Abstract
Circulating tumor cells (CTCs), defined by capture from blood with anti-EpCAM antibodies, have established prognostic value in specific epithelial cancers, but less is known about their utility for assessing patient response to molecularly targeted agents via measurement of pharmacodynamic (PD) endpoints. We discuss the use of CellSearch CTC isolation technology for monitoring PD response in early phase trials. We present representative data from three clinical trials with the PARP inhibitor veliparib (ABT-888) suggesting that CTCs can be used to measure PD effects, but our experience points to the difficulty in obtaining sufficient EpCAM-expressing CTCs from patients with advanced disease to reach statistically significant conclusions about PD effects from each trial, instead often leading to hypothesis generating information. Overall, the level of phenotypic heterogeneity observed in specimens from patients with advanced carcinomas suggests caution in the use of cell-surface differentiation marker-based methods for isolating CTCs.
Introduction
The ability to capture and identify circulating tumor cells (CTCs) from blood specimens of patients with epithelial cancers using anti-EpCAM antibodies has opened a new field of cancer diagnostics. Development and use of the Veridex automated CellSearch® platform (Janssen Diagnostics, LLC, Raritan, NJ) in clinical trials has demonstrated the prognostic value of EpCAM-defined CTC enumeration across a broad variety of epithelial cancers. It is important to emphasize the difference between prognostic indicators (i.e., characteristics that forecast patient outcomes without treatment) and predictive factors (i.e., characteristics that estimate the likely benefit of a given treatment to a patient). No predictive value for CTCs has been established. The cells isolated by EpCAM purification and further classified as CD45−/cytokeratin+/DAPI+ have been shown to have prognostic value in clinical studies;1–4 however, the phenotype these markers describe is an epithelial phenotype, not a tumor-specific or a mesenchymal phenotype. The presence of EpCAM+/cytokeratin+ cells in blood is a valid starting point for the definition of a CTC of an epithelial tumor, but when evaluating a pharmacodynamic (PD) response to a targeted or cytotoxic therapy in a patient, these criteria are inadequate because of the likely confounding event of normal epithelial (and mesenchymal) cells also being shed into the vasculature due to drug toxicity. Because the intent of a PD study is to determine drug effect on the molecular target in tumor cells, the ability to discriminate tumor cells from normal epithelia is important.
CTCs as Prognostic Indicators
In the initial demonstration of CTC isolation from the blood of patients with metastatic carcinomas using the CellSearch system,1 a wide range of CTC numbers (0–23,618 CTCs with a mean±SD of 60±693 CTCs per 7.5 mL whole blood) was present in the 2,183 blood samples from 964 metastatic carcinoma patients tested; 36% (781 of 2,183) of the specimens had ≥2 CTCs. In contrast, only 1 of the 344 (0.3%) healthy or nonmalignant disease subjects had ≥2 EpCAM-defined CTCs per 7.5 mL of blood. The frequency of patients with ≥2 CTCs was subsequently used as the cutoff value to analyze the frequency of CTCs in various metastatic carcinomas. These data showed that the proportion of positive specimens was 57% (107 of 188) of prostate cancers, 37% (489 of 1,316) of breast cancers, 37% (20 of 53) of ovarian cancers, 30% (99 of 333) of colorectal cancers, 20% (34 of 168) of lung cancers, and 26% (32 of 125) of other cancers.
After this initial study, several multicenter, prospective clinical trials were conducted to evaluate the number of Cell Search-detected CTCs associated with progression and survival in patients with metastatic breast cancer (MBC),2 metastatic castration-resistant prostate cancer (mCRPC),3 and metastatic colorectal cancer (mCRC).4 A total of 177 patients with MBC were recruited; 47% were starting their first line of therapy for metastatic disease, 30% were starting hormonal treatment or immunotherapy, and 67% were starting chemotherapy. Additionally, 18% of patients had nonvisceral metastatic sites; 68% of all tumors were positive for estrogen and/or progesterone receptor protein, 26% of the tumors were HER2/neu 2+ or 3+ and 63% of the patients were alive at the time of the Kaplan-Meier analysis. The number of baseline CTCs (≥ 2 CTCs: 61%; ≥ 4 CTCs: 53%; ≥ 5 CTCs: 49%) detected in these patients with MBC was used to assign the cutoff levels associated with longer survivals. In the mCRPC trial, 219 out of 231 patients enrolled were evaluable; 43% of evaluable patients (94 out of 219) had favorable CTCs (<5 CTCs per 7.5 mL blood at baseline) and 57% (125 out of 219) were unfavorable (≥ 5 CTCs at baseline).3 Of the 481 patients enrolled in the mCRC trial, 430 met the inclusion and exclusion criteria. Of these 430 patients, 26% had unfavorable CTC counts (≥ 3 CTCs per 7.5 mL of blood) at baseline, less than what was reported in other epithelial malignancies.4 Based on the data obtained from serial testing for CTCs in conjunction with other clinical methods, it was concluded that in patients with metastatic cancer, unfavorable results (≥5 CTCs per 7.5 mL blood for MBC and mCRPC; ≥3 CTCs per 7.5 mL blood for mCRC) were correlated with shorter progression-free survival and shorter overall survival after the conclusion of therapy. It was subsequently demonstrated that the number of CTCs in the circulation of MBC patients on therapy at monitoring time points from 3 to 20 weeks after the initiation of therapy also had prognostic significance for both progression-free and overall survival.5 Additionally, a prospective clinical trial of abiratarone acetate demonstrated the utility of CTC enumeration in prostate cancer using a tumor marker (RT-PCR detection of the TMPRSS2-ERG gene fusion) to positively identify tumor cells in the population.6–8 The limitation of these study results was that only 41 patients were evaluable; however, CTC counts below 5 per 7.5 mL after treatment were prognostic for longer survival, and 15 of the 41 patients were confirmed as TMPRSS2-ERG positive, proving the identity of those captured CTCs as prostate cancer.
CTCs as Predictive and Pharmacodynamic Indicators
CTCs identified by CellSearch and other technologies have also been studied for their utility in molecular profiling of drug targets and predicting drug responsiveness/resistance during the development of both approved and experimental agents. Returning to the clinical trial discussed in the previous section, further evaluation indicated that CTC TMPRSS2-ERG status did not have predictive value as a biomarker of abiratarone acetate response, in contrast to previous suggestions.7, 9 EpCAM-based CTC capture on microparticles has also been employed to enable RNA sequencing of the eluted cells for molecular profiling. Evaluating ABC drug transporter proteins, ERalpha, HER2/neu, and ALDH1 expression in a two year study, progression free survival was significantly correlated to baseline CTC counts and to expression of the multidrug resistance protein panel members.10 CTCs detected with a fiberoptic array scanning technology from patients with metastatic non-small cell lung cancer undergoing treatment with platinum showed a correlation between increased ERCC1 expression and decreased progression-free survival (PFS),11 and ERCC1-positive CTCs detected in the blood of ovarian cancer patients were found to predict platinum resistance.12
In one of the few CTC PD studies reported, IGF1R was employed as a PD marker on EpCAM-defined CTCs for analysis of response to anti-IGF1R Mab therapy in a Phase I clinical trial.13 The number of CTCs found in these patients was variable and low; however, a decrease in both the total number of CTCs and IGF1R-positive CTCs was observed during the first cycle of drug administration, followed by rebounding IGF1R-bearing CTCs by the end of the cycle. Importantly, the changes in CTC numbers correlated with changes in serum PSA levels in patients with hormone-resistant prostate cancer. The pharmaceutical industry may have collected additional data on the use of CTC biomarkers for assessing molecular drug action using commercial CLIA lab services such as CTC-based biomarker testing offered by Janssen Diagnostics and ApoCell, but the results of these studies are only infrequently published.14 This remains an active area of research, with Janssen Diagnostics recently moving advanced development of their platforms towards DNA sequence analysis applications, focusing on important pharmaceutical targets like ABL1, ALK1, EGFR, ERBB2, FLT3, and others.
One focus of the Pharmacodynamic Assay Development and Implementation Section (PADIS) of the Frederick National Laboratory for Cancer Research has been to establish the application of the clinically validated CellSearch instrument for the experimental study of PD changes using isolated CTCs as a replacement for biopsies in early phase clinical trials. The advanced state of development of the CellSearch system was attractive because of its documented performance criteria, automated data collection system, and manufacture of the device under engineering controls. In addition, methods of specimen stabilization and transport had already been developed, and an additional analysis channel was available for use in PD biomarker measurements.
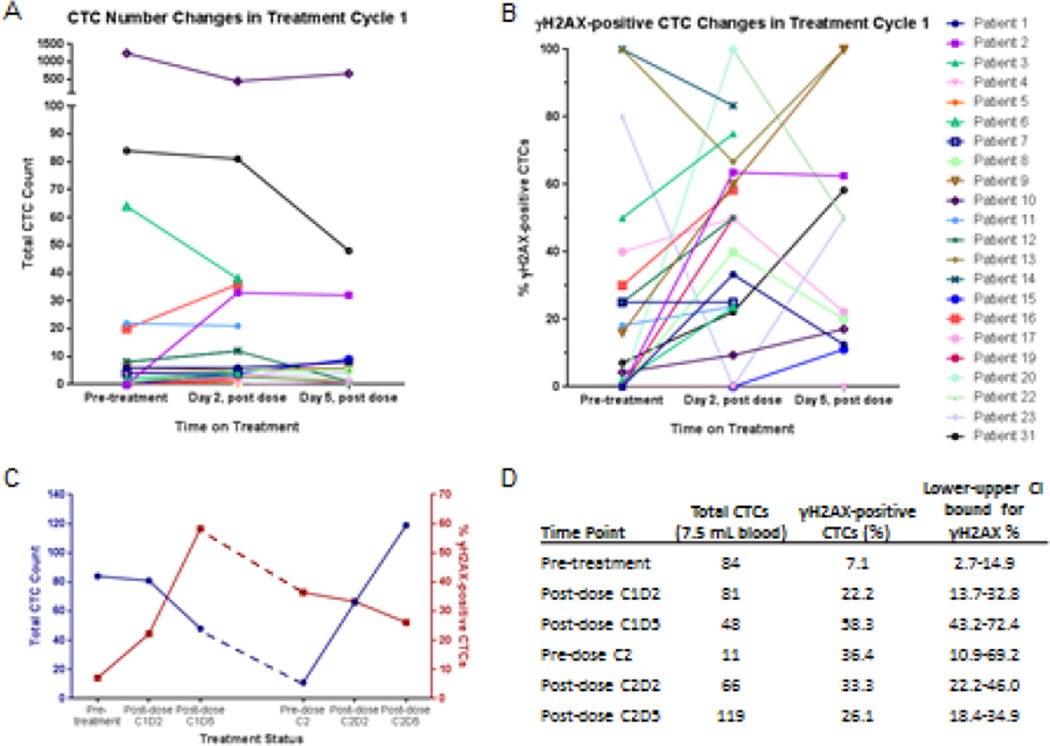
We therefore initiated an investigational study of the CellSearch device for assessment of the activity of DNA damaging agents in putative CTCs, including demonstrating fitness-for-purpose of an assay for the Ser139-phosphorylated histone H2AX (γH2AX) biomarker of DNA damage in CellSearch-isolated cells.15 γH2AX was chosen based on its previous validation as a PD biomarker reporting drug activity for the topoisomerase I inhibitors topotecan and three investigational indenoisoquinolines in tumor tissues.16 Although biomarker fitness-for-purpose studies can be done in xenografts to model patient biopsies, attempting to validate a CTC biomarker in a mouse model is quite difficult; therefore, we took the approach of testing the fitness of the biomarker in patient specimens. A series of 20 patients with epithelial cancers under early phase treatment protocols at the NCI consented to provide a 7.5 mL tube of blood before and after treatment with investigational or established agents known to induce DNA double-strand breaks or agents known not to do so.15 The results clearly demonstrated the specificity of the method to report DNA damage in CTCs, suggested appropriate CTC sampling intervals, and illustrated the wide range of CTC counts in patients enrolled in early phase trials (Figure 1). While CTC counts were generally low, the inter-patient variability was great. In addition, an increased proportion of γH2AX-positive CTCs were generally seen following drug exposure. This appears to be a real effect rather than an artifact of γH2AX-positive CTC selection, as the increase is too great to be attributable to a selective loss of γH2AX-negative CTCs.
Figure 1.
Representative data on (A) total CTC count (normalized to 7.5 mL blood volume) and (B) proportion of γH2AX-positive CTCs enumerated from patients before and after treatment with topotecan (patient #1), topotecan and veliparib (patients #4, 5, and 6), or cyclophosphamide and veliparib (patients #2, 3, and 7–20). Grouped analysis indicated a significant difference in the proportion of γH2AX-positive CTCs before and 2 days after treatment (P = 0.0027). (C-D) Individual data for a patient with neuroendocrine prostate cancer treated with topotecan for 2 cycles. Adapted from Wang et al.15
We have continued investigating the application of the CellSearch device to determine the utility of CTC PD in assessing patient responses to molecularly targeted agents in early stage clinical trials. In support of NCI-sponsored clinical trials at the Developmental Therapeutics Clinic of the NCI Division of Cancer Treatment and Diagnosis (DCTD) and throughout the NCI Clinical Trials Network, PADIS has performed CTC analysis on the CellSearch platform for 28 early-stage clinical trials (mostly phase I trials) over the past 3.5 years. Phase I trials tend to have a limited number of patients per trial, a variable number of specimens per patient, a heavily pretreated patient population that generally has advanced metastatic disease, and involve a wide range of tumor types. These variables have a major impact on the ability to deliver statistically significant results for biomarker analysis, regardless of whether the tissues tested are tumor biopsies or blood specimens; however, as new compounds advance through clinical drug development, it can become more challenging to incorporate correlative PD sampling into later phase clinical trials. Phase I trials offer the ability to collect biopsy specimens from patients and to correlate these measures with evidence of patient response, providing a method to validate CTC readouts, one of the long-term goals of the PADIS CTC program. If CTC-based PD measurements can be validated against bone fide PD responses in tumor biopsy specimens during phase I trials, subsequent phase II and III clinical trials can also include PD evaluations, which is not often possible if the PD measurements are limited to tumor biopsies. Another benefit to correlative PD sampling in phase I trials is that the dose escalation phase allows for the analysis of the effect of different doses on PD biomarker response.
Here, we will review technical considerations for the use of the CellSearch CTC-isolation technology for PD measurements and the epidemiological data on CTCs we have generated from supporting a number of clinical trials. We will then consider specific lessons learned from several recent PD studies of the investigational poly(ADP-ribose) polymerase (PARP) inhibitor veliparib (ABT-888) using the CellSearch device.
CellSearch Technical Considerations
Validation of Methods and Materials to Increase Reproducibility of Tests Based on R&D-grade Reagents
The CellSearch System, when used according to manufacturer’s specifications, is intended to enumerate putative CTCs based on a common epithelial phenotype to provide prognostic information for patients with certain epithelial cancers; this is the FDA-approved use. For applications in evaluating PD responses in early stage clinical trials, the CellSearch Epithelial cell kit (catalog # 7900000) is our preferred choice for CTC capture and enumeration, allowing the available green fluorescence channel to be used to detect PD biomarker signals in nuclei. In our γH2AX application, only fluorescence signal that is concordant with the DAPI nuclear signal is scored as positive to assure that the assay is reporting the nuclear DNA damage response.
As for all antibody-based assays, antibody validation in the system is critical. We employ several methods of antibody validation, and this step is required even for antibodies that have been clinically validated and reported in the literature. This is critical because of inconsistent lot-to-lot performance of the antibodies released for sale by the manufacturers.17 Our current focus is on biomarkers of drug activity, but the same principles apply for phenotyping or the use of tumor marker antibodies such as PSA. We begin, whenever possible, with antibodies that have been previously validated on clinical specimens. Our requirements are:
The signal is pan nuclear (when appropriate, such as for antibodies to measure DNA damage and repair biomarkers).
The required concentration of conjugated primary antibody has controllable autofluorescence in the cytosol.
The primary antibody binds to a single band at appropriate molecular weight on a Western blot, preferably tested on both positive control tissues and on tumor tissues; because of specimen limitations, xenografts are usually used in place of clinical tumor specimens.
Signal is observed in positive control tissue with constant exposure settings.
Signal is greater than an isotype control run at the same concentration on the positive control tissue.
No signal is observed in negative control tissue with the conjugated primary antibody.
Antibody competition with cognate antigen successfully removes nuclear signal in the positive control tissues.
New lots of primary antibody can be run at concentrations comparable to previously used lots.
When validating biomarkers, our rule of thumb is to optimize for specificity for phenotyping markers and sensitivity for PD biomarkers to decrease the probability of disqualifying a useful marker due to unnecessary stringency.
In practice, image quality is the limiting factor in the assay if it is not high enough to allow morphological analysis of the cells. In most specimens, there are a significant number of cells that are captured by the EpCAM antibody but that are negative for cytokeratin (and CD45) and therefore uncalled by the Veridex specimen report. In the specimens that we have evaluated, most of these are single cells, and were not likely co-purified by adherence to EpCAM positive cells.
A reproducible supply of quality reagents is necessary to support any clinical assay that will be performed on multiple specimens over a prolonged period of time. Measurement of our primary PD marker in CTCs, γH2AX, has been possible due to a reliable source of the antibody JBW301, purchased in bulk lots from EMD Millipore (Merck KGaA, Darmstadt, Germany) and custom conjugated to AlexaFluor 488, and institution of an standard operating procedure (SOP)-driven assay methodology for reagent validation.18 Each lot of labelled antibody is pre-qualified by testing on tumor cell lines (principally HT-29) treated in vitro with topotecan to generate the γH2AX signal. The exact method is described in our antibody qualification and laboratory proficiency testing SOP, available on the NCI website.18 A lot-to-lot comparison of γH2AX induction in topotecan-treated HT-29 cells is performed with each new lot of antibody received. To date we have received and validated four lots of Alexa Fluor 488-conjugated JBW301 successfully.
Control of Pre-Analytical Variables via Compliant Specimen Acquisition and Shipping
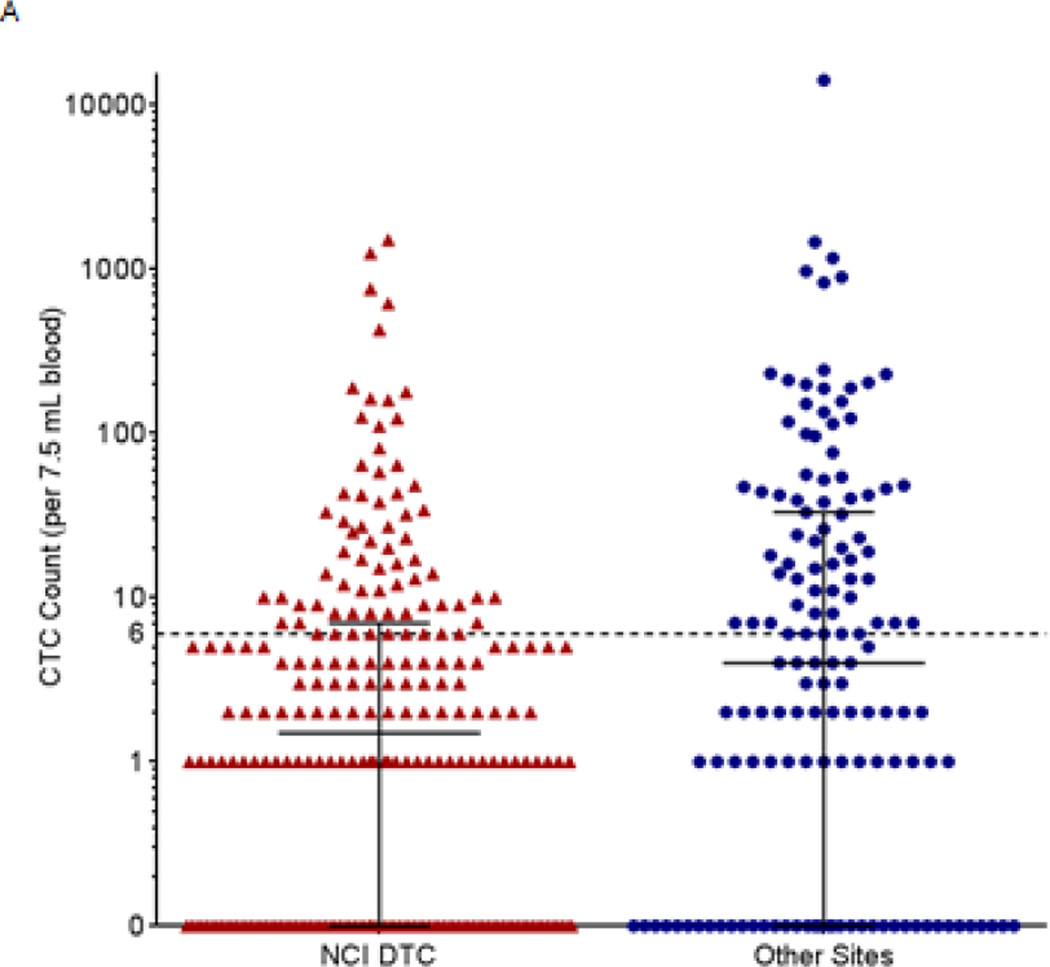
As with any clinical trial, the ability to provide useful biomarker data starts with the ability to deliver analyzable specimens to the laboratory. Daily scheduling of blood collections is influenced by treatment schedules as well as clinical and PD objectives; blood specimens are often collected multiple times during cycle 1 and 2 and then every or every other cycle with continued treatment. Variations in blood collection schedules can result in irregular specimen delivery, which can cause specimen quality control (QC) failures. The CellSearch package insert specifies that blood must be analyzed within 4 days of collection in CellSave vacutainers for the analysis to be valid. Because we used the CellSearch system for correlative PD biomarker studies on early-phase clinical trials (not as a prognostic measurement), we were able to reproducibly analyze specimens up to 5 days (120 hours) after collection as long as hemolyzed or clotted specimens were excluded. Veridex provided validated collection and shipping methods as part of their diagnostic submission to the FDA, and stabilization of blood specimens and transcontinental shipping for CellSearch analysis using CellSave vacutainer tubes has worked well in our experience. Analysis failures are minimal and generally result from either delayed weekend shipments arriving after the 5-day limit for analysis or collection volumes of <4 mL. Despite the variability of shipping times (24 to 96 hours), comparable CTC recoveries were demonstrated from blood specimens shipped from multiple sites in the NCI Clinical Trials Network and from those received within 3–6 hours of collection from the local NCI DCTD Developmental Therapeutics Clinic in Bethesda, MD (Figure 2A).
Figure 2.
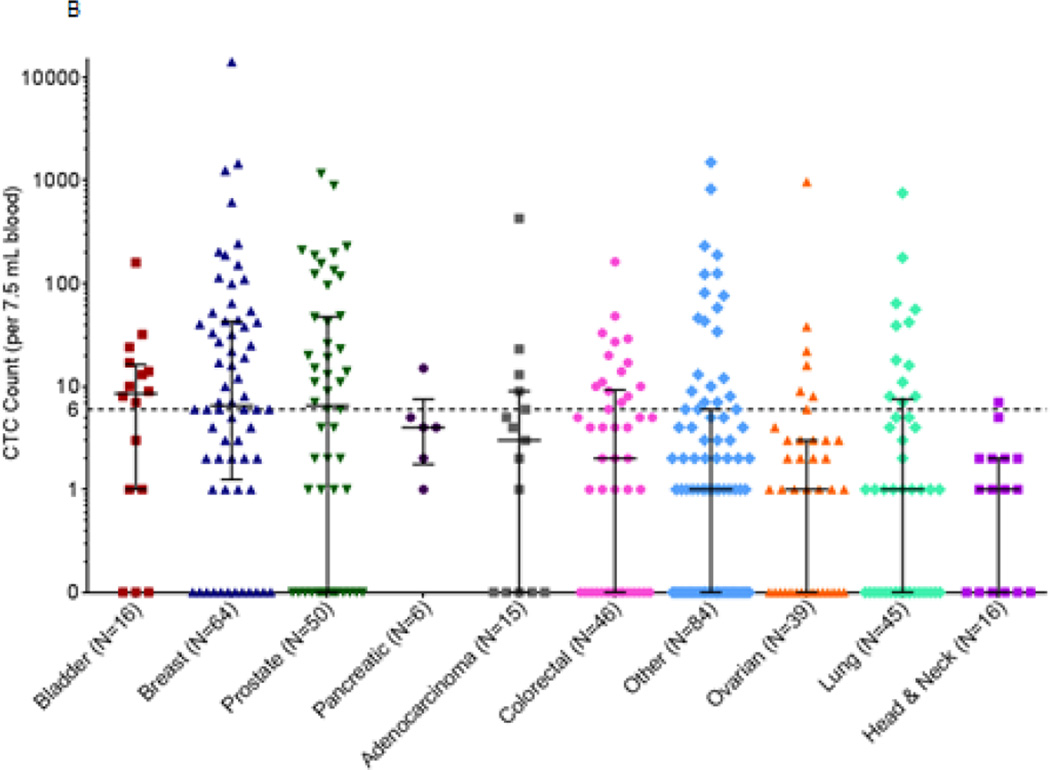
Baseline CTC counts in patients with advanced disease from the NCI Developmental Therapeutics Clinic (DTC) and other sites in the NCI clinical trials network. All patients from the NCI DTC had advanced refractory disease and were enrolled in the following clinical trials: NCT01306032, NCT01051635, NCT00978250, NCT01534598, NCT01748825, NCT01851369, NCT00923481, and NCT00900198. Patients from other sites were enrolled in NCT01264432, NCT01434316, NCT00034216, and NCT00576654. Whole blood samples (7.5 mL) were drawn from 381 patients with a variety of advanced malignancies enrolled in phase I or phase II clinical trials and CTC enumeration was performed by the CellSearch system. (A) Patients at the NCI DTC (N=238) had a mean of 29.1 (standard deviation, 144) and a median of 1.5 baseline CTC, while patients at other sites (N=143) had a mean of 162.5 (standard deviation, 1191) and median of 4 baseline CTCs per 7.5 mL blood. Median and interquartile range are plotted for each. (B) The frequencies of baseline CTC counts for patients at both the NCI DTC and other sites in the NCI clinical trials network (N=381) are shown grouped by cancer type. Median and interquartile range are plotted for each.
QC Metrics for Monitoring Assay Performance over Time
Reliable instrumentation is critical to support a multi-site clinical trial effort. Two early CellTracks AutoPrep instrument failures, resulting in a halt to clinical specimen processing, prompted the utilization of a second device to allow for continued clinical sample analysis even if one system was undergoing repair. We encountered no such issues with the CellTracks Analyzer or with the cell cassette systems, which have performed well over the course of the trials. Actual run failures caused by failure of the High or Low CellSearch Kit Controls were rare across 17 Control lots over the 3.5-year course of these trials (Table 1). Of 549 control runs, 548 low controls and 546 high controls passed the test. In addition, collaboration with our colleagues and fellow CellSearch users at Georgetown University Medical Center (Washington, DC) and MedImmune (Gaithersburg, MD) has allowed us extra instrument capacity to keep trial specimen processing on schedule during times of peak need or instrument failures. The availability of a user’s support network such as this greatly improves the probability of success in supporting multiple clinical sites over time.
Table 1.
The performance of the CellSearch CTC Control Kit over 3.5 years.
| Lot # | Start Use Date |
End Use Date |
Number of Tests Run |
Low Control | High Control | ||
|---|---|---|---|---|---|---|---|
| Pass | Fail | Pass | Fail | ||||
| C913B | 11/26/2010 | 02/28/2011 | 24 | 24 | – | 24 | – |
| D162B | 03/03/2011 | 04/29/2011 | 24 | 24 | – | 24 | – |
| D198B | 05/05/2011 | 07/19/2011 | 22 | 22 | – | 21 | 1 |
| D280B | 07/21/2011 | 09/21/2011 | 18 | 18 | – | 18 | – |
| D388B | 09/29/2011 | 10/11/2011 | 12 | 12 | – | 12 | – |
| D280A | 09/22/2011 | 11/10/2011 | 24 | 24 | – | 24 | – |
| D486A | 11/14/2011 | 01/18/2012 | 24 | 24 | – | 23 | 1 |
| D635A | 01/18/2012 | 03/05/2012 | 24 | 24 | – | 24 | – |
| 0780A | 03/06/2012 | 05/21/2012 | 23 | 23 | – | 23 | – |
| D867A | 05/23/2012 | 08/03/2012 | 24 | 24 | – | 24 | – |
| E086A | 08/07/2012 | 10/01/2012 | 24 | 24 | – | 24 | – |
| E156A | 10/05/2012 | 01/29/2013 | 61 | 60 | 1 | 61 | – |
| E435A | 01/30/2013 | 03/22/2013 | 24 | 24 | – | 24 | – |
| E532A | 03/22/2013 | 09/05/2013 | 72 | 72 | – | 71 | 1 |
| E743A | 06/28/2013 | 08/01/2013 | 24 | 24 | – | 24 | – |
| E902A | 09/06/2013 | 01/30/2014 | 69 | 69 | – | 69 | – |
| E928A | 02/12/2014 | 06/23/2014 | 56 | 56 | – | 56 | – |
| Summary of 17 Lots | 549 | 548 | 1 | 546 | 3 | ||
| percentage of total | 99.8% | 0.18% | 99.5% | 0.55% | |||
Causes of failures over the course of five related clinical trials were highly informative and speak to the value of a properly validated and maintained instrument system and good logistics: 792 blood specimens were received, of which 730 were successfully processed. Reasons for failure in the other 62 analyses included: instrument-aborted run (13 specimens, of which 2 were salvageable), hemolyzed or clotted blood that could not be processed (9 specimens), and specimens expired upon receipt (≥120 hours; 42 specimens). Logistics of specimen shipping and handling therefore accounted for two-thirds of all failures.
Clinical Trial Data
Grouped Epidemiological Data
An analysis of data from all 28 clinical trials we have supported with the CellSearch platform for CTC isolation over the past 3.5 years revealed that the majority (71%) of specimens collected from patients enrolled in trials conducted at the NCI Developmental Therapeutics Clinic (DTC) had too few CTCs (<6 per 7.5 mL blood) to be evaluable (Figure 2A) for PD responses. The DTC is a phase I trials clinic, and the majority of patients enrolled at this site therefore have disseminated cancers and have progressed after multiple rounds of therapy prior to recruitment. Specimens collected from patients at enrollment in early phase trials within the NCI clinical trials network, principally from the Karmanos Cancer Center, City of Hope Comprehensive Cancer Center, Dana Farber Cancer Institute, and Sidney Kimmel Cancer Center, generally had comparable CTC numbers, with the majority of specimens (74/143, 52%) below the minimum 6 CTCs required for evaluation (Figure 2A). When CTC numerical data from all sites were pooled and analyzed by tumor type, a similar statistical distribution appeared, but with some surprises (Figure 2B): only 1 of 6 specimens from patients with pancreatic cancer and 1 of the 19 specimens from patients with head and neck cancer were evaluable based on the minimum CTC requirement of 6 cells. Given that these patients had advanced metastatic disease and should have had significant numbers of tumor cells in circulation, an immediate conclusion from these data is that the EpCAM capture method is likely missing a number of CTCs in these patient populations.
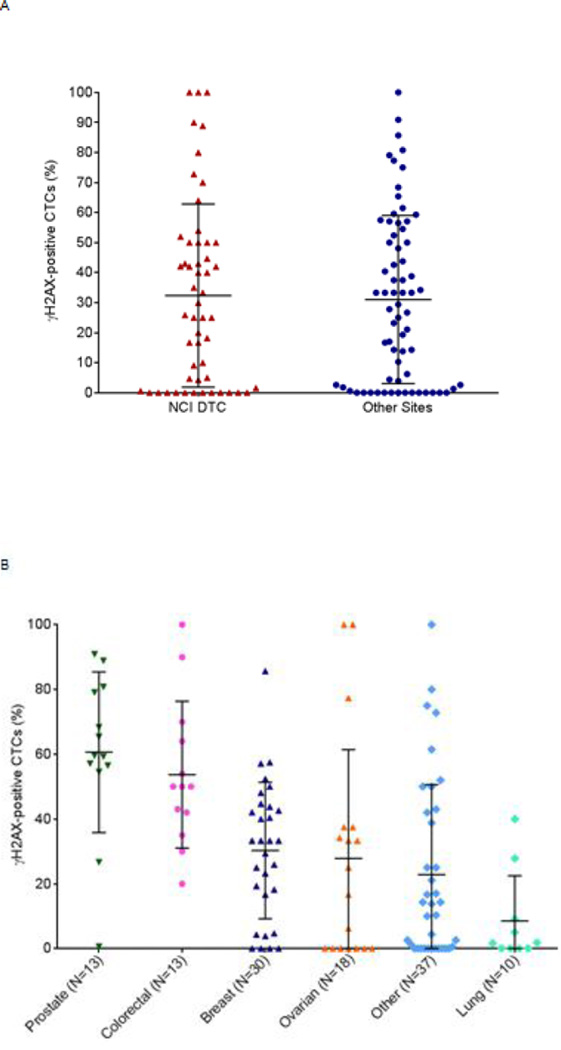
Baseline biomarker positivity was common in these patient populations. Because γH2AX is a marker of DNA damage, it is also a marker of active apoptosis, and indicates that DNA double-strand breaks, which are generated during apoptosis, are present.19 Therefore, a limitation of this biomarker for PD studies is that γH2AX-positive CTCs can be caused by events independent of drug treatment, and thus there is a baseline level of biomarker-positive CTCs in the blood at any given time. The ability to find a drug-induced signal is absolutely dependent on the variability of this baseline and the total number of cells that are measured. We have previously noted that both the total number of CTCs and the fraction that are γH2AX-positive show limited variability over a short time course (1 day to 6 weeks) in patients not undergoing therapy.15 In the patient populations described above, γH2AX-positive CTCs were present in most specimens. In Figure 3, the proportion of baseline γH2AX-positive CTCs for samples with 3 or more CTCs (in 7.5 mL blood) is reported. Specimens from the NCI DTC (N=53) and the NCI clinical trials network (N=68) had 32% and 31% γH2AX-positive CTCs, respectively (Figure 3 A). When specimens from all sites were pooled for analysis according to tumor type, there was a significant fraction of γH2AX-positive cells at baseline for all histologies (Figure 3B). The highest fraction of putative apoptotic cells at baseline was found in prostate and colorectal cancers. Other investigators have previously reported significant numbers of apoptotic CTCs in patients with breast cancer and other diseases using Annexin V or M30 staining.20, 21 This high γH2AX basline positivity raised a statistical challenge in detecting a drug effect associated with DNA damaging agents in this patient population.
Figure 3.
The percentage of baseline γH2AX-positive CTCs in patients with advanced disease from the NCI Developmental Therapeutics Clinic (DTC) and other sites in the NCI clinical trials network who exhibited 3 or more CTCs. Patient specimens with less than 3 baseline CTCs were not graphed because the fraction of γH2AX-positive CTCs in those samples is not a reliable measurement. All patients from the NCI DTC had advanced, refractory disease and were enrolled in trials NCT01306032, NCT01051635, NCT01748825, NCT01851369 and NCT00900198. Patients from other sites were enrolled in trials NCT01264432, NCT01434316, and NCT00576654. CTC enumeration by the CellSearch system on whole blood samples (7.5 mL) found 121 patients with a variety of advanced malignancies enrolled in phase I or phase II clinical trials. (A) At baseline, eligible patients at the NCI DTC (N=53) had, on average, 32% γH2AX-positive CTCs (standard deviation ±31%) and those from other sites (N=68) had, on average, 31% γH2AX-positive CTCs (standard deviation ±28%). Mean and standard deviation are plotted for each. (B) Baseline γH2AX-positive CTC frequencies grouped by cancer type. Mean and standard deviation are plotted for each.
During the first clinical trials in which we evaluated γH2AX in CTCs, we assayed CTCs from patient blood collected 24 hours after the first drug administration, but found that this time point necessarily skews the γH2AX results towards reporting apoptosis, drug-induced or not, rather than the immediate induction of DNA double strand breaks. DNA damaging agents first induce γH2AX as part of the repair process, but if the damage overwhelms the cell, more γH2AX is recruited to the site as apoptosis occurs. In xenograft models, treatment with cytotoxic agents such as topotecan or gemcitabine induces γH2AX expression in tumor tissues within 2–4 hours of treatment, indicative of the increase in DNA double strand breaks.16 In measuring γH2AX in CTCs to document a PD effect, the clearest effect would then seem to be one observed within a few hours of treatment; however, this creates a dilemma because the CTCs with the most predictive value will likely be those that are mobilized from the tumor in response to treatment rather than those already in the blood stream and exposed to plasma concentrations of the drug. Based on the time scale of drug action in the tumor and the short half-life of CTCs (estimated to be between 1 and 2.5 hours),22 time points between 4 and 12 hours after treatment are thought to provide the greatest chance of observing the true drug effect on γH2AX levels in CTCs. Ideally, a rise in γH2AX-positive CTCs would occur during the same time that plasma drug levels decline to reinforce this interpretation.
From the results of these trials, it is evident that EpCAM-defined CTC numbers were not high enough to evaluate drug response in a majority of patients enrolled, even employing a PD biomarker. Despite the fact that these patients have disseminated disease, no EpCAM positive cells that fit the description of CTCs could be detected in the majority of patients enrolled. Application of the γH2AX marker showed that the majority of patients had variable number of CTCs that may have been apoptotic at baseline, prior to drug administration. These results required us to devise a new set of reporting criteria, which were set to be conservative because individual patient CTC numbers are coupled to measurements of drug effect on biomarker response, using each patient as his or her own baseline. For clinical specimens analyzed at PADIS, while all raw numbers are provided, CTCs are only considered clinically reportable if there are a minimum of 6 CTCs detected in the patient specimen by CellSearch, irrespective of blood volume (Table 2). Those specimens that meet this criterion are then normalized to a blood volume of 7.5 mL, the standard blood collection volume in CellSearch tubes. Biomarker response is assessed if either 4 of 5 time points or 3 consecutive time points have reportable CTC numbers; blood specimens are often collected multiple times during cycle 1 and 2 and then every or every other cycle with continued dosing. A post-treatment sample is considered to have a PD biomarker response if there is at least a 3-fold increase at any follow-up time point over the baseline biomarker-positive CTC proportion. For example, 3 biomarker-positive CTCs out of 10 total CTCs at baseline would require ≥9 biomarker-positive CTCs out of 10 total CTCs following treatment (or ≥18 biomarker-positive out of 20 total) to be considered a PD response. In addition, any deviations to the assay SOPs are logged and reported to the clinic in case they may affect the interpretation of the data.
Table 2.
Example of a clinical report of a PD biomarker-positive CTC assessment based on lessons-learned from using the CellSearch system to evaluate specimens from multiple clinical trials.
| Raw CTCsa | Normalized CTCsb | Biomarker:Total CTC Ratioc | |||||
|---|---|---|---|---|---|---|---|
| Time Point |
Total CTCs |
Biomarker -positive CTCs |
Blood Volume |
Total CTCs |
Biomarker -positive CTCs |
Biomarker:Total CTC Ratio |
Specimen Response from Baseline |
| C1D 1 pre |
12 | 4 | 7.5 | 12 | 4 | 0.25 | Baseline |
| C1D 1 4 hours |
8 | 7 | 7.5 | 8 | 7 | 0.88 | Response |
| C1D 1 8 hours |
7 | 6 | 7.5 | 7 | 6 | 0.86 | Response |
| C1D 2 24 hours |
5a | 4 | 7.5 | – | – | – | N/A |
| C2D 2 24 hours |
8 | 3 | 5.5 | 11 | 4 | 0.38 | No Response |
A minimum recovery of 6 CTCs is required for reporting. Samples with less than 6 CTCs are not normalized or compared to other values.
Raw CTC counts are normalized to CTCs per 7.5 mL blood based on the blood volume collected.
To be considered a response over baseline, the biomarker to total CTC ratio must be greater than 3 times that observed at baseline.
In summary, much of the collected data resulted in CTC numbers that were below the eligibility criteria for interpretation and reporting due to the low numbers of EpCAM-positive CTCs in specimens from patients with disseminated metastatic cancers enrolled in early phase clinical trials. Such trials also face the challenge of highly diverse patient populations resulting in a limited number of patients with sufficient time points evaluable for the PD biomarker, and the low probability of drug activity via a documented effect on response (RECIST criteria). Not surprisingly, therefore, the results of PD biomarker evaluation of CTCs using the CellSearch system during early phase clinical trials have been limited to anecdotal data. Despite not rising to the level of statistical significance, the PADIS experience has acquired data regarding the enumeration of CTCs and the fraction of γH2AX-positive CTCs that can inform the planning of future trials. To illustrate this, several clinical trials of the PARP inhibitor veliparib that included evaluation of γH2AX in CTCs have been summarized in the next section.
Individual Clinical Trial Data
Our goal in the early phase trials described here was not to determine if CTC measurements could predict therapeutic outcome; instead, we focused on the detection and measurement of PD biomarker responses to treatment using CTCs. Importantly, there is no evidence to date that the quantitative level of a PD effect in CTCs correlates with a PD effect in the patient tumor. Thus, while CTCs may become useful in evaluating on-target drug activity in CTCs themselves, considerable additional study is required before CTC-based measurements can be considered as a possible surrogate for drug effect in a patient’s tumor.
Veliparib in Combination with Radiation Therapy
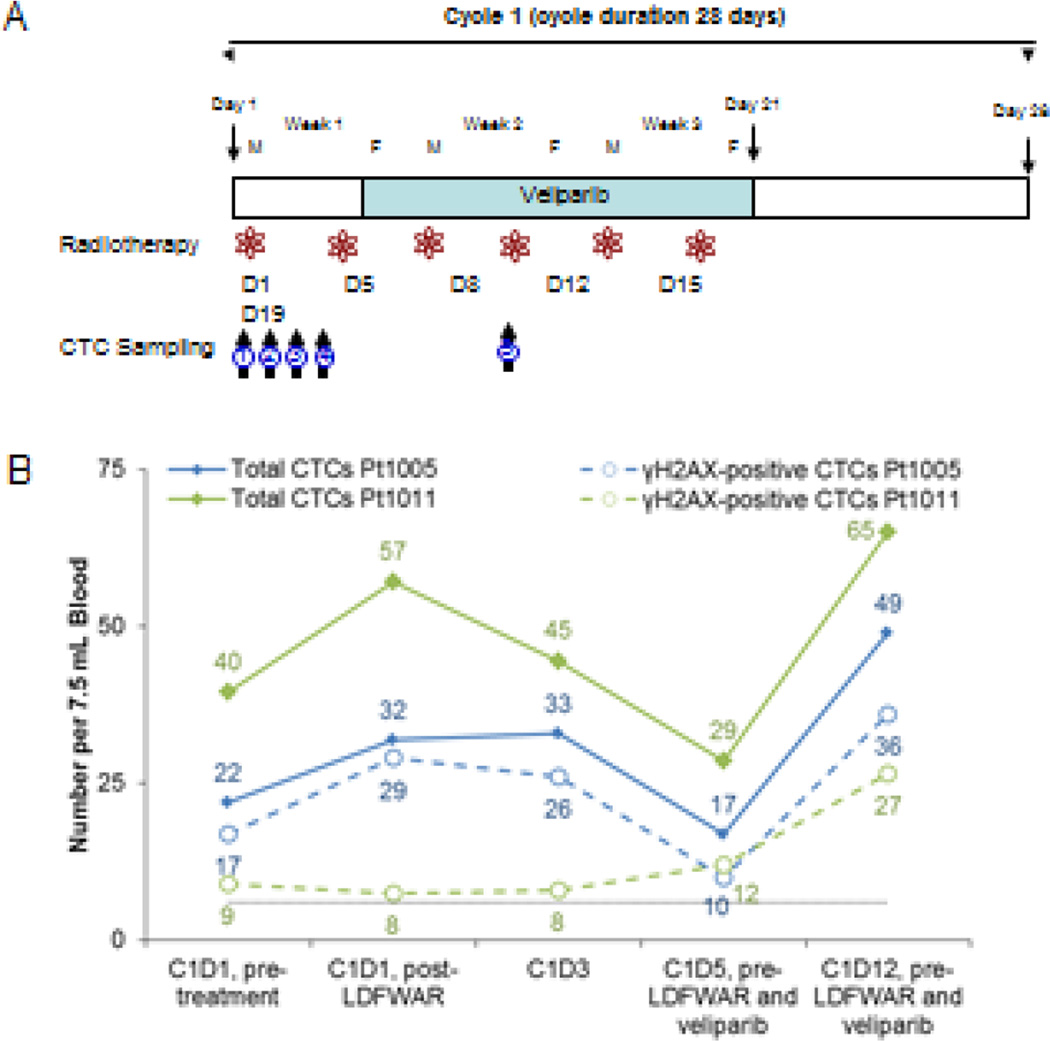
CTCs were collected from patients on a phase I trial (ClinicalTrials.gov: NCT01264432) of veliparib (ABT-888) plus low-dose fractionated whole abdominal radiation therapy (LDFWAR) led by Johns Hopkins.23, 24 In this phase I evaluation of veliparib and radiation, CTCs were evaluated to provide evidence of veliparib-enhanced radiation-induced DNA damage. This study was open to patients with all advanced solid tumors with documented evidence of peritoneal carcinomatosis. Veliparib was administered twice daily (BID) on days 5–21 of 28 day cycles, and LDFWAR was administered in two fractions at a dose of 60 cGy on days 1 and 5 for weeks 1–3 of each cycle (Figure 4A). Tolerable doses of veliparib were achieved up to 160 mg BID.
Figure 4.
Phase I Study of veliparib (ABT-888) in combination with LDFWAR therapy in patients with advanced solid malignancies with peritoneal carcinomatosis. (A) Schema of trial drug administration and 5 separate blood collection time points in cycle 1 of treatment used for CTC analysis. Blood for CTC analysis was collected at (1) C1D1, prior to LDFWAR, (2) C1D1, 6–8 hrs after LDFWAR, (3) C1D3, (4) C1D5, prior to veliparib and LDFWAR, and (5) C1D12 prior to veliparib and LDFWAR (B) CellSearch results for total CTC and γH2AX-positive CTCs over the course of therapy for two different patients. Black dotted line indicates recommended threshold of 6 CTCs for reporting biomarker-positive CTCs.
An initial concern in the evaluation of veliparib plus LDFWAR was that radiotherapy would induce nuclear γH2AX in the specimens at a level that would interfere with or obscure the ability to measure the enhancement of γH2AX response by veliparib. To separate the individual contributions of veliparib and LDFWAR, the first 4 blood collections were prior to veliparib administration: blood was collected at baseline prior to beginning of therapy (cycle 1 day 1; C1D1, pre-treatment), 6 hours after radiotherapy on day 1 (C1D1-, post-LDFWAR), on day 3 (C1D3), and on day 5 before both radiotherapy and the first oral veliparib administration (C1D5). A fifth blood specimen was also collected on day 12 before veliparib or radiotherapy treatment, but following 7 days of veliparib treatment (C1D12). Figure 4B shows a representative CTC profile from two patients on treatment; we observed no significant impact on baseline γH2AX-positive CTC levels immediately following radiotherapy on day 1. For patient 1005, there was a 45% increase in the number of CTCs after the first LDFWAR treatment, but the fraction of CTCs positive for γH2AX signal increased only from 77% to 90% (not statistically significant). At the same point, patient 1011 also had a 42% increase in total CTC number and a small decrease from 22% to 14% in the fraction of CTCs positive for γH2AX signal. In both patients, the total number of CTCs and fraction γH2AX-positive CTCs decreased until the second LDFWAR treatment on C1D5. The next time point collected on C1D12 reflected the effects of both the radiotherapy and the veliparib treatment, and patient 1005 demonstrated a 2.9-fold rise in CTC counts from the pre-veliparib measurement (C1D5) and a 2.2-fold rise from the initial baseline measurement. Patient 1011 had comparable increases of 2.2-fold and 1.6-fold, respectively. Changes in the proportion of γH2AX-positive CTCs were not statistically significant for either patient after the addition of veliparib, but the absolute number of γH2AX-positive CTCs did increase with veliparib treatment. Our interpretation is that the veliparib therapy mobilized CTCs into the bloodstream. There was no evidence for an increase of γH2 AX-positive CTCs on the first day of LDFWAR therapy in either patient, indicating that CTCs already in the peripheral circulation were not affected by the radiotherapy.
Veliparib in Combination with Irinotecan
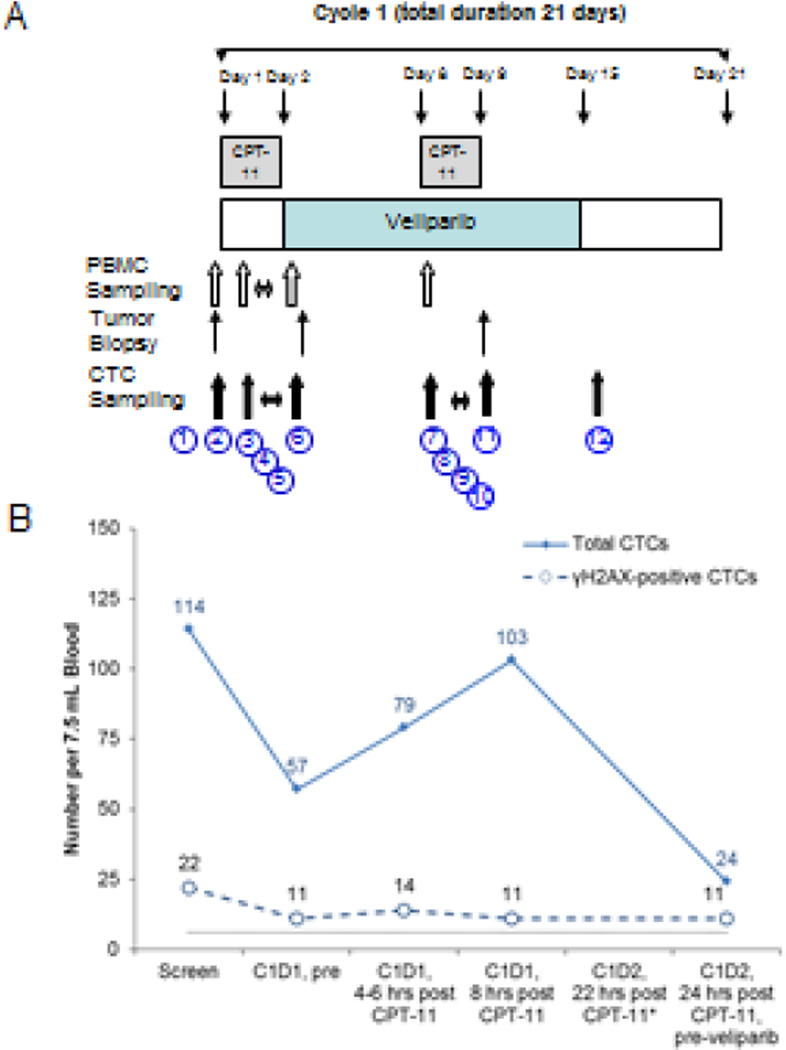
One of the earliest combination trials to test the tolerability of veliparib with chemotherapy was a phase I study of veliparib with irinotecan (CPT-11) led by the Karmanos Cancer Center (ClinicalTrials.gov: NCT00576654).25 Veliparib was given BID from days 1–14 and irinotecan was administered on days 1 and 8 of a 21 day cycle. To assess the PD effects of the PARP inhibitor, veliparib dosing was not begun until day 3 of cycle 1 to allow collection of a tumor biopsy and CTCs on day 1–2, prior to and following irinotecan administration; samples were also collected on day 8–9 after approximately a week of veliparib administration for comparison (Figure 5 A). In this phase I trial, the PD objectives related to CTC collection were to determine (1) if irinotecan administration resulted in an immediate increase in γH2AX-levels in both currently circulating CTCs and those released over the next 8–10 hours (so-called “tumor-exposed” CTCs based on the estimated CTC half-life of 2 hours), (2) if PARP inhibition by veliparib increased the γH2AX response to irinotecan in CTCs 4 hours after treatment, (3) if PARP inhibition by veliparib increased the γH2AX response of tumor cells to tissue irinotecan levels, as indicated by the proportion of γH2AX-positive CTCs at 12–18 hours after treatment, and (4) when the peak γH2AX response in CTCs occurred and if there was a correlation in γH2AX response between CTC and tumor biopsy specimens from the same patient.
Figure 5.
Phase I dose-escalation study of oral veliparib (ABT-888) plus intravenous irinotecan (CPT-11) administered in patients with advanced solid tumors. (A) Clinical trial drug administration schema indicating the 12 separate blood collection time points during treatment cycle 1 for CTC analysis. Blood for CTC analysis was collected at (1) screening; (2) C1D1 prior to irinotecan or veliparib; (3–5) 4–6 hrs, 8 hrs, and 22 hrs after single-agent irinotecan; and (6) C1D2, 24 hrs after irinotecan but before the first veliparib dose. After starting veliparib treatment, blood was also collected at (7–10) C1D8 prior to irinotecan, 4–6 hrs, 8 hrs, and 22 hrs after irinotecan; (11) C1D9, 24 hrs after irinotecan; and (12) C1D15. (B) Longitudinal tracking of total CTC numbers and proportion of γH2AX-positive CTCs in a treated patient. Blood from the C1D2 22 hr time point could not be analyzed due to hemolysis and clotting of the specimen (*). Black dotted line indicates recommended cut-off of 6 CTCs for reporting biomarker-positive CTCs.
Preliminary analysis of the trial data indicates that there was evidence of additive response of veliparib to irinotecan in 5 patients, as indicated by increasing γH2AX-positive CTC counts combined with decreasing total CTC numbers. There was also evidence that irinotecan increased the number of γH2AX-positive CTCs in some patients. An example of patient data from this trial is shown in Figure 5B. Before treatment this patient demonstrated variable numbers of total CTCs (114 to 57) but a consistent proportion of γH2AX-positive CTCs (19%); these counts remained similar up to 8 hours after the first irinotecan treatment. On average, 16% of CTCs over these four time points were γH2AX-positive, reflecting the baseline proportion of apoptotic CTCs, so for this patient there was no immediate effect of the initial high irinotecan levels on the fraction of γH2AX-positive CTCs in circulation. By C1D2 (24 hours post-irinotecan), the number of CTCs had dropped significantly to 27% of the mean value of the four previous time points and the fraction of γH2AX-positive CTCs increased to 46%. The increase in the proportion of γH2AX-positive CTCs was likely driven by the decrease in overall numbers of non-apoptotic CTCs, because the number of γH2AX-positive CTCs was nearly constant between collections.
Veliparib in Combination with Cyclophosphamide
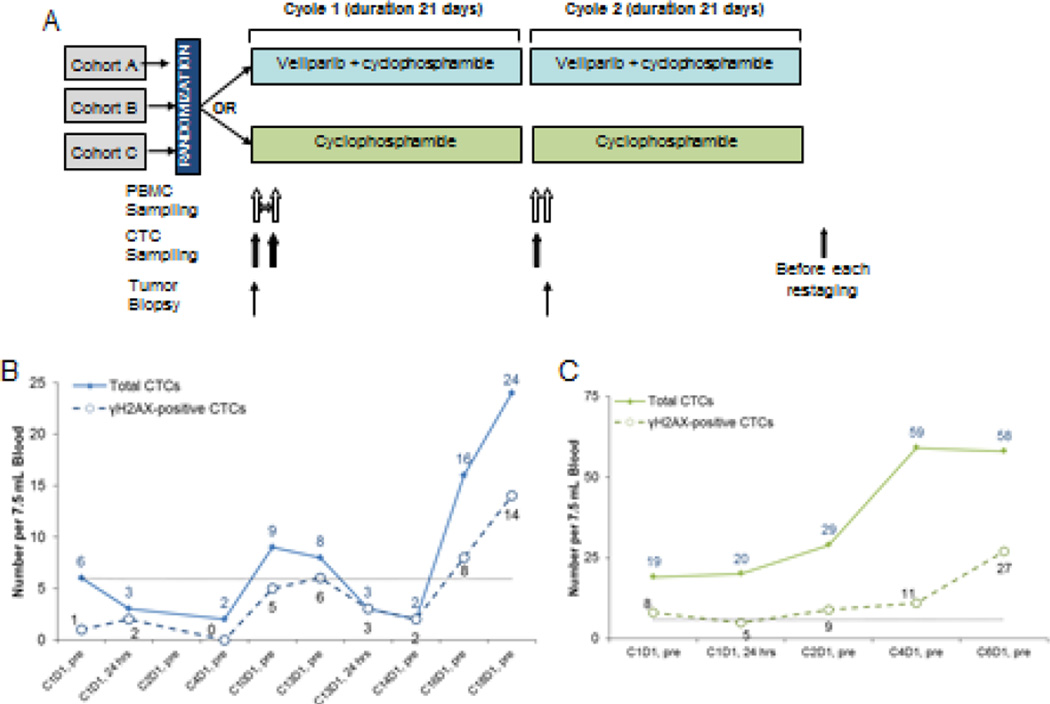
A phase I trial of the combination of veliparib and cyclophosphamide (Cytoxan) reported by the NCI DCTD Developmental Therapeutics Clinic in 2012 (ClinicalTrials.gov: NCT00576654) tested a daily schedule of veliparib rather than BID dosing to allow for better patient compliance when given with daily cyclophosphamide.26 Clinical responses, predominately in patients with BRCA mutations, were seen. A follow-up randomized phase II trial of oral cyclophosphamide with and without veliparib (ClinicalTrials.gov: NCT01306032) was conducted in which veliparib was given at the recommended phase II dose of 60 mg daily with 50 mg cyclophosphamide daily compared to the control group given 50 mg daily cyclophosphamide alone (Figure 6A). There were individual arms to evaluate three different histologies: A) high-grade serous ovarian cancer, B) triple negative breast cancer, and C) low-grade non-Hodgkin lymphoma arm (closed due to poor accrual). In the largest, high-grade serous ovarian cancer cohort, there were 4 responders on the combination arm, as compared to 7 in the single-agent cyclophosphamide arm.27
Figure 6.
Veliparib (ABT-888) with cyclophosphamide in refractory BRCA-positive ovarian, primary peritoneal or ovarian high-grade serous carcinoma, fallopian tube cancer, triple-negative breast cancer, and low-grade non-Hodgkin lymphoma. (A) Schema for clinical trial drug administration and CTC collection. (B-C) Total CTCs and γH2AX-positive CTCs for two patients initially on cyclophosphamide alone who then crossed over to the veliparib plus cyclophosphamide arm. Patient in panel B had breast cancer and crossed over to combination arm at cycle 13; patient in panel C had breast cancer and crossed over to combination arm at cycle 3. Both patients were taken off-study due to disease progression. Black dotted line indicates recommended cut-off of 6 CTCs for reporting biomarker-positive CTCs.
CTC analysis in this trial had several limitations: there were a low number of CTCs per patient blood specimen and patients with non-Hodgkin lymphoma could not be analyzed by CellSearch due to lack of EpCAM positivity. Figure 6B demonstrates the time-course of CTC analysis for a patient with ovarian cancer on the combination drug arm. In this patient, the fraction of γH2AX-positive CTCs increased over baseline by cycle 10 and remained elevated until the patient came off-study in cycle 18 due to disease progression. Several time points were not evaluable for γH2AX because of our established clinical reporting criteria of ≥6 EpCAM-defined CTCs. The low numbers of CTCs found in earlier cycles confound interpretation for statistical significance, but the overall picture from the 5 analyzable specimens support the drug combination being active in this patient’s CTCs. There was a wide range of total and γH2AX-positive CTCs in the patient population. For example, two other patients with breast cancer had 6 and 27 CTCs at baseline with 1 and 7 γH2AX-positive CTCs, respectively; 24 hours later 44 and 30 CTCs were detected with 3 and 11 γH2AX-positive CTCs, respectively (data not shown). The time course of total CTCs and γH2AX-positive CTCs from a patient on the breast cancer arm of the trial is represented in Figure 6C. Interestingly, both patients in Figure 6B and 6C were initially on the cyclophosphamide alone arm and crossed-over to the combination arm at disease progression; the patient with ovarian cancer crossed over at cycle 14 and the patient with breast cancer crossed over at cycle 3. In both cases, the total number of CTCs and the number of γH2AX- positive CTCs did not change significantly during the single agent cyclophosphamide treatment, and the change in treatment regimens was associated with an increase in total CTCs concordant with the clinical observation that the cross-over treatment did not restore tumor growth control after the failure of cyclophosphamide alone.
Advantages and Limitations of the CellSearch System
The compatibility of the CellSearch analysis with specialized vacutainers capable of preserving cellular biomarkers for up to 72 hours during transport (CellSave tubes) is an important advantage in the clinical laboratory setting, especially in the context of central laboratory testing. Additionally, the CellSearch device gives inherently quantitative results; therefore, if cell enumeration is extended to those CTCs that have the desired biomarker signal, that result will also be quantitative. This approach circumvents the fundamental difficulties in attempting to score the intensity of a biomarker signal in a cell being evaluated.
Unlike technologies that capture CTCs for subsequent analysis, the CellSearch device does not create a permanent specimen or derivative slide to use downstream; only electronic results (cell images) are recorded, limiting analysis to the initial biomarker choice. Imaging and image analysis are also particularly limited with this system. Although it would be desirable to evaluate changes in signal intensity of important and commonly used biomarkers, such as phosphorylated ERK for MAP kinase pathway activation or the activity of kinase inhibitors, generating intensity-based image analysis is not feasible in this system because the image resolution is relatively low and the user cannot control or adjust for changes of intensity in the image analyzer (which is controlled by the imaging program software and optimized for cytokeratin channel intensity). Therefore, the preferred method of analysis is evaluation of positive changes in biomarker levels above the detection threshold of the imaging system, and the system is unable to quantify changes in biomarker levels within individual CTCs. Thus, we believe that the best option for CTC quantitation in this system is cell enumeration, consistent with the CellSearch system’s approved use for patient specimen analysis.
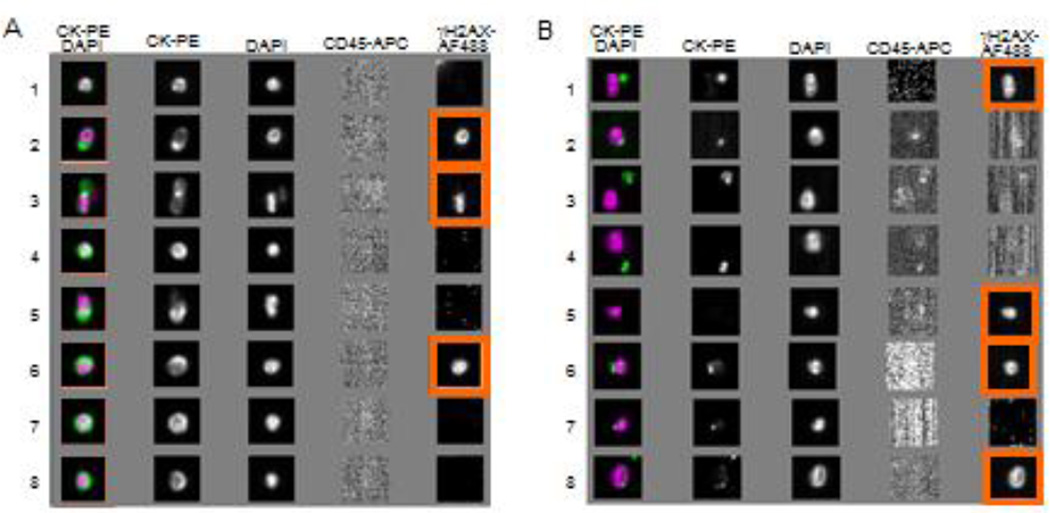
The cells isolated for CellSearch analysis by EpCAM purification and further classified as CD45−/cytokeratin+/DAPI+ have been shown to have prognostic values in clinical studies; however, the phenotype these markers describe is an epithelial phenotype and not a tumor-specific phenotype. As our data indicate, the phenotype described is not sensitive in patients with advanced metastatic disease, and there are significant numbers of EpCAM+ cells that are not scored by the CTC phenotyping criteria used. We re-analyzed the image galleries generated in the CellSearch system to determine if the “events” that are not reported as CTCs by the CellSearch device contained EpCAM-captured cells that were CD45−/cytokeratin−/DAPI+ (nucleated). The galleries for one representative patient are illustrated in Figure 7. There were significant numbers of cells captured by the CellSearch system that would not be classified as epithelial based on the lack of cytokeratin, but may represent a non-epithelial subpopulation of CTCs. It would have been informative to have examined a vimentin stain on these cells to confirm a mesenchymal phenotype, but this was not possible with the instrumentation available.
Figure 7.
Images of CTCs and unassigned events by CellSearch. EpCAM antibody-enriched components from a blood specimen from a patient with advanced breast cancer were stained with DAPI and fluorescence antibodies CK-PE, CD45-APC, and γH2AX-AF488, and analyzed using the CellSearch system. For this specimen, 27 CTCs (EpCAM+/CK+/CD45−/DAPI+) were detected and 7 of them were γH2AX-positive. Additionally, 906 unassigned events were observed in this specimen, among which 43 were CK−/CD45−/DAPI+ and, of those, 9 were γH2AX-positive. Images highlighted in orange show γH2AX-AF488 staining, indicating a γH2AX-positive CTC. (A) Images for 8 representative CTCs and (B) 8 unassigned events (not typical CTCs) are presented. Unassigned events (CK−/CD45−/DAPI+) were also captured by the EpCAM antibody in the specimen from the same patient.
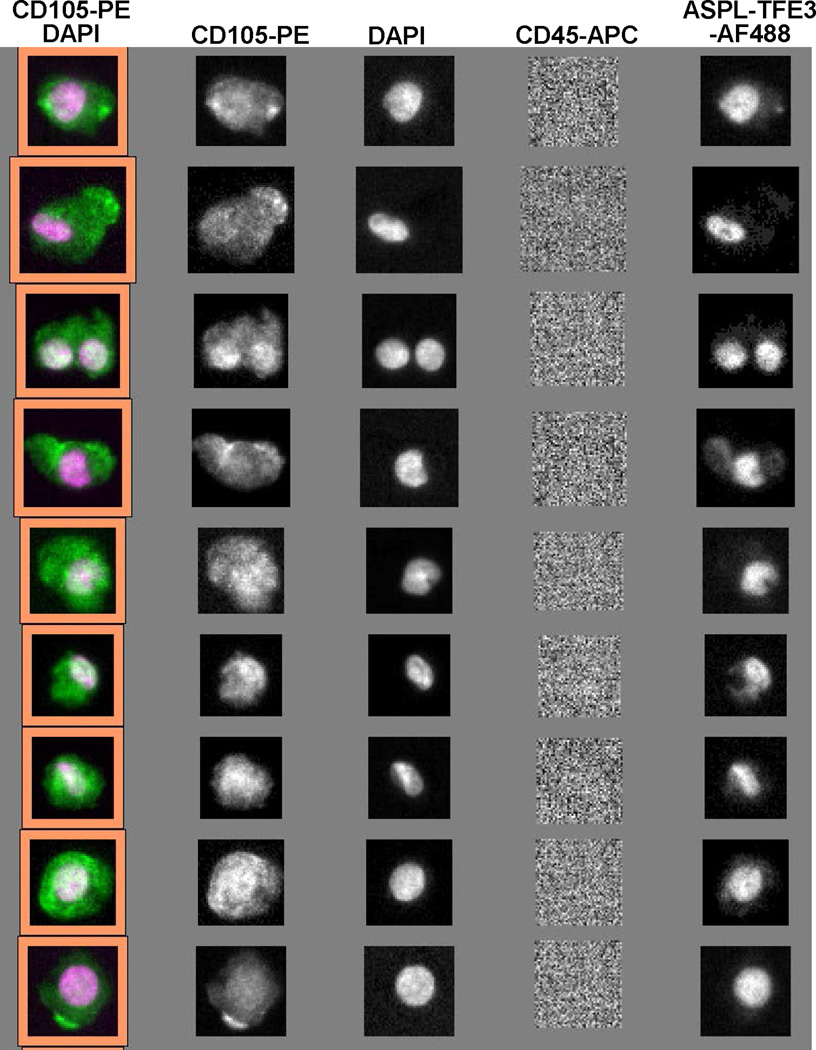
Recently, Veridex has made a research-use only kit available for purifying circulating endothelial cells (CECs) from patient blood. The capture antibody for the ferrofluid in this kit is CD146 (MCAM, MUC18); however, because this marker is found on some leukocyte populations, the specificity of this antibody for CECs is not perfect, so CD105 is used as an additional phenotyping marker. Pharmacologists and toxicologists may view this as a potentially informative marker for drug toxicity, and also as a marker for melanoma and certain sarcomas. We investigated potential use of this capture kit in blood samples from patients with alveolar soft part sarcoma (ASPS), a disease characterized by a genetic rearrangement of the ASPL and TFE3 genes. There are both FISH detection kits (Vysis, Abbot Laboratories, Abbott Park, IL) and specific monoclonal antibodies to detect the fusion protein generated by the rearrangement.28 Figure 8 shows a CellSearch image gallery for CECs from an ASPS patient labelled with the monoclonal antibodies to the fusion protein, demonstrating the capture and identification of CTCs derived from ASPS and suggesting expanded uses for the CellSearch technology. To this end, we are currently testing a next generation CellSearch system that allows this positive tumor marker antibody phenotyping approach to be combined with a labelled antibody for evaluating a PD biomarker such as γH2AX (via the addition of a fifth analysis channel) to expand CTC PD measurements to a greater variety of patient populations.
Figure 8.
CTCs detected from an ASPS patient blood sample using the CellSearch Circulating Endothelial Cell kit with anti-CD146 capture (and anti-CD105 phenotyping confirmation) on the CellSearch device. Staining with an antibody against the ASPL-TFE3 fusion protein in the last column authenticates the malignant origin of the cells.
Conclusion
EpCAM-positive CTCs have been found in advanced cases of all major tumor histologies, yet we detect EpCAM-positive CTCs in only 20–40% of early phase trial patients, a low frequency that is counterintuitive in the context of these cases of widely disseminated, metastatic disease. The presence of EpCAM-positive CTCs in only a subset of advanced cancer cases therefore significantly hinders the application of the CellSearch platform in early clinical trials, despite its several clinical advantages such as multi-day specimen preservation and proven suitability for central laboratory testing. This limitation forces an untenable decision during early clinical trials: either accept sample sizes smaller than those needed to reach statistically significant conclusions about pharmacological effects, or screen higher than needed numbers of patients to select only those with evaluable EpCAM-positive CTC numbers at baseline while risking selection bias in the trial population. Perhaps CellSearch evaluations will instead find niche applications in early stage clinical trials using smaller sample sizes than those required for statistical significance, such as guiding and informing other evaluations scheduled during a clinical trial. For example, instead of using a threshold plasma exposure or PD response in circulating blood mononuclear cells to justify performing core needle biopsies for PD evaluation in phase 0 or I trials, one could envision using CellSearch CTC monitoring of a PD biomarker until a drug effect appears in two patients as evidence that biopsy-based PD assessment will most likely be successful. In this scenario, CTC analysis is essentially providing early, less-invasive evidence for a PD effect that then justifies more invasive, biopsy-based procedures to confirm the molecular drug action in tumor.
In the setting of advanced disease, the accumulating CTC data from phase 1 solid tumor clinics point to the important conclusion that a universal cell surface marker for circulating carcinoma cells has not been identified, and that either a universal marker or an alternative strategy will be needed for the CellSearch platform to realize its full potential as a tool for developmental therapeutics. Because commonly expressed epithelial markers such as EpCAM, MUC1, and others have not proven to be universal cell surface differentiation markers for identifying CTCs, it seems highly unlikely that any other marker of this type would be more successful in detecting CTCs in ≥80% of all carcinoma cases. Thus, the path forward to increase the utility of the CellSearch platform will likely involve a three-pronged strategy: (1) moving to more homogeneous phase I/II patient populations, perhaps of the same histology or sub-type, or earlier stages of disease where tumor phenotypic heterogeneity may not be so pronounced so the need for a universal cell surface epitope for CTC capture is replaced by the need for a tissue-specific or cell lineage-specific biomarker; (2) using a cell surface marker that identifies subpopulations of cells based on shared biological function of importance in oncology, such as cancer stem cells, because shared function implies common molecular pathways and therefore cell surface receptors in common that could provide useful epitopes for CTC capture; and (3) a biomarker test to distinguish malignant from non-malignant cells, because normal epithelial stem cells may also circulate in blood, especially if mobilized by chemotherapy, confounding CTC isolation based on tissue, lineage, and/or function -specific biomarkers.
Even with the addition of a fifth analysis channel, the current configuration of the CellSearch platform cannot accommodate such a multi-marker analysis, and pursuit of the three-prong strategy would require additional modification of the instrument and the validated analysis. However, if such modifications were successful, the payoff for developmental therapeutics would be quite large, because the longitudinal assessment of drug action on molecular targets required to characterize the PD of a new drug and set its optimal scheduling cannot be achieved using tumor biopsies, but must rely on less invasive assessment strategies such as the CTC “liquid biopsy”. The main competition for cell surface biomarker-based capture of CTCs comes from EpCAM-independent platforms for isolating and evaluating CTCs, including filtration approaches, 29, 30 depletion of normal blood cells (negative selection),31, 32 microchip technology,33 and dielectrophoresis field-flow.34, 35
The application of the CellSearch technology in monitoring drug responses in patients with advanced cancers within the NCI clinical trials network has yielded several key insights including demonstrating that patients with a variety of advanced stage carcinomas have low numbers of EpCAM-defined CTCs, independent of disease type or participating clinical site. This was a major limiting factor in our efforts to statistically quantitate CTCs and CTC-associated biomarkers. It also serves as a caution for the use of individual cell-surface differentiation markers to isolate and characterize CTCs from patients with advanced carcinomas, as there could well be additional populations of important but currently unidentified circulating cells to be discovered and categorized. This is a double-edged sword, providing an exciting area of future research as well as cautioning scientists and clinicians about the extent and validity of CTC results reported from advanced-stage patients.
Acknowledgments
We thank Dr. Andrea Regier Voth, Leidos Biomedical Research, Inc., for medical writing support in the preparation of this manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: None
Research Performed At: The Frederick National Laboratory for Cancer Research
References
- 1.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 2.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 4.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 5.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 6.Reid AH, Attard G, Danila DC, Oommen NB, Olmos D, Fong PC, et al. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J Clin Oncol. 2010;28:1489–1495. doi: 10.1200/JCO.2009.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danila DC, Anand A, Sung CC, Heller G, Leversha MA, Cao L, et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol. 2011;60:897–904. doi: 10.1016/j.eururo.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helo P, Cronin AM, Danila DC, Wenske S, Gonzalez-Espinoza R, Anand A, et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 10.Gradilone A, Naso G, Raimondi C, Cortesi E, Gandini O, Vincenzi B, et al. Circulating tumor cells (CTCs) in metastatic breast cancer (MBC): prognosis, drug resistance and phenotypic characterization. Ann Oncol. 2011;22:86–892. doi: 10.1093/annonc/mdq323. [DOI] [PubMed] [Google Scholar]
- 11.Das M, Riess JW, Frankel P, Schwartz E, Bennis R, Hsieh HB, et al. ERCC1 expression in circulating tumor cells (CTCs) using a novel detection platform correlates with progression-free survival (PFS) in patients with metastatic non-small-cell lung cancer (NSCLC) receiving platinum chemotherapy. Lung Cancer. 2012;77:421–426. doi: 10.1016/j.lungcan.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Kuhlmann JD, Wimberger P, Bankfalvi A, Keller T, Scholer S, Aktas B, et al. ERCC1-positive circulating tumor cells in the blood of ovarian cancer patients as a predictive biomarker for platinum resistance. Clin Chem. 2014;60:1282–1289. doi: 10.1373/clinchem.2014.224808. [DOI] [PubMed] [Google Scholar]
- 13.de Bono JS, Attard G, Adjei A, Pollak MN, Fong PC, Haluska P, et al. Potential applications for circulating tumor cells expressing the insulin-like growth factor-I receptor. Clin Cancer Res. 2007;13:3611–3616. doi: 10.1158/1078-0432.CCR-07-0268. [DOI] [PubMed] [Google Scholar]
- 14.Peek VL, Um SLS, Betty Yan SB, Yan L, Dempsey J, Barnard D, et al. Circulating tumor cell (CTC) assay development for detection of H2AX as a clinical pharmacodynamic (PD) marker for Chk1 kinase inhibitors [abstract] Mol Cancer Ther (Meeting Abstracts) 2011;10:A49. [Google Scholar]
- 15.Wang LH, Pfister TD, Parchment RE, Kummar S, Rubinstein L, Evrard YA, et al. Monitoring drug-induced gammaH2AX as a pharmacodynamic biomarker in individual circulating tumor cells. Clin Cancer Res. 2010;16:1073–1084. doi: 10.1158/1078-0432.CCR-09-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinders RJ, Hollingshead M, Lawrence S, Ji J, Tabb B, Bonner WM, et al. Development of a validated immunofluorescence assay for gammaH2AX as a pharmacodynamic marker of topoisomerase I inhibitor activity. Clin Cancer Res. 2010;16:5447–5457. doi: 10.1158/1078-0432.CCR-09-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 18. [accessed October 1, 2014];NCI Division of Cancer Treatment and Diagnosis Validated Biomarker Assays. Assays Available from: http://dctd.cancer.gov/ResearchResources/ResearchResources-biomarkers.htm.
- 19.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 20.Kallergi G, Konstantinidis G, Markomanolaki H, Papadaki MA, Mavroudis D, Stournaras C, et al. Apoptotic circulating tumor cells in early and metastatic breast cancer patients. Mol Cancer Ther. 2013;12:1886–1895. doi: 10.1158/1535-7163.MCT-12-1167. [DOI] [PubMed] [Google Scholar]
- 21.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 22.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 23.Reiss KA, Herman JM, Zahurak M, Brade AM, Dawson LA, Scardina A, et al. A phase I study of veliparib (ABT-888) in combination with low-dose fractionated whole abdominal radiation therapy (LDFWAR) in patients with advanced solid malignancies and peritoneal carcinomatosis [abstract] J Clin Oncol (Meeting Abstracts) 2014;32:4139. [Google Scholar]
- 24.Heitz F, du Bois A, Rochon J, Scheil-Bertram S, Hils R, Fisseler-Eckhoff A, et al. Requirements to assess feasibility of phase 0 trials during major abdominal surgery: variability of PARP activity. Clin Cancer Res. 2012;18:2632–2637. doi: 10.1158/1078-0432.CCR-12-0021. [DOI] [PubMed] [Google Scholar]
- 25.LoRusso P, Ji JJ, Li J, Heilbrun LK, Shapiro G, Sausville EA, et al. Phase I study of the safety, pharmacokinetics (PK), and pharmacodynamics (PD) of the poly(ADP-ribose) polymerase (PARP) inhibitor veliparib (ABT-888; V) in combination with irinotecan (CPT-11; Ir) in patients (pts) with advanced solid tumors [abstract] J Clin Oncol (Meeting Abstracts) 2011;29:3000. [Google Scholar]
- 26.Kummar S, Ji J, Morgan R, Lenz HJ, Puhalla SL, Belani CP, et al. A phase I study of veliparib in combination with metronomic cyclophosphamide in adults with refractory solid tumors and lymphomas. Clin Cancer Res. 2012;18:1726–1734. doi: 10.1158/1078-0432.CCR-11-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kummar S, Oza AM, Fleming GF, Sullivan DM, Gandara DR, Naughton MJ, et al. Randomized Trial of Oral Cyclophosphamide and Veliparib in High-Grade Serous Ovarian, Primary Peritoneal, or Fallopian Tube Cancers, or BRCA-Mutant Ovarian Cancer. Clin Cancer Res. 2015;21:1574–1582. doi: 10.1158/1078-0432.CCR-14-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vistica DT, Krosky PM, Kenney S, Raffeld M, Shoemaker RH. Immunohistochemical discrimination between the ASPL-TFE3 fusion proteins of alveolar soft part sarcoma. J Pediatr Hematol Oncol. 2008;30:46–52. doi: 10.1097/MPH.0b013e31815d1d6f. [DOI] [PubMed] [Google Scholar]
- 29.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu T, Lu B, Tai YC, Goldkorn A. A cancer detection platform which measures telomerase activity from live circulating tumor cells captured on a microfilter. Cancer Res. 2010;70:6420–6426. doi: 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jatana KR, Balasubramanian P, Lang JC, Yang L, Jatana CA, White E, et al. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: initial results. Arch Otolaryngol Head Neck Surg. 2010;136:1274–1279. doi: 10.1001/archoto.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, et al. ApoStream(TM), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gascoyne PR, Shim S. Isolation of circulating tumor cells by dielectrophoresis. Cancers (Basel) 2014;6:545–579. doi: 10.3390/cancers6010545. [DOI] [PMC free article] [PubMed] [Google Scholar]