Abstract
Rationale: Obstructive sleep apnea is a common disorder associated with increased risk for cardiovascular disease, diabetes, and premature mortality. Although there is strong clinical and epidemiologic evidence supporting the importance of genetic factors in influencing obstructive sleep apnea, its genetic basis is still largely unknown. Prior genetic studies focused on traits defined using the apnea–hypopnea index, which contains limited information on potentially important genetically determined physiologic factors, such as propensity for hypoxemia and respiratory arousability.
Objectives: To define novel obstructive sleep apnea genetic risk loci for obstructive sleep apnea, we conducted genome-wide association studies of quantitative traits in Hispanic/Latino Americans from three cohorts.
Methods: Genome-wide data from as many as 12,558 participants in the Hispanic Community Health Study/Study of Latinos, Multi-Ethnic Study of Atherosclerosis, and Starr County Health Studies population-based cohorts were metaanalyzed for association with the apnea–hypopnea index, average oxygen saturation during sleep, and average respiratory event duration.
Measurements and Main Results: Two novel loci were identified at genome-level significance (rs11691765, GPR83, P = 1.90 × 10−8 for the apnea–hypopnea index, and rs35424364; C6ORF183/CCDC162P, P = 4.88 × 10−8 for respiratory event duration) and seven additional loci were identified with suggestive significance (P < 5 × 10−7). Secondary sex-stratified analyses also identified one significant and several suggestive associations. Multiple loci overlapped genes with biologic plausibility.
Conclusions: These are the first genome-level significant findings reported for obstructive sleep apnea–related physiologic traits in any population. These findings identify novel associations in inflammatory, hypoxia signaling, and sleep pathways.
Keywords: sleep apnea, hypoxia, genetics, GWAS, Prader–Willi syndrome
At a Glance Commentary
Scientific Knowledge on the Subject
Previous genetic studies of obstructive sleep apnea (OSA) have largely focused on candidate genes and have not found any associations at commonly accepted levels of significance. Previous studies have also focused on the apnea–hypopnea index, which may not fully reflect information on key OSA physiologic parameters. No studies have systematically examined genetic associations with OSA in Hispanics/Latinos.
What This Study Adds to the Field
In the largest genetic study of OSA to date, we investigate the genetics of the apnea–hypopnea index, average oxyhemoglobin saturation across the sleep episode, and average apnea/hypopnea event duration length in Hispanic/Latino Americans, a growing population with a high risk of OSA and related comorbidities. We identify three loci significantly associated with OSA traits, and several suggestively associated loci. Several of the identified loci contain genes with plausible OSA-related functions.
Obstructive sleep apnea (OSA) is a common disorder characterized by recurrent episodes of partial or complete upper airway obstruction that result in intermittent hypoxemia and sleep disruption. Individuals with OSA are at increased risk for cardiovascular disease, diabetes, and other disorders that carry significant morbidity and premature mortality, underscoring the need to better understand the etiology of this disorder and to identify targets for novel interventions (1–3). Although there is strong evidence supporting the importance of familial, and specifically genetic factors in influencing OSA susceptibility (4), its genetic basis is not well understood. Moreover, the genetic risk of OSA is known to vary by continental ancestries; a study of 31 global ancestral informative markers in Brazilians has indicated ancestry-related differences of risk for OSA (5). No prior study has studied the genetic associations of OSA in Hispanic/Latino Americans, a growing demographic group with a high prevalence of OSA and associated cardiovascular disease risk factors (6–8).
The physiologic components underlying OSA include contributions from energy balance/obesity, craniofacial structure, upper airway neuronal control, ventilatory control, and inflammation (4). In this paper, we studied the apnea–hypopnea index (AHI) as the primary measure of OSA severity given its heritability (h2 = 0.37) (9) and widespread clinical use. We also examined two other traits. First, the average percentage of oxyhemoglobin saturation (SpO2) during the sleep period, a measure widely available and easily collected in large numbers of individuals using pulse oximetry. Mean sleep SpO2 correlates with AHI while also providing information on a key physiologic stress associated with OSA. Second, the average respiratory disturbance (apnea or hypopnea) length, a measure reflecting the respiratory arousal threshold, which has been postulated to be a phenotype informative for targeted interventions addressing respiratory stability (10).
In this study, we conducted the largest genome-wide association study (GWAS) of three OSA traits to date. Primary analyses were adjusted for body mass index (BMI) to increase the likelihood of identifying variants that influence OSA through obesity-independent pathways. We focus on Hispanic/Latino Americans, a growing population at high risk for OSA and related comorbidities. We also conducted sex-stratified analyses given differences in OSA clinical manifestations (6).
Methods
Study Sample
Data were derived from three community-based cohort studies using in-home overnight sleep studies: Hispanic Community Health Study/Study of Latinos (HCHS/SOL), Multi-Ethnic Study of Atherosclerosis (MESA), and the Starr County Health Studies (Starr) (11–13). These studies comprise all known Hispanic/Latino OSA studies with available genome-wide data. Demographic and basic clinical parameters are presented in Table 1. Additional characteristics of each study are provided in the Methods section of the online supplement.
Table 1.
Sample Description
| HCHS/SOL | MESA | Starr County | |
|---|---|---|---|
| N, AHI/sleep O2 saturation/event duration | 11,317/11,317/9,791 | 459/458/447 | 782/782/0 |
| Age, yr, mean (SD) | 46.17 (13.79) | 68.34 (9.20) | 52.34 (11.29) |
| Percent female | 59.1 | 52.8 | 71.9 |
| BMI, kg/m2, mean (SD) | 29.79 (6.00) | 30.07 (5.52) | 32.15 (6.78) |
| AHI, events/h, median (IQR) | 1.97 (0.42–6.62) | 17.00 (7.50–30.53) | 10.35 (3.60–20.78) |
| Sleep O2 saturation, %, mean (SD) | 96.45 (0.95) | 94.33 (1.56) | 94.65 (2.09) |
| Waking O2 saturation, %, mean (SD) | 96.94 (3.13) | 96.12 (1.37) | 95.93 (2.43) |
| Event duration, s, mean (SD) | 23.62 (7.98) | 21.21 (5.92) | NA |
| Percent sleep under 90% saturation, mean (SD) | 0.85 (3.14) | 3.84 (7.35) | 2.83 (8.79) |
| Percent asthma | 7.70 | 5.25 | NA |
| Percent COPD | 2.78 | 0.44 | NA |
| Percent diabetes | 19.55 | 27.47 | 47.90 |
| Percent hypertensive | 27.60 | 58.30 | 57.25 |
| Percent Central American | 10.37 | NA | 0.00 |
| Percent Cuban | 14.63 | 3.05 | 0.00 |
| Percent Dominican | 9.44 | 18.54 | 0.00 |
| Percent Mexican | 39.51 | 49.77 | 100.00 |
| Percent Puerto Rican | 16.48 | 14.55 | 0.00 |
| Percent South American | 6.49 | NA | 0.00 |
| Percent other Hispanic | 3.08 | 14.08 | 0.00 |
Definition of abbreviations: AHI = apnea–hypopnea index; BMI = body mass index; COPD = chronic obstructive pulmonary disease; HCHS/SOL = Hispanic Community Health Study/Study of Latinos; IQR = interquartile range; MESA = Multi-Ethnic Study of Atherosclerosis; NA = not available; Starr = Starr County Health Studies.
Sample sizes are listed for AHI (12,558)/average O2 saturation (12,557)/average event duration (10,238) individuals with genotypes and phenotypes. Event duration sample size is reduced because of availability in Starr County and the exclusion of individuals without an apnea or hypopnea event. Population groups are self-identified for Hispanics/Latinos residing in the United States.
Phenotype and Covariate Definitions
The quantitative phenotypic outcomes were (1) AHI (defined by events associated with a ≥3% desaturation); (2) average hemoglobin oxygen saturation across the sleep period (SpO2; excluding intermittent waking episodes); and (3) average length of apnea/hypopnea events (“event duration,” defined as the average length of respiratory disturbances lasting 10 s or more). The sleep apnea assessments were based on type 3 home sleep apnea testing monitors for HCHS/SOL (ARES Unicorder 5.2; Advanced Brain Monitoring, Inc. [formerly B-Alert], Carlsbad, CA) and Starr (WatchPAT-200; Itamar Medical Ltd., Caesarea, Israel); and with 14-channel at-home polysomnography for MESA (Compumedics Somte; Compumedics, Ltd., Abbotsford, VIC, Australia). Further descriptions of equipment and more detailed definition of phenotypes are provided in the Methods section of the online supplement. All sleep records were scored by a central Sleep Reading Center at Brigham and Women's Hospital with high levels of established reliability (14). Covariates were obtained by questionnaires, direct measurement (BMI), and oximetry (waking oxygen saturation was measured before the sleep recording).
Genotyping and Quality Control
HCHS/SOL participants were assayed with an Illumina Omni 2.5M chip (Illumina, Inc., San Diego, CA). MESA and Starr participants were assayed with an Affymetrix 6.0 chip (Affymetrix, Inc., Santa Clara, CA). Imputation based on the 1000 Genomes Project (15) and quality control is described in the online supplement.
All single-nucleotide polymorphisms (SNPs) and insertion/deletions with an IMPUTE2 (16) Info score less than 0.88, minor allele frequency less than 1%, or fewer than 20 minor allele counts within a cohort were removed from applicable analyses.
Statistical Analysis
AHI and SpO2 values were inverse-normal rank-normalized and event duration values were log-transformed (see Methods section of the online supplement) to meet statistical assumptions for normality. Residuals were constructed for each trait using identical covariates. The primary model adjusted for age, age2, sex, age × sex, BMI, and BMI2. Population stratification and cryptic relatedness were controlled for using a linear mixed model or population-specific principal components, as described in the online supplement.
GWAS were performed on the residuals using an additive model for SNP effects in linear mixed models (17, 18). A fixed effect, inverse variance weighted metaanalysis was performed using METAL with genomic control applied to each cohort's results (19). Statistically significant and suggestive thresholds were set at P less than 5.0 × 10−8 and P less than 5.0 × 10−7, respectively. The choice of P less than 5 × 10−8 reflects a Bonferroni correction of 1 million independent tests, following a long-standing tradition in genome-wide studies (20, 21). Other choices, including calculating the number of independent linkage disequilbrium blocks, may be less effective when considering differences in admixture patterns. Secondary models were stratified by sex. To reduce the possibility of false-positives caused by small sample sizes, we only report results for SNPs where there was a minimum sample size of 1,000. We defined a locus region to include all SNPs with P less than 1.0 × 10−6 that were within 500 kb of the lead SNP. LocusZoom (22) was used for regional visualization with SNP linkage disequilibrium calculated within HCHS/SOL (comprised of six Hispanic/Latino population background groups).
Results
Study Sample
An overview of sample characteristics is shown in Table 1. As many as 12,558 individuals contributed to the analyses of AHI and average SpO2 in sleep. Fewer participants contributed to the analysis of average event duration (10,238) because of the exclusion of individuals with no respiratory disturbances and an absence of this phenotype in the Starr County sample. The mean ages across cohorts were 46–68 (HCHS/SOL, 46.1 [SD = 13.8]; Starr, 52.3 [SD = 9.2]; MESA, 68.3 [SD = 11.3]). The percentage of female participants ranged from 53 to 72% (MESA, 52.8%; HCHS/SOL, 59.1%; Starr, 71.9%). The largest demographic subgroup was of Mexican background (including all of the Starr participants), with additional self-identified Caribbean, Central American, and South American backgrounds. Average BMI varied from 29.8 to 32.2 across cohorts (HCHS/SOL, 29.8; MESA, 30.1; Starr, 32.2). Type 2 diabetes was common in the cohorts (HCHS/SOL, 19.6%; MESA, 27.5%; Starr, 47.9%). The median AHI was lower in HCHS/SOL relative to other cohorts (HCHS/SOL, 1.97; Starr, 10.35; MESA, 17.00), likely reflecting the younger age and lower BMI of this cohort.
Pairwise Correlations between OSA Phenotypes
Pairwise correlations between OSA phenotypes, calculated across all three cohorts, are presented in Table 2. AHI was strongly correlated with average sleep SpO2 and modestly correlated with event duration (pooled Spearman ρ = −0.608 and 0.220, respectively). A weak correlation between sleep SpO2 and event duration was also observed (ρ = −0.089). Waking SpO2 was modestly correlated with average sleep SpO2 and with AHI (ρ = 0.303 and −0.155, respectively). Both age and BMI were most strongly correlated with AHI, followed by sleep SpO2 and event duration, respectively. Cohort-specific cross-phenotype correlations results are shown in Table E1 in the online supplement.
Table 2.
Pairwise Correlations between OSA Phenotypes and Select Covariates
| AHI | Sleep SpO2 | Event Duration | Wake SpO2 | Age | |
|---|---|---|---|---|---|
| Sleep SpO2 | −0.608 (−0.619 to −0.596) | — | — | — | — |
| Event duration | 0.220 (0.201 to 0.240) | −0.089 (−0.109 to −0.068) | — | — | — |
| Wake SpO2 | −0.155 (−0.173 to −0.137) | 0.303 (0.286 to 0.319) | 0.003 (−0.018 to 0.023) | — | — |
| Age | 0.427 (0.411 to 0.442) | −0.391 (−0.406 to −0.375) | 0.220 (0.200 to 0.239) | −0.128 (−0.146 to −0.109) | — |
| BMI | 0.375 (0.359 to 0.391) | −0.300 (−0.317 to −0.283) | −0.127 (−0.147 to −0.106) | −0.063 (−0.081 to −0.044) | 0.083 (0.064 to 0.101) |
Definition of abbreviations: AHI = apnea–hypopnea index; BMI = body mass index; OSA = obstructive sleep apnea; SpO2 = average percentage of oxyhemoglobin saturation.
Each table cell lists Spearman ρ correlations metaanalyzed using Fisher Z-transformations weighted by sample size. Ninety-five percent confidence intervals are listed in parentheses.
GWAS Results
We performed metaanalyses of GWAS for AHI, average sleep SpO2, and respiratory (apnea and/or hypopnea) event duration. Manhattan and QQ plots for analyses across both sexes and stratified by sex are presented in Figures E1–E3. The QQ plots and genomic inflation factors showed little inflation (range, 0.980–1.015). Lead SNPs for statistically significant or suggestive associations with AHI, sleep SpO2, and event duration are shown in Tables 3–5.
Table 3.
Genome-Level Significant and Suggestive Associations with AHI
| SNP | B37 Region | SNPs in Region | Genes in Region | N | CAF | β (SE) BMI Adjusted | β (SE) BMI Unadjusted | P Value | Direction |
|---|---|---|---|---|---|---|---|---|---|
| rs116791765 T | 11q21:94,074,914–94,242,697 | 28 | GPR83, LINC01171, MRE11A | 11,774 | 0.989/0.988/NA | −0.323 (0.057) | −0.345 (0.061) | 1.90 × 10−8 (1.84 × 10−8) | −+? |
| rs999944 A | 2p14:65,049,853 | 1 | 12,557 | 0.112/0.106/0.073 | −0.095 (0.019) | −0.104 (0.020) | 4.53 × 10−7 (2.48 × 10−7) | −+− |
Definition of abbreviations: AHI = apnea–hypopnea index; BMI = body mass index; CAF = coded allele frequency; NA = not available; SNP = single-nucleotide polymorphism.
Lead SNPs for regions with significant (P < 5.0 × 10−8) P values. Individual regions were minimally 500 kb apart. Minimum n = 1,000. SNPs in region denotes the count of regional SNPs with P less than 1.0 × 10−6. Genes in region indicates overlapping nonpseudogene Ensembl genes within 5 kb of the P less than 1 × 10−6 SNPs. P values in parentheses were obtained from equivalent BMI-unadjusted model results. Individual regional SNP results are provided in Table E2. BMI-adjusted and unadjusted directions are equivalent except where noted. CAF and direction columns are ordered from left to right as Hispanic Community Health Study/Study of Latinos, Multi-Ethnic Study of Atherosclerosis, and Starr County Health Studies.
Table 5.
Genome-Level Significant and Suggestive Associations with Average Event Duration
| SNP | B37 Region | SNPs in Region | Genes in Region | N | CAF | β (SE) BMI Adjusted | β (SE) BMI Unadjusted | P Value | Direction |
|---|---|---|---|---|---|---|---|---|---|
| rs35424364 A | 6q21:109,584,988–109,643,605 | 3 | C6ORF183, CCDC162P | 10,240 | 0.129/0.097/NA | 0.030 (0.006) | 0.032 (0.006) | 4.88 × 10−8 (1.82 × 10−8) | ++ |
| rs74472562 T | 11p11:44,762,755 | 1 | RP11-45A12.2, TSPAN18 | 10,240 | 0.086/0.092/NA | −0.034 (0.006) | −0.035 (0.006) | 1.05 × 10−7 (6.47 × 10−8) | −− |
| rs2743173 T | 20p12:8,245,293 | 1 | PLCB1 | 10,240 | 0.567/0.570/NA | 0.019 (0.004) | 0.019 (0.004) | 3.85 × 10−7 (2.23 × 10−7) | ++ |
| rs72699765 A | 15q12:25,809,781–25,829,197 | 4 | AC124997.1 | 10,240 | 0.110/0.112/NA | 0.029 (0.006) | 0.029 (0.006) | 3.93 × 10−7 (5.20 × 10−7) | ++ |
| rs2033354 T* | 2p24:15,212,995–15,218,152 | 5 | 10,240 | 0.784/0.779/NA | −0.022 (0.004) | −0.022 (0.004) | 4.77 × 10−7 (3.65 × 10−7) | −− |
Definition of abbreviations: BMI = body mass index; CAF = coded allele frequency; NA = not available; SNP = single-nucleotide polymorphism.
P values in parentheses were obtained from equivalent BMI-unadjusted model results. CAF and direction columns are ordered from left to right as Hispanic Community Health Study/Study of Latinos and Multi-Ethnic Study of Atherosclerosis.
Heterogeneity P = 0.012.
Table 4.
Suggestive Associations with Sleep SpO2
| SNP | B37 Region | SNPs in Region | Genes in Region | N | CAF | β (SE) BMI Adjusted | β (SE) BMI Unadjusted | P Value | Direction |
|---|---|---|---|---|---|---|---|---|---|
| rs75108997 A | 1q42:233,947,565–233,954,231 | 4 | 11,809 | 0.013/0.014/NA | −0.291 (0.055) | −0.296 (0.058) | 1.40 × 10−7 (3.52 × 10−7) | −−? | |
| rs116133558 T | 1q32:203,716,843 | 1 | ATP2B4 | 11,351 | 0.014/NA/NA | 0.291 (0.056) | 0.231 (0.059) | 2.18 × 10−7 (9.09 × 10−5) | +?? |
Definition of abbreviations: BMI = body mass index; CAF = coded allele frequency; NA = not available; SNP = single-nucleotide polymorphism; SpO2 = average percentage of oxyhemoglobin saturation.
P values in parentheses were obtained from equivalent BMI-unadjusted model results. CAF and direction columns are ordered from left to right as Hispanic Community Health Study/Study of Latinos, Multi-Ethnic Study of Atherosclerosis, and Starr County Health Studies.
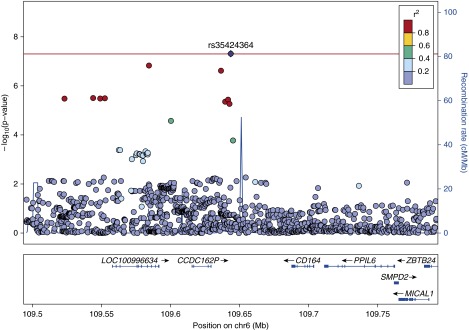
A significant association with AHI was found at chr 11q21 (lead SNP rs116791765, P = 1.90 × 10−8). The locus included 28 SNPs with P less than 1 × 10−6 and spanned several genes (GPR83, LINC01171/C11ORF97, MRE11A) (Table 3, Figure 1). A significant association with event duration was found at chr 6q21 (lead SNP rs35424364, P = 4.88 × 10−8). This locus included three SNPs with P less than 1 × 10−6 that spanned the pseudogenes CCDC162P and C6ORF183/LOC100996694 (Table 5, Figure 2; see Table E2).
Figure 1.
rs116791765 apnea–hypopnea index regional association plot. Physical positions (Build 37 coordinates) are shown on the x-axis. Log-transformed P values are shown on the y-axis. Single-nucleotide polymorphism (SNP) colors indicate the degree of linkage disequilibrium with the lead SNP rs116791765 (based on Hispanic Community Health Study/Study of Latinos genotypes). Regional recombination rates are denoted with the blue line, with recombination hotspots indicated by large increases in the recombination rates. Although rs116791765 is located in an intergenic region, the entire locus with P values < 1 × 10−6 spans three genes (GPR83, MRE11A, and ANKRD49). A fourth gene (ANKRD49) is within the region but does not contain any SNPs with P values < 1 × 10−6.
Figure 2.
rs35424364 event duration regional association plot.
Additionally, there were one AHI, two SpO2, and four event duration signals with suggestive evidence of association (P < 5 × 10−7) (Tables 3–5). These signals included SNPs that overlap biologically plausible genes, including rs2743173 (event duration; PLCB1) (see Figure E8) and rs116133558 (sleep SpO2; ATP2B4/PMCA4B) (see Figure E6). Additional details and results for SNPs at each locus with P less than 1 × 10−6 are listed in Table E2. Regional association plots are shown in Figures E4–E21. Regional plots for the significant associations are provided in Figures E22–E29. Forest plots for all lead loci effect sizes and standard errors within individual cohorts are shown in Figures E30–E36.
OSA likely reflects genetic factors that operate through BMI-dependent and BMI-independent pathways (9). Results for the top BMI-adjusted model regions were broadly similar using a BMI-unadjusted model (Tables 3–5), suggesting that these novel loci do not contribute to OSA through obesity.
Sex-stratified analyses identified six additional suggestive AHI regions, one suggestive sleep episode SpO2 region, and one significant and four suggestive event duration regions (Table 6; see Table E3).
Table 6.
Suggestive Sex-stratified Associations
| Model | SNP | B37 Region | SNPs in Region | Genes in Region | N | CAF | β (SE) BMI Adjusted | β (SE) BMI Unadjusted | P Value | Direction |
|---|---|---|---|---|---|---|---|---|---|---|
| AHI females | rs199803244 A | 12p11:27,645,082–27,659,326 | 3 | SMCO2 | 6,691 | 0.945/NA/NA | 0.182 (0.035) | 0.187 (0.037) | 1.64 × 10−7 (4.98 × 10−7) | +?? |
| rs11897825 A | 2p24:21,694,451 | 1 | AC011752.1, AC067959.1 | 6,691 | 0.606/NA/NA | −0.085 (0.017) | −0.090 (0.018) | 4.34 × 10−7 (5.26 × 10−7) | −?? | |
| AHI males | rs11588454 T | 1q31:191,618,878–191,808,947 | 14 | 5,062 | 0.750/0.710/0.701 | −0.114 (0.021) | −0.124 (0.023) | 6.97 × 10−8 (8.04 × 10−8) | −+− | |
| rs140743827 A | 1q23:164,985,756 | 1 | 4,626 | 0.018/NA/NA | 0.392 (0.076) | 0.428 (0.083) | 2.26 × 10−7 (2.27 × 10−7) | +?? | ||
| rs4796285 A | 17q12:35,181,996 | 1 | 5,062 | 0.581/0.556/0.593 | −0.096 (0.019) | −0.092 (0.020) | 3.09 × 10−7 (7.15 × 10−6) | −+− | ||
| rs111942351 A | 5q31:143,073,550 | 1 | CTB-57H20.1 | 4,842 | 0.981/0.984/NA | 0.363 (0.071) | 0.338 (0.078) | 3.75 × 10−7 (1.34 × 10−5) | ++? | |
| SpO2 males | rs11074782 T | 16p12:26,505,011 | 1 | 5,076 | 0.230/0.205/0.263 | −0.112 (0.022) | −0.113 (0.024) | 2.95 × 10−7 (1.55 × 10−6) | −+− | |
| Event duration females | rs78897171 A | 20q13:60,443,251 | 1 | CDH4 | 5,589 | 0.023/NA/NA | 0.106 (0.020) | 0.108 (0.020) | 1.63 × 10−7 (1.12 × 10−7) | +? |
| rs35329661 T | 11q13:74,974,990 | 1 | ARRB1, CTD-2562J17.7, CTD-2562J17.9 | 5,589 | 0.013/NA/NA | 0.132 (0.026) | 0.134 (0.026) | 2.80 × 10−7 (2.06 × 10−7) | +? | |
| Event duration males | rs148024591 T | 15q26:93,906,988–93,927,482 | 10 | RP11-266O8.1 | 4,414 | 0.017/0.015/NA | 0.119 (0.021) | 0.117 (0.022) | 2.81 × 10−8 (6.72 × 10−8) | ++ |
| rs4849682 T | 2q14:118,911,223–118,942,883 | 2 | AC093901.1 | 4,414 | 0.290/0.320/NA | −0.032 (0.006) | −0.030 (0.006) | 1.07 × 10−7 (4.56 × 10−7) | −− | |
| rs11610782 A | 12q24:126,196,289–126,256,105 | 2 | 4,204 | 0.024/NA/NA | 0.109 (0.021) | 0.108 (0.021) | 3.01 × 10−7 (3.69 × 10−7) | +? |
Definition of abbreviations: AHI = apnea–hypopnea index; BMI = body mass index; CAF = coded allele frequency; NA = not available; SNP = single-nucleotide polymorphism; SpO2 = average percentage of oxyhemoglobin saturation.
P values in parentheses were obtained from equivalent BMI-unadjusted model results. CAF and direction columns are ordered from left to right as Hispanic Community Health Study/Study of Latinos, Multi-Ethnic Study of Atherosclerosis, and Starr County. Event duration was not collected in Starr County.
For AHI, rs11588454 was marginally above genome-level significance for association in males (P = 6.97 × 10−8). The 190-kb locus region with SNP P values less than 1 × 10−6 is intergenic. RGS18 is the nearest protein-coding gene (see Figure E13).
For event duration, rs148024591 was significantly associated in males (P = 2.81 × 10−8; 10 regional SNPs P < 1 × 10−6) (Figure 3). rs148024591 is within the lincRNA gene RP11-226O8.1 (Ensembl ID ENSG00000257060) and 282 kb away from the closest protein-coding gene RGMA. Interestingly, among the suggestive loci SNPs in males was rs4849682 (P = 1.07 × 10−7). This SNP is located in the lincRNA AC093901.1 (Ensembl ID ENSG00000226856) and downstream of the candidate gene INSIG2 (see Figure E20) (23). Among the suggestive loci in females was an association (rs35329661, P = 2.80 × 10−7) near ARRB1, a gene known to interact with HIF1A (see Discussion). A comparison of sex-stratified results of the lead SNPs identified by combined and stratified analyses is provided in Table E3.
Figure 3.
rs148024591 event duration in males regional association plot. Note the two recombination hotspots that bound the P value < 1 × 10−6 region. Although rs148024591 is distant from the closest protein-coding gene, RGMA, it is physically located within the lincRNA gene RP11-226O8.1 (Ensembl ID ENSG00000257060).
We tested if all 21 loci significantly or suggestively associated with AHI, average SpO2, or event duration were associated with the other two OSA traits. Consistent with the crude cross-phenotype correlations reported previously, all lead AHI and sleep SpO2 SNPs showed the expected inverse effect directionality and significance levels of P less than 0.05 when compared with the estimates for the other OSA trait (see Table E4). The lead event duration SNPs showed no consistent evidence of association for signals identified for the other two traits, with the exception of rs78897171 in females (AHI, P = 3.28 × 10−3; sleep SpO2, P = 5.78 × 10−3).
Association Signal Generalizability into Other Populations
To our knowledge, no additional genome-wide studies of OSA in Hispanics/Latinos exist apart from our cohorts, limiting our ability to replicate. We examined the possible generalizability of our signals into Asian Americans (minimal, n = 214), African Americans (minimal, n = 1,183), and European Americans (minimal, n = 5,638) from the Atherosclerosis Risk In Communities, Cleveland Family Study, Framingham Heart Study, MESA, and Osteoporotic Fractures in Men studies (see Methods section of the online supplement). No consistent evidence of association in other ethnic groups for the top SNPs was observed (see Table E5). The Asian American and African American sample sizes, however, were relatively small, providing limited ability to assess generalizability.
We also examined generalizability of previously reported AHI candidate gene results to our Hispanic/Latino cohorts (see Table E6) (23–31). Of 17 SNPs tested for association with AHI, only rs10097555 in NRG1 was nominally significant (P = 0.034; A allele, β [SE] = −0.025 [0.012]). rs1421085, a FTO region variant that is causative in obesity-related traits (32), was nominally significant using a BMI-unadjusted model (P = 0.012; T allele, β [SE] = −0.036 [0.015]).
Bioinformatics Data Mining: Correlated Functional SNPs and Mouse Phenotypes
We searched for SNPs with supporting ENCODE or Roadmap Epigenomics regulatory evidence as calculated by HaploReg (33). A total of 33 SNPs with an association P value less than 1 × 10−6 and located within a significant or suggestive region overlapped promoter, enhancer, or DNase I hypersensitivity regions (see Table E7). Notable SNPs include rs74472562 (event duration, P = 1.05 × 10−7), which overlaps enhancer marks in 60 cell lines; rs4849682 (event duration in men, P = 1.07 × 10−7), which overlaps promoter marks in 82 cell lines and enhancer marks in 45 cell lines; and rs35329661 (event duration in females, P = 2.80 × 10−7), which overlaps enhancers in 54 cell lines. All three SNPs and 10 others overlapped marks in central nervous system–related cell lines. Two genome-level significant SNPs displayed regulatory evidence: rs116791765 (one promoter mark in embryonic stem cells) and rs35424364 (five enhancer marks in blood cell lines).
Four genes in suggestive regions have been analyzed in ongoing work by the International Knockout Mouse Consortium: Arrb1, Cdh4, Smco2, and Tspan18 (see Table E8) (34). Potentially relevant phenotypes for these associations were reported for body fat (Arrb1 and Smco2); and fasting glucose, immune system functioning, and mean corpuscular hemoglobin (Tspan18).
Discussion
In the present study, we identified several significant genetic associations with OSA-related phenotypes, using the largest objectively measured sample assembled to date in any ethnic group, and the first in Hispanic/Latino Americans. In total, we detected two significant and nine suggestive regions (P < 5 × 10−8; 5 × 10−7) in the primary analyses and one significant and 11 suggestive regions in sex-stratified analyses. Multiple genes in associated regions have plausible OSA-related functions, including GPR83, ATP2B4, ARRB1, INSIG2, and PLCB1.
The genetics of OSA remain largely unknown. Research in this area has been limited by the relatively small numbers of participants that have undergone overnight sleep studies and have genome-wide assays. Prior research has consisted predominantly of candidate gene studies and has had limited power. That body of work identified potential associations of variants in genes including CRP, GDNF, and HTR2A; LPAR1, PLEK, and PTGER3; PPARGC1B; NRG1; FTO; TRABD2B; SLC64A; LEPR; and TNF (23–31). To our knowledge, however, no loci have been identified at the generally accepted significance threshold of 5.0 × 10−8 and most results have not been replicated. Large samples are usually required to identify common variants with individually small effects that have been typical of most complex traits (35).
GWAS can help identify genomic regions containing causal variants or variants influencing the expression of genes outside of the identified region (e.g., enhancers and/or expression quantitative trait loci). Although the current study cannot establish causal mechanisms, several genes in our identified regions have published biologic relationships that suggest prioritization in future studies. The rs116791765 locus on chr 11q21 that associated with AHI includes the G-protein receptor GPR83 (formerly Gir). Rodent studies show that this protein is expressed in multiple brain regions of relevance to OSA, including the hypoglossal nuclei, dorsal motor nucleus of vagus nerve, and the nucleus of solitary tract (36). Gpr83 down-regulation in mice leads to decreased core body temperature and increases in weight and circulating adiponectin (37). Gpr83 knockout mice display differences in expression of the sleep- and wake-related genes Npy and Hcrt in the hypothalamus (38). GPR83 is also highly expressed in Fox3p regulatory T (Treg) cells, which are important in modulating inflammatory responses, and seem to be important in Treg cell development. Overexpression of a Gpr83 isoform in T cells interferes with inflammatory responses in vivo (39). Hypoxia impacts the relative abundance of Treg cells compared with Th17 cells, both in vivo and in vitro (40). Alterations in the balance of Treg to Th17 cells, an indication of autoimmunity and inflammation, have been associated with OSA severity. Following tonsillectomy, a significant reduction in the AHI was accompanied by a significant Treg/Th17 ratio reversal (41, 42). Inflammation may weaken upper airway muscle and exacerbate OSA through effects on muscle fibers and stimulating nerves (43). Hypoxia-induced chemoreceptor responses are muted with antiinflammatory drugs (44).
In addition to AHI, the most common clinical metric of OSA severity, we examined additional measures that provide complementary information on relevant physiologic pathways. We studied average oxygen saturation across the sleep period, an index of hypoxemia, one of the key physiologic stresses associated with OSA. Variation in oxyhemoglobin saturation levels may reflect frequency and duration of respiratory disturbances, the magnitude of work of breathing, baseline oxygen saturation and pulmonary function, and possibly pulmonary vascular and parenchymal responses to the recurrent “stress” associated with hypoxemia. SpO2 is measured with a simple oximeter and is easily scalable to the sample sizes required for genetic epidemiologic studies.
The relevance of the average sleep SpO2 phenotype to genetics of OSA is supported by the high phenotypic correlation between SpO2 and AHI, and evidence of overlapping genetic signals for this trait and for the AHI (see Table E4). The sleep SpO2-associated locus at chr 1q32 includes ATP2B4 (formerly PMCA4B), the main cellular membrane calcium pump in erythrocytes (45). ATP2B4 also negatively regulates nitric oxide bioavailability in endothelial cells (46), impacts blood pressure and regulates vascular tone (47), and thus may have pleiotropic effects relevant to gas exchange during sleep.
Wellman and colleagues have proposed that four physiologic traits affect OSA susceptibility, including the respiratory arousal threshold, defined as the intensity of respiratory stimuli (e.g., negative pharyngeal pressure and increase of carbon dioxide concentration) required to invoke awakening (10, 48). As a surrogate for respiratory arousability, we analyzed average apnea and hypopnea event duration length, with shorter duration events indicating greater arousability (49). This phenotype was only moderately correlated with AHI (ρ = 0.220). Analysis of this unique phenotype identified multiple regions containing biologically plausible genes, including ARRB1, which physically interacts with and regulates HIF1A, which plays a central role in hypoxia sensing. ARRB1 affects HIF1A stabilization in normoxia and impacts the expression of HIF1A target genes, including VEGFA (50). rs35329661, the lead SNP in the region, overlaps enhancer histone marks in 54 cell lines (33). The suggestive locus at 15q12 (see Figure E9) is located within the Prader–Willi syndrome region (15q11–15q13). Patients with Prader–Willi have an altered hypercapnic arousal response. The prevalence of OSA in pediatric patients across 14 studies was 79% (51, 52).
rs4849682, showing suggestive significance with event duration, is located downstream of the insulin signaling gene INSIG2 (see Figure E20), a candidate OSA gene known to block the actions of sterol regulatory element binding protein (SREBP), which in turn mediates intermittent hypoxia-induced hyperlipidemia (21, 53, 54). rs4849682 may also impact the lincRNA AC093901.1 (Ensembl ID ENSG00000226856) caused by overlaps with promoter histone marks in multiple cell lines (33).
rs2743173, also showing association with event duration, is located within PLCB1. This gene has been shown to positively regulate SREBP signaling and to affect insulin secretion (55, 56). Future work is required to determine if multiple loci invoking SREBP signaling and related pathways may contribute to variation in OSA because of alterations in insulin signaling, fat deposition, or ventilatory control effects. PLCB1 has also been shown to directly affect sleep. Plcb1 binds to muscarinic acetylcholine receptors in mice (57), and knockout mice have altered theta rhythms (58). A locus including PLCB1 was among the strongest (albeit nonsignificant) regions in a GWAS of insomnia in Koreans (59). The lead locus SNP rs6140722 displayed modest association with event duration in our population (P = 0.0030).
Several of the SNPs identified in the current analysis are located in known regulatory regions. Other loci overlap RNA genes. Future research is required to understand these regions and the event duration associated region near rs35424364 (with the pseudogenes CCDC162P and C6ORF183/LOC100996694). This region seems to be biologically active and is associated with red blood cell traits (60).
Our study contains multiple strengths when compared with prior genetic analyses of OSA. Most notably, the sample of 12,558 individuals with objective measures of OSA and genotype data represents the largest study to date on OSA genetics, and is over triple the sample size of our prior largest analysis of objectively phenotyped OSA (24). This boost in power has improved our ability to obtain statistically significant results, in which variants have small effect sizes, in line with other complex phenotypes. We used a stringent imputation quality score threshold to reduce random error. The use of dense imputation allowed us to measure SNPs that were poorly tagged by European-centric gene chip SNPs. For the first time, we have conducted genome-wide studies of sleep SpO2 and respiratory event duration. These traits may facilitate the genetic dissection of OSA, providing additional information on OSA-related physiology using relatively easily scalable measurements derived from ambulatory recordings.
Our study also has some weaknesses. Although this is the largest OSA study to date, identification of variants with small effects may require much larger sample sizes. We did not have an independent sample of Hispanic/Latino individuals for replication and were unable to detect generalization of Hispanic/Latino SNP associations in our much smaller sample of African American, Asian American, and European American ancestry samples. These small sample sizes make it difficult to distinguish lack of power from differences in allele frequency and/or mutations private to a subset of continental populations, or true allelic heterogeneity across populations. Opposite directionality may also have been observed because the top SNPs may be in linkage disequilibrium with one or more common or rare causal variants that vary by population and degree of admixture. We also identified multiple loci with opposite directionality among our cohorts. The three cohorts also varied in ancestral backgrounds, including a large proportion of HCHS/SOL individuals with Caribbean ancestry (with higher recent African admixture contributions). This seems to be of particular relevance for our AHI association near GPR83, with lead SNPs specifically enriched in African 1,000 Genomes Project populations with rare or nonexistent minor alleles in the American, Asian, and European populations. This metaanalysis focused on common genetic variants. Additional rare variants may be uncovered by future investigations that incorporate exome or whole-genome sequencing, particularly in a family-based context. Future replication is required to validate our findings, which incorporated all known Hispanic/Latino studies with available genotypes and phenotypes.
In summary, we have identified several genome-level significant associations with OSA-related traits in Hispanic/Latino Americans in the largest genetic analysis conducted to date in any population, and also identify the potential utility of two novel OSA phenotypes for use in future genetic studies.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the staff and participants of the Hispanic Community Health Study/Study of Latinos for their important contributions. A complete list of staff and investigators is available on the study website http://www.cscc.unc.edu/hchs/. The authors thank the investigators, the staff, and the participants of the Multi-Ethnic Study of Atherosclerosis for their valuable contributions. A full list of participating Multi-Ethnic Study of Atherosclerosis investigators and institutions can be found at http://www.mesa-nhlbi.org. We also thank the field staff in Starr County for their careful collection of these data and are especially grateful to the participants who so graciously cooperated and gave of their time. The authors thank the staff and participants of the Atherosclerosis Risk In Communities (ARIC) study for their important contribution. This manuscript was not prepared in collaboration with investigators of the ARIC Study and does not necessarily reflect the opinions or conclusions of the ARIC Study or the NHLBI.
Footnotes
Supported by National Institutes of Health grants T32-HL007901-16 and R01-HL113338-04 (B.E.C.). The Sleep Reading Center of Brigham and Women's Hospital has been supported by National Institutes of Health grants 5-R01-HL046380-15 and 5-KL2-RR024990-05. Hispanic Community Health Study/Study of Latinos (HCHS/SOL): baseline examination of HCHS/SOL was performed as a collaborative study supported by contracts from the NHLBI to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following institutes/centers/offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, and National Institutes of Health Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and National Institute of Dental and Craniofacial Research contracts (HHSN268201300005C AM03 and MOD03). Provision of genotyping services supported in part by National Center for Advancing Translational Sciences (NCATS) CTSI grant UL1TR000124 and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) DRC grant DK063491. Multi-Ethnic Study of Atherosclerosis (MESA): MESA is conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. Funding for SHARe genotyping was provided by NHLBI contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and Massachusetts Institute of Technology (Boston, MA). Funding support for the Sleep Polysomnography dataset was provided by grant HL56984. Provision of genotyping services supported in part by NCATS CTSI grant UL1TR000124 and NIDDK DRC grant DK063491. Starr County Health Studies (Starr): Starr is supported in part by grants R01 DK073541, U01 DK085501, R01 AI085014, and R01 HL102830 from the National Institutes of Health, and funds from the University of Texas Health Science Center at Houston. G.I.B. supported in part by grant P30 DK020595 and a gift from the Kovler Family Foundation. The Atherosclerosis Risk in Communities Study is conducted and supported by the NHLBI in collaboration with the University of North Carolina (N01-HC-55015, N01-HC-55018), Baylor College of Medicine (N01-HC-55016), University of Minnesota (N01-HC-55019), Johns Hopkins University (N01-HC-55020), and University of Mississippi Medical Center (N01-HC-55021).
Author Contributions: Conception and design, S.A.-I., R.A., J.S.L., K.L.S., G.J.T., P.C.Z., C.L.H., S.R.P., C.C.L., and S.R. Data acquisition, B.E.C., H.C., A.M.S., K.J.G., T.S., G.I.B., J.E.B., A.C.B., S.C., M.P.C., D.S.E., W.C.J., A.C.F.-W., J.M.L., E.K.L., W.S.P., A.R.R., K.R., J.I.R., N.A.S., K.D.T., T.A.T., C.W., S.R.S., X.Z., R.S., X.L., and S.R. Analysis, all authors. Interpretation, draft and review, and final approval, all authors. B.E.C. and S.R. had full access to the study data and take responsibility for the integrity of the data and accuracy of analyses.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201512-2431OC on March 15, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Marin JM, Carrizo SJ, Vicente E, Agusti AG. Long-term cardiovascular outcomes in men with obstructive sleep apnoea-hypopnoea with or without treatment with continuous positive airway pressure: an observational study. Lancet. 2005;365:1046–1053. doi: 10.1016/S0140-6736(05)71141-7. [DOI] [PubMed] [Google Scholar]
- 2.Gottlieb DJ, Yenokyan G, Newman AB, O’Connor GT, Punjabi NM, Quan SF, Redline S, Resnick HE, Tong EK, Diener-West M, et al. Prospective study of obstructive sleep apnea and incident coronary heart disease and heart failure: the Sleep Heart Health study. Circulation. 2010;122:352–360. doi: 10.1161/CIRCULATIONAHA.109.901801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kendzerska T, Gershon AS, Hawker G, Tomlinson G, Leung RS. Obstructive sleep apnea and incident diabetes: a historical cohort study. Am J Respir Crit Care Med. 2014;190:218–225. doi: 10.1164/rccm.201312-2209OC. [DOI] [PubMed] [Google Scholar]
- 4.Redline S. Genetics of obstructive sleep apnea. In: Kryger MH, Roth T, Dement WC, editors. Principles and practice of sleep medicine. 5th ed. St. Louis, MO: Saunders; 2011. pp. 1183–1193. [Google Scholar]
- 5.Guindalini C, Colugnati FA, Pellegrino R, Santos-Silva R, Bittencourt LR, Tufik S. Influence of genetic ancestry on the risk of obstructive sleep apnoea syndrome. Eur Respir J. 2010;36:834–841. doi: 10.1183/09031936.00146809. [DOI] [PubMed] [Google Scholar]
- 6.Redline S, Sotres-Alvarez D, Loredo J, Hall M, Patel SR, Ramos A, Shah N, Ries A, Arens R, Barnhart J, et al. The Hispanic Community Health Study/Study of Latinos. Sleep-disordered breathing in Hispanic/Latino individuals of diverse backgrounds. Am J Respir Crit Care Med. 2014;189:335–344. doi: 10.1164/rccm.201309-1735OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daviglus ML, Talavera GA, Avilés-Santa ML, Allison M, Cai J, Criqui MH, Gellman M, Giachello AL, Gouskova N, Kaplan RC, et al. Prevalence of major cardiovascular risk factors and cardiovascular diseases among Hispanic/Latino individuals of diverse backgrounds in the United States. JAMA. 2012;308:1775–1784. doi: 10.1001/jama.2012.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneiderman N, Llabre M, Cowie CC, Barnhart J, Carnethon M, Gallo LC, Giachello AL, Heiss G, Kaplan RC, LaVange LM, et al. Prevalence of diabetes among Hispanics/Latinos from diverse backgrounds: the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) Diabetes Care. 2014;37:2233–2239. doi: 10.2337/dc13-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel SR, Larkin EK, Redline S. Shared genetic basis for obstructive sleep apnea and adiposity measures. Int J Obes (Lond) 2008;32:795–800. doi: 10.1038/sj.ijo.0803803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellman A, Eckert DJ, Jordan AS, Edwards BA, Passaglia CL, Jackson AC, Gautam S, Owens RL, Malhotra A, White DP. A method for measuring and modeling the physiological traits causing obstructive sleep apnea. J Appl Physiol (1985) 2011;110:1627–1637. doi: 10.1152/japplphysiol.00972.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, Schneiderman N, Raij L, Talavera G, Allison M, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20:629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 13.Hanis CL, Ferrell RE, Barton SA, Aguilar L, Garza-Ibarra A, Tulloch BR, Garcia CA, Schull WJ. Diabetes among Mexican Americans in Starr County, Texas. Am J Epidemiol. 1983;118:659–672. doi: 10.1093/oxfordjournals.aje.a113677. [DOI] [PubMed] [Google Scholar]
- 14.Whitney CW, Gottlieb DJ, Redline S, Norman RG, Dodge RR, Shahar E, Surovec S, Nieto FJ. Reliability of scoring respiratory disturbance indices and sleep staging. Sleep. 1998;21:749–757. doi: 10.1093/sleep/21.7.749. [DOI] [PubMed] [Google Scholar]
- 15.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conomos MP.Inferring, estimating and accounting for population and pedigree structure in genetic analysesPh.D. thesis 2014. Seattle: University of Washington) [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 21.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 22.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin EK, Patel SR, Goodloe RJ, Li Y, Zhu X, Gray-McGuire C, Adams MD, Redline S. A candidate gene study of obstructive sleep apnea in European Americans and African Americans. Am J Respir Crit Care Med. 2010;182:947–953. doi: 10.1164/rccm.201002-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SR, Goodloe R, De G, Kowgier M, Weng J, Buxbaum SG, Cade B, Fulop T, Gharib SA, Gottlieb DJ, et al. Association of genetic loci with sleep apnea in European Americans and African-Americans: the Candidate Gene Association Resource (CARe) PLoS One. 2012;7:e48836. doi: 10.1371/journal.pone.0048836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kripke DF, Kline LE, Nievergelt CM, Murray SS, Shadan FF, Dawson A, Poceta JS, Cronin J, Jamil SM, Tranah GJ, et al. Genetic variants associated with sleep disorders. Sleep Med. 2015;16:217–224. doi: 10.1016/j.sleep.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baik I, Seo HS, Yoon D, Kim SH, Shin C. Associations of sleep apnea, NRG1 polymorphisms, alcohol consumption, and cerebral white matter hyperintensities: analysis with genome-wide association data. Sleep. 2015;38:1137–1143. doi: 10.5665/sleep.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cronin RM, Field JR, Bradford Y, Shaffer CM, Carroll RJ, Mosley JD, Bastarache L, Edwards TL, Hebbring SJ, Lin S, et al. Phenome-wide association studies demonstrating pleiotropy of genetic variants within FTO with and without adjustment for body mass index. Front Genet. 2014;5:250. doi: 10.3389/fgene.2014.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grilo A, Ruiz-Granados ES, Moreno-Rey C, Rivera JM, Ruiz A, Real LM, Sáez ME. Genetic analysis of candidate SNPs for metabolic syndrome in obstructive sleep apnea (OSA) Gene. 2013;521:150–154. doi: 10.1016/j.gene.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Guan J, Yi H, Yin S. A systematic review and meta-analysis of the association between serotonergic gene polymorphisms and obstructive sleep apnea syndrome. PLoS One. 2014;9:e86460. doi: 10.1371/journal.pone.0086460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin B, Sun Z, Liang Y, Yang Z, Zhong R. The association of 5-HT2A, 5-HTT, and LEPR polymorphisms with obstructive sleep apnea syndrome: a systematic review and meta-analysis. PLoS One. 2014;9:e95856. doi: 10.1371/journal.pone.0095856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varvarigou V, Dahabreh IJ, Malhotra A, Kales SN. A review of genetic association studies of obstructive sleep apnea: field synopsis and meta-analysis. Sleep. 2011;34:1461–1468. doi: 10.5665/sleep.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sah R, Pritchard LM, Richtand NM, Ahlbrand R, Eaton K, Sallee FR, Herman JP. Expression of the glucocorticoid-induced receptor mRNA in rat brain. Neuroscience. 2005;133:281–292. doi: 10.1016/j.neuroscience.2005.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubins JS, Sanchez-Alavez M, Zhukov V, Sanchez-Gonzalez A, Moroncini G, Carvajal-Gonzalez S, Hadcock JR, Bartfai T, Conti B. Downregulation of GPR83 in the hypothalamic preoptic area reduces core body temperature and elevates circulating levels of adiponectin. Metabolism. 2012;61:1486–1493. doi: 10.1016/j.metabol.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller TD, Müller A, Yi CX, Habegger KM, Meyer CW, Gaylinn BD, Finan B, Heppner K, Trivedi C, Bielohuby M, et al. The orphan receptor Gpr83 regulates systemic energy metabolism via ghrelin-dependent and ghrelin-independent mechanisms. Nat Commun. 2013;4:1968. doi: 10.1038/ncomms2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen W, Westendorf AM, Toepfer T, Mauel S, Geffers R, Gruber AD, Buer J. Inflammation in vivo is modulated by GPR83 isoform-4 but not GPR83 isoform-1 expression in regulatory T cells. Genes Immun. 2010;11:357–361. doi: 10.1038/gene.2010.5. [DOI] [PubMed] [Google Scholar]
- 40.Clambey ET, McNamee EN, Westrich JA, Glover LE, Campbell EL, Jedlicka P, de Zoeten EF, Cambier JC, Stenmark KR, Colgan SP, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci USA. 2012;109:E2784–E2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenstein EM, Williams CB. The T(reg)/Th17 cell balance: a new paradigm for autoimmunity. Pediatr Res. 2009;65:26R–31R. doi: 10.1203/PDR.0b013e31819e76c7. [DOI] [PubMed] [Google Scholar]
- 42.Ye J, Liu H, Li P, Chen ZG, Zhang GH, Yang QT, Li Y. CD4(+)T-lymphocyte subsets in nonobese children with obstructive sleep apnea syndrome. Pediatr Res. 2015;78:165–173. doi: 10.1038/pr.2015.76. [DOI] [PubMed] [Google Scholar]
- 43.Boyd JH, Petrof BJ, Hamid Q, Fraser R, Kimoff RJ. Upper airway muscle inflammation and denervation changes in obstructive sleep apnea. Am J Respir Crit Care Med. 2004;170:541–546. doi: 10.1164/rccm.200308-1100OC. [DOI] [PubMed] [Google Scholar]
- 44.Liu X, He L, Stensaas L, Dinger B, Fidone S. Adaptation to chronic hypoxia involves immune cell invasion and increased expression of inflammatory cytokines in rat carotid body. Am J Physiol Lung Cell Mol Physiol. 2009;296:L158–L166. doi: 10.1152/ajplung.90383.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauffer TP, Guerini D, Carafoli E. Tissue distribution of the four gene products of the plasma membrane Ca2+ pump. A study using specific antibodies. J Biol Chem. 1995;270:12184–12190. doi: 10.1074/jbc.270.20.12184. [DOI] [PubMed] [Google Scholar]
- 46.Holton M, Mohamed TM, Oceandy D, Wang W, Lamas S, Emerson M, Neyses L, Armesilla AL. Endothelial nitric oxide synthase activity is inhibited by the plasma membrane calcium ATPase in human endothelial cells. Cardiovasc Res. 2010;87:440–448. doi: 10.1093/cvr/cvq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuh K, Quaschning T, Knauer S, Hu K, Kocak S, Roethlein N, Neyses L. Regulation of vascular tone in animals overexpressing the sarcolemmal calcium pump. J Biol Chem. 2003;278:41246–41252. doi: 10.1074/jbc.M307606200. [DOI] [PubMed] [Google Scholar]
- 48.Eckert DJ, White DP, Jordan AS, Malhotra A, Wellman A. Defining phenotypic causes of obstructive sleep apnea. Identification of novel therapeutic targets. Am J Respir Crit Care Med. 2013;188:996–1004. doi: 10.1164/rccm.201303-0448OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berry RB, Gleeson K. Respiratory arousal from sleep: mechanisms and significance. Sleep. 1997;20:654–675. doi: 10.1093/sleep/20.8.654. [DOI] [PubMed] [Google Scholar]
- 50.Zecchini V, Madhu B, Russell R, Pértega-Gomes N, Warren A, Gaude E, Borlido J, Stark R, Ireland-Zecchini H, Rao R, et al. Nuclear ARRB1 induces pseudohypoxia and cellular metabolism reprogramming in prostate cancer. EMBO J. 2014;33:1365–1382. doi: 10.15252/embj.201386874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livingston FR, Arens R, Bailey SL, Keens TG, Ward SL. Hypercapnic arousal responses in Prader-Willi syndrome. Chest. 1995;108:1627–1631. doi: 10.1378/chest.108.6.1627. [DOI] [PubMed] [Google Scholar]
- 52.Sedky K, Bennett DS, Pumariega A. Prader Willi syndrome and obstructive sleep apnea: co-occurrence in the pediatric population. J Clin Sleep Med. 2014;10:403–409. doi: 10.5664/jcsm.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Nanayakkara A, Jun J, Savransky V, Polotsky VY. Effect of deficiency in SREBP cleavage-activating protein on lipid metabolism during intermittent hypoxia. Physiol Genomics. 2007;31:273–280. doi: 10.1152/physiolgenomics.00082.2007. [DOI] [PubMed] [Google Scholar]
- 55.Chatterjee S, Szustakowski JD, Nanguneri NR, Mickanin C, Labow MA, Nohturfft A, Dev KK, Sivasankaran R. Identification of novel genes and pathways regulating SREBP transcriptional activity. PLoS One. 2009;4:e5197. doi: 10.1371/journal.pone.0005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishihara H, Wada T, Kizuki N, Asano T, Yazaki Y, Kikuchi M, Oka Y. Enhanced phosphoinositide hydrolysis via overexpression of phospholipase C beta1 or delta1 inhibits stimulus-induced insulin release in insulinoma MIN6 cells. Biochem Biophys Res Commun. 1999;254:77–82. doi: 10.1006/bbrc.1998.9468. [DOI] [PubMed] [Google Scholar]
- 57.Kim D, Jun KS, Lee SB, Kang NG, Min DS, Kim YH, Ryu SH, Suh PG, Shin HS. Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature. 1997;389:290–293. doi: 10.1038/38508. [DOI] [PubMed] [Google Scholar]
- 58.Shin J, Kim D, Bianchi R, Wong RK, Shin HS. Genetic dissection of theta rhythm heterogeneity in mice. Proc Natl Acad Sci USA. 2005;102:18165–18170. doi: 10.1073/pnas.0505498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ban HJ, Kim SC, Seo J, Kang HB, Choi JK. Genetic and metabolic characterization of insomnia. PLoS One. 2011;6:e18455. doi: 10.1371/journal.pone.0018455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, Paul DS, Elling U, Allayee H, Li X, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.