinc. J Am Chem Soc 118: 5752–5763 [Google Scholar]
- Kopycki JG, Rauh D, Chumanevich AA, Neumann P, Vogt T, Stubbs MT (2008) Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferases. J Mol Biol 378: 154–164 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Leu SY, Zhu JY (2013) Substrate-related factors affecting enzymatic saccharification of lignocelluloses: our recent understanding. Bioenerg Res 6: 405–415 [Google Scholar]
- Makita Y, Shimada S, Kawashima M, Kondou-Kuriyama T, Toyoda T, Matsui M (2015) MOROKOSHI: transcriptome database in Sorghum bicolor. Plant Cell Physiol 56: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD (2016) Designer lignins: harnessing the plasticity of lignification. Curr Opin Biotechnol 37: 190–200 [DOI] [PubMed] [Google Scholar]
- Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol 179: 738–750 [DOI] [PubMed] [Google Scholar]
- Noel JP, Dixon RA, Pichersky E, Zubieta C, Ferrer JL (2003) Structural, functional, and evolutionary basis for methylation of plant small molecules. Recent Advances in Phytochemistry 37: 37–58 [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo D, Blount JW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pinçon G, Maury S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2001) Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Propheter JL, Staggenborg SA, Wu X, Wang D (2010) Performance of annual and perennial biofuel crops: yield during the first two years. Agron J 102: 806–814 [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Rautengarten C, Baidoo E, Keasling JD, Scheller HV (2010) A simple method for enzymatic synthesis of unlabeled and radiolabeled hydroxycinnamate-CoA. Bioenerg Res 3: 115–122 [Google Scholar]
- Regassa TH, Wortmann CS (2014) Sweet sorghum as a bioenergy crop: literature review. Biomass Bioenergy 64: 348–355 [Google Scholar]
- Rooney WL, Blumenthal J, Bean B, Mullet JE (2007) Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod Biorefin 1: 147–157 [Google Scholar]
- Russell WR, Forrester AR, Chesson A, Burkitt MJ (1996) Oxidative coupling during lignin polymerization is determined by unpaired electron delocalization within parent phenylpropanoid radicals. Arch Biochem Biophys 332: 357–366 [DOI] [PubMed] [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70: 818–830 [DOI] [PubMed] [Google Scholar]
- Saballos A, Vermerris W, Rivera L, Ejeta G (2008) Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenerg Res 1: 193–204 [Google Scholar]
- Sattler SE, Palmer NA, Saballos A, Greene AM, Xin ZG, Sarath G, Vermerris W, Pedersen JF (2012) Identification and characterization of four missense mutations in brown midrib12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenerg Res 5: 855–865 [Google Scholar]
- Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol 150: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Saballos A, Xin Z, Funnell-Harris DL, Vermerris W, Pedersen JF (2014) Characterization of novel Sorghum brown midrib mutants from an EMS-mutagenized population. G3 (Bethesda) 4: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully ED, Gries T, Funnell-Harris DL, Xin Z, Kovacs FA, Vermerris W, Sattler SE (2016) Characterization of novel Brown midrib 6 mutations affecting lignin biosynthesis in sorghum. J Integr Plant Biol 58: 136–149 [DOI] [PubMed] [Google Scholar]
- Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE (2011) Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Sci USA 108: 6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, Abril A (2015) Enhancing cellulose utilization for fuels and chemicals by genetic modification of plant cell wall architecture. Curr Opin Biotechnol 32: 104–112 [DOI] [PubMed] [Google Scholar]
- Vidgren J, Svensson LA, Liljas A (1994) Crystal structure of catechol O-methyltransferase. Nature 368: 354–358 [DOI] [PubMed] [Google Scholar]
- Walker AM, Hayes RP, Youn B, Vermerris W, Sattler SE, Kang C (2013) Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol 162: 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengenmayer H, Ebel J, Grisebach H (1976) Enzymic synthesis of lignin precursors: purification and properties of a cinnamoyl-CoA: NADPH reductase from cell suspension cultures of soybean (Glycine max). Eur J Biochem 65: 529–536 [DOI] [PubMed] [Google Scholar]
- Wils CR, Brandt W, Manke K, Vogt T (2013) A single amino acid determines position specificity of an Arabidopsis thaliana CCoAOMT-like O-methyltransferase. FEBS Lett 587: 683–689 [DOI] [PubMed] [Google Scholar]
- Ximenes E, Kim Y, Mosier N, Dien B, Ladisch M (2011) Deactivation of cellulases by phenols. Enzyme Microb Technol 48: 54–60 [DOI] [PubMed] [Google Scholar]
- Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Wang ML, Burow G, Burke J (2009) An induced sorghum mutant population suitable for bioenergy research. Bioenerg Res 2: 10–16 [Google Scholar]
- Zeng Y, Zhao S, Yang S, Ding SY (2014) Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr Opin Biotechnol 27: 38–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.