Chemical screening for novel inhibitors of lignification uncovered a compound processed in the plant into two such inhibitors, one of which, p-iodobenzoic acid, inhibits CINNAMATE-4-HYDROXYLASE activity.
Abstract
Plant secondary-thickened cell walls are characterized by the presence of lignin, a recalcitrant and hydrophobic polymer that provides mechanical strength and ensures long-distance water transport. Exactly the recalcitrance and hydrophobicity of lignin put a burden on the industrial processing efficiency of lignocellulosic biomass. Both forward and reverse genetic strategies have been used intensively to unravel the molecular mechanism of lignin deposition. As an alternative strategy, we introduce here a forward chemical genetic approach to find candidate inhibitors of lignification. A high-throughput assay to assess lignification in Arabidopsis (Arabidopsis thaliana) seedlings was developed and used to screen a 10-k library of structurally diverse, synthetic molecules. Of the 73 compounds that reduced lignin deposition, 39 that had a major impact were retained and classified into five clusters based on the shift they induced in the phenolic profile of Arabidopsis seedlings. One representative compound of each cluster was selected for further lignin-specific assays, leading to the identification of an aromatic compound that is processed in the plant into two fragments, both having inhibitory activity against lignification. One fragment, p-iodobenzoic acid, was further characterized as a new inhibitor of CINNAMATE 4-HYDROXYLASE, a key enzyme of the phenylpropanoid pathway synthesizing the building blocks of the lignin polymer. As such, we provide proof of concept of this chemical biology approach to screen for inhibitors of lignification and present a broad array of putative inhibitors of lignin deposition for further characterization.
Lignin is a phenolic polymer deposited predominantly in the plant’s secondary-thickened cell walls together with cellulose and other matrix polysaccharides (Vanholme et al., 2012a). By rendering the cell wall impermeable, lignin allows long-distance transport of water and dissolved nutrients along the vascular system. A lignin-impregnated cell wall also constitutes a physical barrier against insects and pathogens (Miedes et al., 2014). In addition, the mechanical strength it provides to the plant ensures stability, erect growth, and resistance to the negative pressure caused by transpiration. Therefore, the emergence of lignin is considered a crucial evolutionary adaptation of plants for successful land colonization (Weng and Chapple, 2010).
In addition to its essential physiological role during plant growth, the lignin polymer has an intrinsic economic significance. For example, it can be processed into important food additives (e.g. vanillin) or foam stabilizers (Agrawal et al., 2014), and some lignans (i.e. dimers derived from monolignols) possess pharmacological properties (Vogt, 2010). On the other hand, lignin is a major factor causing cell wall recalcitrance for industrial processing of lignocellulosic biomass (Baucher et al., 2003). For example, the extraction of fermentable sugars for the production of biofuels and other bio-based chemicals is largely hampered by the presence of lignin (Vanholme et al., 2012a). To improve downstream processing, the biomass needs to be pretreated using harsh physical and chemical procedures. These additional processing costs give bio-based products a commercial disadvantage compared with conventional fossil-based alternatives (Lynd et al., 2008).
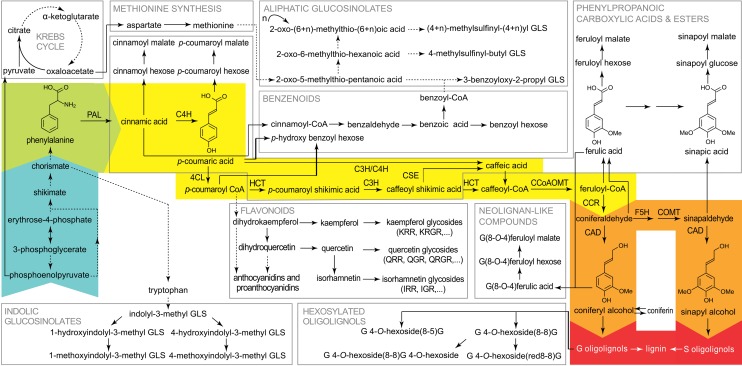
Because of its economic importance, lignin has been the subject of intense research (Baucher et al., 2003). Forward and reverse genetic approaches have significantly broadened our understanding of the role of many lignin biosynthetic enzymes (Bonawitz and Chapple, 2010). In the last decade, profiling of the secondary metabolism via gas chromatography or ultra-high-pressure liquid chromatography (UHPLC)-mass spectrometry (MS) has facilitated further elucidation of the pathway (Gross, 2008; Vanholme et al., 2013), and this scientific research condensed into the pathway we know today (Fig. 1). The phenylpropanoid pathway uses the aromatic amino acid Phe as an entry substrate and converts it into a plethora of secondary metabolites. Phe is deaminated by PAL. The produced cinnamic acid is converted subsequently to p-coumaric acid by C4H and further into p-coumaroyl-CoA by 4CL (Boerjan et al., 2003). From this point on, the phenylpropanoid pathway branches into several routes ultimately leading to the formation of monolignols and other secondary metabolites, like phenylpropanoic esters, benzenoids, and flavonoids (Vogt, 2010). The monolignol-specific pathway departs from the general phenylpropanoid pathway from feruloyl-CoA on and converts it into coniferyl and sinapyl alcohols, the two main lignin monomers (Vanholme et al., 2012b). After their synthesis in the cytosol, monolignols are either combinatorially coupled in the cytoplasm, glycosylated, and stored in the vacuole (Dima et al., 2015) or transported to the apoplast, where they are polymerized into lignin, mainly forming 8-O-4, 8-8, and 8-5 bonds (Vanholme et al., 2012a).
Figure 1.
The phenylpropanoid pathway with a focus on lignin metabolism. Phe (green), derived from the shikimate pathway (blue), is converted via the general phenylpropanoid (yellow) and the monolignol-specific (orange) pathways to the monolignols coniferyl and sinapyl alcohol, which can polymerize into G and S oligolignols (red). Other pathways are intertwined with or branch off from the general phenylpropanoid pathway (in gray squares). Dashed arrows represent multiple enzymatic steps, and solid arrows stand for a single conversion. This figure was adapted from Vanholme et al. (2012b). Proteins: CAD, CINNAMOYL ALCOHOL DEHYDROGENASE; CCoAOMT, CAFFEOYL-COENZYME A O-METHYLTRANSFERASE; CCR, CINNAMOYL-COENZYME A REDUCTASE; C3H, p-COUMARATE 3-HYDROXYLASE; C4H, CINNAMATE 4-HYDROXYLASE; 4CL, 4-COUMARATE:COENZYME A LIGASE; COMT, CAFFEIC ACID O-METHYLTRANSFERASE; CSE, CAFFEOYL SHIKIMATE ESTERASE; F5H, FERULATE 5-HYDROXYLASE; HCT, p-HYDROXYCINNAMOYL-COENZYME A:QUINATE SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE; PAL, PHENYLALANINE AMMONIA LYASE. Rhamnosides: GLS, glucosinolate; IGR, isorhamnetin-3-O-glucosyl-7-O-rhamnoside; IRR, isorhamnetin-3-O-rhamnosyl-7-O-rhamnoside; KRGR, kaempferol-3-O-rhamnosyl(1→2)β-glucopyranosyl-7-O-α-rhamnoside; KRR, kaempferol-3-O-rhamnosyl-7-O-rhamnoside; QGR, quercetin-3-O-β-glucopyranosyl-7-O-α-rhamnopyranoside; QRGR, quercetin-3-O-rhamnosyl(1→2)β-glucopyranosyl-7-O-α-rhamnopyranoside; QRR, quercetin-3-O-rhamnosyl-7-O-rhamnoside.
Perturbations provoked in a biosynthetic pathway can be useful to study the mechanism steering the flux through the pathway or to identify the enzymes embedded in the metabolic network (Vanholme et al., 2012b). The response to pathway perturbations also can reveal novel genes closely associated with the studied pathway (Vanholme et al., 2012b, 2013; Sundin et al., 2014). A complementary strategy to study biological processes is chemical biology. This research strategy, whereby small molecules are used to elucidate molecular pathways or assign functions to proteins, has notable advantages compared with the classical forward and reverse genetic approaches (Serrano et al., 2015). For example, the concentration and timing of a treatment with chemicals are flexible parameters, allowing one to specifically target vital processes that would result in lethality if affected using a genetic approach. Small molecules also can target multiple members of a protein family, circumventing to some extent the genetic redundancy typically encountered using genetic approaches. Furthermore, multiple biological synthesis routes or multiple parts of a biological process can be dissected via the simultaneous application of multiple compounds. Finally, compounds can be applied easily to essentially any plant model system or crop, whereas a genetic perturbation needs to be introduced in one genotype at a time (Tóth and van der Hoorn, 2010). Thanks to these characteristics, chemical biology has been successful for drug discovery (Cong et al., 2012), and it is becoming increasingly more popular to discover molecular mechanisms of physiological processes in plants (De Rybel et al., 2009, 2012; Dejonghe and Russinova, 2014; Hicks and Raikhel, 2014).
In this study, we performed a high-throughput screen of a diverse set of 10,000 synthetic molecules to identify inhibitors of the lignin biosynthetic pathway. Of the 73 positive hits, 39 putative inhibitors that caused major perturbations in the phenylpropanoid pathway were divided into five functional classes based on the phenolic profiles they provoked in seedlings. One representative compound of each functional class was then further subjected to a range of assays to quantify the extent of inhibition of lignin deposition and its effect on phenolic metabolism. Our approach discovered p-iodobenzoic acid (pIBA) as a new inhibitor of C4H, a key enzyme of the phenylpropanoid pathway.
RESULTS
Development of a High-Throughput Screening Assay
We devised a high-throughput assay in a 96-well plate format to enable the quick screening of the impact of a chemically diverse collection of 10,000 molecules on the extent of lignin deposition in Arabidopsis (Arabidopsis thaliana) seedlings. In this setup, three to seven Columbia-0 (Col-0) seeds were added per well in liquid plant growth medium. Three days after germination (DAG), library compounds were supplied to the seedlings at a final concentration of 50 µm. Because at this young seedling stage lignin deposition is not yet extensive, making it difficult to screen for altered lignin deposition, isoxaben was added to the medium 3 h after the addition of the library compound. Isoxaben inhibits cellulose synthesis and causes ectopic lignin deposition in seedlings (Caño-Delgado et al., 2003). Three days after the addition of the library compound and isoxaben, the seedlings were screened for perturbations in lignin deposition using the Wiesner reagent (1% phloroglucinol in ethanol and HCl), a dye that turns red upon reaction with O-4-linked coniferyl and sinapyl aldehydes that are typical parts of the lignin polymer (Pomar et al., 2002; Boerjan et al., 2003; Ralph et al., 2004). The screen aimed to identify compounds that reduced the staining compared with that of mock-treated plants or plants treated with an inactive compound.
To validate this experimental setup, a proof-of-concept experiment was set up with 3,4-methylenedioxycinnamic acid (MDCA). MDCA is a known competitive inhibitor of 4CL and has been shown to reduce lignin content in the cell wall (Funk and Brodelius, 1990). Initially, the assay suffered from severe background staining with the Wiesner reagent. Because a reddish color also was observed in the plant tissue upon HCl treatment in the absence of phloroglucinol, this artifact could be attributed to the Suc used as a carbon source in the plant medium (Supplemental Fig. S1). It is known that Suc can lead to the accumulation of anthocyanins in Arabidopsis (Teng et al., 2005; Solfanelli et al., 2006; Poustka et al., 2007), which turn red in an acidic environment (Castañeda-Ovando et al., 2009). This undesired effect was circumvented by replacing Suc by Glc in the growth medium. Under these conditions, a clear red vasculature was visible in isoxaben-treated control seedlings upon Wiesner staining (Fig. 2A). This coloration was absent or largely reduced in intensity when seedlings were cotreated with 50 µm MDCA (Fig. 2B), proving the efficacy of this assay to report lignin reduction. As an additional validation, the 4cl1-1 mutant was analyzed as a genetic positive control for lignification inhibition. This mutant is considerably impaired in the phenylpropanoid pathway (Vanholme et al., 2012b), resulting in a final lignin content in senesced stems that is 26% lower than that in wild-type plants (Van Acker et al., 2013). Based on Wiesner staining, isoxaben treatment of these plants did not induce lignin deposition to a similar level to that in wild-type plants (Fig. 2C), but the vasculature of the 4cl1-1 mutant was lignified to a higher extent compared with that of MDCA-treated seedlings.
Figure 2.
Wiesner staining of Arabidopsis seedlings (6 DAG). A, Wild-type seedlings treated with isoxaben. B, Wild-type seedlings cotreated with isoxaben and the 4CL inhibitor MDCA. C, 4cl1-1 seedlings treated with isoxaben. Bar = 1 mm.
Identification of 73 Putative Lignification Inhibitors
After assay optimization and validation, a full-scale library screen was performed using the ChemBridge DIVERSet library, which consists of a collection of 10,000 diverse synthetic molecules. Seedlings cotreated with 50 µm MDCA and isoxaben were used as positive controls for the inhibition of lignification; dimethyl sulfoxide (DMSO)- and isoxaben-treated (mock) seedlings were used as negative controls. The first screen resulted in 156 compounds causing a visual reduction in Wiesner staining in seedlings. These 156 compounds were retested under the same conditions, and for 73 compounds, the reduced staining was confirmed. All 73 compounds were given a unique number to facilitate tracking in follow-up experiments (Supplemental Table S1). Their structures were uploaded to the ChemMine Web site (http://chemmine.ucr.edu/), and a hierarchical tree was constructed based on structural similarity (Supplemental Fig. S2; Backman et al., 2011). Seven known inhibitors of the phenylpropanoid pathway were included as reference compounds: cinnamaldehyde (CAld) as an inhibitor of PAL (Fujita et al., 2006); 2,4-dinitrophenol (DNP) and menadione (MD) as inhibitors of C4H (Billett and Smith, 1978); and caffeic acid (CA), ferulic acid (FA), MDCA, and N-(3,4-dichlorophenyl)propanamide (propanil) as inhibitors of 4CL (Funk and Brodelius, 1990; Harding et al., 2002; Yun et al., 2006). The structural clustering revealed four prominent structural classes and one class with remaining compounds (Supplemental Fig. S2). Structural class 1 consisted of these 16 remaining compounds (7, 8, 9, 10, 11, 22, 26, 28, 30, 32, 37, 44, 57, 68, 69, and 71) and three known inhibitors (CAld, DNP, and propanil). The known inhibitors CA, FA, and MDCA clustered together in structural class 2, which is in line with their shared phenylpropanoid structure. None of the 73 compounds clustered in this group of canonical phenylpropanoid-like compounds. The remaining known inhibitor (i.e. MD) resided in the third structural class. This was the largest cluster, consisting of 52 compounds (MD, 1, 3, 4, 5, 6, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 23, 24, 25, 29, 31, 34, 35, 36, 38, 39, 40, 41, 42, 43, 45, 46, 48, 49, 50, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 63, 65, 66, 67, 70, 72, and 73). Structural class 4 contained four compounds (2, 27, 47, and 64), and structural class 5 contained two compounds (33 and 60).
Phenolic Profiling Reveals Functional Classes
To validate the effect of the 73 retained compounds on the phenylpropanoid pathway, the phenolic profile of compound-treated plants was assessed. Arabidopsis seedlings (3 DAG) were treated for 3 d with the compounds and harvested, and their metabolites were methanol extracted for phenolic profiling by UHPLC-MS. CAld, MD, DNP, CA, FA, MDCA, and propanil were included as reference inhibitors. Of the 80 assessed compounds, three were lethal to the developing seedlings (i.e. compounds 20, 30, and 31). The seedlings treated with the 77 remaining compounds and, as controls, eight Arabidopsis mutants perturbed in specific steps of the phenylpropanoid biosynthetic pathway (pal1-2, pal2-2, c4h-3, 4cl1-1, 4cl2-1, ccoaomt1-5, ccr1-6, and ccr2-1) were subjected to metabolic profiling.
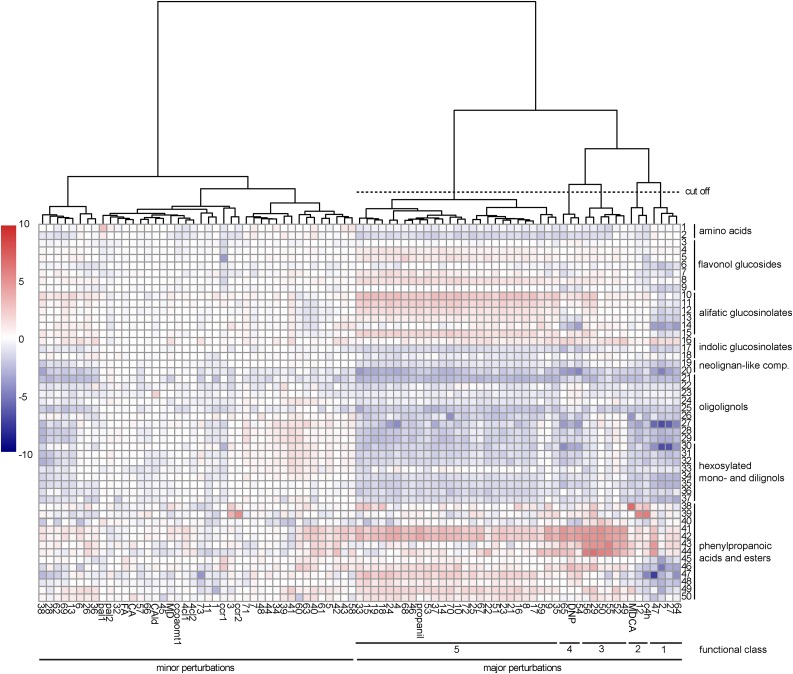
A total of 20,973 peaks were detected over the different samples (Supplemental Table S2). A targeted search was performed on these peaks, annotating 50 lignin-related metabolites. This subset was used to produce a heat map visualizing the average normalized peak area of these metabolites over the 85 samples (Fig. 3). The heat map revealed two big clusters: compounds or mutations causing only minor shifts in the phenolic profile of seedlings (31 compounds, seven mutants, and four known inhibitors) versus those having a major impact (39 compounds, one mutant, and three known inhibitors). The latter were considered for further research and classified into five functional classes through hierarchical clustering of the phenolic profiles (Fig. 3).
Figure 3.
Phenolic profile of Arabidopsis seedlings treated with putative inhibitors of lignification. Targeted analysis was performed on phenylpropanoids (numbered from 1 to 50 at right) in Arabidopsis seedlings treated with 70 putative inhibitors (listed below the heat map). Seven known inhibitors of lignin biosynthesis and eight lignin biosynthesis mutants were included as positive controls (below the heat map). Peak areas were divided by the relative total peak area (total peak area in the sample/maximum total peak area of all samples) to be able to compare the relative peak areas across samples. Then, the peak area was divided by the average control peak area and log2 transformed. Finally, values were converted to color codes to visualize the increase (red) or decrease (blue) of metabolite levels compared with mock-treated controls. The resulting heat map was clustered in R using the pheatmap package (clustering distance set to Euclidean, and Ward was used as the clustering method). Based on this clustering, compounds with a major impact on the phenolic profile were divided into five functional classes as indicated below the heat map. 1, Phe; 2, Trp; 3, kaempferol-3-rhamnose-7-rhamnose; 4, kaempferol-3-O-rhamnosyl(1→2)β-glucopyranosyl-7-O-α-rhamnopyranoside; 5, quercetin-3-rhamnose-7-rhamnose; 6, quercetin-3-O-β-glucopyranosyl-7-O-α-rhamnopyranoside; 7, quercetin-3-O-rhamnosyl(1→2)β-glucopyranosyl-7-O-α-rhamnopyranoside; 8, isorhamnetin-3,7-dirhamnose; 9, isorhamnetin-3-Glc-7-rhamnose; 10, 5-methylsulfinyl-pentyl glucosinolate; 11, 6-methylsulfinyl-hexyl glucosinolate; 12, 7-methylsulfinyl-heptyl glucosinolate; 13, 8-methylsulfinyl-octyl glucosinolate; 14, 9-methylsulfinyl-nonyl glucosinolate; 15, 3-benzoyloxy-n-propyl glucosinolate; 16, indolyl-3-methylglucosinolate; 17, 1-methoxyindolyl-3-methyl glucosinolate; 18, 4-methoxyindolyl-3-methyl glucosinolate; 19, G(8-O-4)FA; 20, G(8-O-4)FA-hexoside; 21, G(8-O-4)G(8-O-4)G; 22, G(8-5)G (dehydrodiconiferyl alcohol); 23, G(8-5)G fragment; 24, G(8-5)G′; 25, G(8-8)G (pinoresinol); 26, G(8-5)G glucoside (dehydrodiconiferyl alcohol glucoside); 27, G(8-8)G hexoside (pinoresinol hexoside); 28, G(8-8)G hexoside fragment 1; 29, G(8-8)G hexoside fragment 2; 30, coniferin; 31, G(8-8)G dihexoside; 32, G(8-8)G dihexoside fragment; 33, G(8-8)G dihexoside isomer; 34, G(red8-8)G hexoside; 35, G(red8-8)G hexoside isomer; 36, G(red8-8)G hexoside fragment; 37, G(red8-8)G hexoside isomer fragment; 38, cinnamoyl hexose; 39, cinnamoyl malate; 40, p-coumaric acid; 41, p-coumaroyl hexose 1; 42, p-coumaroyl hexose 2; 43, p-coumaroyl malate 1; 44, p-coumaroyl malate 2; 45, cis-feruloyl Glc; 46, trans-feruloyl Glc; 47, trans-sinapoyl Glc; 48, cis-sinapoyl Glc; 49, trans-sinapoyl malate; 50, cis-sinapoyl malate.
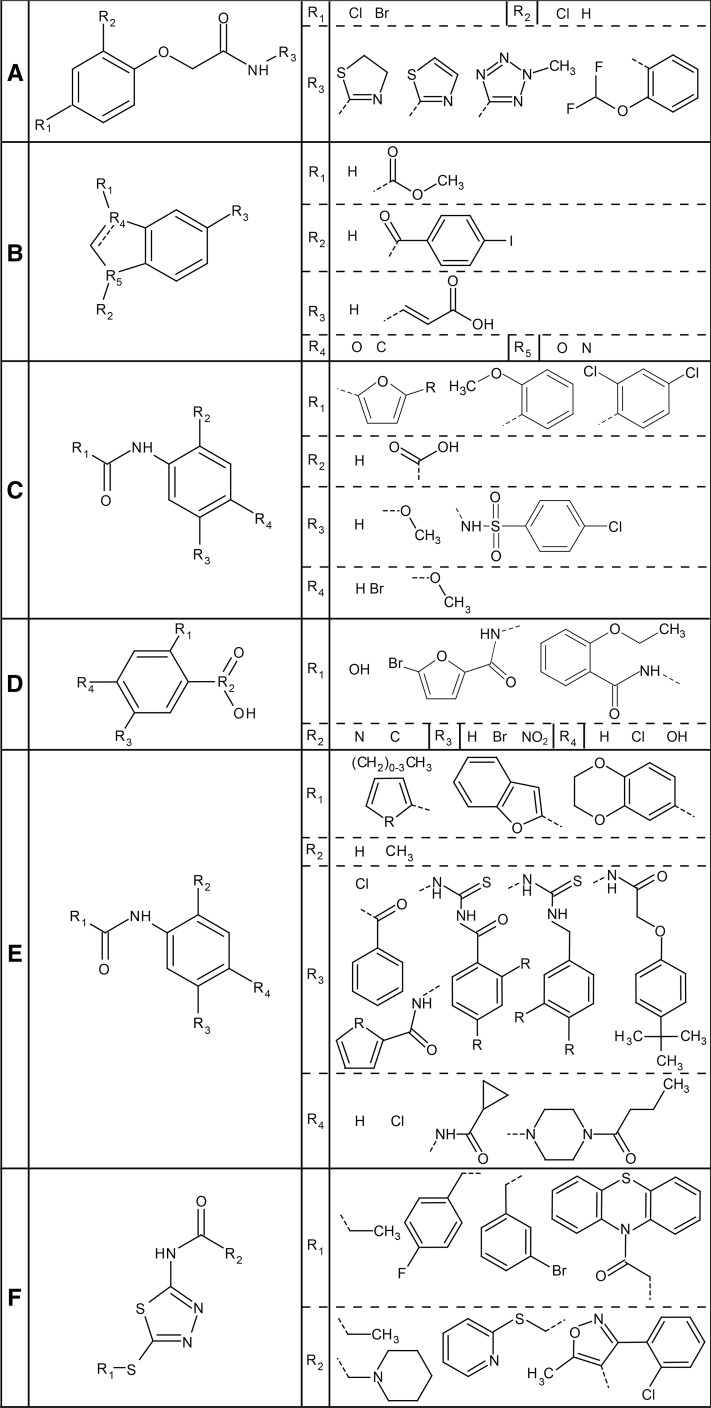
The first functional class contained compounds 2, 27, 47, and 64, all belonging to structural class 4, being acetamides with a halogen-substituted phenoxyl group on the α-position of the carbonyl (Fig. 4A). A second, more variable ring structure is attached to the nitrogen atom. The phenolic profile of seedlings treated with these compounds showed a major decrease in abundance of two coniferyl alcohol derivatives: hexosylated coniferyl alcohol (i.e. coniferin) and a hexosylated dilignol of coniferyl alcohol [i.e. G(8-8)G hexoside]. Furthermore, the intermediates of the first part of the phenylpropanoid pathway had a tendency to accumulate to higher levels (cinnamoyl malate and p-coumaric acid with and without hexose or malate), whereas intermediates appearing farther downstream in the pathway had rather decreased in abundance (feruloyl Glc, sinapoyl Glc, and sinapoyl malate).
Figure 4.
Consensus chemical pattern of each functional class. A, The four compounds of functional class 1 (compounds 2, 27, 47, and 64) are all acetamides with a halogen-substituted phenoxyl group. At the R3 position, a ring structure is attached, usually a five-membered ring. B, The two compounds of functional class 2 (MDCA and compound 12) contain a benzene ring with a five-membered ring attached to it. The double bond is dashed because it is present only in compound 12 and not in MDCA. Also, the heteroatoms of the five-membered ring are not common between the two compounds, as are neither of the substitutions. C, The third functional class (compounds 29, 49, 50, 52, 55, and 56) is characterized by an amide attached to a benzene ring. This benzene ring often is substituted with a carboxyl group (R2), except in compound 29. R1 is always a ring structure, while R3 and R4 vary more. The R in the first structure of R1 can be a Br or an H. D, The compounds of functional class 4 (54, 65, and DNP) are structurally similar to the compounds of class 3 but have no amide connecting their rings. E to F, Functional class 5 is the largest cluster and can be further divided into two structural classes with two different consensus patterns (except 13, 22, 37, and 59). E, The first one is similar to the one in C but with different side chains. Most compounds (4, 14, 15, 16, 17, 18, 19, 21, 23, 24, 25, 33, 35, 46, 51, 53, 67, 70, and 72) contain this consensus pattern, including propanil. Side chains can have variable substitutions (R), which can be consulted in Supplemental Table S1. F, Four compounds (8, 9, 10, and 68) contain the second consensus pattern of class 5. It consists of a thiadiazole amide with the R1 chain attached to it with a sulfide bond. The substitutions generally are single or even multiple ring structures.
The second functional class contained only the known 4CL inhibitor MDCA and compound 12. Both compounds belong to different structural classes (structural classes 2 and 3, respectively), suggesting that there is little structural similarity between the two compounds (Fig. 4B). Interestingly, functional class 2 was the only cluster in which a lignin mutant was included (i.e. c4h). In this cluster, a clear accumulation of cinnamoyl hexoside (profile of MDCA-treated seedlings) or cinnamoyl malate (profiles of seedlings treated with compound 12 and c4h) was detected.
The six compounds of the third functional class (29, 49, 50, 52, 55, and 56) belong to structural class 3 and share two aromatic rings, linked via an amide (Fig. 4C). The aromatic ring at the N-side is a substituted phenyl group, whereas the aromatic ring at the carbonyl side is variable. All compounds, except 29, have a carboxylic acid ortho of the amide. The para position with respect to the amide group is substituted with bromide in compounds 49 and 55, whereas the para and meta positions are methoxy groups in compounds 50 and 52. Compound 29 has a p-chlorobenzenesulfonamide group attached to the aromatic ring at the N-side. The compounds of this functional class caused a profound increase of p-coumaroyl hexose and p-coumaroyl malate.
The fourth functional class contained compounds 54 and 65 as well as the reference inhibitor DNP (Fig. 4D). Compounds 54 and 65 both belong to the same structural class as the compounds of functional class 3 (Fig. 4C; i.e. structural class 3). Compound 65 shares the same structure as compounds 49 and 55, in addition to the variable ring at the carbonyl side of the amide bond. The same holds true for compound 54, but instead of a bromide para to the amide, it has a meta-substituted chlorine. DNP belongs to structural class 1 and has a more simple structure: two nitro-substituents ortho and para of the hydroxyl group of the phenol. Functional class 4 compounds caused an increase in p-coumaroyl hexose, resembling the metabolic shift induced by functional class 3 compounds, but p-coumaroyl malate did not accumulate to a similar extent.
The fifth and last functional class consisted of 27 compounds, including the known inhibitor propanil. Based on structural analysis, most compounds of this functional group could be divided in two subgroups, belonging to structural classes 3 and 1 (Fig. 4, E and F). The majority of the compounds of this functional class contained the same amide group flanked by ring structures as functional classes 3 and 4 (compounds 4, 14, 15, 16, 17, 18, 19, 21, 23, 24, 25, 33, 35, 46, 51, 53, 67, 70, 72, and propanil; Fig. 4E). A smaller number of compounds of this functional class shared another substructure (8, 9, 10, and 68; Fig. 4F): a thiadiazole amide substituted by a single or multiple ring structures. Although two consensus structures could be made for this functional class, still not all compounds could be included (compounds 22, 37, and 59), underlining the structural diversity of this class. Seedlings treated with compounds of this functional class showed an accumulation of p-coumaroyl hexose, but to a lesser extent to that observed after treatment with functional class 4 compounds. Besides the shift in the phenylpropanoid profile, the level of some glucosinolates also was altered. Aliphatic glucosinolates tended to accumulate to higher levels in propanil-treated plants when their aliphatic chains were shorter. For instance, 5-methylsulfinylpentyl glucosinolate clearly accumulated in all samples, whereas 9-methylsulfinylnonyl glucosinolate accumulated in a few samples only.
To obtain more insight into the mode of action of the putative inhibitors, one representative compound of each functional class was selected (i.e. compounds 47, 12, 56, 54, and 72 for functional class 1, 2, 3, 4, and 5, respectively), and these were subjected to further research.
Phenotypes of Plants Treated with the Five Representative Compounds
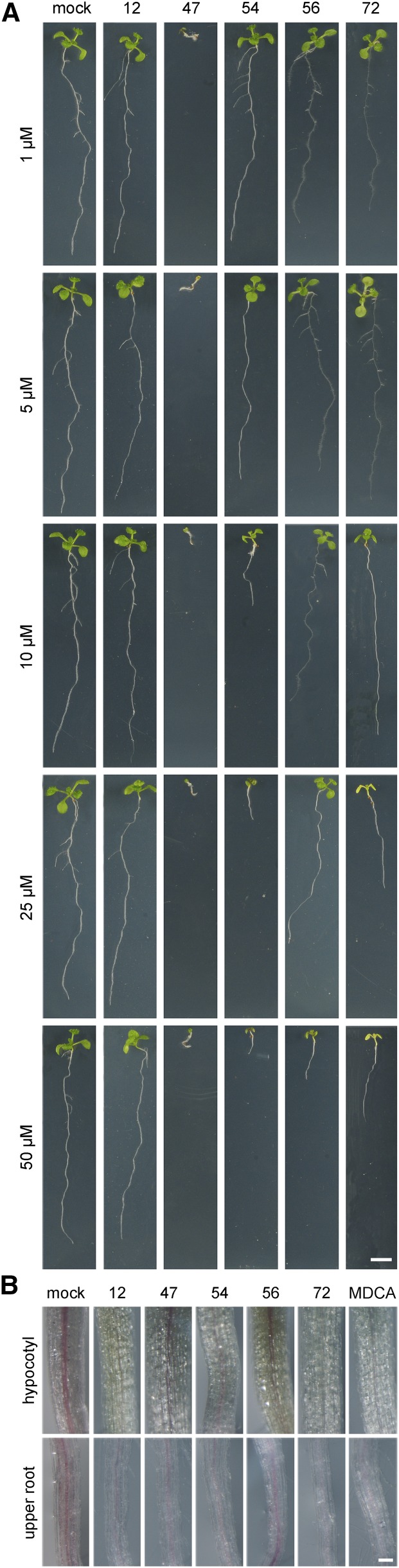
Based on the screening method used, we could not exclude that some of the selected compounds interfered with growth or with the isoxaben treatment rather than with lignification. To test the ability of the compounds to inhibit lignification without interfering with isoxaben, seedlings were grown for 9 d in the absence of isoxaben on solid medium supplemented with one of the five representative compounds. Upon Wiesner staining, a reduction in the intensity of the red vasculature of both hypocotyl and root became apparent for all compounds tested, indicating that these compounds were not retained from the library based on their putative obstruction of the action of isoxaben (Supplemental Fig. S3). However, in this experiment, growth defects (see below) were observed when seedlings were treated with compounds 47, 54, 56, and 72 at 50 µm; for compound 12, a mild growth inhibition was observed only when the concentration exceeded a threshold of 100 µm. Therefore, at this point, we could not exclude that the reduction in the intensity of the Wiesner staining was merely the result of an overall developmental shift (postponing differentiation) rather than a direct inhibition of monolignol biosynthesis or polymerization itself.
To determine the highest compound concentration that did not provoke a dramatic growth defect, seedlings were grown on Murashige and Skoog medium supplemented with the five compounds at different concentrations. Compounds 54, 56, and 72 affected plant growth as visualized by a reduction in primary root length at concentrations ranging from 1 to 50 µm (Fig. 5A). Compound 47 caused severe root growth defects already at 50 nm (Supplemental Fig. S4A) and undifferentiated cell proliferation in both the roots (0.1–1 µm) and leaves (5–25 µm; Supplemental Fig. S4B). When applied at 50 nm, compound 47 induced lateral roots at irregular positions along the primary root. The severe growth defects induced by this compound when applied at low concentrations suggested a hormonal activity. In line with this hypothesis, compound 47 as well as the other compounds belonging to functional class 1 (i.e. 2, 27, and 64) are derivatives of the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D). In fact, 2,4-D could be traced back in the metabolic profile of seedlings treated with compound 47, suggesting that compound 47 is chemically unstable or processed by the plant to 2,4-D (Supplemental Fig. S5).
Figure 5.
Plant phenotypes caused by compounds representing the five functional classes. A, Effect of the five representative compounds (12, 47, 54, 56, and 72) on the growth and development of in vitro-grown Arabidopsis (9 DAG). Bar = 5 mm. B, Wiesner staining of seedlings (9 DAG) chemically treated with the following compounds at the concentration that resulted in a close to wild-type phenotype: 100 µm 12, 0.01 µm 47, 10 µm 54, 25 µm 56, and 10 µm 72. MDCA (10 µm) was included as a positive control. Bar = 0.1 mm.
Based on these growth experiments, the working concentrations for the different compounds were set at 100 µm for compound 12, 25 µm for compound 56, 10 µm for compounds 54, 72, and MDCA, and 10 nm for compound 47. At these concentrations, residual lignification was found for all compounds (Fig. 5B), but clear differences in inhibitory activity were observed. Based on the reduction in the Wiesner stain intensity, we concluded that compounds 12, 54, 72, and MDCA reduced lignification in the hypocotyl and the root, whereas compound 47 reduced lignin in the root only. For compound 56, local patches of lignin inhibition were observed together with the occasional induction of ectopic lignification in some areas of the vascular tissue in both the hypocotyl and the root.
Inhibition of Lignification by Compound 12
In order to quantify the inhibitory potential of the five representative compounds, they were used at the above-defined working concentrations to inhibit lignification in the Casparian strip. In contrast to a previously generally accepted model, lignin is essential for the barrier function of the Casparian strip in Arabidopsis (Naseer et al., 2012). In an assay described by Naseer et al. (2012), Arabidopsis seedlings are stained with propidium iodide (PI) at 5 DAG. This dye diffuses apoplastically in the root tissue but cannot penetrate the lignin-rich Casparian strip, leaving the cell walls of the stele unstained. Counting the number of endodermal cells from the elongation zone in the root tip to the region where PI is excluded from the stele gives a measure for the efficiency of Casparian strip formation and, hence, lignification. In plants treated with an inhibitor of lignification (e.g. the C4H inhibitor piperonylic acid [PA]; Schalk et al., 1998), the number of endodermal cells with a defective Casparian strip is higher than in mock-treated plants due to reduced lignin deposition. The lignification of the Casparian strip can be restored by supplying coniferyl and sinapyl alcohols to the PA-treated plants (Naseer et al., 2012).
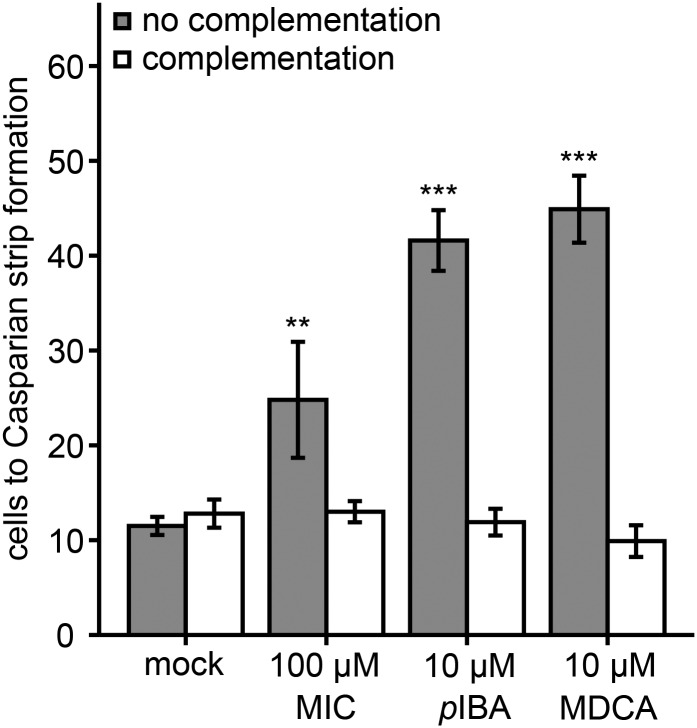
To test the five selected compounds in the Casparian strip assay, 5-d-old seedlings were subjected to a 24-h incubation with each of the compounds (Fig. 6). MDCA and PA were included as positive controls, and the solvent DMSO was used as a negative control. Of the five compounds tested, only compound 12 caused a significant and reversible inhibition of the Casparian strip lignification. The number of endodermal cells up to the PI exclusion zone was increased by 57% compared with the wild type. For MDCA- and PA-treated roots, this number was increased by 104% and 134%, respectively.
Figure 6.

Casparian strip assay of the five representative compounds. Five-day-old Arabidopsis seedlings were subjected to a 24-h treatment with 100 µm compound 12 (A), 0.01 µm compound 47 (B), 10 µm compound 54 (C), 25 µm compound 56 (D), or 10 µm compound 72 (E) either noncomplemented (gray bars) or complemented (white bars) with the monolignols coniferyl and sinapyl alcohol (20 µm each). As positive controls, 10 µm MDCA and PA were included, whereas mock-treated plants served as negative controls. Endodermal cells were counted from the root meristem to the point where Casparian strip formation prevented PI from diffusing into the vascular tissue. Averages were calculated from 10 seedlings per condition. Statistical differences were estimated using Student’s t test compared with the respective controls. Error bars represent sd, and differences are significant with P values below 0.05 (*) and 0.001 (***).
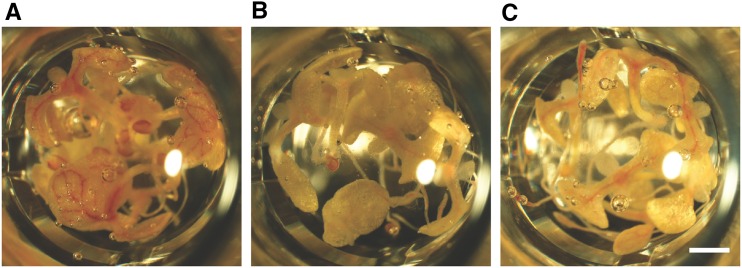
To further explore the impact of the five representative compounds on lignification independent of other developmental processes, their activity was tested on in vitro Arabidopsis pluripotent cell cultures induced to differentiate into lignified tracheary elements (TEs). The lignin content in the cell walls of these specialized xylem cells can be reduced by treating the cell cultures with known inhibitors of lignification, such as PA, and reverted by adding coniferyl or sinapyl alcohol to the PA-treated cells (Pesquet et al., 2013). Cell suspension cultures complement the Casparian strip assay in testing the potential of the candidate lignin inhibitors.
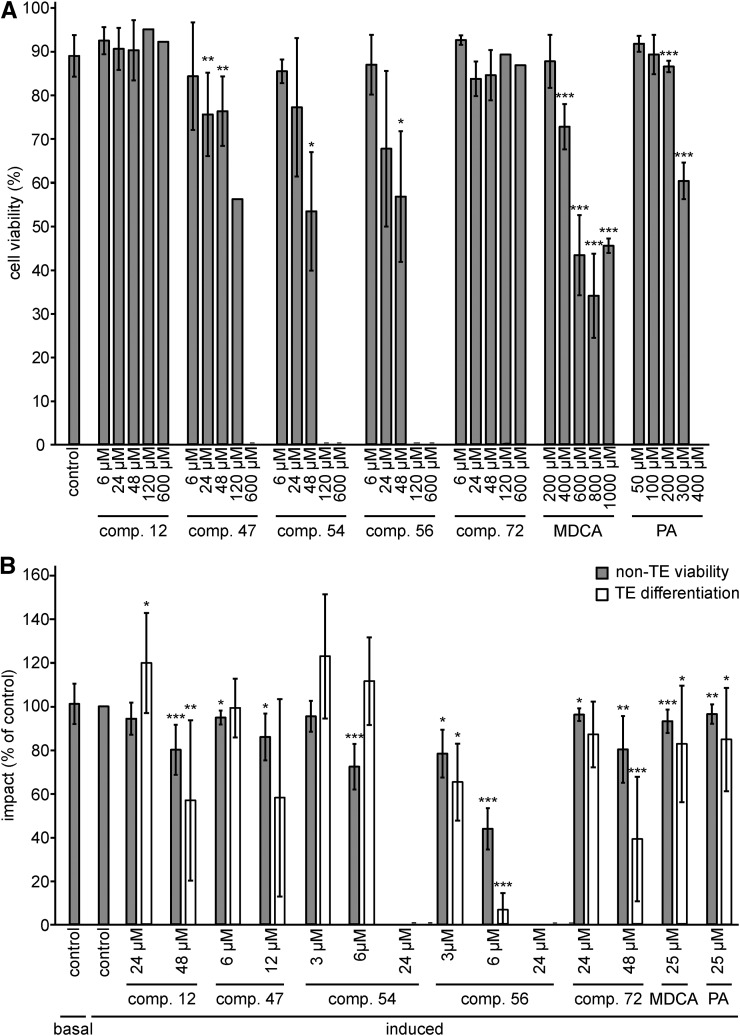
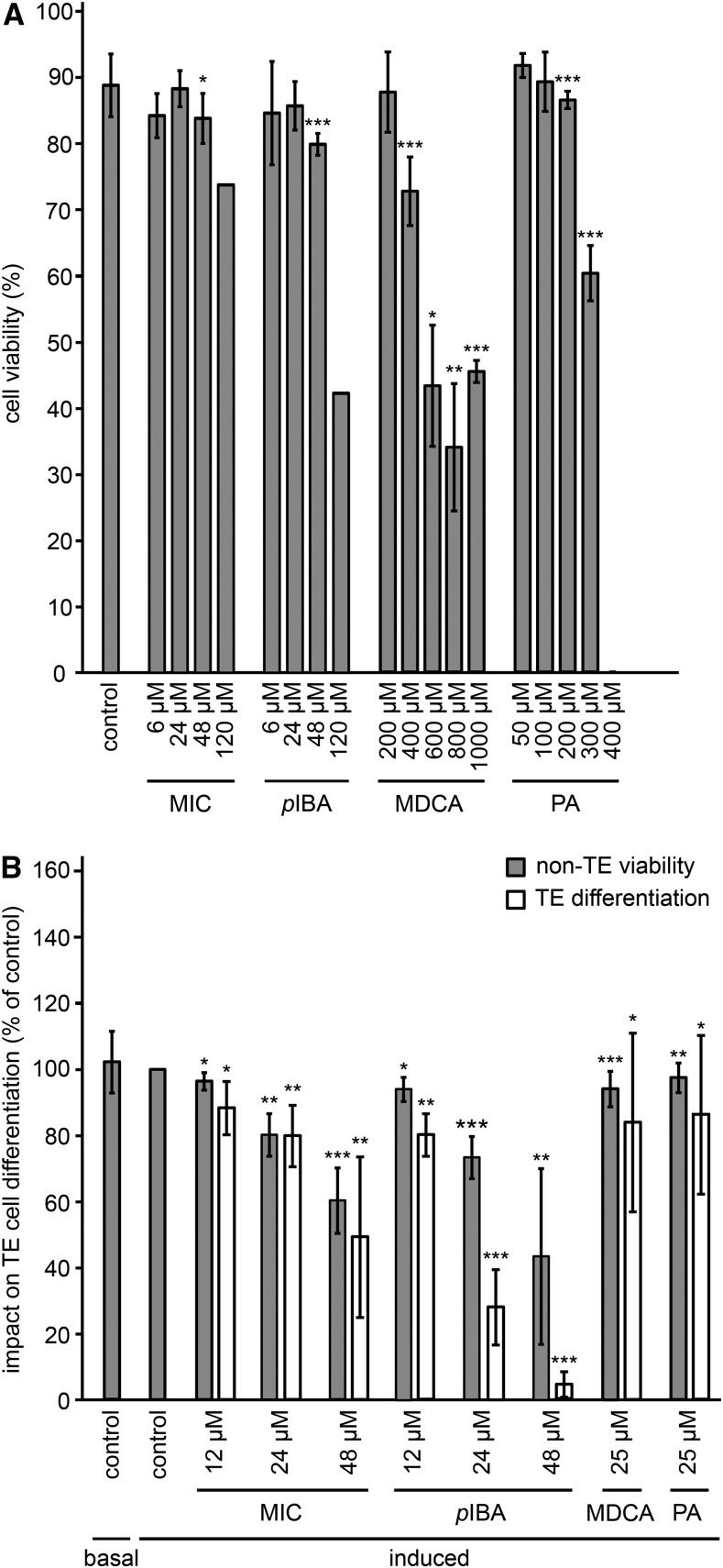
First, the cytotoxic concentration was defined for each compound by determining the cell mortality in a cell culture after a 24-h treatment (Fig. 7A; Supplemental Fig. S6). Compound 12 exhibited no toxicity when used in a concentration range from 6 to 600 µm, which was in line with the absence of toxic effects when applied to Arabidopsis seedlings. The effect of the other compounds on the cell cultures was different from that observed on root growth. For example, in contrast to the deleterious effect of compound 47 on Arabidopsis seedlings at nanomolar concentrations, this compound was less toxic in cell cultures, with a mild reduction in cell viability observed only at 24 µm or higher. Similarly, none of the concentrations tested for compound 72 affected cell viability (up to 600 µm), whereas in seedlings, a mild toxic effect was observed at concentrations above 10 µm. In conclusion, the highest nontoxic concentration of each compound was set to 6 µm for 47 and 24 µm for both 54 and 56. For the nontoxic compounds (i.e. compounds 12 and 72), the highest concentration was set at 48 µm.
Figure 7.
Impact of the five representative compounds on cell viability and TE differentiation. A, Impact of compounds 12, 47, 54, 56, and 72 on the cell viability of an Arabidopsis (Col-0) pluripotent cell suspension culture after 24 h of treatment. Error bars indicate sd from 18 independent biological replicates for the control and three to four independent replicates for each compound (200–400 cells counted per replicate sample), except for the concentrations at 120 and 600 μm, which represented only one replicate. B, Proportion of viable parenchyma cells not differentiated into TEs (non-TE viability) and changes in TE differentiation efficiency (TE differentiation) after long-term (9-d) treatment with nontoxic concentrations of each representative compound. Results are expressed as percentages of the 2% DMSO-treated controls. Error bars indicate sd of three to 24 independent biological replicates (200–400 cells counted per replicate sample). Statistical differences were estimated using Student’s t test compared with the respective controls. Differences are significant with P values below 0.05 (*), 0.01 (**), and 0.001 (***).
Next, the impact of each compound on TE cell differentiation and the viability of the surrounding, nondifferentiated cells (non-TE viability) was evaluated (Pesquet et al., 2010) after a long-term (9-d) incubation period (Fig. 7B; Supplemental Fig. S6). In addition to the above-defined concentrations, one or two additional subtoxic concentrations were evaluated to extend the concentration range at which the compounds were tested. Overall, the non-TE viability was reduced compared with that in mock-treated samples for all conditions tested, except for the treatments with compounds 12 and 54 used at concentrations of 24 and 3 µm, respectively. However, the observed reduction was, in general, mild and not exceeding 30% (Fig. 7B). Based on these observations, the following concentrations were chosen for further experiments: 48, 12, 6, 3, and 48 µm for compounds 12, 47, 54, 56, and 72, respectively. At these concentrations, compounds 12, 56, and 72 reduced TE differentiation by 42%, 34%, and 61%, respectively, whereas for compounds 47 and 54, no significant change was observed compared with mock-treated samples. Treatments with nontoxic concentrations of MDCA or PA in the cell viability assay (i.e. 25 µm) reduced surrounding cell viability by 6% and 3%, respectively, and TE differentiation efficiency by 17% and 14%, respectively (Fig. 7B).
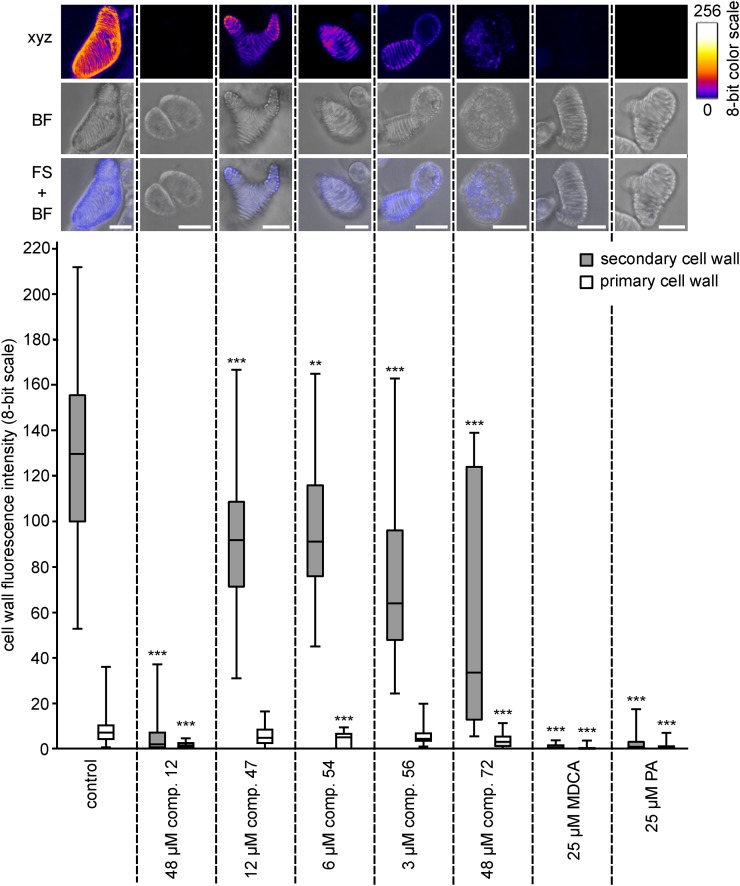
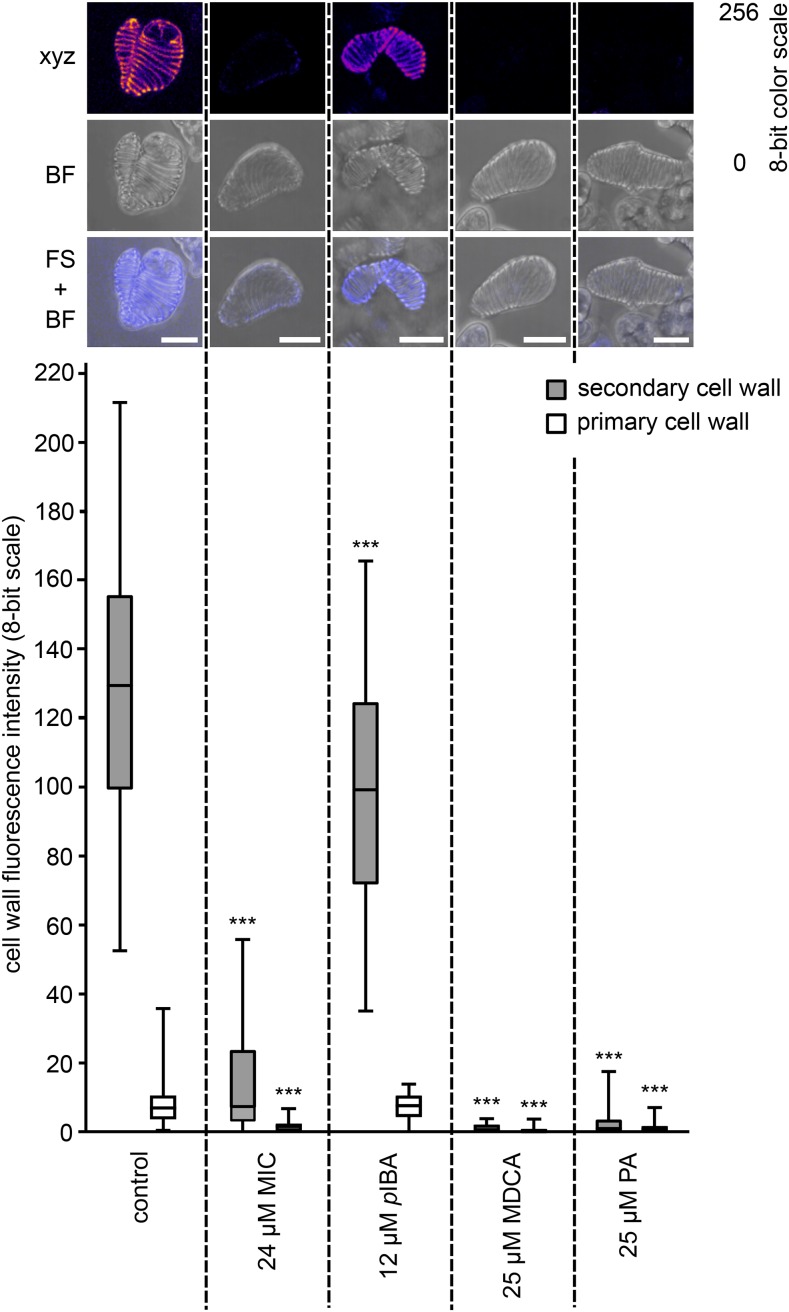
The impact of each compound on TE lignification was quantified using UV confocal microspectroscopy, which allows one to measure lignin autofluorescence in the TE cell walls (Decou et al., 2016). Autofluorescence was restricted to the lignin-rich TE secondary cell walls (SCWs), leaving the primary cell walls (PCWs) as a background that could be used as a reference for each isolated cell in each independent replicate (Fig. 8). Quantifying autofluorescence confirmed that the lignin fluorescence intensity (represented on an eight-bit scale) was conserved between independent replicates (Supplemental Fig. S7). Treatment with the positive controls (i.e. MDCA and PA) completely abolished lignin fluorescence in TE SCWs (Fig. 8). Compared with the mock-treated control, treatments with nontoxic concentrations of compounds 12, 47, 54, 56, and 72 resulted in 98%, 29%, 30%, 51%, and 74% reductions of lignin autofluorescence, respectively (Fig. 8; Supplemental Fig. S7). This confirmed the inhibitory activity of the identified compounds on lignification. Of the different compounds and concentrations tested, compound 12 used at 48 µm gave the largest reduction in lignin deposition, although not to the levels achieved by MDCA or PA. The superiority of compound 12 as an inhibitor of lignification is in line with the results obtained in the Casparian strip assay.
Figure 8.
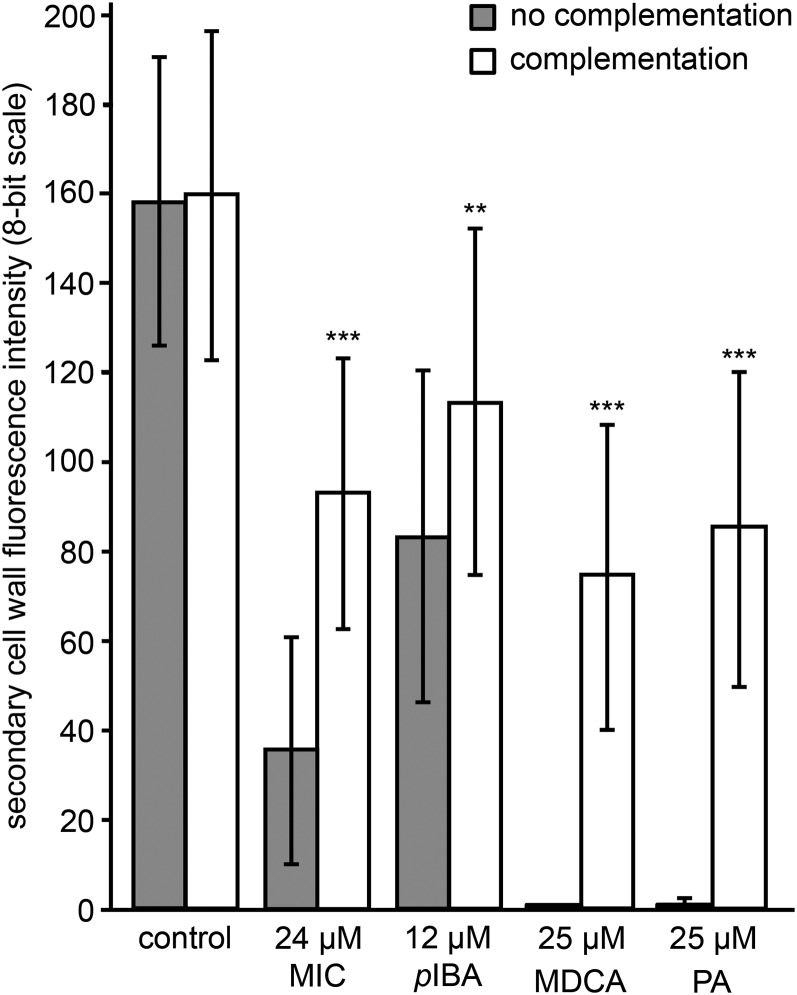
Effects of the five selected compounds on TE lignification. UV confocal microspectroscopy was used to observe TE lignin autofluorescence. Representative TEs are presented for each treatment, with a maximal projection of confocal xyz stacks in an eight-bit artificial color scale (xyz), bright-field transmission images (BF), as well as merged images between the fluorescence signal and the bright-field projection (FS + BF). Bars = 25 µm. Below the images, box plots show the range of lignin fluorescence intensity (eight-bit scale) in TE SCWs (gray bars) compared with TE PCWs (white bars) in response to nontoxic concentrations of each representative compound compared with the control treated with DMSO. Per condition, three to 12 independent biological replicates were analyzed (five to 10 individual TEs analyzed per treatment per replicate). Statistics were performed via Student’s t test, comparing the autofluorescence of the SCWs and PCWs of each treatment with the autofluorescence of the SCWs and PCWs of the control, respectively. Differences are significant with P values below 0.01 (**) and 0.001 (***).
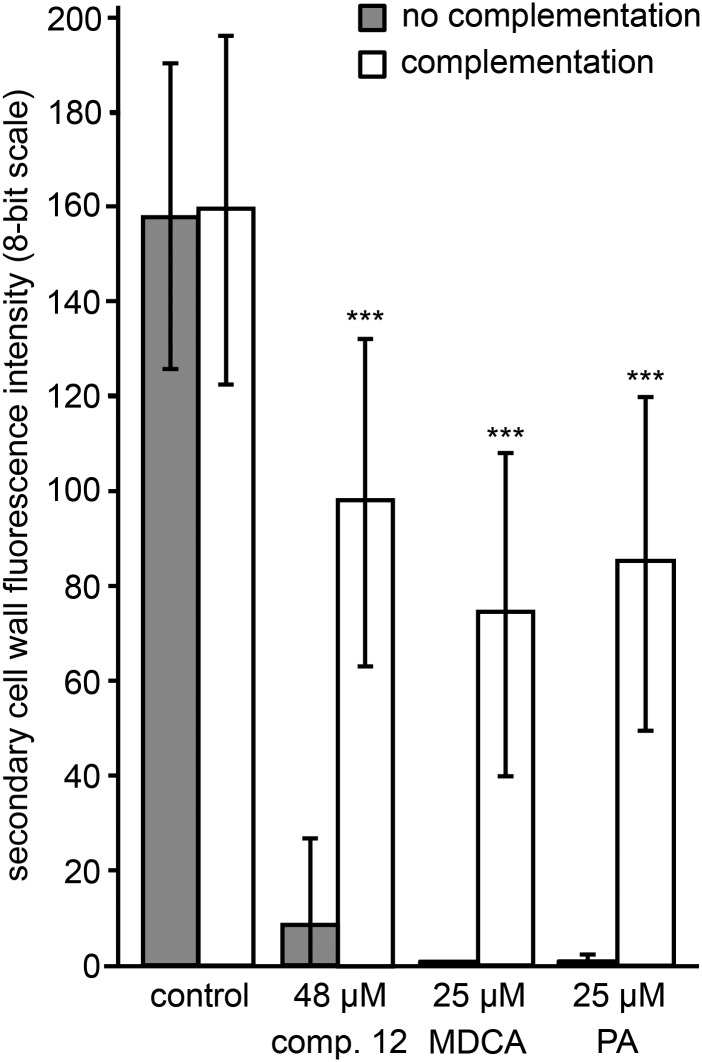
To further confirm the inhibitory effect of compound 12 on lignin biosynthesis in TE-induced cell cultures, the potential for postmortem lignification was verified by adding 120 µm coniferyl alcohol as a lignin monomer to 9-d-old, compound 12-treated, dead and hypolignified cells. After a 120-h incubation period, UV confocal microspectroscopy was used to measure the intensity of SCW lignin autofluorescence (Fig. 9). This treatment led to a more than 50% recovery of lignification in dead, hypolignified TE SCWs, which is similar to the recovery observed for the MDCA- and PA-treated samples used as positive controls. In line with the results obtained with the Casparian strip assay, this observation further supports the idea that compound 12 affects lignin monomer biosynthesis rather than lignin polymerization.
Figure 9.
Postmortem lignification of TEs inhibited by compound 12. UV confocal microspectroscopy observations are shown for the SCW lignin autofluorescence intensity of 14-d-old TE-induced cell cultures and treated with different compounds and either complemented at 9 d with coniferyl alcohol (white bars) or not (gray bars). Error bars indicate sd of three independent biological replicates (five to 10 individual TEs analyzed per treatment per replicate). Statistical differences were estimated using Student’s t test compared with the respective controls. Only the significances between no complementation and complementation of a particular treatment are shown and are significant with P values below 0.001 (***).
Phenolic Profiling Suggests in Planta Processing of Compound 12
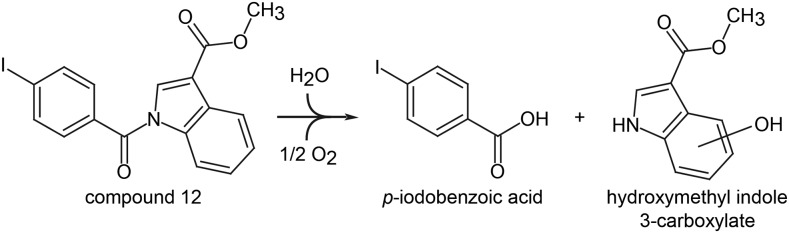
Compound 12 scored positive in both bioassays for lignin inhibition and, for this reason, was selected for a more detailed analysis. To obtain further insight into its mode of action, we tested its effect on actively lignifying tissue. To this end, 15- to 20-cm-tall Arabidopsis stems were collected from soil-grown plants and treated with 100 µm compound 12 diluted in liquid Murashige and Skoog medium. MDCA and PA were included as positive controls, and DMSO was used as a negative control. After a 48-h incubation period, stems were snap frozen in liquid nitrogen and ground to powder. The metabolites were extracted by methanol, and phenolic profiling was performed using UHPLC-MS, resulting in the detection of 4,695 peaks that were further filtered based on their presence in all replicates of at least one treatment and that had an average peak area higher than 500 in at least one treatment (Supplemental Table S3). Next, we selected peaks for which the area differed by at least 10-fold between the treatments. This resulted in a list of 30 peaks, of which the 10 largest peaks were selected for identification. Because three peaks were identified as in-source fragments of one of the metabolites in this top-10 list, the analysis resulted in the characterization of seven metabolites (Table I). Six of the seven highly accumulating metabolites detected in the extracts of plants treated with compound 12 were not found in MDCA- or PA-treated controls or in the mock-treated controls (Supplemental Table S3). Interestingly, their m/z values corresponded to the monoisotopic masses of compounds that could theoretically be obtained by the hydrolysis of compound 12, hydroxy-methylindole carboxylic acid (hydroxy-MIC; m/z 190.05) and pIBA (m/z 246.92) and derivatives thereof, suggesting that compound 12 is processed in the plant (Fig. 10). The only peak in the list that could not be linked to the processing of compound 12 was cinnamoyl hexose (metabolite 7). The accumulation of this metabolite might point toward the inhibition of an enzymatic step at the beginning of the phenylpropanoid pathway.
Table I. Phenolic profiling of stems treated with compound 12.
The top-10 highest peaks of compounds of which the abundance differed by at least 10-fold in compound 12-treated plants compared with mock-treated plants were selected for identification and were derived from seven metabolites. Three were hydroxymethyl indole 3-carboxylate (hydroxy-MIC) derivatives, three others were pIBA derivatives, and the last was a cinnamoyl hexose derivative. For detailed information about the listed metabolites, see Supplemental Table S3 and Supplemental Figure S9. Peak areas are given as average ± sd. RT, retention time.
| Metabolite Name | Metabolite No. | m/z | RT (min) | Peak Area, DMSO | Peak Area, Compound 12 |
|---|---|---|---|---|---|
| Hydroxy-MIC | 1 | 190.05 | 13.84 | 5 ± 9 | 4,186 ± 1,136 |
| Hydroxy-MIC hexose | 2 | 352.10 | 9.23 | 0 ± 0 | 8,036 ± 2,358 |
| Hydroxy-MIC acetyl hexose | 3 | 394.11 | 11.48 | 0 ± 0 | 7,680 ± 4,691 |
| pIBA + malate | 4 | 362.93 | 16.61 | 0 ± 0 | 8,217 ± 5,393 |
| pIBA + malate (-dehydrated malate) | 4 | 246.92 | 16.61 | 0 ± 0 | 42,225 ± 25,722 |
| pIBA + malate (-dehydrated malate -CO2) | 4 | 202.93 | 16.61 | 0 ± 0 | 4,758 ± 2,947 |
| pIBA + hexose (-dehydrated hexose) | 5 | 246.92 | 12.29 | 22 ± 10 | 9,751 ± 2,252 |
| pIBA + hexose + 224 D | 6 | 633.05 | 19.09 | 0 ± 0 | 23,202 ± 6,369 |
| pIBA + hexose + 224 D | 6 | 246.92 | 19.05 | 0 ± 0 | 8,315 ± 2,229 |
| Cinnamoyl hexose + 224 D | 7 | 533.17 | 17.45 | 74 ± 106 | 7,260 ± 3,450 |
Figure 10.
Presumed processing of compound 12 in Arabidopsis plants. The amide bond in compound 12 is probably hydrolyzed in planta, resulting in the formation of two fragments: pIBA and hydroxy-MIC. The exact position of the hydroxyl as well as the moment of hydroxylation (prehydrolysis or posthydrolysis) is so far unknown.
Both Methylindole Carboxylic Acid and pIBA Inhibit Lignification
Because compound 12 is processed in the plant, the question arose whether the compound itself or one of its fragments (pIBA or hydroxy-MIC) or a corresponding derivative inhibited in planta lignification. For one of the fragments, hydroxy-MIC, the exact structure could not be identified, because the hydroxyl group could not be unequivocally positioned on the benzene ring based on the MS data. Because none of the possible isomers was commercially available, its nonhydroxylated derivative methylindole carboxylic acid (MIC) was used. Both MIC and pIBA were tested for their putative inhibitory activity in the Casparian strip assay (Fig. 11). Initially, they were tested at the same concentration (100 µm) as for compound 12, but because 100 µm pIBA was toxic, the concentration was lowered to 10 µm pIBA. The effect on Casparian strip formation was determined by counting endodermal cells from the meristem to the point where PI was excluded from the stele, as described before. The cell count increased significantly by 116% and 262% upon treatment with 100 µm MIC and 10 µm pIBA, respectively. In both cases, the inhibition was rescued by the addition of coniferyl and sinapyl alcohols (Fig. 11).
Figure 11.
Casparian strip assay with MIC and pIBA. Five-day-old Arabidopsis seedlings were subjected to 100 µm MIC or 10 µm pIBA either noncomplemented (gray bars) or complemented (white bars) with the monolignols coniferyl and sinapyl alcohol (100 µm). MDCA (10 µm) was used as a positive control, whereas mock-treated plants served as negative controls. Endodermal cells surrounded by PI staining were counted until Casparian strip formation prevented PI from diffusing beyond the apoplast. Statistical differences were estimated using Student’s t test compared with the respective controls. Error bars indicate sd of 10 seedlings per condition and are significant with P values below 0.01 (**) and 0.001 (***).
In parallel, the impacts of both compounds on plant cell viability, TE differentiation efficiency, and TE SCW lignin deposition were tested as described above. Although compound 12 showed no toxic effect at the concentrations tested (up to 600 µm) after a 24-h incubation, both MIC and pIBA turned out to be more toxic at 120 µm, leading to 17% and 52% dead cells compared with the control, respectively (Fig. 12A). The highest nontoxic concentrations tested were 24 µm for both MIC and pIBA. However, at this concentration, both compounds still exhibited significant long-term toxicity, reducing cell viability in the 9-d incubation experiment used to study TE differentiation (Fig. 12B). Nevertheless, 24 µm MIC and 12 µm pIBA were used to determine the TE lignification level: in these conditions, the non-TE viability was reduced by less than 30% and TE differentiation efficiency was 79% and 80%, respectively, compared with DMSO-treated controls (Fig. 12B). Although pIBA clearly outperformed both compound 12 and MIC in the Casparian strip assay, MIC abolished TE SCW lignin autofluorescence by 94%, whereas pIBA could only reduce autofluorescence by 23% at the tested concentration (Fig. 13; Supplemental Movie S1). Similar to the postmortem restoration of lignification in compound 12-treated cells, lignification was restored in the SCW of MIC-treated (58%) and pIBA-treated (71%) dead TEs complemented with 120 µm coniferyl alcohol (Fig. 14).
Figure 12.
Impact of MIC and pIBA fragments on cell viability and TE differentiation. A, Impact of MIC and pIBA on the cell viability of Arabidopsis (Col-0) pluripotent cell suspension cultures after 24 h of treatment. Error bars indicate sd from 15 independent biological replicates for the control and three to four independent biological replicates for each compound (200–400 cells counted per replicate sample), except at the concentration of 120 µm, which represented only one replicate. B, Proportion of viable parenchyma cells not differentiated into TEs (non-TE viability) and changes in TE differentiation efficiency (TE differentiation) after long-term (9-d) treatment in response to 12, 24, and 48 µm of each compound. Results are expressed as percentages of the 2% DMSO-treated controls. Error bars represent sd of three to 22 independent biological replicates (200–400 cells counted per replicate sample). Statistical differences were estimated using Student’s t test compared with the respective controls. Differences are significant with P values below 0.05 (*), 0.01 (**), and 0.001 (***).
Figure 13.
Effects of MIC and pIBA on TE lignification. UV confocal microspectroscopy was used to observe TE lignin autofluorescence. Representative TEs are presented for each treatment, with a maximal projection of confocal xyz stacks in an eight-bit artificial color scale (xyz), bright-field transmission images (BF), as well as merged images between the fluorescence signal and the bright-field projection (FS + BF). Bars = 25 µm. Below the images, box plots show the range of lignin fluorescence intensity (eight-bit scale) in TE SCWs (gray bars) compared with TE PCWs (white bars) in response to nontoxic concentrations of MIC and pIBA compared with the control treated with DMSO. MDCA and PA were included as positive controls. Per condition, three to 12 independent biological replicates were analyzed (five to 10 individual TEs analyzed per treatment per replicate). Statistics were performed via Student’s t test, comparing the autofluorescence of the SCWs and PCWs of each treatment with that of the SCWs and PCWs of the control, respectively. Differences are significant with P values below 0.001 (***).
Figure 14.
Postmortem lignification of TEs inhibited by MIC and pIBA. UV confocal microspectroscopy observations are shown for the SCW lignin autofluorescence intensity on 14-d-old TE-induced cell cultures treated with different compounds and either supplemented at 9 d with coniferyl alcohol (white bars) or not (gray bars). Error bars indicate sd of three independent biological replicates (five to 10 individual TEs analyzed per treatment per replicate). Statistical differences were estimated using Student’s t test compared with the respective controls. Only the significances between no complementation and complementation of a particular treatment are shown and are significant with P values below 0.01 (**) and 0.001 (***).
C4H Is the Target of pIBA
To obtain further insight into the mode of action of MIC and pIBA, stems were treated with both compounds using a similar approach to that described above. A total of 4,793 compounds were detected over the different samples, and subsequent analysis was performed on peaks present in all replicates of at least one treatment and with an average peak intensity higher than 500 in at least one of the treatments (Supplemental Table S4). We further selected those peaks for which the area was significantly different in MIC- and pIBA-treated samples compared with that in the DMSO-treated control samples, using a 10-fold change as threshold. This resulted in lists of 64 peaks with increased peak areas and 101 peaks with reduced peak areas upon MIC treatment. The 20 largest peaks in both lists were selected for further characterization (Table II). Based on the characterized metabolites, we were not able to pinpoint a specific target for MIC, because most accumulating metabolites were MIC itself (metabolite 8) or hydroxylated derivatives of MIC (metabolites 1, 2, 3, 9, 10, 11, 12, 13, 14, 15, 16, and 17). Notably, metabolites 1, 2, and 3 also were detected in stems treated with compound 12. Metabolites that were decreased in abundance upon treatment with MIC were hexosylated phenolic dimers made by oxidative coupling of p-coumaryl alcohol with another unit of p-coumaryl alcohol, coniferyl alcohol, ferulic acid, or p-coumaric acid (metabolites 20, 21, 22, and 23, respectively), two hydroxyphenyl (H)- and guaiacyl (G)-unit-containing trilignols (metabolites 24 and 25), and two G- and syringyl (S)-unit-containing trilignols (metabolites 26 and 27). A targeted approach showed that not only trilignols 24, 25, 26, and 27 were reduced in abundance in MIC-treated plants compared with mock controls, but that all oligolignols detected in the stems showed reduced abundance (Supplemental Table S5).
Table II. Phenolic profiling of stems treated with MIC.
The top-20 highest peaks of both accumulating and decreasing compounds in MIC-treated plants compared with mock-treated plants were selected for identification and were derived from 35 metabolites. For detailed information about the listed metabolites, see Supplemental Table S4 and Supplemental Figure S9. *, No tandem MS spectrum available. Peak areas are given as average ± sd. RT, Retention time.
| Metabolite Name | Metabolite No. | m/z | RT (min) | Peak Area, DMSO | Peak Area, MIC | Peak Area, MDCA | Peak Area, PA | Up/Down |
|---|---|---|---|---|---|---|---|---|
| MIC (confirmed with standard) | 8 | 174.05 | 16.14 | 0 ± 0 | 26,000 ± 7,466 | 0 ± 0 | 1 ± 3 | Up |
| Hydroxy-MIC | 1 | 190.05 | 13.98 | 25 ± 21 | 28,272 ± 4,716 | 761 ± 348 | 14 ± 14 | Up |
| Hydroxy-MIC + hexose 1 | 2 | 352.10 | 9.34 | 0 ± 0 | 96,066 ± 12,137 | 15,874 ± 4,458 | 6 ± 16 | Up |
| Hydroxy-MIC + hexose 1 (-dehydrated hexose) | 2 | 190.05 | 9.34 | 0 ± 1 | 7,574 ± 1,134 | 1 ± 2 | 0 ± 0 | Up |
| Hydroxy-MIC + hexose 2 | 11 | 352.10 | 6.45 | 3 ± 5 | 78,902 ± 10,218 | 3 ± 6 | 24 ± 62 | Up |
| Hydroxy-MIC + hexose 2 (dimer) | 11 | 705.21 | 6.44 | 0 ± 0 | 20,355 ± 3,045 | 0 ± 0 | 0 ± 0 | Up |
| Hydroxy-MIC + hexose 3 (formic acid adduct) | 12 | 398.11 | 12.06 | 0 ± 0 | 3,896 ± 1,085 | 0 ± 0 | 0 ± 0 | Up |
| Hydroxy-MIC + acetyl hexose 1 | 3 | 394.11 | 11.60 | 0 ± 0 | 55,801 ± 6,711 | 64 ± 73 | 2 ± 5 | Up |
| Hydroxy-MIC + acetyl hexose 2 | 13 | 394.11 | 9.26 | 0 ± 0 | 4,199 ± 954 | 0 ± 0 | 0 ± 0 | Up |
| Hydroxy-MIC + acetyl hexose 3 | 14 | 394.11 | 11.33 | 0 ± 0 | 4,865 ± 601 | 56 ± 95 | 0 ± 0 | Up |
| Hydroxy-MIC + 687 D | 9 | 877.22 | 13.97 | 0 ± 0 | 30,781 ± 9,647 | 427 ± 1,208 | 0 ± 0 | Up |
| Hydroxy-MIC + 687 D | 10 | 877.22 | 11.60 | 0 ± 0 | 12,437 ± 1,581 | 417 ± 684 | 0 ± 0 | Up |
| Hydroxy-MIC (-CH3OH) | 1 | 158.02 | 13.97 | 0 ± 0 | 11,542 ± 2,100 | 48 ± 134 | 1 ± 3 | Up |
| Hydroxy-MIC [G(8-5)feruloyl malate adduct] | 1 | 926.24 | 14.00 | 0 ± 0 | 12,716 ± 1,496 | 0 ± 0 | 0 ± 0 | Up |
| Dihydroxy-MIC + hexose | 15 | 368.10 | 9.06 | 1 ± 3 | 20,790 ± 8,160 | 28 ± 20 | 2,164 ± 1,733 | Up |
| Dihydroxy-MIC + acetyl hexose | 16 | 410.11 | 10.85 | 0 ± 0 | 9,430 ± 4,040 | 0 ± 0 | 0 ± 0 | Up |
| Hexosylated hydroxy-MIC (-dehydrated hexose) | 11 | 190.05 | 6.45 | 2 ± 5 | 12,428 ± 1,690 | 34 ± 70 | 11 ± 11 | Up |
| Methoxy-hydroxy-MIC + hexose | 17 | 382.11 | 7.22 | 3 ± 9 | 4,414 ± 739 | 5 ± 12 | 0 ± 0 | Up |
| Unknown 1 | 18 | 487.10 | 4.20 | 0 ± 0 | 7,182 ± 1,862 | 0 ± 0 | 165 ± 265 | Up |
| Unknown 2 | 19 | 673.17 | 4.90 | 0 ± 0 | 4,911 ± 2,373 | 0 ± 0 | 0 ± 0 | Up |
| H(8-8)H 4-O-hexoside | 20 | 459.17 | 9.24 | 9,801 ± 3,211 | 621 ± 350 | 499 ± 965 | 40 ± 88 | Down |
| H/G(8-5)G/H hexoside (formic acid adduct) | 21 | 535.18 | 8.75 | 6,649 ± 1,860 | 223 ± 173 | 1,674 ± 919 | 256 ± 378 | Down |
| H(8-O-4)G(8-5)H | 24 | 493.19 | 14.45 | 6,004 ± 1,129 | 5 ± 11 | 572 ± 556 | 35 ± 37 | Down |
| H(8-O-4)H/G(8-8)G/H | 25 | 493.19 | 14.61 | 4,277 ± 956 | 8 ± 22 | 606 ± 465 | 27 ± 44 | Down |
| G(8-O-4)S(8-5)G | 26 | 583.22 | 14.95 | 8,258 ± 3,523 | 670 ± 663 | 3,310 ± 2,832 | 1,128 ± 905 | Down |
| G(8-O-4)G/S(8-8)S/G | 27 | 583.22 | 17.16 | 4,411 ± 2,209 | 0 ± 0 | 1,575 ± 1,327 | 191 ± 164 | Down |
| H 4/9-O-hexoside (8-5)p-coumaric acid | 23 | 473.15 | 11.06 | 3,211 ± 1,769 | 81 ± 131 | 182 ± 202 | 10 ± 16 | Down |
| H(8-O-4)ferulic acid + hexose (-dehydrated hexose) | 22 | 359.11 | 6.26 | 3,134 ± 1,688 | 55 ± 73 | 781 ± 609 | 238 ± 268 | Down |
| Unknown 3 | 28 | 391.05 | 12.76 | 8,034 ± 3,604 | 776 ± 684 | 558 ± 304 | 153 ± 264 | Down |
| Unknown 4 | 29 | 517.14 | 5.51 | 6,951 ± 2,477 | 16 ± 31 | 1,367 ± 1,494 | 43 ± 47 | Down |
| Unknown 5 | 30 | 695.20 | 9.83 | 5,710 ± 2,416 | 198 ± 127 | 875 ± 830 | 54 ± 72 | Down |
| Unknown 6 | 31 | 517.13 | 5.16 | 4,326 ± 1,769 | 3 ± 5 | 864 ± 768 | 45 ± 37 | Down |
| Unknown 7 | 32 | 829.26 | 6.26 | 3,457 ± 2,017 | 105 ± 271 | 1,280 ± 686 | 185 ± 190 | Down |
| Unknown 8 | 33 | 379.09 | 12.53 | 3,344 ± 1,301 | 10 ± 24 | 314 ± 548 | 0 ± 0 | Down |
| Unknown 9 | 34 | 499.13 | 8.54 | 3,340 ± 1,120 | 10 ± 17 | 732 ± 355 | 90 ± 84 | Down |
| Unknown 10 | 35 | 267.10 | 13.45 | 3,117 ± 1,143 | 157 ± 199 | 425 ± 350 | 163 ± 229 | Down |
| Unknown 11* | 36 | 789.18 | 3.93 | 9,499 ± 2,625 | 698 ± 1,016 | 13,711 ± 5,399 | 5,609 ± 4,596 | Down |
| Unknown 12* | 37 | 799.24 | 9.22 | 6,400 ± 2,120 | 410 ± 223 | 447 ± 672 | 64 ± 119 | Down |
| Unknown 13* | 38 | 969.23 | 5.79 | 4,224 ± 2,521 | 44 ± 90 | 312 ± 427 | 19 ± 40 | Down |
| Unknown 14* | 39 | 359.11 | 9.40 | 3,258 ± 877 | 322 ± 287 | 919 ± 1131 | 533 ± 399 | Down |
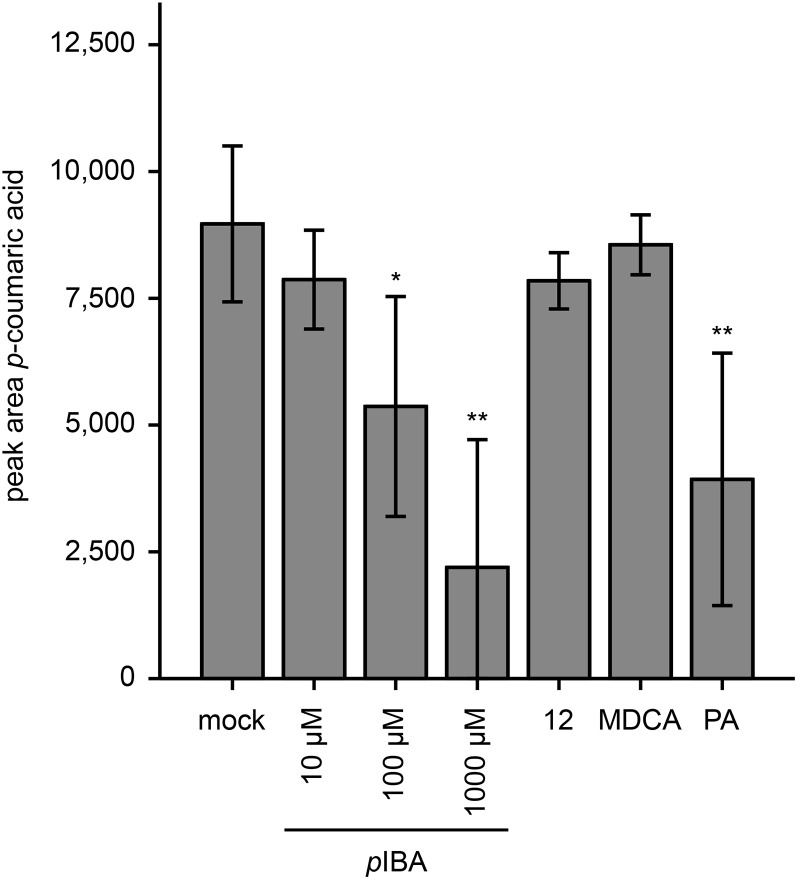
For pIBA, a similar approach was followed: the top 20 of the 174 accumulating and 95 decreasing peaks upon pIBA treatment were selected for further characterization (Table III). In line with its suggested role as an inhibitor of lignification, several hexosylated dimers of p-coumaryl alcohol with another unit of p-coumaryl alcohol or p-coumaric acid (metabolites 20, 23, 49, 50, 51, 52, 53, and 54) were reduced in abundance, as well as two H- and G-unit-containing trilignols (metabolites 24 and 25) and one hexosylated trilignol with G and S units (metabolite 55). As for MIC, a reduction in the abundance of additional oligolignols was found via a targeted search in pIBA-treated seedlings (Supplemental Table S5). The highest accumulating compounds were pIBA (metabolite 40), pIBA derivatives (metabolites 4, 5, 6, 41, 42, and 43), and compounds derived from cinnamic acid (metabolites 7, 44, 45, 46, and 47), which is the substrate of C4H, suggesting an inhibition at the level of C4H. Intriguingly, p-coumaroyl hexose (metabolite 48), derived from the C4H product p-coumaric acid, also accumulated. However, this was also the case in plants treated with the C4H inhibitor PA and might be explained by the fact that C4H also plays a role in the conversion of p-coumaric acid to caffeic acid when in complex with C3H (Chen et al., 2011), perhaps only in certain cell types. Nonetheless, the level of several other p-coumaric acid-containing metabolites was reduced in pIBA-treated plants (metabolites 23, 50, 51, 52, 53, and 54), indicating a classical C4H inhibition.
Table III. Phenolic profiling of stems treated with pIBA.
The top-20 highest peaks of both accumulating and decreasing compounds in pIBA-treated plants compared with mock-treated plants were selected for identification and were derived from 31 metabolites. For detailed information about the listed metabolites, see Supplemental Table S4 and Supplemental Figure S9. *, No tandem MS spectrum available. Peak areas are given as average ± sd. RT, retention time.
| Metabolite Name | Metabolite No. | m/z | RT (min) | Peak Area, DMSO | Peak Area, pIBA | Peak Area, MDCA | Peak Area, PA | Up/Down |
|---|---|---|---|---|---|---|---|---|
| pIBA (confirmed by standard) | 40 | 246.93 | 19.30 | 5 ± 7 | 56,081 ± 16,654 | 4 ± 10 | 7 ± 9 | Up |
| pIBA + hexose | 5 | 408.98 | 12.42 | 0 ± 0 | 61,727 ± 9,841 | 0 ± 0 | 0 ± 0 | Up |
| pIBA + hexose (formic acid adduct) | 5 | 454.98 | 12.43 | 0 ± 0 | 65,818 ± 12,662 | 0 ± 0 | 0 ± 0 | Up |
| pIBA + hexose (-dehydrated hexose) | 5 | 246.93 | 12.43 | 98 ± 59 | 146,491 ± 2,0301 | 158 ± 169 | 62 ± 48 | Up |
| pIBA + hexose + 224 D | 6 | 633.05 | 19.25 | 0 ± 0 | 78,911 ± 18,448 | 13 ± 25 | 500 ± 899 | Up |
| pIBA + malate | 4 | 362.94 | 16.85 | 0 ± 0 | 287,442 ± 4,6250 | 5 ± 10 | 0 ± 0 | Up |
| pIBA + malate (iodine ion) | 4 | 126.91 | 16.85 | 1 ± 4 | 30,709 ± 4,462 | 0 ± 1 | 0 ± 1 | Up |
| pIBA + malate (-pIBA) | 4 | 115.00 | 16.84 | 14 ± 6 | 12,591 ± 2,088 | 29 ± 13 | 21 ± 10 | Up |
| pIBA + malate (-dehydrated malate) | 4 | 246.93 | 16.84 | 37 ± 105 | 803,651 ± 130,376 | 133 ± 190 | 29 ± 51 | Up |
| pIBA + malate (-dehydrated malate -CO2) | 4 | 202.94 | 16.85 | 0 ± 0 | 61,073 ± 8,733 | 0 ± 0 | 0 ± 1 | Up |
| pIBA + Asp | 41 | 361.95 | 12.64 | 0 ± 0 | 18,848 ± 10,907 | 0 ± 0 | 11 ± 22 | Up |
| pIBA + 248 D (-248 D) | 42 | 246.93 | 15.45 | 0 ± 0 | 16,460 ± 5,254 | 0 ± 0 | 0 ± 0 | Up |
| pIBA + 248 D (dimer) | 42 | 990.97 | 15.45 | 0 ± 0 | 19,200 ± 11,369 | 0 ± 0 | 0 ± 0 | Up |
| Vaniloylhexose + pIBA + hexose (formic acid adduct) | 43 | 767.07 | 13.59 | 0 ± 0 | 21,262 ± 7,872 | 0 ± 0 | 0 ± 0 | Up |
| Cinnamoyl hexose (formic acid adduct) | 44 | 355.10 | 9.81 | 291 ± 174 | 17,461 ± 6,158 | 29,580 ± 14,804 | 99,938 ± 17,296 | Up |
| Cinnamoyl hexose + 224 D | 7 | 533.17 | 17.59 | 133 ± 196 | 18,345 ± 6,831 | 19,586 ± 7,221 | 29,632 ± 14,439 | Up |
| Cinnamoyl malate (-dehydrated malate) | 47 | 147.04 | 14.00 | 16 ± 18 | 34,977 ± 5,266 | 53,877 ± 33,222 | 174,560 ± 53,114 | Up |
| Cinnamoyl Asp | 45 | 262.07 | 10.17 | 17 ± 37 | 16,206 ± 5,358 | 29,826 ± 30,603 | 76,685 ± 18,056 | Up |
| Cinnamoyl Glu | 46 | 276.09 | 10.93 | 0 ± 0 | 12,497 ± 4,314 | 22,740 ± 14,510 | 184,756 ± 52,274 | Up |
| p-Coumaroyl hexose | 48 | 325.09 | 4.65 | 572 ± 462 | 16,238 ± 2,088 | 12,335 ± 6,987 | 22,265 ± 14,124 | Up |
| H(8-8)H 4-O-hexoside | 20 | 459.17 | 9.24 | 9,801 ± 3,211 | 37 ± 43 | 499 ± 965 | 40 ± 88 | Down |
| H 4-O-hexoside(8-5)H (formic acid adduct) | 49 | 505.17 | 9.37 | 31,717 ± 8,101 | 2,769 ± 1,220 | 3,821 ± 2,852 | 683 ± 477 | Down |
| H 4/9-O hexoside (8-5)p-coumaric acid | 23 | 473.15 | 11.06 | 3,211 ± 1,769 | 76 ± 75 | 182 ± 202 | 10 ± 16 | Down |
| H(8-O-4)p-coumaroyl hexose + 224 D 1 | 50 | 715.23 | 11.43 | 17,677 ± 5,991 | 1,490 ± 1,271 | 1,685 ± 2,838 | 247 ± 358 | Down |
| H(8-O-4)p-coumaroyl hexose + 224 D 2 | 51 | 715.23 | 11.81 | 11,527 ± 3,882 | 956 ± 1,116 | 1,061 ± 1,839 | 79 ± 147 | Down |
| H(8-5)p-coumaroyl hexose + 224 D | 52 | 697.21 | 16.00 | 15,108 ± 6,568 | 200 ± 201 | 575 ± 1,567 | 0 ± 0 | Down |
| H(8-5)p-coumaroyl malate 1 (-dehydrated malate -CH2O) | 53 | 281.08 | 13.95 | 6,757 ± 1,199 | 396 ± 225 | 821 ± 1,193 | 1 ± 2 | Down |
| H(8-5)p-coumaroyl malate 1 (-dehydrated malate -H2O) | 53 | 293.08 | 13.96 | 8,247 ± 1,507 | 589 ± 278 | 1,083 ± 1,519 | 11 ± 20 | Down |
| H(8-5)p-coumaroyl malate 1 (-dehydrated malate) | 53 | 311.09 | 13.96 | 3,779 ± 677 | 126 ± 132 | 502 ± 874 | 0 ± 0 | Down |
| H(8-5)p-coumaroyl malate 2 | 54 | 427.10 | 14.23 | 3,682 ± 1,890 | 257 ± 333 | 851 ± 1,194 | 133 ± 146 | Down |
| H(8-O-4)G(8-5)H | 24 | 493.19 | 14.45 | 6,004 ± 1,129 | 154 ± 134 | 572 ± 556 | 35 ± 37 | Down |
| H(8-O-4)H/G(8-8)G/H | 25 | 493.19 | 14.61 | 4,277 ± 956 | 346 ± 257 | 606 ± 465 | 27 ± 44 | Down |
| G 4-O-hexoside(8-O-4)S(8-5)G (formic acid adduct) | 55 | 791.23 | 11.92 | 5,830 ± 1,622 | 256 ± 240 | 1,370 ± 730 | 13,685 ± 13,933 | Down |
| Unknown 3 | 28 | 391.05 | 12.76 | 8,034 ± 3,604 | 138 ± 158 | 558 ± 304 | 153 ± 264 | Down |
| Unknown 4 | 29 | 517.14 | 5.51 | 6,951 ± 2,477 | 454 ± 522 | 1,367 ± 1,494 | 43 ± 47 | Down |
| Unknown 6 | 31 | 517.13 | 5.16 | 4,326 ± 1,769 | 179 ± 238 | 864 ± 768 | 45 ± 37 | Down |
| Unknown 8 | 33 | 379.09 | 12.53 | 3,344 ± 1,301 | 25 ± 62 | 314 ± 548 | 0 ± 0 | Down |
| Unknown 12* | 37 | 799.24 | 9.22 | 6,400 ± 2,120 | 84 ± 108 | 447 ± 672 | 64 ± 119 | Down |
| Unknown 13* | 38 | 969.23 | 5.79 | 4,224 ± 2,521 | 196 ± 241 | 312 ± 427 | 19 ± 40 | Down |
| Unknown 15* | 56 | 745.27 | 13.51 | 3,851 ± 1,143 | 93 ± 103 | 521 ± 577 | 235 ± 274 | Down |
In conclusion, the phenolic profiles of pIBA-treated plants provided supporting evidence that this fragment of compound 12 inhibits lignin biosynthesis at the level of C4H, whereas the profiles of MIC-treated plants were inconclusive. To further support the mode of pIBA action, C4H of Arabidopsis was heterologously expressed in yeast, and purified microsomes were incubated with cinnamic acid in the presence or absence of pIBA or compound 12 (Fig. 15). Mock- and MDCA-treated (100 µm) microsomes were used as negative controls, whereas treatment with 100 µm PA served as a positive control. After treatment with the 4CL inhibitor MDCA, p-coumaric acid levels were similar to those of the mock-treated microsomes, whereas treatment with PA significantly reduced product synthesis. pIBA significantly inhibited C4H at 100 and 1,000 µm. The inhibition at 100 µm was as strong as that caused by PA at equimolar concentrations. In contrast to pIBA, compound 12 did not significantly inhibit C4H in the in vitro assay, not even at 1 mm, indicating that compound 12 by itself does not inhibit C4H and that its activity as an inhibitor is obtained only upon in planta processing.
Figure 15.
Effects of pIBA on C4H activity. Microsomal preparations from C4H-overexpressing yeast cells were incubated with cinnamic acid in the presence of NADPH. C4H activity was followed by measuring p-coumaric acid levels. Compound 12 was used at 1,000 µm, and pIBA was added in different concentrations (10, 100, and 1,000 µm). MDCA (100 µm) and a mock treatment were used as negative controls, whereas PA (100 µm) was used as a positive control. Statistical differences were estimated using Student’s t test compared with the respective controls. Error bars represent sd of four independent biological replicates per condition and are significant with P values below 0.05 (*) and 0.01 (**).
As a final proof for the inhibition of lignification, the effect of pIBA on the lignin monomeric composition of 6-d-old Arabidopsis seedlings was evaluated. This was done by thioacidolysis, a procedure that breaks β-O-4 ether bounds within the lignin polymer, followed by analysis of the H, G, and S monomers by gas chromatography-MS. The most abundant lignin monomers (i.e. G and S) were clearly present in mock-treated plants, confirming the presence of lignin in young Arabidopsis seedlings. Isoxaben increased the concentrations of the three different lignin units, which is in line with the suggested increase in lignin deposition (Caño-Delgado et al., 2003). Both PA and pIBA reduced the levels of the lignin units in isoxaben-treated seedlings, confirming that both PA and pIBA effectively reduce lignin amount in plant cell walls (Supplemental Fig. S8).
DISCUSSION
We used a phenotype-based chemical screening approach to discover novel inhibitors of lignification. To this end, a screening-compatible assay was developed based on histochemical staining of the lignin-enriched vasculature of Arabidopsis seedlings. Despite the relatively low amount of lignin in seedlings, this developmental stage has been used successfully as a model for lignin deposition (Naseer et al., 2012). A main advantage of using seedlings in the screen is their small size, allowing one to perform a whole-plant assay in a 96-well microplate format, which considerably improved the throughput.
Once optimized, the assay was used to screen 10,000 compounds, which resulted in the identification of 73 putative inhibitors of lignification. The number of hits in a chemical genetic screen depends on the setting of the selection threshold. Stringent selection criteria will result in only a few hits, but these few hits will have high potential. Lowering the selection threshold will increase the number of hits but also the risk of selecting false positives. The relatively high number of hits we retained was indicative for the presence of a subset of false positives. The latter could be a direct consequence of the fact that all compounds were screened at a relatively high concentration (i.e. 50 µm). Indeed, some compounds will be toxic at this concentration, resulting in growth inhibition or even lethality. As lignin deposition in the cell wall is known to be highly intertwined with plant growth and development, the developmental shift could phenocopy the reduction in lignin deposition we screened for (Grbić and Bleecker, 1995; Mattsson et al., 2003). The dramatic developmental defects observed in compound 47-treated plants clearly illustrate that compounds affecting development passed the screen. These drawbacks are inherent to a high-throughput screening approach and make follow-up screens a necessity to confirm the activity of the compounds on the target process and to weed out false positives.
For this project, we implemented phenolic profiling of seedlings treated with the 73 retained compounds as a secondary screen. Thirty-four of the compounds caused no dramatic shift in the phenolic profile of seedlings. The other 39 compounds caused significant alterations in the phenolic profiles, suggesting a direct perturbation of the phenylpropanoid biosynthetic pathway. Here, clear shifts in both the end products as well as the intermediates of the lignin biosynthetic pathway and derivatives thereof were seen. This pattern was in striking contrast to the profiles of most of the included monolignol biosynthesis mutants, which showed only mild shifts in the abundance of these compounds. The only exception among the mutants was c4h, for which a drastic reduction in the detectable oligolignols was found, accompanied by an increase in two cinnamic acid derivatives, cinnamoyl hexose and malate. This pattern is in line with a perturbation at the level of C4H, which catalyzes the conversion of cinnamic acid into p-coumaric acid. Notably, C4H is encoded by a single-copy gene in Arabidopsis, whereas the genes affected in the other mutants are members of multigene families, underlining the buffering effect provided by gene redundancy. In line with this observation, 4cl1-1 seedlings were found to have considerably higher residual lignin levels as visualized by Wiesner staining, compared with wild-type Arabidopsis plants of the same developmental stage, treated with the 4CL inhibitor MDCA. Together, these results support the idea that gene redundancy is at the base of the observed differences between a mutant defective in a particular gene of a biosynthetic pathway and a wild-type plant treated with an inhibitor of the corresponding enzymatic step that would target all isozymes.
The shift in the phenolic profile induced by the different compounds was used to guide the selection of compounds for further characterization. The profiles were used to classify the compounds into five functional classes, and for each of the classes, one representative compound was selected. To validate the inhibitory effect on lignification, we analyzed the effects of the five selected representative compounds in the Casparian strip and the TE assay. Only compound 12 scored positive in both assays, and its inhibitory activity could be complemented by the addition of monolignols. Together with the particular shift in the phenolic profile observed in compound 12-treated seedlings, this indicated that this compound inhibits the biosynthesis of monolignols rather than their polymerization. The observation that the other putative inhibitors scored negative in one of the assays could be the result of a different uptake, stability, or effectiveness, dependent on the conditions in the different assays (e.g. shorter incubation times, lower concentrations, etc.). On the other hand, these compounds could specifically inhibit one particular isozyme of an enzyme family, resulting in a visible reduction in Wiesner staining of seedlings, whereas the inhibition might not be sufficient to inhibit lignification in Casparian strips of roots or in TE-induced cell cultures. Although overcoming gene redundancy is one of the most cited advantages of chemical genetics, precedents exist of inhibitors that do not target all members of a conserved family (Hicks and Raikhel, 2014). Jarin-1, for instance, selectively inhibits jasmonate-Ile formation by specifically interacting with JASMONATE RESISTANT1 of the GH3 enzyme family (Meesters et al., 2014). Another example is bikinin, which inhibits only two of the four groups of the Arabidopsis SHAGGY-like kinase family (ASK; De Rybel et al., 2009). Moreover, bikinin only moderately inhibits ASKθ of group 3 ASKs, illustrating that putative inhibitors can inhibit an enzyme or an entire family only moderately, which also could explain residual activity.
Further experiments on compound 12 revealed its in planta processing into pIBA and hydroxy-MIC. Although the exact position of the hydroxyl group of hydroxy-MIC could not be determined, treating stems with MIC yielded hydroxy-MIC derivatives (metabolites 1, 2, and 3), suggesting that compound 12 is hydrolyzed in planta into MIC, which is subsequently hydroxylated. MIC, therefore, is a good compound to inhibit lignification, regardless of whether MIC, hydroxy-MIC, or yet another derivative is the active compound. Intriguingly, both MIC and pIBA (or their corresponding derivatives) inhibit lignification, although not at the same efficiency. In the Casparian strip assay, pIBA performed better than MIC, whereas the opposite was the case in the TE assay. The observed differences are possibly a direct consequence of the higher impact of pIBA on TE differentiation compared with MIC. Again, it could also be due to the different experimental setup of the two assays, involving different cell types, compound concentrations, timing, and readout for lignification. Unfortunately, metabolomics on MIC-treated plants did not provide insight into the mode of action of MIC. This could indicate that MIC does not target a specific enzyme of the biosynthetic pathway but rather reduces lignin deposition in an indirect way. In contrast to MIC, the profiles of pIBA-treated plants hinted toward a perturbation in one of the first steps of the pathway, which was supported by identifying C4H as the molecular target in a heterologous yeast expression system.
The functional classification together with the structural information of each potential inhibitor gave a perfect setting to explore the structure-activity causality. This key concept in drug discovery, chemical genetics, and cheminformatics assumes that structural similarity reflects similar biological activity (Martin et al., 2002). Although the assumed causality was found to some extent (e.g. functional class 1 and structural class 4 aligned completely), some structurally related compounds ended up in different functional classes (e.g. compound 54 and 56 differ only by a single chlorine atom but were classified in functional classes 4 and 3, respectively). The other way around, structurally different compounds ended up in the same functional class, illustrated by compound 12, which clustered together with the structurally unrelated 4CL inhibitor MDCA in functional class 2. An explanation for weak correlations between the structural and functional classification is in planta processing of compounds. Our data on compounds 12 and 47 illustrate that processing indeed occurs and is even crucial to lead to active compounds. Interestingly, recent findings also showed in planta processing of MDCA to PA (Steenackers et al., 2016). Hence, the processing of both compound 12 and MDCA toward a C4H inhibitor (pIBA and PA, respectively) explains their clustering based on the induced shift in the phenolic profile. The fact that these compounds do not cluster in the structural tree is simply because the latter was generated on the nonprocessed compounds. When looking at the corresponding processed (and hence bioactive) compounds, there is closer structural similarity. PA differs from pIBA only in a methylenedioxy group, replacing the iodo group at the para position of the aromatic ring. A similar structural replacement of a methylenedioxy and iodo group with conservation of the bioactivity has been described before (Coppola and Mondola, 2013). The in planta processing of compounds and resulting variability in structure-activity causality imply that the selected representative compounds will not always reflect the activity of every individual member of that class. Ideally, each of the 39 compounds needs to be characterized as was done for the five representative compounds. To efficiently screen larger numbers of compounds in different concentrations, further development of high-throughput methods to quantify the lignin amount will be necessary.
In conclusion, a chemical genetics screen was developed to find novel inhibitors of lignification, and one compound (compound 12) was studied in detail. Using phenolic profiling, we showed that compound 12 is hydrolyzed in the plant into hydroxy-MIC and pIBA. We demonstrated that pIBA affects lignification by inhibiting C4H activity. The finding of pIBA as a novel inhibitor of C4H validates chemical genetics as a successful approach to discover novel inhibitors of lignification.
MATERIALS AND METHODS
Chemicals
Chemicals were purchased from Sigma-Aldrich unless stated otherwise. All solvents used were UHPLC-MS grade (Biosolve). Water was produced by a DirectQ-UV water purification system (Millipore).
Chemical Genetics Screening
The chemical genetics screening was performed by the VIB Compound Screening Facility using the 10-k DIVERSet of ChemBridge. Arabidopsis (Arabidopsis thaliana) Col-0 seeds were surface sterilized with 70% ethanol for 2 min followed by 15 min of bleach solution (4.6% NaOCl) and stratified in the dark at 4°C for 48 h. Three to seven seeds were distributed per well of a 96-well plate (353072; Falcon) in 150 µL of liquid 0.5× Murashige and Skoog medium (Duchefa Biochemie), pH 5.7, supplemented with 0.5 g L−1 MES monohydrate (Duchefa Biochemie), 0.1 g L−1 myoinositol, and 2% Glc. The plates were put on a shaker in continuous light at 21°C. At 3 DAG, each well was supplied with a final concentration of 50 µm of a library chemical (in DMSO). Wells serving as controls were supplied with DMSO only. Two hours later, isoxaben (in DMSO) was added to all wells in a final concentration of 80 nm per well. Robotics were implemented for automatic liquid handling to allow fast and reproducible plate loading. At 6 DAG, the seedlings were stained with Wiesner reagent (1% phloroglucinol in 100 mL of 95% ethanol and 16 mL of 37% HCl), and lignification was assessed visually.
Hierarchical Clustering
All 73 selected compounds and the known inhibitors of the lignification pathway were clustered by single linkage using the ChemMine clustering algorithm (Backman et al., 2011).
Plant Growth and Metabolite Extraction for Phenolic Profiling
For phenolic profiling of the confirmed hits, the same conditions were used as for the chemical genetics screening. This time, however, seeds were sterilized overnight by chlorine gas (5.5 mL of HCl in 150 mL of NaClO) and germinated on petri plates with 0.5× Murashige and Skoog medium with 0.8% plant tissue culture agar (Lab M). At the time of radicle protrusion (0 DAG), exactly six seedlings were transferred to each individual well of a 96-well plate. Arabidopsis mutants defective in the lignin biosynthetic pathway (pal1-2, pal2-2, c4h-3, 4cl1-1, 4cl2-1, ccoaomt1-5, and ccr1-6 [all described in Vanholme et al., 2012b] as well as ccr2-1 [SAIL_207_G05], a T-DNA insertion mutant for the gene with accession number AT1G80820 from the SAIL collections [McElver et al., 2001] and ordered via the European Arabidopsis Stock Centre) were used as controls for phenolic profiling. In addition, known inhibitors of the lignification pathway were included: CAld, DNP, MD, MDCA (ACROS Organics), CA, FA (Janssen Chimica), and propanil. The outer borders of each 96-well plate were filled with medium but were not used to screen compounds. Wells serving as controls and wells with seeds defective in lignin biosynthesis were supplied only with DMSO. The experiment was done with two replicates per condition. Subsequently, metabolites were extracted by incubating the seedlings in Eppendorf tubes with 350 µL of methanol for 15 min at 70°C (850 rpm). The samples were purified by solid-phase extraction using 30-mg Varian Bond-Elut Plexa columns (Agilent) pretreated with 400 µL of methanol. After loading of the sample, each column was washed with another 100 µL of methanol to elute the phenolics. The purified samples were vaporized in the Savant SpeedVac Concentrator (Thermo Electron), and the pellets were resuspended in 25 µL of sterile water. The metabolite solutions were transferred to LCMS Certified Vials (Waters) and analyzed by UHPLC-MS.
Phenolic Profiling
UHPLC-MS was performed on a Waters Acquity UHPLC system connected to a SYNAPT HDMS Q-TOF mass spectrometer (Micromass). UHPLC-MS settings were as described previously (Vanholme et al., 2013). For the analysis of the chromatograms of the screening experiment, integration and alignment were done using MZmine (Katajamaa et al., 2006). The chromatograms of the samples treated with compounds 20, 30, and 31 were not taken into account, because they appeared to be lethal to the plants, at least in the concentration used. The chromatograms were cropped from RT 0 to 30 min. Peak detection was done using the exact mass option for mass detection with absolute noise level set at 15. The FTML shoulder peaks filter was used at a mass resolution of 10,000, and the peak model function was set on Lorentzian extended. For chromatogram construction, a time frame of 4 s was taken with a minimum height of 2 and an m/z tolerance of 0.04. During deconvolution, peaks were recognized by taking the local minimum search with chromatographic threshold, minimum relative height, and minimum ratio of peak edge set at zero, while the minimum RT interval was 10 s and the minimum absolute height was 20. The alignment was done using an m/z tolerance of 0.04, an m/z weight of 10, an absolute RT tolerance of 20 s, a relative RT tolerance of 15%, an RT weight of 10, and a threshold level of 65% for isotope pattern score. Finally, gap filling was done with a 20% intensity tolerance, an m/z tolerance of 0.04, an absolute RT tolerance of 15 s, and a relative RT tolerance of 15%. The RT correction also was activated. The list of peak areas resulting from these parameters was imported in a digital spreadsheet (Excel) for further analysis. All peaks were normalized by dividing the peak area by the relative total peak area of the respective sample. Subsequently, all values were divided by the averages of the corresponding controls, to set the average control values at 1. Finally, these values were log2 transformed. The functional clustering was done using Euclidean clustering distance and Ward as clustering method in R studio 3.2.2 (R Core Team, 2015) via the pheatmap package. For analysis of the chromatograms from the experiments with Arabidopsis inflorescence stems treated with compound 12, MIC, and pIBA, chromatogram integration and alignment was done using ProgenesisQI (Nonlinear) as described previously (Vanholme et al., 2013).
In Vitro Plant Growth Conditions for Phenotyping
Arabidopsis Col-0 seeds were stratified in the dark at 4°C and germinated on petri plates with 0.5× Murashige and Skoog medium. Germinated seeds were transferred to 0.5× Murashige and Skoog agar plates containing the desirable concentration of each of the five selected candidate compounds (12, 47, 54, 56, and 72). An equal amount of DMSO was added to all control plates. The plants were grown vertically for 9 d in long-day conditions (16 h of light and 8 h of dark) at 21°C. Seedlings treated with 100 µm compound 12, 0.01 µm compound 47, 10 µm compound 54, 25 µm compound 56, and 10 µm compound 72 were additionally incubated for 30 min in Wiesner reagent and photographed using a Leica MZ16 stereomicroscope (Meyer Instruments).
Casparian Strip Assay
The Casparian strip assay was performed as described by Naseer and coworkers (2012) with minor adjustments. Arabidopsis Col-0 seeds were germinated on 0.5× Murashige and Skoog agar plates after 2 d in the dark at 4°C. Then, seedlings were grown vertically under long-day conditions (16 h of light and 8 h of dark) at 21°C. Five days after the shift to room temperature, the seedlings were incubated for 24 h in the dark in liquid 0.5× Murashige and Skoog medium supplemented with the right inhibitor and a 40 µm mixture of coniferyl alcohol (50%) and sinapyl alcohol (50%) as monolignols. For visualization of the apoplastic barrier, seedlings were incubated in the dark for 2 min in a fresh solution of 10 µg mL−1 PI and rinsed in water twice. Confocal laser scanning microscopy was performed on a Zeiss LSM5 Exciter confocal microscope (Carl Zeiss). An excitation window of 543 nm was used, and detection was done using LP560. For quantification, onset of elongation was defined as the point where an endodermal cell in a median optical section was more than twice its width. From this point on, cells in the file were counted until PI could not diffuse through the apoplastic barrier anymore, indicating that the Casparian strip was formed. The endodermal cells of 10 seedlings per condition were counted, and P values were calculated using one-way ANOVA in SPSS (IBM).
Short-Time Cytotoxicity Analysis on Arabidopsis Cell Cultures
Arabidopsis Col-0 cells were cultured in 50 mL of Murashige and Skoog medium at pH 5.7 and were placed on a shaker at 120 rpm in a dark culture room at 24°C. Induction of TEs was performed according to Pesquet et al. (2010). Cytotoxicity analyses were performed on six-well plates (4 mL of cells per well). Seven-day-old noninduced cells adjusted to a density of 60 mg fresh weight mL−1 in Murashige and Skoog medium at pH 5.7 were treated with the different compounds (DMSO, MDCA, PA, MIC, pIBA, and compounds 12, 47, 54, 56, and 72). After 24 h of culture in a dark room, viability staining of the cells was performed with 0.01% fluorescein diacetate, and the cells were observed with a fluorescence microscope. Viability quantification was done for three to 18 replicate samples with 200 to 400 cells in each sample and was expressed as percentage of cell viability.
Xylogenic Cell Cultures and Chemical Treatments for Cell Viability, TE Differentiation, and Lignification Rate Analysis
To analyze the inhibitor’s effect on non-TE viability, TE differentiation, and lignification, 9-d-old cell cultures were calibrated at 30 mg cells mL−1 in Murashige and Skoog medium supplemented with MES at pH 6. The cells were immediately induced by hormone supplementation according to Pesquet et al. (2010). The treatments were performed 3 d after induction in order to respect the TE differentiation period. During the treatment, the plates were placed in the culture room as described previously. Nine days after induction, the cells were stained with 0.01% fluorescein diacetate. A total of 200 to 400 cells were counted using the fluorescence microscope (three to 22 replicates), and the non-TE viability/TE differentiation rates were defined as percentage of the control (i.e. induced cells treated with DMSO).
UV Confocal Microspectroscopy
For the 9-d-old treated cells, confocal UV lignin imaging was performed with the confocal Leica TCS SP2 AOBS spectral system on a Leica DM IRE2 inverted microscope. Lignin autofluorescence emission spectra were obtained in a 411- to 472-nm emission window with excitation at 405 nm. The lignin distribution between PCW and SCW was measured by five independent measurements for each cell wall in 12 to 89 independent images and analyzed with ImageJ software (eight-bit scale quantification; Schneider et al., 2012). The results were expressed as cell wall lignin fluorescence intensity. Movies including confocal flow through and 3D half-cell reconstruction were made according to Derbyshire and coworkers (2015).
Recovery of Postmortem Lignification in TEs Inhibited to Lignify
For each biological replicate, induced cells were treated with the different compounds (two technical replicates per compound treatment). At 9 d, the initial chemical treatment was repeated for each sample, and only one of the two technical replicates was supplemented with 120 µm coniferyl alcohol. After 5 d, both technical replicates were observed by UV confocal microspectroscopy as described above.
In Vivo Plant Growth Conditions for Inflorescence Stem Treatments
Wild-type Arabidopsis Col-0 seeds were sown in soil and stratified in the dark at 4°C. After 2 d, the trays were transferred to short-day conditions (8 h of light and 16 h of dark) at 21°C for 8 weeks. Then, the plants were transferred to long-day conditions (16 h of light and 8 h of dark) at 21°C until the stems were between 15 and 20 cm in height. Eight stems per condition were cut at the base while submerged in water and were treated with liquid 0.5× Murashige and Skoog medium supplemented with the appropriate amount of compound (100 µm compound 12, 50 µm MDCA, 50 µm PA, 100 µm MIC, and 50 µm pIBA). After 48 h of incubation, stems were snap frozen in liquid nitrogen. The lower 11 cm was selected, and the bottom 1 cm was cut off, leaving a stem piece of 10 cm that was ground for analysis. Phenolic compounds were extracted by incubation in 1 mL of 90% methanol for 15 min at 70°C (850 rpm). After centrifugation, the supernatant was vaporized in the Savant SpeedVac Concentrator (Thermo Electron), and the pellets were used for liquid-liquid extraction using a 1:1 ratio of water and cyclohexane. The lower watery phase was transferred to LCMS Certified Vials (Waters) and analyzed by UHPLC-MS, as described above.
Microsome Assay
The C4H gene of Arabidopsis (AT2G30490) was ordered from The Arabidopsis Information Resource as a Gateway-compatible open reading frame clone (DKLAT2G30490). Heterologous expression in Saccharomyces cerevisiae strain WAT21 (Pompon et al., 1996) was done using the destination vector pAg424GAL-ccdB (Addgene plasmid 14151; Alberti et al., 2007). After harvesting and washing the yeast cells, they were incubated in dropout medium containing Gal to induce transcription, supplemented with the appropriate dropout supplement. Microsomal fractions were isolated by differential centrifugation after mechanical disruption of the yeast cells with glass beads of 0.3 to 0.5 mm according to Pompon et al. (1996). Microsomes containing the C4H enzyme were dissolved in ice-cold sodium phosphate buffer (PBS; 20 mm, pH 7.4) together with the appropriate inhibitor (10, 100, and 1,000 µm pIBA; 1,000 µm compound 12; 100 µm MDCA; 100 µm PA). The substrate of C4H (i.e. cinnamic acid) was added in an end concentration of 200 µm, and to start the reaction, 5 µg µL−1 NADPH+ solution was added. The samples were incubated at 28°C for 10 min (750 rpm), and to stop the reaction, 100 µL of boiling PBS was added and samples were boiled for 10 min. To remove lipids, liquid-liquid extraction was performed by adding 200 µL of cyclohexane, vortexing, and centrifugation, and metabolites were retrieved by transferring the watery phase to UHPLC-grade vials. Ten microliters of the aqueous phase was subjected to UHPLC-MS with the same settings as for the phenolic profiling experiments and as described previously (Vanholme et al., 2013). Peak detection and integration of p-coumaric acid (whose identity was confirmed by the use of an authentic standard) was done using MassLynx software (Waters), and P values were calculated using unpaired Student’s t test.
Thioacidolysis
Arabidopsis seeds were gas sterilized and germinated on 0.5× Murashige and Skoog medium under long-day growth conditions. At 3 DAG, seedlings were transferred to 100 mL of liquid Murashige and Skoog medium. Here, care was taken not to transfer the seed coat to avoid contamination with lignin not derived from the developing seedling. After transfer, chemicals (isoxaben, PA, and pIBA) were added as described for the initial screen. For the mock treatment, DMSO was used. After another 3 d in the growth chamber, seedlings were harvested and thioacidolysis was performed on purified cell wall residue as described by Van Acker et al. (2013).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. High-throughput chemical genetics screen toward inhibitors of lignification.
Supplemental Figure S2. Hierarchical clustering of compounds according to their chemical structure.
Supplemental Figure S3. Effects of the compounds representing the five functional classes.
Supplemental Figure S4. Effects of compound 47 at lower concentrations.
Supplemental Figure S5. Evidence for 2,4-D fragments in seedlings treated with compound 47.
Supplemental Figure S6. Cell viability and TE differentiation rates after a long-term (9-d) treatment with the representative compounds.
Supplemental Figure S7. Distribution of lignin autofluorescence intensity of SCWs in TEs.
Supplemental Figure S8. H/G/S monomeric composition of seedlings treated with pIBA.
Supplemental Figure S9. Tandem MS spectra of structurally characterized and unknown metabolites.
Supplemental Table S1. Overview of selected ChemBridge compounds and their characteristics.
Supplemental Table S2. Metabolite data of seedlings treated with candidate lignin inhibitors.
Supplemental Table S3. Phenolic profiling of stems treated with compound 12.
Supplemental Table S4. Phenolic profiling of stems treated with MIC and pIBA.
Supplemental Table S5. Targeted search for oligolignols isolated from stems treated with MIC and pIBA.
Supplemental Movie S1. XYZ confocal image series of TEs produced in cell cultures.
Supplementary Material
Acknowledgments
We thank Annick Bleys for help in preparing the article.
Glossary
- UHPLC
ultra-high-pressure liquid chromatography
- MS
mass spectrometry
- Col-0
Columbia-0
- DAG
days after germination
- MDCA
3,4-methylenedioxycinnamic acid
- DMSO
dimethyl sulfoxide
- CAld
cinnamaldehyde
- DNP
2,4-dinitrophenol
- MD
menadione
- CA
caffeic acid
- FA
ferulic acid
- 2,4-D
2,4-dichlorophenoxyacetic acid
- PI
propidium iodide
- PA
piperonylic acid
- TE
tracheary element
- SCW
secondary cell wall
- PCW
primary cell wall
- hydroxy-MIC
hydroxymethyl indole 3-carboxylate
- pIBA
p-iodobenzoic acid
- MIC
methylindole carboxylic acid
- S
syringyl
- G
guaiacyl
- H
hydroxyphenyl
- RT
retention time
Footnotes
This work was supported by the Hercules Foundation for the SYNAPT Q-TOF (grant no. AUGE/014) and by the Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy (grant no. 01MRB510W) of Ghent University; by the Agency for Innovation by Science and Technology in Flanders (to D.V.d.W.) and the Research Foundation Flanders (postdoctoral fellowship to R.V.); by an OMICS@VIB Marie Curie COFUND fellowship to R.H.; by the Vetenskapsrådet Swedish Research Council (grant no. 2010–4620 to E.P.) and the Kempe Foundation (Gunnar Öquist Fellowship to E.P.); and by Stanford University’s Global Climate and Energy Projects Towards New Degradable Lignin Types and Lignin Management: Optimizing Yield and Composition in Lignin-Modified Plants to W.B.
Articles can be viewed without a subscription.
References
- Agrawal A, Kaushik N, Biswas S (2014) Derivatives and applications of lignin: an insight. The SciTech Journal 1: 30–36 [Google Scholar]
- Alberti S, Gitler AD, Lindquist S (2007) A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman TWH, Cao Y, Girke T (2011) ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res (Suppl 2) 39: W486–W491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Halpin C, Petit-Conil M, Boerjan W (2003) Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol 38: 305–350 [DOI] [PubMed] [Google Scholar]
- Billett EE, Smith H (1978) Cinnamic acid 4-hydroxylase from gherkin tissues. Phytochemistry 17: 1511–1516 [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Castañeda-Ovando A, de Lourdes Pacheco-Hernández M, Páez-Hernández ME, Rodríguez JA, Galán-Vidal CA (2009) Chemical studies of anthocyanins: a review. Food Chem 113: 859–871 [Google Scholar]
- Chen HC, Li Q, Shuford CM, Liu J, Muddiman DC, Sederoff RR, Chiang VL (2011) Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F, Cheung AK, Huang SMA (2012) Chemical genetics-based target identification in drug discovery. Annu Rev Pharmacol Toxicol 52: 57–78 [DOI] [PubMed] [Google Scholar]
- Coppola M, Mondola R (2013) 5-Iodo-2-aminoindan (5-IAI): chemistry, pharmacology, and toxicology of a research chemical producing MDMA-like effects. Toxicol Lett 218: 24–29 [DOI] [PubMed] [Google Scholar]
- Decou R, Serk H, Ménard D, Pesquet E (2016) Analysis of lignin composition and distribution using fluorescence laser confocal microspectroscopy. Methods Mol Biol (in press) [DOI] [PubMed] [Google Scholar]
- Dejonghe W, Russinova E (2014) Target identification strategies in plant chemical biology. Front Plant Sci 5: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P, Ménard D, Green P, Saalbach G, Buschmann H, Lloyd CW, Pesquet E (2015) Proteomic analysis of microtubule interacting proteins over the course of xylem tracheary element formation in Arabidopsis. Plant Cell 27: 2709–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L, et al. (2009) Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol 16: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Xuan W, Overvoorde P, Strader LC, Kepinski S, Hoye R, Brisbois R, Parizot B, Vanneste S, et al. (2012) A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat Chem Biol 8: 798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W (2015) Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27: 695–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Tanaka E, Murata M (2006) Cinnamaldehyde inhibits phenylalanine ammonia-lyase and enzymatic browning of cut lettuce. Biosci Biotechnol Biochem 70: 672–676 [DOI] [PubMed] [Google Scholar]
- Funk C, Brodelius PE (1990) Phenylpropanoid metabolism in suspension cultures of Vanilla planifolia Andr. II. Effects of precursor feeding and metabolic inhibitors. Plant Physiol 94: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V, Bleecker AB (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J 8: 595–602 [Google Scholar]
- Gross GG. (2008) From lignins to tannins: forty years of enzyme studies on the biosynthesis of phenolic compounds. Phytochemistry 69: 3018–3031 [DOI] [PubMed] [Google Scholar]
- Harding SA, Leshkevich J, Chiang VL, Tsai CJ (2002) Differential substrate inhibition couples kinetically distinct 4-coumarate:coenzyme A ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol 128: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV (2014) Plant chemical biology: are we meeting the promise? Front Plant Sci 5: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajamaa M, Miettinen J, Orešič M (2006) MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 22: 634–636 [DOI] [PubMed] [Google Scholar]
- Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J, et al. (2008) How biotech can transform biofuels. Nat Biotechnol 26: 169–172 [DOI] [PubMed] [Google Scholar]
- Martin YC, Kofron JL, Traphagen LM (2002) Do structurally similar molecules have similar biological activity? J Med Chem 45: 4350–4358 [DOI] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meesters C, Mönig T, Oeljeklaus J, Krahn D, Westfall CS, Hause B, Jez JM, Kaiser M, Kombrink E (2014) A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat Chem Biol 10: 830–836 [DOI] [PubMed] [Google Scholar]
- Miedes E, Vanholme R, Boerjan W, Molina A (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Korolev AV, Calder G, Lloyd CW (2010) The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr Biol 20: 744–749 [DOI] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar F, Merino F, Barceló AR (2002) O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220: 17–28 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Poustka F, Irani NG, Feller A, Lu Y, Pourcel L, Frame K, Grotewold E (2007) A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol 145: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, http://www.R-project.org [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Schalk M, Cabello-Hurtado F, Pierrel MA, Atanossova R, Saindrenan P, Werck-Reichhart D (1998) Piperonylic acid, a selective, mechanism-based inactivator of the trans-cinnamate 4-hydroxylase: a new tool to control the flux of metabolites in the phenylpropanoid pathway. Plant Physiol 118: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Kombrink E, Meesters C (2015) Considerations for designing chemical screening strategies in plant biology. Front Plant Sci 6: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers W, Cesarino I, Klíma P, Quareshy M, Vanholme R, Corneillie S, Kumpf RP, Van de Wouwer D, Ljung K, Goeminne G, et al. (2016) The allelochemical MDCA inhibits lignification and affects auxin homeostasis. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin L, Vanholme R, Geerinck J, Goeminne G, Höfer R, Kim H, Ralph J, Boerjan W (2014) Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol 166: 1956–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, van der Hoorn RAI (2010) Emerging principles in plant chemical genetics. Trends Plant Sci 15: 81–88 [DOI] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012a) Metabolic engineering of novel lignin in biomass crops. New Phytol 196: 978–1000 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012b) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Yun MS, Chen W, Deng F, Kiyokawa T, Mametsuka K, Yogo Y (2006) An in vitro screening assay to discover novel inhibitors of 4-coumarate:CoA ligase. Pest Manag Sci 62: 1065–1071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.