Abstract
Please cite this paper as: WHO/OIE/FAO. (2012) Continued evolution of highly pathogenic avian influenza A(H5N1): Updated nomenclature. Influenza and Other Respiratory Viruses 6(1), 1–5.
Background Continued evolution of highly pathogenic avian influenza A (H5N1) throughout many regions of the eastern hemisphere has led to the emergence of new phylogenetic groups. A total of 1637 new H5N1 hemagglutinin (HA) sequences have become available since the previous nomenclature recommendations described in 2009 by the WHO/OIE/FAO H5N1 Evolution Working Group. A comprehensive analysis including all the new data is needed to update HA clade nomenclature.
Methods Phylogenetic trees were constructed from data sets of all available H5N1 HA sequences. New clades were designated on the basis of phylogeny and p‐distance using the pre‐established nomenclature system (Emerg Infec Dis 2008; 14:e1). Each circulating H5N1 clade was subjected to further phylogenetic analysis and nucleotide sequence divergence calculations.
Results All recently circulating clades (clade 1 in the Mekong River Delta, 2.1.3 in Indonesia, 2.2 in India/Bangladesh, 2.2.1 in Egypt, 2.3.2, 2.3.4 and 7 in Asia) required assignment of divergent HA genes to new second‐, third‐, and/or fourth‐order clades. At the same time, clades 0, 3, 4, 5, 6, 8, 9, and several second‐ and third‐order groups from clade 2 have not been detected since 2008 or earlier.
Conclusions New designations are recommended for 12 HA clades, named according to previously defined criteria. In addition, viruses from 13 clades have not been detected since 2008 or earlier. The periodic updating of this dynamic classification system allows continued use of a unified nomenclature in all H5N1 studies.
Keywords: H5N1, hemagglutinin, highly pathogenic avian influenza, molecular epidemiology, nomenclature, phylogenetics, viral evolution
Introduction
The nomenclature system designed by the WHO/OIE/FAO H5N1 Evolution Working Group to classify the A/goose/Guangdong/1/1996 lineage of Eurasian highly pathogenic H5N1 avian influenza viruses has been in place since early 2008. Based on the criteria used to distinguish variant groups of the H5 hemagglutinin (HA) gene, the system has formally identified 20 distinct clades of the virus. 1 , 2 Using phylogenetic characterization and nucleotide sequence divergence, H5N1 virus clades have been defined based on sharing of a common ancestral node and monophyletic evolution with a bootstrap value of ≥60 at the clade‐defining node (after 1000 neighbor‐joining bootstrap replicates) and maintaining the average percentage pairwise nucleotide distances between and within clades of >1·5% and <1·5%, respectively. 2 Since the inception of this approach to H5N1 classification, clade 2.2 viruses (first detected in the Qinghai Lake area) introduced into Egypt diverged into a third‐order clade designated 2.2.1. 1 As expected, however, the continued evolution of H5N1 has led to the periodic emergence of new phylogenetic groups of H5N1 in several regions of the world. Therefore, the WHO/OIE/FAO H5N1 Evolution Working Group has examined the HA sequence data available as of January 2011 in the context of previous nomenclature assignments to update this classification system.
Materials and methods
Data sets and phylogenetic analysis
Hemagglutinin sequences of the highly pathogenic avian influenza A (H5N1) viruses within the Eurasian A/goose/Guangdong/1/1996 lineage were obtained from the Epiflu Database in GISAID, which contains all of the H5N1 HA sequences available in the Influenza Virus Resource of NCBI, as well as shared data from individual laboratories within the WHO Global Influenza Surveillance and Response System (GISRS) and OIE/FAO Global Network of Expertise on Animal Influenza (OFFLU). Only sequences with at least 1600 nucleotides of the HA gene were included (>90% of the full‐length open reading frame). A nucleotide alignment containing 2947 HA sequences was generated using MUSCLE. 3 A large neighbor‐joining tree was initially constructed from the alignment using a GTR+I+G model in PAUP* v4.0b10, 4 and clade assignments (using previously established nomenclature parameters) were made for new sequences based on their location within a clade (Figure S1). Clade‐specific alignments were then used to generate neighbor‐joining trees with 1000 bootstraps and rooted to either A/goose/Guangdong/1/96 or a common outlier to that clade (Figure S2A–G). Monophyletic groups within each of the clades defined in the 2009 update 1 containing more than four non‐redundant sequences with bootstrap values of ≥60 were selected as potential candidates for new order clade designation.
Genetic distance calculations
To determine whether a clade had undergone a sufficient divergence to exceed the pre‐established limit (i.e. required splitting), the average within‐group nucleotide p‐distance for sequences was determined using MEGA4 5 ; clades having within‐group distances ≥1·5% were analyzed further. Only groups containing samples with a collection date of 2007 or later were subjected to this analysis. New order clade designations were made for those groups that had between‐group divergences of ≥1·5% and within‐group divergences of ≤1·5%. As described previously, the new clade number consisted of the name of the originating clade followed by a period and a single numeric digit added on the right (Figure S2A–G). Following the identification of new higher‐order clades, neighbor‐joining and Bayesian trees of 196 representative sequences were constructed using PAUP* v4.0b10 4 and MrBayes, 6 to verify the overall H5N1 HA tree topology and for statistical inferences (Figure 1).
Figure 1.
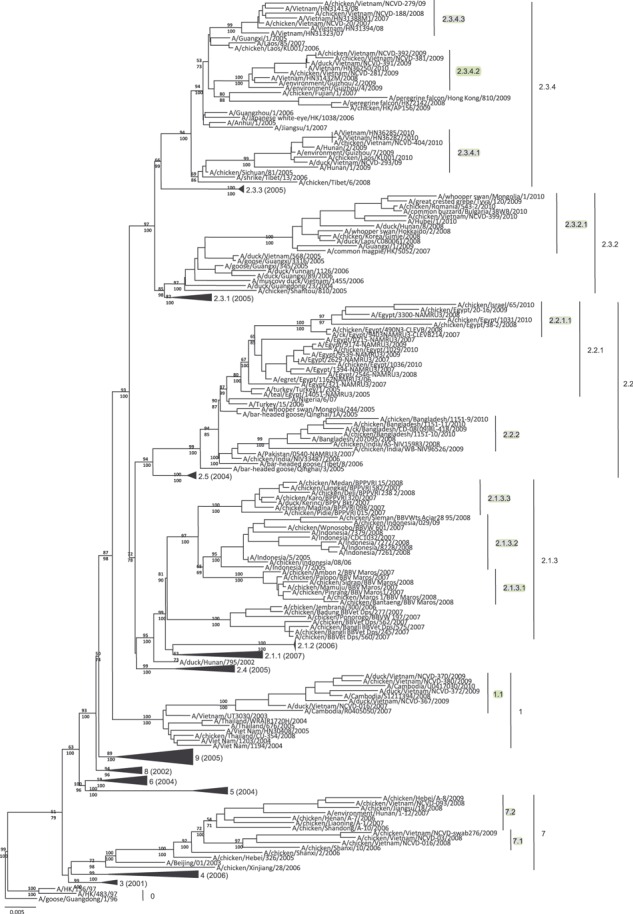
Phylogenetic relationships of recently diverged A/goose/Guangdong/1/1996‐like H5 hemagglutinin (HA) genes. Neighbor‐joining tree of 196 HA gene sequences from H5N1 viruses constructed with 1000 bootstrap replicates (above branches) and Bayesian posterior probabilities (below branches). The tree was constructed using PAUP* v4.0b10 4 with a GTR+I+G model and rooted to A/goose/Guangdong/1/1996. Newly designated clades are highlighted. Solid triangles denote HA clades of viruses that have not been in circulation since 2008 or earlier; the year of the most recent isolation is shown in parenthesis. Scale bar denotes nucleotide substitutions per site. The sequence data set used to generate this figure is provided as Supporting Information in FASTA format (Supplementary Data S2).
Results
Further divergence of the H5N1 HA gene
Surveillance for the continuing circulation of highly pathogenic avian influenza A (H5N1) in poultry and wild birds in Asia, the Middle East, Europe, and Africa has led to an increase in the number of sequences available for analysis. The current analysis was applied to 2947 HA gene sequences consisting of at least 1600 nucleotides, of which 1637 sequences (primarily from 2008 to 2010) were added since the previous nomenclature update. Phylogenetic analysis of the neighbor‐joining tree generated from this data set revealed that the addition of these new sequence data resulted in the generation of one or more monophyletic groups with high bootstrap support within each of the currently circulating clades of H5N1 (Figure S1). In addition, many of these groups had long branches separating them from the nearest node in the tree, indicating further nucleotide divergence.
Each of the currently circulating clades (1, 2.1.3, 2.2, 2.2.1, 2.3.2, 2.3.4, and 7) was then examined independently, to measure the average within‐group pairwise nucleotide distances. All were found to have >1·5% within‐group divergence indicating the need to split these groups into new order clades. After generation of clade‐specific trees, monophyletic groups with bootstrap values more than 60 were selected for further testing (Figure S2A–G). Each monophyletic group identified was then tested to determine the within‐ and between‐group p‐distances. Based on the previously defined nomenclature, 12 new second‐, third‐, and fourth‐order clades were identified (Table 1).
Table 1.
Updates to the H5N1 highly pathogenic avian influenza virus clade nomenclature system
| New order clade | Country | Isolate dates | Average between‐group divergence (%) | Average within‐group divergence (%) |
|---|---|---|---|---|
| 7.2 | Vietnam/China | 2006–2009 | 4·5 | 1·2 |
| 7.1 | Vietnam/China | 2006–2009 | 4·5 | 1·1 |
| 1.1 | Vietnam/Cambodia | 2007–2010 | 2·8 | 1·4 |
| 2.1.3.3 | Indonesia | 2005–2008 | 2·9 | 0·8 |
| 2.1.3.2 | Indonesia | 2005–2009 | 3·2 | 1·4 |
| 2.1.3.1 | Indonesia | 2005–2008 | 2·7 | 1·1 |
| 2.3.4.3 | Vietnam/China | 2007–2009 | 2·7 | 0·7 |
| 2.3.4.2 | Vietnam/China | 2008–2010 | 2·7 | 1·2 |
| 2.3.4.1 | Vietnam/China/Laos | 2009–2010 | 3·4 | 0·9 |
| 2.3.2.1 | Southeast Asia/Eastern Europe/Far East | 2007–2010 | 3·6 | 1·7 |
| 2.2.1.1 | Egypt/Israel | 2007–2010 | 2·9 | 1·5 |
| 2.2.2 | Bangladesh/India | 2008–2010 | 2·3 | 1·3 |
To summarize these findings and to confirm the consistency of clade topology using a smaller data set, a small tree containing 196 HA sequences was generated using both NJ and Bayesian methods (Figure 1). This tree is annotated with the 12 new clades presented here and displays older, non‐circulating, and putatively non‐circulating clades as collapsed nodes (Figure 1). Both NJ bootstrap support and Bayesian posterior probabilities were calculated to demonstrate confidence in clade assignments when using smaller data sets.
Discussion
The highly pathogenic avian influenza virus A (H5N1) lineage was first identified in 1996 and has subsequently spread over much of the eastern hemisphere. As these viruses have established geographically and ecologically isolated enzootic foci of infection, new clades of viruses continuously evolve. The WHO/OIE/FAO H5N1 Evolution Working Group has previously identified 20 such clades and established specific criteria for naming H5N1 clades. Since the most recent recommendations of the H5N1 Evolution Working Group in 2009, H5N1 viruses have continued to spread and evolve locally and regionally.
To address the question of whether this continued evolution has led to the appearance of new clades, we initially constructed a phylogenetic tree composed of nearly 3000 HA sequences from either public databases or ongoing surveillance programs. As expected, the addition of more than 1500 new sequences to the previous H5N1 nomenclature update tree produced a number of monophyletic groups within each of the currently circulating clades of H5N1. After measuring within‐ and between‐group average nucleotide p‐distances, it was observed that all extant clades of H5N1 required splitting into one or more newly defined second‐, third‐, and/or fourth‐order clades. The current analysis, therefore, proposes the designation of 12 new clades that meet the criteria of the H5N1 nomenclature system.
The designation of 12 new H5N1 clades (in contrast with the previous nomenclature update in which only one new clade was identified 1 ) is not surprising, considering the length of time since the last nomenclature update and improved surveillance/reporting of viruses in recent years, increasing the number of sequences available for analysis. Interestingly, the clades that had already diverged into third‐order groups in previous analyses required the largest number of new clade designations. Previous assignment to third‐order clades indicates that the viruses had already evolved considerably, presumably because of short generation times and large population sizes in enzootic areas. The subdivision of these clades suggests the continued evolution and divergence in regions where H5N1 is considered entrenched in poultry. 7 For example, clade 2.3.4 viruses circulating in China, Vietnam, Laos, and neighboring regions were split into 2.3.4.1/2.3.4.2/2.3.4.3, whereas clade 2.1.3 in Indonesia was split into 2.1.3.1/2.1.3.2/2.1.3.3. The Egyptian third‐order clade 2.2.1 also required splitting into new clade 2.2.1.1, indicating further divergence of contemporary strains of H5N1 circulating in Egyptian poultry. 8 In addition, clade 2.3.2 enzootic in China was subdivided to accommodate the new clade 2.3.2.1, which has been identified in poultry populations as well as wild birds in many diverse regions outside of China. 9 , 10 While these groups have been identified as sufficiently divergent from the previously defined clades, their ancestral populations have not been detected in recent years and are likely to no longer circulate.
One new third‐order clade, termed 2.2.2, comprising viruses from India and Bangladesh was identified. This clade diverged from clade 2.2 after its introduction into this region in approximately 2006. 7 Finally, first‐order clades 1 and 7 required designation of second‐order clades as a result of their continued divergence in the Mekong Delta regions of southern Vietnam/Cambodia and China, respectively. While clade 1.1 appears to represent a continuum of evolution in the relatively well‐sampled Mekong Delta region, 11 , 12 clade 7.1 and 7.2 viruses remain widely diverse groups (average between‐group divergence of 4·5%), suggesting that these groups represent under‐sampled virus populations with only sporadic detection (Figure S2A–G). 13 , 14 Antigenic analysis of viruses from each of these new clades will inform decisions on whether to update the library of pre‐pandemic vaccine candidates (see http://www.who.int/influenza/resources/documents/characteristics_virus_vaccines/en/index.html).
In addition to the emergence of novel clades of H5N1, a number of previously circulating clades of H5N1 have not been detected for several years (since at least 2008 or earlier). Although we cannot rule out the possibility that the lack of detection of viruses in these clades may in some cases be related to gaps in surveillance, it is more likely that many of them have been supplanted by new clades and become inactive. As shown in Figure 1, 13 clades (shown as collapsed nodes in the tree with the last year of detection shown in parentheses) have not been detected since 2008 or earlier (0, 2.1.1, 2.1.2, 2.3.1, 2.3.3, 2.4, 2.6, 3, 4, 5, 6, 8, and 9). Therefore, although 12 new clades have been identified and characterized in this study, 13 older clades may now be considered out of circulation. Again, this is not unexpected given the time since the last nomenclature update.
As observed previously, the majority of viruses cluster with others of similar geographic and temporal origin, with few exceptions that might indicate viral spread over longer distances (i.e., poultry trade and/or bird migration). 1 , 2 For example, clade 2.3.2.1 viruses have now been detected in 9 countries since 2007. In addition, while the majority of the novel HA gene clades have approximately similar levels of within‐ (<1·5% nucleotide differences) and between‐ (>1·5%) group diversity, the clade 2.3.2.1 viruses have a higher within‐group mean nucleotide divergence (1·7%). This result suggests that the clade 2.3.2.1 HA genes have diversified to a greater extent than those in other clades, perhaps owing to the adaptation to different bird species and introduction into multiple geographic regions by migratory birds. Although this average within‐group nucleotide p‐distance exceeds the <1·5% within‐group mean criterion originally proposed as a cutoff for clade determination, 1 , 2 designation of a single fifth‐order clade awaits additional data to determine its significance.
Despite very successful efforts to eliminate or control the spread of H5N1 in poultry, viruses persisting in enzootic regions seed virus epizootics in new areas via poultry trade or wild birds. Periodic population sweeps in enzootic and epizootic areas with sustained viral circulation may lead to the extinction of some clades and extensive divergence of viruses within more successful clades. As a consequence, the classification of H5N1 viruses based on HA evolution requires periodic updating, making classification dynamic as the virus has expanded within several disparate ecosystems and along distinct evolutionary trajectories. Although this report focuses strictly on genetic divergence as a measure of H5N1 classification, clearly additional studies on H5N1 antigenicity and cross‐clade protective immunity are needed to inform pre‐pandemic vaccine strain selection as well as risk management policies for both veterinary and human health. As always, continued surveillance for H5N1 in animals and humans is crucial for this process to be most effective.
Supporting information
Data S1. H5N1 evolution working group members.
Data S2. The nucleotide sequence alignment used to generate the tree shown in Figure 1 is provided as an electronic text file in FASTA format.
Figure S1. Neighbor‐joining tree of 2947 H5N1 HA sequences constructed using PAUP* v4.0b10. The tree was rooted using AturkeyEngland50921991
Figure S2. (A–G) Clade‐specific neighbor‐joining trees with 1000 bootstrap replicates were generated for each clade and rooted to either AgooseGuangdong196 or a common outlier to that clade.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Acknowledgements
We thank Justin Bahl, Ian H. Brown, Giovanni Cattoli, Todd Davis, Ruben O. Donis, Ron A.M. Fouchier, Elizabeth Mumford, Pierre Rivailler, Gavin J.D. Smith, and Ian A. York for drafting the manuscript on behalf of the H5N1 Evolution Working Group. We also thank Justin Bahl, Todd Davis, Pierre Rivailler, and Gavin Smith for performing sequence and phylogenetic analyses. We acknowledge access to sequence information from the GISAID database. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. This publication contains the collective views of an international group of experts and does not necessarily represent the decisions or the stated policy of the Food and Agriculture Organization of the United Nations (FAO), the World Organisation for Animal Health (OIE), or the World Health Organization (WHO).
© 2011 Blackwell Publishing Ltd. The World Health Organization retains copyright and all other rights in the manuscript of this article as submitted for publication.
References
- 1. WHO/OIE/FAO H5N1 Evolution Working Group . Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: divergence of clade 2.2 viruses. Influenza Other Respi Viruses 2009; 3:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization H5N1 Evolution Working Group . Towards a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infec Dis 2008; 14:e1. doi: 10.3201/eid1407.071681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta. Sunderland, MA: Sinauer Associates, 2001. [Google Scholar]
- 5. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 6. Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- 7. FAO . Approaches to controlling, preventing and eliminating H5N1 Highly Pathogenic Avian Influenza in endemic countries. Animal Production and Health Paper. No. 171. Rome, 2011.
- 8. Kayali G, Webby RJ, Ducatez MF et al. The epidemiological and molecular aspects of influenza H5N1 viruses at the human‐animal interface in Egypt. PLoS ONE 2011; 6:e17730. doi:10.1371/journal.pone.0017730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakoda Y, Sugar S, Batchluun D et al. Characterization of H5N1 highly pathogenic avian influenza virus strains isolated from migratory waterfowl in Mongolia on the way back from the southern Asia to their northern territory. Virology 2010; 406:88–94. [DOI] [PubMed] [Google Scholar]
- 10. L’Vov DK, Zhchelkanov M, Vlasov NA et al. The first break‐through of the genotype 2.3.2 of high‐virulence influenza A virus A/H5N1, which is new for Russia, in the Far East. Vopr Virusol 2008; 53:4–8. [PubMed] [Google Scholar]
- 11. Buchy P, Fourment M, Mardy S et al. Molecular epidemiology of clade 1 influenza A viruses (H5N1), Southern Indochina Peninsula, 2004–2007. Emerg Infect Dis 2009; 15:1641–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wan XF, Nguyen T, Davis CT et al. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS ONE 2008; 3:e3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis CT, Balish AL, O’Neill E et al. Detection and characterization of clade 7 high pathogenicity avian influenza H5N1 viruses in chickens seized at ports of entry and live poultry markets in Vietnam. Avian Dis 2010; 54(1 Suppl):307–312. [DOI] [PubMed] [Google Scholar]
- 14. Li Y, Shi J, Zhong G et al. Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J Virol 2010; 84:8389–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. H5N1 evolution working group members.
Data S2. The nucleotide sequence alignment used to generate the tree shown in Figure 1 is provided as an electronic text file in FASTA format.
Figure S1. Neighbor‐joining tree of 2947 H5N1 HA sequences constructed using PAUP* v4.0b10. The tree was rooted using AturkeyEngland50921991
Figure S2. (A–G) Clade‐specific neighbor‐joining trees with 1000 bootstrap replicates were generated for each clade and rooted to either AgooseGuangdong196 or a common outlier to that clade.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


