SUMMARY
The inflammasome is an intracellular signaling complex, which on recognition of pathogens and physiological aberration, drives activation of caspase-1, pyroptosis, and the release of the pro-inflammatory cytokines IL-1β and IL-18. Bacterial ligands must secure entry into the cytoplasm to activate inflammasomes, however, the mechanism by which concealed ligands are liberated in the cytoplasm have remained unclear. Here, we showed that the interferon-inducible protein IRGB10 is essential for activation of the DNA-sensing AIM2 inflammasome by Francisella novicida, and contributed to the activation of the LPS-sensing caspase-11 and NLRP3 inflammasome by Gram-negative bacteria. IRGB10 directly targeted cytoplasmic bacteria through a mechanism requiring guanylate-binding proteins. Localization of IRGB10 to the bacterial cell membrane compromised bacterial structural integrity and mediated cytosolic release of ligands for recognition by inflammasome sensors. Overall, our results reveal IRGB10 as part of a conserved signaling hub at the interface between cell-autonomous immunity and innate immune sensing pathways.
Keywords: Innate immunity, cell-autonomous immunity, sensors, immunity-related GTPases, GBPs, LPS, caspase-1, interferons
Graphical Abstract
eTOC
Finding the route to the cytoplasm: an interferon-inducible protein localizes to the cell membrane of bacteria, compromising its structural integrity and mediating release of ligands, otherwise inaccessible, for sensing by inflammasomes.
INTROUDCTION
Inflammasomes are innate immune signaling complexes which contribute to the host protection against infectious agents and have a critical role in regulating the development of cancer, autoinflammation, and metabolic disorders (Lamkanfi and Dixit, 2014). The identity of an inflammasome is defined by the sensor initiating its formation, which could either be a nucleotide-binding domain, leucine-rich repeat containing protein (NLR), an AIM2-like receptor (ALR), or pyrin (Man and Kanneganti, 2016). These cytoplasmic sensors detect pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS) and flagellin from invading microbes, but also respond to host-derived danger-associated molecular patterns (DAMPs) released from damaged or dying cells (Latz et al., 2013). On recognition of PAMPs and DAMPs, inflammasome sensors initiate assembly of an inflammasome complex containing caspase-1. Caspase-1 is activated and cleaves the pro-inflammatory cytokine precursors pro-IL-1β and pro-IL-18. Activated caspase-1 also drives a form of inflammatory cell death called pyroptosis. Bioactive IL-1β and IL-18 released from pyroptotic cells unleash their pleiotropic effects in the extracellular space.
The ability of the pathogen to reach, and/or inject virulence factors into the cytoplasm is a fundamental requirement for activation of the inflammasome. For example, LPS introduced into the cytoplasm during infection by Gram-negative bacteria engages non-canonical activation of the NLRP3 inflammasome via caspase-11 (Kayagaki et al., 2011; Kayagaki et al., 2013). In this pathway, extracellular LPS is detected by Toll-like receptor 4 (TLR4), inducing TRIF-dependent type I interferon (IFN) production and upregulation of caspase-11 expression (Broz et al., 2012; Gurung et al., 2012; Rathinam et al., 2012). Guanylate-binding proteins (GBPs) mediate entry of Gram-negative bacteria into the cytoplasm, where LPS binds and activates caspase-11 (Hagar et al., 2013; Meunier et al., 2014; Shi et al., 2014). Activated caspase-11 directly cleaves the pro-pyroptotic factor gasdermin D, whereby the N-terminal fragment of gasdermin D induces pyroptosis and activation of the NLRP3 inflammasome (Kayagaki et al., 2015; Shi et al., 2015).
Unlike NLRP3 which does not appear to interact with any ligands directly, the DNA sensor AIM2 binds dsDNA and activates the inflammasome (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). AIM2 recognizes the Gram-negative bacterium Francisella novicida, DNA viruses mouse cytomegalovirus (MCMV) and vaccinia virus, as well as mammalian DNA (Fernandes-Alnemri et al., 2010; Jones et al., 2010; Muruve et al., 2008; Rathinam et al., 2010). Of these, F. novicida infection engages activation of an IRF1–dependent AIM2 inflammasome pathway (Man et al., 2015). Activation of this pathway requires type I IFN-potentiated expression of GBPs via the transcription factor IRF1 (Man et al., 2015). GBPs mediate the exposure of DNA from F. novicida that ultimately leads to activation of the AIM2 inflammasome (Man et al., 2015; Meunier et al., 2015). Although it is established that bacterial ligands must gain entry into the cytoplasm to activate inflammasomes, the mechanism governing how ligands are liberated and presented to inflammasome-initiating sensors have remained unclear.
Here, we showed that the IFN-inducible protein IRGB10 is essential for the activation of the IRF1–dependent AIM2 and caspase-11–dependent NLRP3 inflammasomes in response to bacterial infection. IRGB10 directly targeted Gram-negative bacteria decorated by GBPs. Localization of IRGB10 to the bacterial cell membrane conferred anti-bacterial activity and liberation of DNA and LPS for sensing by inflammasome sensors. Therefore, our results provide insights into the mechanism by which concealed ligands are liberated in the cytoplasm to activate inflammasomes and establish a mechanistic nexus between cell-autonomous immunity and innate immune sensing pathways.
RESULTS
IRGB10 is expressed in response to infection by F. novicida
We previously demonstrated that the transcription factor IRF1 induces the expression of a family of IFN-inducible proteins called GBPs in response to Francisella novicida infection (Man et al., 2015). In addition to GBPs, IRF1 induces upregulation of hundreds of IFN-stimulated genes in response to F. novicida infection, many of which are poorly characterized. We analyzed our entire microarray dataset and identified 20 genes which had the lowest levels of expression in F. novicida-infected Irf1−/− BMDMs compared with infected WT BMDMs (Figure S1A). Among the 20 included genes encoding GBPs (Figure S1A), confirming our previous analysis (Man et al., 2015).
The gene encoding IRGB10 had the lowest level of expression in Irf1−/− BMDMs relative to WT BMDMs infected with F. novicida (Figure S1A). The reduced gene and protein expression of IRGB10 in F. novicida-infected Irf1−/− and Ifnar1−/− BMDMs compared with that of WT BMDMs was further confirmed by immunoblotting and real-time quantitative RT-PCR analysis (Figures 1A and 1B). We also observed reduced expression of guanylate-binding protein 2 (GBP2) and GBP5 in F. novicida-infected Irf1−/− and Ifnar1−/− BMDMs compared with that of WT BMDMs (Figure 1A). We also observed a rapid and dynamic expression of IRGB10 in WT BMDMs treated with recombinant mouse IFN-β and IFN-γ, LPS and poly(I:C) (Figure S1B). Together, these results suggested that IRGB10 is an IFN-inducible protein and is produced in response to F. novicida infection through a mechanism requiring type I IFN signaling and IRF1.
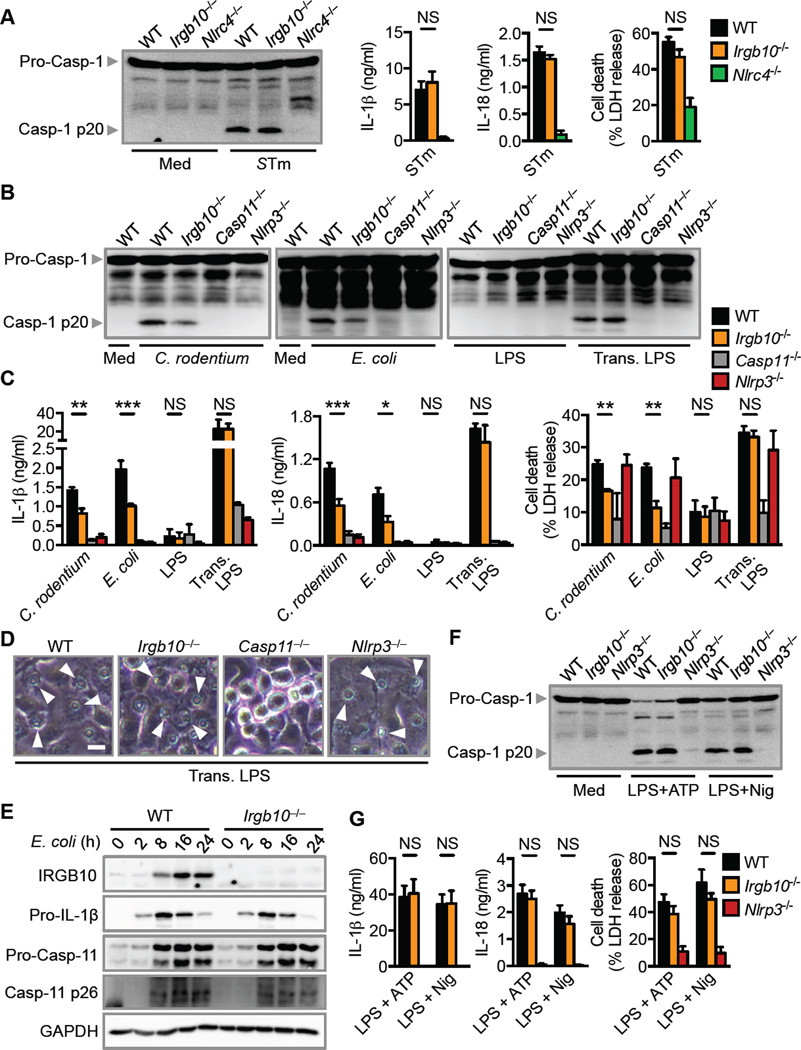
Figure 1. IRGB10 is required for activation of the AIM2 inflammasome by F. novicida (See also Figures S1–S3).
(A) Immunoblot analysis of IRF1, IRGB10, GBP2, GBP5 and GAPDH (loading control) in unprimed wild-type (WT) or mutant BMDMs at various times (above lane) after infection with F. novicida (multiplicity of infection [MOI], 50).
(B) Real-time quantitative RT-PCR analysis of the gene encoding IRGB10 in BMDMs 8 h after infection with F. novicida, presented relative to that of the gene encoding GAPDH.
(C) Immunoblot analysis of pro-caspase-1 (Pro-Casp-1) and the caspase-1 subunit p20 (Casp-1 p20) in unprimed WT or mutant BMDMs left uninfected or untreated (medium alone [Med]) or assessed 20 h after infection with F. novicida (MOI, 100; left) or 5 h after transfection with poly(dA:dT) and pcDNA (middle) or 10 h after infection with mouse cytomegalovirus (MCMV; right).
(D) Release of IL-1β (top), IL-18 (middle) and death (bottom) of unprimed BMDMs after treatment as in (C).
(E) Microscopy analysis of the death of unprimed BMDMs after treatment as in (C).
(F) Confocal microscopy analysis of ASC in unprimed BMDMs infected with F. novicida (MOI, 100) for 20 h or 5 h after transfection with poly(dA:dT). Quantification of the prevalence of ASC inflammasome speck. At least 200 BMDMs from each genotype were analyzed.
Scale bars, 10 µm (E and F). Arrowheads indicate dead cells (E) or inflammasome specks (F). NS, not statistically significant, ***P < 0.001 and ****P < 0.0001 (two-tailed t-test [D]); one-way analysis of variance [ANOVA] with Dunnett’s multiple-comparisons test [F]). Data are representative of two (B and F) or three independent experiments (A, C–E; mean and s.e.m. in B, D and F).
IRGB10 is required for activation of the AIM2 inflammasome by F. novicida
The IRF1-dependent pathway is essential for the activation of the AIM2 inflammasome induced by F. novicida infection, however, how this pathway results in the cytosolic liberation of otherwise hidden bacterial DNA for sensing by AIM2 is still unclear. We generated a mouse strain with a deletion of IRGB10 to investigate whether IRGB10 contributed to the activation of the AIM2 inflammasome in response to F. novicida infection. Irgb10−/− mice were viable and gained similar body weight over time compared with WT mice (Figure S1C). BMDMs generated from Irgb10−/− mice displayed similar expression of the myeloid cell markers F4/80 and CD11c compared with that of WT mice (Figure S1D). Irgb10−/− mice also had a similar prevalence of neutrophils, macrophages, DCs, CD4+ T cells, CD8+ T cells and B cells in the spleen, CD4+ T cells and CD8+ T cells in the thymus, and neutrophils, monocytes, CD4+ T cells, CD8+ T cells and B cells in the bone marrow compared with WT mice (Figure S2A–C).
We infected unprimed WT and Irgb10−/− BMDMs with F. novicida and analyzed caspase-1 activation, release of IL-1β and IL-18 and induction of cell death mediated by inflammasome activation. Activation of caspase-1, release of IL-1β and IL-18 and induction of cell death were impaired in Irgb10−/− BMDMs infected with F. novicida compared with WT BMDMs infected with F. novicida (Figures 1C–1E). Aim2−/− BMDMs infected with F. novicida also failed to engage inflammasome activities, confirming specificity of F. novicida infection to the AIM2 inflammasome. Expression of the gene encoding IL-1β or IL-18 and secretion of the pro-inflammatory cytokines tumor-necrosis factor (TNF) and IL-6 and IFN-β were similar between WT and Irgb10−/−BMDMs infected with F. novicida (Figure S3A and S3B), suggesting there were no global defects in Irgb10−/−BMDMs.
Engagement of AIM2 by transfection of dsDNA ligands poly(dA:dT) and plasmid pcDNA3.1 DNA into the cytoplasm or infection by the DNA virus MCMV induced maturation of caspase-1, release of IL-18 and cell death in both WT and Irgb10−/− BMDMs, but not in Aim2−/− BMDMs (Figures 1C–1E), suggesting that the requirement for IRGB10 in the activation of the AIM2 inflammasome was specific to F. novicida infection. Furthermore, we knocked down the gene encoding IRGB10 in primary WT BMDMs using small interfering RNA and found that silencing of Irgb10 led to a reduction in secretion of IL-1β and activation of caspase-1 in response to infection with F. novicida, whereas the expression of GBP2, IL-1β or TNF was not affected (Figure S3C–E). Irgb10−/− BMDMs also had an impaired ability to generate inflammasome specks in response to infection with F. novicida, but not in response to transfected poly(dA:dT) (Figure 1F). These results indicated that cytoplasmic recognition of bacteria, viruses or transfected dsDNA by AIM2 is governed by the differential requirement of IRGB10. Identification of a role for IRGB10 in the activation of the DNA-sensing AIM2 inflammasome by F. novicida adds another layer of complexity underpinning the regulation of inflammasome activity.
IRGB10 contributes to activation of the caspase-11–NLRP3 inflammasome
Gram-negative bacteria other than F. novicida can activate the inflammasome via alternative cytosolic sensors, including NLRC4 and NLRP3 (Man and Kanneganti, 2015). To investigate whether IRGB10 is required for activation of the inflammasome in response to other Gram-negative bacteria, we stimulated unprimed WT and Irgb10−/− BMDMs with Salmonella enterica serovar Typhimurium (S. Typhimurium) which had been grown to a log-phase and can activate the NAIP-NLRC4 inflammasome via its flagellin or the Salmonella Pathogenicity Island-1 type III secretion system (Broz et al., 2010; Man et al., 2014). We found similar levels of activation of caspase-1, release of IL-1β and IL-18 and cell death in WT and Irgb10−/− BMDMs infected with S. Typhimurium grown to a log-phase (Figure 2A).
Figure 2. IRGB10 contributes to activation of the caspase-11–NLRP3 inflammasome (See also Figures S3 and S4).
(A) Immunoblot analysis of caspase-1, the release of IL-1β and IL-18, and death of unprimed BMDMs left untreated or assessed 4 h after infection with Salmonella Typhimurium (STm; MOI, 1).
(B) Immunoblot analysis of caspase-1 in unprimed BMDMs left untreated or assessed 20 h after infection with C. rodentium, E. coli or 10 h after LPS transfection.
(C) Release of IL-1β (left), IL-18 (middle) and death (right) of unprimed BMDMs after treatment as in (B).
(D) Microscopy analysis of the death of primed BMDMs 10 h after LPS transfection.
(E) Immunoblot analysis of IRGB10, pro-IL-1β, pro-caspase-11 (Pro-Casp-11) and the caspase-11 subunit p26 (Casp-11 p26) and GAPDH (loading control) in unprimed WT or mutant BMDMs at various times (above lane) after infection with E. coli (MOI, 20).
(F) Immunoblot analysis of caspase-1 in unprimed BMDMs (Med) or LPS-primed BMDMs stimulated with ATP (LPS + ATP) or nigericin (LPS + Nig).
(G) Release of IL-1β and IL-18 and death of BMDMs analyzed as in (F).
Scale bar, 10 µm (D). Arrowheads indicate dead cells (D). NS, not statistically significant, *P < 0.05, **P < 0.01 and ***P < 0.001 (two-tailed t-test [A, C and G]). Data are representative of three independent experiments (A–G; mean and s.e.m. in A, C and G).
We next infected WT and Irgb10−/− BMDMs with the Gram-negative bacteria C. rodentium and E. coli, both of which can engage the caspase-11–NLRP3 inflammasome (Kayagaki et al., 2011). We found a reduced level of caspase-1 activation and partial, but statistically significant reduction in the release of IL-1β and IL-18 and cell death in Irgb10−/− BMDMs compared with infected WT BMDMs (Figures 2B–2C). The levels of TNF and IL-6 release were similar in WT and Irgb10−/− BMDMs infected with C. rodentium or E. coli (Figure S3F). In addition, we found similar levels of pro-IL-1β and caspase-11 expression in WT and Irgb10−/− BMDMs infected with E. coli (Figure 2E), demonstrating that the ‘priming’ signal necessitating activation of the NLRP3 inflammasome was not compromised in the absence of IRGB10. Silencing of Irgb10 using small interfering RNA also led to a reduction in secretion of IL-1β in response to infection with E. coli (Figure S3E). We also observed reduced secretion of IL-1β and IL-18 in Irgb10−/− BMDMs infected with S. Typhimurium lacking the flagellin subunits fliC and fljB – a strain which preferentially activates the non-canonical NLRP3 inflammasome (Broz et al., 2012; Man et al., 2014) – compared with infected WT BMDMs (Figure S3G).
Engagement of caspase-11 activity leads to non-canonical activation of the NLRP3 inflammasome in response to Gram-negative bacteria (Kayagaki et al., 2011). We observed reduced levels of caspase-11 activation in Irgb10−/− BMDMs infected with E. coli compared with infected WT BMDMs (Figure 2E), suggesting that IRGB10 was apical to caspase-11-mediated binding of LPS. We transfected Salmonella LPS into the cytoplasm of BMDMs to investigate whether IRGB10 was directly involved in LPS sensing. We found that transfection of LPS activated caspase-1 and induced release of IL-1β and IL-18 in an IRGB10-independent and caspase-11- and NLRP3-dependent manner (Figures 2B–2D). In addition, LPS-induced pyroptosis was independent of IRGB10 but required caspase-11 (Figures 2C and 2D). These data demonstrated that IRGB10 contributed to activation of the caspase-11–NLRP3 inflammasome in response to Gram-negative bacteria, but not when LPS is directly transfected into the cytoplasm. This important observation suggested that IRGB10 could have a role in liberating LPS from Gram-negative bacteria during the course of infection to enhance cytosolic accessibility by caspase-11.
We further observed that activators of the canonical NLRP3 inflammasome, LPS plus ATP or nigericin, engaged activation of caspase-1, release of IL-1β and IL-18, and cell death in both WT and Irgb10−/− BMDMs, but not in Nlrp3−/− BMDMs (Figures 2F and 2G). These results excluded a role for IRGB10 in the canonical NLRP3 inflammasome pathway and provided further evidence to support the specificity of IRGB10 to the activation of inflammasomes by Gram-negative bacteria.
Recruitment of IRGB10 to bacteria requires guanylate-binding proteins
Members of the IFN-inducible GTPase family are executioners of cell-autonomous immunity and have the ability to target the vacuolar membrane encapsulating intracellular parasites and bacteria (Kim et al., 2016; Man et al., 2016). We used confocal microscopy to investigate the spatial distribution of IRGB10 relative to F. novicida in BMDMs. Endogenous IRGB10 recruited to and engulfed F. novicida bacteria in WT BMDMs (Figure 3A). In addition, we observed localization of endogenous IRGB10 to E. coli and C. rodentium bacteria (Figure 3A and Figure S4A). Recruitment of IRGB10 to the bacterial structures is reminiscent of the spatial localization of GBPs to Gram-negative bacteria (Man et al., 2015; Meunier et al., 2014; Meunier et al., 2015). Indeed, IRGB10 is a phylogenetic relative of GBPs in the IFN-inducible protein family.
Figure 3. Recruitment of IRGB10 to bacteria requires guanylate-binding proteins (See also Figure S4).
(A) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with F. novicida (left) or E. coli (right).
(B) Immunofluorescence staining as in (A) in unprimed WT, Irgb10−/− and Gbpchr3-KO BMDMs 16 h after infection with F. novicida (top) or E. coli (bottom).
(C) Immunoblot analysis of IRGB10 in cross-linked fraction or the lysate of unprimed BMDMs left untreated or assessed 16 h after stimulation with IFN-β (200 U/ml) or infection with F. novicida (MOI, 100).
(D) Release of IL-1β, IL-18 and death of unprimed BMDMs assessed 16 h after infection as in (A).
(E) Immunoblot analysis of IRGB10, GBP2, GBP5 and GAPDH (loading control) in unprimed WT and Gbpchr3-KO BMDMs infected with F. novicida (MOI, 50).
Scale bar, 10 µm (A) and 5 µm (B). White arrowheads indicate bacteria targeted by IRGB10 (B). Black arrowheads indicate a non-specific band (C). **P < 0.01, ***P < 0.001 and ****P < 0.0001; (two-tailed t-test [D]). Data are representative of two (C) or three independent experiments (A, B, D and E; mean and s.e.m. in D).
We hypothesized that IRGB10 and GBPs synergistically contributed to the liberation of bacterial ligands for sensing by inflammasomes. We immuno-localized endogenous IRGB10 in WT BMDMs and BMDMs lacking GBPs encoded on chromosome 3 (Gbp1, Gbp2, Gbp3, Gbp5 and Gbp7; called Gbpchr3-KO). While endogenous IRGB10 recruited to F. novicida in WT BMDMs, we observed an impaired ability of IRGB10 to localize to the bacteria in Gbpchr3-KO BMDMs (Figure 3B). The accumulation of IRGB10 on bacterial structures suggested that these proteins have the capacity to undergo oligomerization. Indeed, the G domain of IRGB10 mediates homotypic interaction which allows generation of higher order protein oligomers (Haldar et al., 2013). We performed oligomerization assays and cross-linked proteins in the lysates derived from untreated BMDMs, BMDMs treated with recombinant mouse IFN-β, or from BMDMs infected with F. novicida. Consistent with the microscopy analyses, we observed oligomerization of IRGB10 in WT BMDMs infected with F. novicida, but not in that of Irgb10−/− or Gbpchr3-KO BMDMs (Figure 3C). Furthermore, WT BMDMs treated with exogenous IFN-β expressed monomeric IRGB10, but failed to generate large quantities of IRGB10 oligomers (Figure 3C), suggesting that formation of these higher-order protein species requires infection by a pathogen. Similar to the case in F. novicida, we found that recruitment of IRGB10 to E. coli was abrogated in the absence of GBPs encoded on chromosome 3 (Figure 3B).
Activation of the inflammasome driven by F. novicida or E. coli infection was abolished in Gbpchr3-KO BMDMs infected either with F. novicida or E. coli, compared with WT BMDMs (Figure 3D). The reduction in IL-1β and IL-18 secretion and pyroptosis were more pronounced in Gbpchr3-KO BMDMs compared with Irgb10−/− BMDMs (Figure 3D), suggesting that GBPs may recruit additional effectors other than IRGB10 to the bacteria to mediate inflammasome activation. These results also indicated that IRGB10 and GBPs are largely non-redundant in the activation of the AIM2 inflammasome in response to infection with F. novicida.
To investigate whether IRGB10 and GBPs have the ability to regulate the expression of one another, we infected WT and Gbpchr3-KO BMDMs with F. novicida or E. coli and monitored the dynamics of expression of IRGB10 over the course the infection. Both WT and Gbpchr3-KO BMDMs produced similar levels of IRGB10 in response to F. novicida or E. coli infection over time (Figure 3E and Figure S4B). Furthermore, we observed that the expression of GBP2 and GBP5 were intact in Irgb10−/− BMDMs infected with F. novicida or E. coli (Figure S4B and S4C). These results demonstrated that IRGB10 and GBPs do not regulate the expression of one another, but also highlighted that the impaired ability of IRGB10 to localized to F. novicida and E. coli bacteria in Gbpchr3-KO BMDMs was not due to a reduced bioavailability of IRGB10 in these macrophages.
IRGB10 directly targets intracellular bacteria
The mouse IRG family from the IFN-inducible GTPase superfamily contains IRGA, IRGB, IRGC, IRGD and IRGM members (Figure S5A) (Bekpen et al., 2005). The protein sequence of IRGB10 is most closely related to other IRGB members in the family (Figure S5A). We used bioinformatic tools to predict transmembrane regions of IRG proteins, which might provide insights into their ability to recruit to membrane compartments. We found that the IRG family members, including IRGB10, contain two putative myristoylation sites at the amino terminus region (Figure S5B), consistent with previous studies reporting the presence of a myristoylation motif in IRGB10 and IRGA6 (also known as IIGP1) (Haldar et al., 2013; Martens et al., 2004; Papic et al., 2008). We further identified two putative transmembrane helices on the carboxyl terminus of IRGB10 and several other IRG proteins, of which putative transmembrane helix 1 is an amphipathic helix and predicted to show antimicrobial potential (Figures S5C and S5D).
Indeed, the putative transmembrane helix 1 of IRGB10 (residues 284 to 302) contains a highly conserved proline residue, which exists in high frequency in transmembrane α-helices and is essential for inducing hinge or distortion in the membrane helix essential for the function of certain transmembrane channels (Cordes et al., 2002; Jin et al., 2002; Tieleman et al., 2001; Yohannan et al., 2004). All other IRG proteins analyzed harbor a putative amphipathic helix except IRGA2, IRGM1, IRGM2 and IRGM3 (Figure S5C). The second putative transmembrane helix (called transmembrane helix 2) was identified in IRGB10 (residues 370 to 387) and all other IRG proteins analyzed (Figures S5E and S5F), consistent with the identification of a transmembrane helix in the corresponding region of IRGM1 (Martens et al., 2004; Tiwari et al., 2009).
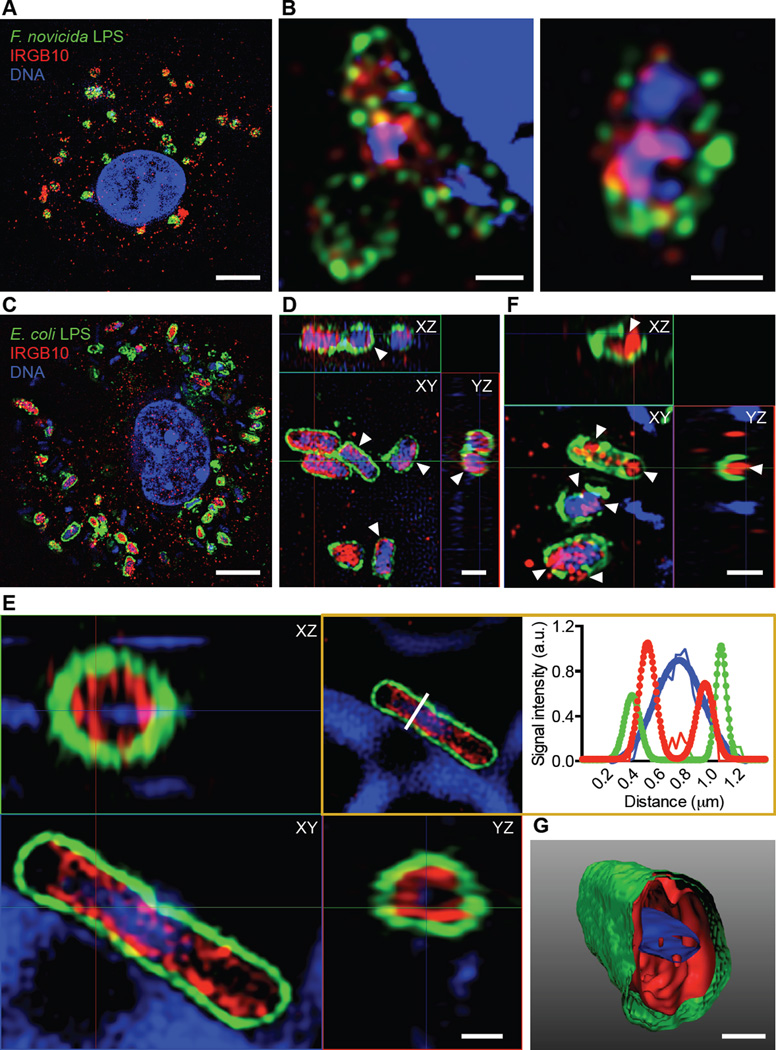
To investigate whether IRGB10 can directly target bacteria, we immunostained endogenous IRGB10 and LPS in primary WT BMDMs infected with F. novicida or E. coli, and examined their relative localization using super resolution and structured illumination microscopy. Remarkably, we observed localization of IRGB10 within a layer of F. novicida LPS (Figures 4A and 4B) and on the surface of the LPS layer of F. novicida (Figure S4D). Structured illumination microscopy also revealed a distinct layer of IRGB10 bound by a layer of E. coli LPS (Figures 4C–4F). In addition, we observed a layer of IRGB10 surrounding the bacterial DNA, both encased by a layer of E. coli LPS (Figures 4D and 4E). The average distance between the peak signal from the E. coli LPS and the peak signal from the IRGB10 layer was 0.12 µm, and the average distance between the signal from the E. coli LPS and the signal from the bacterial DNA was 0.34 µm. These measurements validated that the IRGB10 layer was found between the outer LPS layer and the bacterial DNA (Figure 4E).
Figure 4. IRGB10 directly targets intracellular bacteria (See also Figure S4 and Movie S1).
(A) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with F. novicida. Three-dimensional images were taken using the Structured Illumination Microscopy (SIM) technique. A single z-plane of a 3D image is shown.
(B) A higher resolution image of F. novicida cells taken using SIM as in (A).
(C) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli. Images were taken using SIM.
(D) An enlarged and orthogonal-view image of E. coli cells taken using SIM as in (C).
(E) An enlarged and orthogonal-view image of an E. coli cell taken using SIM. White line (boxed image) indicates a line scan analysis of the SIM image. The line scan values for each signal were plotted and fitted with single and double Gaussian equations (boxed graph).
(F) An enlarged and orthogonal-view image of an E. coli cell taken using SIM.
(G) Three-dimensional surface rendering of an E. coli cell shown in (E).
Scale bars, 5 µm (A and C), 0.5 µm (B, E and G) and 1 µm (D and F). Arrowheads indicate a layer of IRGB10 staining internal to a layer of LPS staining (D) or a disrupted LPS layer (F). Data are representative of two independent experiments (A–G).
Analysis of additional E. coli cells revealed that IRGB10 fully penetrated and integrated into the E. coli cytoplasm (Figures 4E–4G, Figure S4E and S4F). Some bacteria even displayed a compromised structural integrity of their cell wall (Figure 4F, Figure S4E and Movie S1). An initial rupture of the LPS layer might allow IRGB10 to penetrate inside the bacteria (Figure S4G). Consistent with confocal microscopy analysis (Figures 3A and 3B), structured illumination microscopy revealed that IRGB10 had an impaired ability to target bacteria in the absence of GBPs encoded on chromosome 3 (Figure S4H).
We further used immunogold labeling and transmission electron microscopy techniques to immunolocalize endogenous IRGB10 at the subcellular level. We observed localization of IRGB10 on the bacterial cell membrane and within the bacteria (Figure 5A and 5B and Figure S6A), consistent with our confocal and structured illumination microscopy findings. We found a loss of a defined bi-layer cell membrane on the bacteria in regions where IRGB10 staining was found, whereas the bacterial cell membrane remained intact in regions free of IRGB10 staining (Figure 5B). We also observed significantly increased prevalence of IRGB10 proteins in irregular-shaped (22.2 ± 2.9 puncta per bacterium) bacteria than in regular-shaped (1.7 ± 0.3 puncta per bacterium) bacteria (Figure 5A). Furthermore, IRGB10 was often found in vesicle-like structures, with IRGB10-containing vesicles being delivered to and fused with the bacterial cell membrane (Figures 5B and 5C). IRGB10-containing vesicles targeted the bacterial cell membrane of 43% of the IRGB10+ bacteria (Figure S6B). We also observed IRGB10 being targeted to F. novicida (Figure S6C). These findings suggested that IRGB10 proteins might be delivered as a “lethal-hit” to damage cytosolic bacteria in macrophages. The presence of transmembrane helices in IRGB10 and its ability to decorate the cell membrane and internal architecture of bacteria suggested that IRGB10 might have the ability to induce bacteriolysis.
Figure 5. IRGB10 targets the bacterial cell membrane (See also Figures S5 and S6).
(A) Immunogold staining of IRGB10 in unprimed WT BMDMs 16 h after infection with E. coli, acquired using transmission electron microscopy (left). Quantification of the number of IRGB10 puncta in regular-shaped (n = 35) and irregular-shaped (n = 74) bacteria in control samples (secondary antibody only) and regular-shaped (n = 82) and irregular-shaped (n = 79) bacteria in IRGB10-stained samples (right). Images of regular-shaped (top inset) and irregular-shaped (bottom inset) bacteria.
(B) Images of E. coli cells targeted by IRGB10, acquired as in (A). White arrowheads indicate an intact bi-layer bacterial cell membrane. Black arrowheads indicate the presence of IRGB10 and a loss of the bacterial cell membrane. Images of IRGB10-containing vesicles fused with the bacterial cell membrane (insets).
(C) Images of E. coli cells targeted by IRGB10, acquired as in (A). Black arrowheads indicate IRGB10-containing vesicles being delivered to or fused with the bacterial cell membrane.
Scale bars, 500 nm (A), 200 nm (B), and 100 nm (C). NS, not statistically significant, ****P < 0.0001 (one-way ANOVA with Tukey’s multiple-comparisons test [A]). Data are from two (A–C) independent experiments (mean and s.e.m. in A).
IRGB10 and GBPs coordinate bacterial killing and inflammasome activation
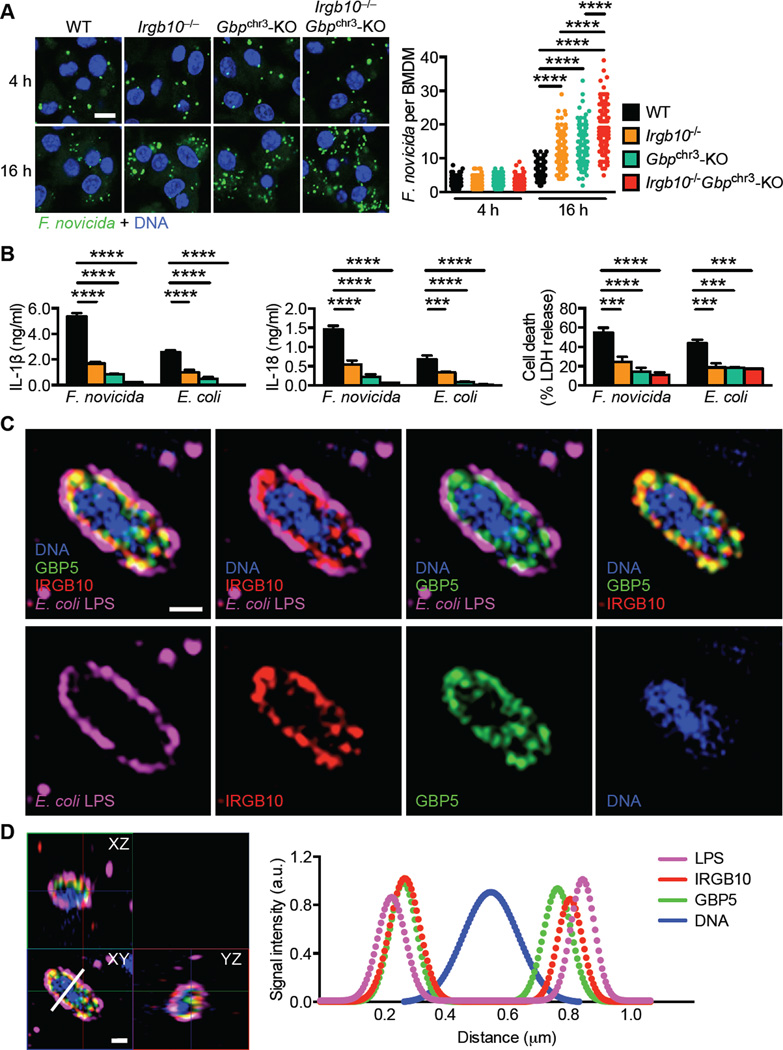
To investigate whether IRGB10 directly affected bacterial viability in macrophages, we infected WT, Irgb10−/− and Gbpchr3-KO BMDMs with F. novicida and quantified bacteria in these cells over time. Single-cell analysis revealed that WT BMDMs restricted bacterial replication from 4 h to 16 h after infection. In contrast, both Irgb10−/− and Gbpchr3-KO BMDMs failed to control bacterial replication and harbored significantly more bacteria than WT BMDMs at 16 h (Figure 6A). Moreover, Irgb10−/− Gbpchr3-KO BMDMs infected with F. novicida harbored significantly more bacteria than did WT, Irgb10−/− or Gbpchr3-KO BMDMs (Figure 6A). The residual secretion of IL-1β and IL-18 in Irgb10−/− or Gbpchr3-KO BMDMs infected with F. novicida or E. coli was abolished in Irgb10−/− Gbpchr3-KO BMDMs (Figure 6B). We further investigated the localization of endogenous IRGB10 and GBPs relative to one another in BMDMs infected with E. coli and found that both were recruited to the same bacteria (Figure S6D). Structured illumination microscopy revealed a layer of IRGB10 co-localized or closely associated with a layer of GBP5, both encased by a layer of LPS (Figures 6C and 6D and Figure S6E).
Figure 6. IRGB10 and GBPs coordinate to mediate bacterial killing (See also Figure S7).
(A) Immunofluorescence staining of F. novicida LPS (green) and DNA (blue) in unprimed BMDMs 4 and 16 h after infection with F. novicida (left). Quantification of F. novicida in WT BMDMs (4 h, n = 177; 16 h, n = 127), Irgb10−/− (4 h, n = 160; 16 h, n = 130), Gbpchr3-KO (4 h, n = 158; 16 h, n = 116), and Irgb10−/− Gbpchr3-KO (4 h, n = 178; 16 h, n = 143) BMDMs, assessed by confocal microscopy (right). Each symbol represents an individual BMDM.
(B) Release of IL-1β (left), IL-18 (middle) and death (right) of unprimed BMDMs and 16 h after infection with F. novicida (MOI, 100) or E. coli (MOI, 20).
(C) Immunofluorescence staining of DNA (blue), GBP5 (green), IRGB10 (red), and LPS (magenta) in unprimed WT BMDMs 16 h after infection with E. coli. 3D images were taken using SIM. A single z-plane of a 3D image of an enlarged image of an E. coli cell is shown (top).
(D) An enlarged and orthogonal-view image of the same E. coli cell as in (C) is shown (left). White line indicates a line scan analysis of the SIM image. The line scan values for each signal were plotted and fitted with single and double Gaussian equations (right).
Scale bars, 10 µm (A) and 0.5 µm (C and D). ***P < 0.001 and ****P < 0.0001 (one-way ANOVA with Dunnett’s multiple-comparisons test [A and B]). Data are from two (A, C and D) or three (B) independent experiments (mean and s.e.m. in B).
Recruitment of IRGB10 to the bacteria to drive bacterial killing may require additional anti-microbial effector molecules. We identified in our microarray dataset, Nos2, the gene encoding inducible nitric oxide synthase (iNOS), as one of the 20 genes with the lowest expression in Irf1−/− BMDMs infected with F. novicida compared with infected WT BMDMs (Figure S1A). This finding was further validated by immunoblotting and confocal microscopy (Figures S7A and S7B). However, both WT and Nos2−/− BMDMs expressed similar levels of IRGB10, GBP2 and GBP5, and were able to undergo activation of caspase-1 and release of IL-1β and IL-18 in response to F. novicida infection (Figures S7C and S7D). Furthermore, we found that the expression of iNOS was not compromised in Irgb10−/− and Gbpchr3-KO BMDMs infected with F. novicida compared with WT BMDMs (Figures S7E and S7F). Collectively, these data suggested that IRGB10-and GBP-dependent killing of F. novicida and activation of the AIM2 inflammasome is uncoupled from iNOS production.
IRGB10 does not affect cytosolic escape by F. novicida
The ability of IRGB10 to target the bacteria directly does not exclude a role for this protein in mediating entry of the bacteria into the cytoplasm. Therefore, we investigated whether IRGB10 is required for driving escape of F. novicida from the vacuole into the cytoplasm. Cytosolic pathogens such as F. novicida escape the vacuole and can be used as a mechanism to deliver ligands into the cytoplasm of a cell, such that ultrapure Salmonella LPS can be delivered to activate the caspase-11–NLRP3 inflammasome (Man et al., 2015).
We took advantage of this cytosolic delivery method and infected WT, Irgb10−/−, Gbpchr3-KO, Aim2−/−, Aim2−/− Nlrp3−/− and Nlrc4−/− BMDMs with F. novicida in the presence of ultrapure LPS from Salmonella to investigate whether Salmonella LPS can be efficiently introduced into the cytoplasm to activate the inflammasome in the absence of IRGB10. In the absence of Salmonella LPS, F. novicida-induced secretion of IL-1β and IL-18 was dependent on IRGB10, AIM2 and GBPs encoded on chromosome 3, but not dependent on NLRC4 (Figure S7G). Importantly, we found that WT, Irgb10−/− and Aim2−/− BMDMs all released similar levels of IL-1β and IL-18 in response to infection with F. novicida in the presence of Salmonella LPS (Figure S7G). These results suggested successful delivery of Salmonella LPS into the cytoplasm and activation of the caspase-11–NLRP3 inflammasome even in the absence of IRGB10. Indeed, Aim2−/− Nlrp3−/− BMDMs infected with F. novicida in the presence of Salmonella LPS failed to release IL-1β and IL-18 (Figure S7G). The partial reduction in IL-1β and IL-18 release in Gbpchr3-KO BMDMs infected with F. novicida in the presence of Salmonella LPS also suggested that GBPs encoded on chromosome 3 had a minor role in promoting escape of F. novicida.
Expression of the gene encoding IL-1β or IL-18 was normal in BMDMs of all genotypes, suggesting that the ‘priming’ signal is intact in these cells (Figure S7H). We also validated our findings and used an isogenic mutant of F. novicida (called F. novicida ΔmglA), which cannot disrupt the vacuole and enter the cytoplasm, as an additional control (Man et al., 2015). Salmonella LPS plus F. novicida ΔmglA was unable to induce IL-1β and IL-18 release from BMDMs of any genotype (Figure S7G). These results further validated the concept that cytosolic entry and exposure of the bacterial ligands is a fundamental requirement for activation of inflammasomes by bacteria. Collectively, our findings indicated that IRGB10 did not interfere with the ability of F. novicida to escape from the vacuole into the cytoplasm and that liberation of F. novicida DNA by IRGB10 is mediated by disruption of the bacteria in the cytoplasm.
IRGB10 provides host protection against F. novicida in vivo
To date, the physiological relevance of IRGB10 in the host defense against pathogens is unknown owing to the lack of a knockout mouse model. We investigated the role of IRGB10 in the host defense against F. novicida infection in vivo. We infected WT, Irgb10−/−, Gbpchr3-KO, Aim2−/− and Casp1Null mice with F. novicida and monitored their susceptibility to infection. All mutant mice lost more body weight compared with WT mice, and these mutant mice succumbed to infection within 8 days, whereas 65% of the WT mice survived beyond day 12 (Figures 7A and 7B). In addition, Irgb10−/− mice harbored significantly more F. novicida bacteria in the liver and spleen compared with WT mice (Figure 7C). Analysis of serum IL-18 showed that Irgb10−/− mice had an impaired ability to produce this cytokine following infection with F. novicida (Figure 7D). Reduced levels of serum IL-18 were also observed in Gbpchr3-KO, Aim2−/− and Casp1Null mice (Figure 7D). Together, these results highlighted a crucial role for IRGB10 in the host defense against F. novicida infection, through a mechanism dependent on GBPs and the AIM2 inflammasome.
Figure 7. IRGB10 provides host protection against infection with F. novicida in vivo.
(A) Survival of 8-week-old WT mice (n = 20), Irgb10−/− mice (n = 30), Gbpchr3-KO mice (n = 13), Aim2−/− (n = 20) and Casp1Null mice (n = 12) infected subcutaneously with 7.5 × 104 CFU of F. novicida.
(B) Body weight of 8-week-old WT mice (n = 12), Irgb10−/− mice (n = 20), Gbpchr3-KO mice (n = 8), Aim2−/− (n = 11) and Casp1Null mice (n = 10) 0–7 d (horizontal axis) after subcutaneous infection with 1.5 × 105 CFU of F. novicida, presented relative to initial body weight at day 0, set as 100%.
(C) Bacterial burden in the liver (left) and spleen (right) of 8-week-old WT mice (n = 18), Irgb10−/− (n = 10) and Aim2−/− mice (n = 8) on day 3 after infection with 1 × 105 CFU of F. novicida.
(D) Concentration of IL-18 in the serum of WT mice (n = 28), Irgb10−/− mice (n = 23), Gbpchr3-KO mice (n = 7), Aim2−/− (n = 14) and Casp1Null mice (n = 14) 24 h after infection with 1.5 × 105 CFU of F. novicida. Each symbol represents an individual mouse (C and D). ***P < 0.0001 and ****P < 0.0001 (log-rank test [A] or one-way ANOVA with Dunnett’s multiple-comparisons test [C and D]). Data are pooled from two independent experiments (A, C and D) or are from one experiment representative of two independent experiments (B; mean and s.e.m. in B, C and D).
DISCUSSION
Pattern-recognition receptors recognize an extraordinarily diverse set of PAMPs and DAMPs. Pathogens can strategically invade the cytoplasm and minimize detection by membrane-bound sensors. In addition, certain ligands are sequestered within the pathogen, and virulence factors produced by pathogens can also mask potential ligands or inhibit innate immune sensing and signaling. Therefore, detection of pathogens heavily relies on the ability of host sensors to gain access to concealed ligands. Here, we identified the first member within the IRG family that contributed to the activation of inflammasomes. In a broader context, the ability of IRGB10 to license activation of multiple inflammasomes highlights its functionality as a conserved signaling hub in innate immunity.
We and others previously demonstrated that the DNA sensor cGAS detects F. novicida infection and triggers a type I IFN and IRF1-dependent signaling cascade leading to the upregulation of hundreds of IFN-stimulated genes (Man et al., 2015; Storek et al., 2015). In this study, we identified IRGB10 as part of this IFN-inducible network regulating activation of inflammasomes. IRGB10 is one of 23 members within the mouse IRG family (Bekpen et al., 2005). The gene encoding IRGB10 was originally identified by fine mapping of 1.2 megabases of DNA on chromosome 11 of mouse embryonic fibroblasts (Bernstein-Hanley et al., 2006b). Differences in this genomic region between inbred mouse strains provided a clue as to why C57BL/6J mice are more resistant to Chlamydia trachomatis infection than do C3H/HeJ mice (Bernstein-Hanley et al., 2006a). Embryonic fibroblasts from C57BL/6J mice treated with IFN-γ express a higher level of IRGB10 and are markedly more resistant to C. trachomatis infection compared with embryonic fibroblasts derived from C3H/HeJ mice (Bernstein-Hanley et al., 2006b). A later study also found that the increased resistance of C57BL/6J mice to Chlamydia psittaci infection relative to the DBA/2J mouse strain is attributed to the increased expression of IRGB10 in C57BL/6J mice (Miyairi et al., 2007).
In this study, we have found that IRGB10 mobilized to the F. novicida bacteria to disrupt bacterial integrity and mediated liberation of otherwise sequestered bacterial DNA. Our identification of putative myristoylation motifs and transmembrane amphipathic helices of IRGB10 indicated that any or all of these could be involved in membrane attachment and permeabilization. A previous study has shown that a myristoylation motif and an amphipathic helix αk region are essential for the recruitment of IRGB10 to the pathogen-containing vacuole of C. trachomatis (Haldar et al., 2013). Furthermore, amphipathic helices are commonly found in anti-microbial peptides, allowing them to induce loss of membrane potential, membrane permeabilization and disruption (Dathe and Wieprecht, 1999). It is intriguing to hypothesize that the myristoylation motifs might bind the bacterial membrane, and the transmembrane helices excavate holes on the membrane to cause bacteriolysis and leakage of DNA into the cytoplasm. It is also possible that other IFN-inducible factors might form, in a hierarchical manner, higher order complexes with IRGB10 and GBPs to orchestrate bacteriolysis. This idea is supported by a previous study showing that IRGA6, IRGB6, IRGD and IRGM2 are all recruited to the same T. gondii pathogen-containing vacuole (Khaminets et al., 2010). However, in cases such as infection by Chlamydia muridarum, GBP-dependent activation of the inflammasome occurs independently of the ability of GBPs or IRGB10 to localize to the pathogen-containing vacuole (Coers et al., 2008; Finethy et al., 2015), suggesting pathogen-specific mechanisms of inflammasome activation by IFN-inducible GTPases.
With respect to the function of IRGB10 in the engagement of caspase-11 activation by Gram-negative bacteria, we observed a partial role for IRGB10 in the activation of the caspase-11–NLRP3 inflammasome. Unlike DNA, LPS is found on the surface of Gram-negative bacteria and is directly exposed to cytoplasmic sensors following entry of the bacteria into the cytoplasm mediated by GBPs or following delivery of LPS by bacterial outer membrane vesicles (Meunier et al., 2014; Vanaja et al., 2016). This observation is consistent with our findings that GBPs were required for the recruitment of IRGB10, where GBPs expose the bacteria in the cytoplasm for subsequent attack by IRGB10 and GBPs themselves. We reason that mechanical disruption of the bacteria by IRGB10 would make LPS more freely accessible for caspase-11, and therefore, drives a more robust inflammasome response.
In addition to the mechanism of ligand release, the biological effects of membrane disruption include reduced bacterial viability and inhibition of bacterial replication. Overall, our results provide insights into how pathogen-associated ligands are released in the cytoplasm to activate inflammasomes. Our work also highlights a functionality of IRGB10 as a signaling hub between cell-autonomous immunity and innate immune sensing pathways.
STAR⋆METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○Mice
-
○Bone Marrow-Derived Macrophages
-
○Bacterial Culture
-
○
- METHOD DETAILS
-
○Stimulation of Bone Marrow-Derived Macrophages
-
○Lactate Dehydrogenase Assay
-
○Immunoblotting Analysis
-
○Immunofluorescence Staining
-
○Super Resolution Microscopy and Structured Illumination Microscopy
-
○Transmission Electron Microscopy
-
○Amino Acid Sequence Analysis
-
○Real Time qRT-PCR Analysis
-
○siRNA Knockdown
-
○Cytokine Analysis
-
○Flow Cytometry
-
○Oligomerization Assay
-
○Animal Infection
-
○
QUANTIFICATION AND STATISTICAL ANALYSIS
- DATA RESOURCES
-
○Microarray
-
○
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one movie and can be found with this article online at XXX.
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Thirumala-Devi Kanneganti (thirumala-devi.kanneganti@stjude.org).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice
Aim2−/−, Nlrp3−/−, Nlrc4−/− , Casp1Null , Casp11−/−, Gbpchr3-KO and Irf1−/− mice have been described previously (Man et al., 2015). Aim2−/− Nlrp3−/− mice were generated by crossing Aim2−/− mice and Nlrp3−/− mice as described previously (Karki et al., 2015). Nos2−/− mice were purchased from Jackson Laboratories (Stock Number 002609). Mice with a genomic deletion of IRGB10 (called Irgb10−/− mice) were generated by Cas9/CRISPR-mediated genome editing. The insert fragment of Irgb10 gRNA was amplified using KODFXNEO (Toyobo) and the following primers; Irgb10_gRNA1_F (5’-TTT CTT GGC TTT ATA TAT CTT GTG GAA AGG ACG AAA CAC Cga aag tga gtc agc tga gag-3’) and Irgb10_gRNA1_R (5’-gaa agt gag tca gct gag agG TTT TAG AGC TAG AAA TAG CAA GTT AAA ATA AGG CTA GTC-3’). The fragment for Irgb10 gRNA was inserted into the gRNA cloning vector (41824, Addgene) using Gibson Assembly Master Mix (E2611S, New England Biolab) to generate Irgb10 gRNA-expressing plasmids. A T7 promoter was added to the Irgb10 gRNA template using KODFXNEO and the following primers; Irgb10_T7gRNA_F (5’-TTA ATA CGA CTC ACT ATA GGa aag tga gtc agc tga gag GTT T-3’) and gRNA_common_R (5’- AAA AGC ACC GAC TCG GTG CCA CTT TT-3’). The T7-Irgb10 gRNA PCR product was gel-purified and used for the subsequent generation of the Irgb10 gRNA. The MEGAshortscript T7 Transcription Kit (AM1354, ThermoFisher Scientific) was used for the generation of the Irgb10 gRNA. Cas9 mRNA was generated by in vitro transcription (IVT) using mMESSAGE mMACHINE T7 ULTRA kit (AM1345, ThermoFisher Scientific). The template was amplified by PCR using pEF6-hCas9-Puro and the primers T7Cas9_IVT_F and Cas9_R as described (Ohshima et al., 2014), and the PCR products gel-purified. The synthesized Irgb10 gRNA and Cas9 mRNA were purified using MEGAclear Transcription Clean-Up Kit (AM1908, ThermoFisher Scientific) and eluted in RNase-free water. Irgb10−/− Gbpchr3-KO mice were generated by crossing Irgb10−/− mice and Gbpchr3-KO mice.
To obtain IRGB10-mutated mice, B6C3F1 (C57BL/6 × C3H) female mice (6 weeks old) were superovulated and mated with B6C3F1 stud males. Fertilized one-cell stage embryos were collected from the oviducts and injected into the pronuclei or the cytoplasm with the Cas9 mRNA (100 ng/µl) and the Irgb10 gRNA (50 ng/µl) in accordance with previous instructions (Wang et al., 2013). The injected live embryos were transferred into the oviducts of pseudopregnant ICR females at 0.5 dpc. The Irgb10 loci of the resulting pups were screened using primers; Irgb10_indel_F (5’- CCG ATG CAA AGG CTC ACA ATA TGG C-3’) and Irgb10_indel_R (5’-GCT GGG CTT CAT TAT CTC TAA AGC G-3’). The male pup harboring the mutation was mated with C57BL/6 female mice and tested for the germ line transmission. Heterozygotic mice for the mutated Irgb10 locus were intercrossed to generate homozygotic IRGB10-deficient mice. All mouse strains were housed separately before and during the experiment. Male and female mice of 6–8 weeks old were used in this study. All mice were bred at the St. Jude Children’s Research Hospital. Animal studies were conducted under protocols approved by the St. Jude Children’s Research Hospital on the Use and Care of Animals.
Bone Marrow-Derived Macrophages
Primary bone marrow–derived macrophages (BMDMs) were grown for 5–6 days in DMEM (11995073, ThermoFisher Scientific) supplemented with 10% FBS (TMS-013-B, Millipore), 30% L929 conditioned media and 1% penicillin and streptomycin (15070-063, ThermoFisher Scientific). BMDMs were seeded in antibiotic–free media at a concentration of 1 × 106 cells onto 12-well plates and incubated overnight.
Bacterial Culture
Francisella novicida strain U112 or its isogenic mutant F. novicida ΔmglA were grown in BBL™ Trypticase™ Soy Broth (TSB) (211768, BD) supplemented with 0.2% L-cysteine (BP376-100, ThermoFisher Scientific) overnight under aerobic conditions at 37°C. Bacteria were subcultured (1:10) in fresh TSB supplemented with 0.2% L-cysteine for 4 h and resuspended in PBS. S. Typhimurium SL1344, an isogenic mutant lacking fliC and fljB (ΔfliCΔfljB STm), Citrobacter rodentium (51459, American Type Culture Collection) and Escherichia coli (11775, American Type Culture Collection) were inoculated into Luria-Bertani media (3002-031, MP Biomedicals) and incubated overnight under aerobic conditions at 37°C. S. Typhimurium SL1344 was subcultured (1:10) into fresh LB media for 3 h at 37°C to generate log-phase grown bacteri a.
METHODS DETAILS
Stimulation of Bone Marrow-Derived Macrophages
The following conditions were used to stimulate BMDMs: F. novicida or F. novicida ΔmglA (MOI 100 and 20 h for caspase-1 activation; MOI 50 for 2, 8, 16 and 24 h for IRF1 or GBP expression), S. Typhimurium (MOI 1, 4 h), ΔfliCΔfljB STm (MOI 20 for 20 h), C. rodentium (MOI 20 for 20 h) and E. coli (MOI 20 for 20 h). 50 µg/ml gentamicin (15750-060, ThermoFisher Scientific) was added after 2 h (S. Typhimurium), 4 h (C. rodentium and ΔfliCΔfljB STm), and 8 h (F. novicida) post-infection to kill extracellular bacteria. The MCMV Smith MSGV strain (VR-1399™, American Type Culture Collection) was obtained from P.G. Thomas (St. Jude Children’s Research Hospital). MCMV was added to unprimed BMDMs at an MOI of 10 for 10 h to activate the AIM2 inflammasome. To activate the canonical NLRP3 inflammasome, BMDMs were primed using 500 ng/ml ultrapure LPS from Salmonella minnesota R595 (tlrl-smlps, InvivoGen) for 4 h and stimulated with 5 mM ATP (10127531001, Roche) or 10 µM nigericin (N7143, Sigma) for 45 min. For DNA transfection, each reaction consisted of 2.5 µg of poly(dA:dT) (tlrl-patn, InvivoGen) or plasmid pcDNA3.1 DNA (V79020, ThermoFisher Scientific) resuspended in PBS and mixed with 0.3 µl of Xfect polymer in Xfect reaction buffer (631318, Clontech Laboratories, Inc.). After 10 min, DNA complexes were added to BMDMs in Opti-MEM (31985-070, ThermoFisher Scientific) and incubated for 5 h.
Lactate Dehydrogenase Assay
Levels of lactate dehydrogenase released by cells were determined using the CytoTox 96 Non-Radioactive Cytotoxicity Assay according to the manufacturer’s instructions (G1780, Promega). Cell culture supernatants were collected for ELISA.
Immunoblotting Analysis
For caspase-1 immunoblotting, BMDMs and supernatant were lysed in RIPA buffer and sample loading buffer containing SDS and 100 mM DTT. For immunoblotting of IRF1 or GBP, supernatant was removed and BMDMs washed once with PBS, followed by lysis in RIPA buffer and sample loading buffer containing SDS and 100 mM DTT. Proteins were separated on 8–12% polyacrylamide gels. Following electrophoretic transfer of protein onto PVDF membranes (IPVH00010, Millipore), membranes were blocked in 5% skim milk and incubated with primary antibodies against caspase-1 (1:3,000 dilution, AG-20B-0042, Adipogen), IRF1 (1:1,000 dilution, #8478S, Cell Signaling Technologies), GBP2 (1:1,000 dilution, 11854-1-AP, Proteintech), GBP5 (1:1,000 dilution, 13220-1-AP, Proteintech), caspase-11 (1:1,000 dilution, NB120-10454, Novus), iNOS (1:1,000 dilution, #13120, Cell Signaling Technologies), IL-1β (1:500 dilution, AF-401-NA, R&D Systems), anti-IRGB10 rabbit serum raised against recombinant full-length IRGB10 (1:10,000 dilution) (Steinfeldt et al., 2010) or GAPDH (1:10,000 dilution, #5174, Cell Signaling Technologies). Membranes were then incubated with HRP-conjugated secondary antibody for 1 h and proteins were visualized using Super Signal Femto substrate (34096, ThermoFisher Scientific).
Immunofluorescence Staining
For visualization of inflammasomes, BMDMs were infected with F. novicida for 20 h, washed three times with PBS and fixed in 4% paraformaldehyde for 15 min at room temperature, followed by blocking in 10% normal goat serum (X090710-8, Dako) supplemented with 0.1% saponin (47036, Sigma) for 1 h. Cells were incubated with a rabbit anti-ASC antibody (1:500 dilution, clone AL177, AG-25B-0006-C100, AdipoGen) overnight at 4°C. For IRGB10 staining, BMDMs were infected for the indicated times and washed three times with PBS. Cells were fixed and blocked as described above. IRGB10 was stained using an anti-IRGB10 rabbit serum (1:5,000 dilution) (Steinfeldt et al., 2010) overnight at 4°C. The antibody was further purified by pre-absorption with cell lysates from IFN-β-stimulated Irgb10−/− BMDMs. Bacteria were stained with antibodies against F. novicida LPS (1:50 dilution, anti-F. novicida 13, ImmunoPrecise), E. coli LPS (1:50 dilution, DS-MB-01267, RayBiotech), or C. rodentium-specific Tir antibody (1:500 dilution, from W. Deng and B.B. Finlay, University of British Columbia, Canada). For co-staining of IRGB10, GBP5 and LPS, cells were first stained with the IRGB10 antibody and an anti-rabbit secondary Alexa Fluor 568 antibody. Cells were washed five times with PBS. Cells were then stained with the antibody to GBP5 (1:50 dilution, 13220-1-AP, Proteintech) which had been conjugated with Alexa Fluor 488 in house, as well as with the antibody to E. coli LPS. Cells were washed five times with PBS. An anti-mouse secondary Alexa Fluor 647 antibody was used to couple the LPS antibody. Cells were counterstained in DAPI mounting medium (H-1200, Vecta Labs). Bacteria, inflammasomes and BMDMs were visualized, counted, and imaged using a Nikon C2 confocal microscope.
Super Resolution Microscopy and Structured Illumination Microscopy
Sample preparation was described above. Super resolved three-dimensional images were taken on a Zeiss Elyra PS.1 microscope with Structured Illumination Microscopy (SIM) technique. In order to show the localization or arrangement of LPS, IRGB10, GBP5 and DNA, a line scan analysis was applied to the SIM images using the ZEN software (410135-1004-230, Zeiss). Raw values from the line scan for each signal were exported to IGOR Pro (Wavemetrics) where they were plotted and fitted with single and double Gaussian equations. The Gaussian peak positions extracted from the fits were used to calculate the distance of separation between each peak signal.
Transmission Electron Microscopy
BMDMs were infected with F. novicida or E. coli for 16 h, washed with PBS, fixed in 4% paraformaldehyde (15710, Electron Microscopy Sciences) for 15 min at room temperature, washed with PBS and blocked with blocking buffer (0.1% saponin, 3% BSA in PBS) for 1 h at room temperature. Primary rabbit antisera against IRGB10 (Coers et al., 2008) (1:100 dilution) in blocking buffer were incubated with BMDMs overnight at 4°C. Cells were washed with blocking buffer and sec ondary anti-rabbit IgG antibody conjugated with ultra-small gold particles (1:50 dilution, 25100, Electron Microscopy Sciences) was incubated overnight at 4°C. Cells wer e washed extensively with blocking buffer followed by PBS and post-fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer 30 min and further washed with PBS. Gold particles were then silver enhanced with the use of the Aurion R-Gent SE-EM kit (25521-90, Electron Microscopy Sciences) following manufacturer’s protocols. Cells were rinsed in 0.1 M sodium cacdodylate buffer, fixed in 0.5% osmium tetroxide in 0.1 M sodium cacodylate buffer for 15 min and dehydrated in a series of alcohol, propylene oxide and embedded in Hard Embed 812. Samples were polymerized in a 70°C oven overnight. Thin section were cut at 80 nm and viewed using a Tecnai Electron Microscope at 80 kv.
Amino Acid Sequence Analysis
Phylogenetic tree from IRG protein sequences was constructed using Pylogeny.fr server (Dereeper et al., 2008). Putative transmembrane regions in IRGB10 are predicted using PredictProtein (Rost et al., 2004), TMpred (Hofmann and Stoffel, 1993), TMHMM (Krogh et al., 2001) servers. Helical wheel diagrams are made using HELIQUEST (Gautier et al., 2008) and anti-microbial propensities of the transmembrane regions are plotted using AMPA algorithm (Torrent et al., 2012). Conservation at each residue position of predicted transmembrane helices graphically plotted using WebLogo3 (Crooks et al., 2004).
Real Time qRT-PCR Analysis
RNA was extracted using TRIzol (15596026, ThermoFisher Scientific) and converted into cDNA using the High capacity cDNA Reverse Transcription kit (4368814, Applied Biosystems). Real-time qPCR was performed on an ABI 7500 real-time PCR instrument with 2× SYBR Green (4368706, Applied Biosystems). Real time qRT-PCR sequences are found in Table S1.
siRNA Knockdown
BMDMs were transfected with siRNA from siGENOME smart pools for 48 h using GenMute siRNA Transfection Reagent and the according to the manufacturer’s instructions (SL100568, SignaGen Laboratories). The siGENOME SMARTpool siRNA specific for the gene encoding mouse IRGB10 (M-077430-00-0005, Dharmacon) and a control siRNA pool were used. Transfected cells were infected with F. novicida as described above.
Cytokine Analysis
Cytokine levels were determined using a multiplex ELISA (MCYTOMAG-70K, Millipore) or IL-18 ELISA (BMS618/3TEN, Affymetrix eBioscience) according to the manufacturers’ instructions.
Flow Cytometry
The following monoclonal antibodies were used for flow cytometry cellular analyses: CD4 (RM4-5, 14-0042-85), CD11b (M1/70, 48-0112-82), CD8a (53-6.7, 48-0081-82) from Affymetrix eBioscience, CD4 (GK1.5, 100408), TCR-β (H57-597, 109222), CD11c (N418, 117306), Gr1 (RB6-815, 108426), F4/80 (BM8, 123109) from BioLegend, and CD19 (1D3, 35-0193-u025) from Tonbo Biosciences. The dilution factor used for these antibodies was 1:300. Flow cytometry data were acquired on FACSCalibur (BD) and were analyzed with FlowJo software (FlowJo, LLC and Illumina, Inc).
Oligomerization Assay
Cells were lysed in buffer A [20 mM HEPES-KOH (pH 7.5), 1.5 mM MgCl2, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 320 mM sucrose, protease (11697498001, Roche) and phosphatase inhibitors (04906837001, Roche)] and syringed 20 times on ice using 1-ml syringes with 25-guage needles. Cell lysates were centrifuged at 1,000 rpm for 8 min. The supernatant was collected and equal volume of buffer A was added to the lysate and centrifuged again at 1,800 rpm for 8 min to further remove the nuclei. The supernatant was collected into a 2 ml tube and 1 ml of CHAPS buffer [20 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 0.5 mM EGTA, 0.1 % CHAPS] was added followed by centrifugation at 5,000 rpm for 8 min at 4°C. The p ellet was washed once with 0.5 ml CHAPS buffer. IRGB10 oligomers were cross-linked using 2 mM disuccinimidyl suberate (21655, ThermoFisher Scientific) for 25 min at room temperature. Proteins were then separated on 4–20% Mini-Protean TGX Precast Gel (456–1096, Biorad) for immunoblotting.
Animal Infection
Francisella novicida strain U112 was grown as described above. For survival and weight change analyses, mice were injected subcutaneously with 7.5 × 104 colony-forming units (CFU) of F. novicida in 200 µl PBS. For colony-forming units (CFU) analysis, mice were injected subcutaneously with 1.5 × 105 CFU of F. novicida in 200 µl PBS. After 3 days, liver and spleen were harvested and homogenized in PBS with metal beads for 2 min using the Qiagen TissueLyser II apparatus. CFU were determined by plating lysates onto TSB agar supplemented with 0.2% L-cysteine and incubated overnight.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 6.0 software was used for data analysis. Data are shown as mean ± s.e.m. Statistical significance was determined by t tests (two-tailed) for two groups or One-way ANOVA (with Dunnett’s or Tukey’s multiple comparisons tests) for three or more groups. Survival curves were compared using the log-rank test. P<0.05 was considered statistically significant.
DATA RESOURCES
Microarray
Differentially expressed genes were analyzed from our previously published microarray dataset (Man et al., 2015). The dataset was deposited under the accession code GSE66461.
Supplementary Material
(A) Microarray analysis of the expression of genes with higher or lower expression in unprimed Irf1−/−, Ifnar1−/− and Aim2−/− BMDMs 8 h after infection with F. novicida, relative to that of their WT counterparts. The 20 genes with the lowest levels of expression in Irf1−/− BMDMs compared with WT BMDMs are shown.
(B) Immunoblot analysis of IRGB10 and GAPDH (loading control) in WT BMDMs stimulated with IFN-β, IFN-γ, LPS and poly(I:C).
(C) Body weight of 3-week-old WT male mice (n = 8) and Irgb10−/− male mice (n = 9), 6–7 week-old WT male mice (n = 35) and Irgb10−/− male mice (n = 26), and 8–9 week-old WT male mice (n = 15) and Irgb10−/− male mice (n = 20).
(D) Flow cytometric analysis of side-scattered light (SSC), forward-scattered light (FSC) and expression of F4/80 and CD11c of bone marrow-derived macrophages generated from WT mice and Irgb10−/− mice.
NS, not statistically significant (one-way ANOVA with Tukey’s multiple-comparisons test [C]). Each symbol represents an individual mouse (C). Data are representative of one experiment with two biological replicates per genotype (A) or are one experiment representative of two (D) or three independent experiments (B).
(A) Total number of neutrophils, macrophages, DCs, CD4+ T cells, CD8+ T cells, and B cells in the spleen of WT mice (n = 5) and Irgb10−/− mice (n = 5).
(B) Total number of CD4+ T cells and CD8+ T cells in the thymus of WT mice (n = 5) and Irgb10−/− mice (n = 5).
(C) Total number of neutrophils, monocytes, CD4+ T cells, CD8+ T cells, and B cells in the bone marrow of WT mice (n = 5) and Irgb10−/− mice (n = 5).
NS, not statically significant (two-tailed t-test [A–C]). Each symbol represents an individual mouse (A–C). Data are one experiment representative of two independent experiments (A–C).
(A) Real-time quantitative RT-PCR analysis of the gene encoding IL-1β, IL-18, TNF and IFN-β in WT and mutant BMDMs 8 h after infection with F. novicida, presented relative to that of the gene encoding GAPDH.
(B) Release of TNF and IL-6 of unprimed WT and mutant BMDMs assessed 16 h after infection with F. novicida (MOI, 100).
(C) Real-time quantitative RT-PCR analysis of the gene encoding IRGB10, GBP2 and IL-1β in WT BMDMs treated with control or Irgb10-specific siRNA molecules, 8 h after infection with F. novicida, presented relative to that of the levels observed with control siRNA, set as 100%.
(D) Immunoblot analysis of caspase-1 in unprimed BMDMs treated with control or Irgb10-specific siRNA molecules and left untreated or assessed 20 h after infection with F. novicida.
(E) Release of IL-1β and TNF of unprimed BMDMs after treatment as in (D) or 20 h after infection with E. coli.
(F) Release of TNF and IL-6 of unprimed WT and mutant BMDMs assessed 16 h after infection with C. rodentium and E. coli (MOI, 20).
(G) Release of IL-1β and IL-18 of unprimed BMDMs assessed 16 h after infection with S. Typhimurium (STm) ΔfliCΔfljB (MOI, 20).
NS, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed t-test [A–C, E and F] and one-way ANOVA with Dunnett’s multiple-comparisons test [G]). Data are from two (A, C) or three (B, D–G) independent experiments (mean and s.e.m. in A–C and E–G).
(A) Immunofluorescence staining of the Citrobacter rodentium Tir protein (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with C. rodentium (MOI, 20).
(B) Immunoblot analysis of IRGB10, GBP2, GBP5 and GAPDH (loading control) in unprimed WT and mutant BMDMs infected with E. coli (MOI, 20).
(C) Immunoblot analysis of IRGB10, GBP2, GBP5 and GAPDH (loading control) in unprimed WT and Irgb10−/− BMDMs infected with F. novicida (MOI, 50).
(D) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with F. novicida. Three-dimensional images were taken using the Structured Illumination Microscopy (SIM) technique. Enlarged and orthogonal-view images of a F. novicida cell taken using SIM.
(E) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli. Three-dimensional images were taken using the Structured Illumination Microscopy (SIM) technique. A single z-plane of a 3D image is shown. White lines indicate a line scan analysis of the SIM image shown in (F).
(F) The line scan values for each signal were plotted and fitted with single and double Gaussian equations for analysis performed in (E).
(G) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli. Images were taken using SIM as in (E).
(H) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT or mutant BMDMs 16 h after infection with E. coli. Images were taken using SIM as in (E).
Scale bar, 10 µm (A), 2 µm (H), 1 µm (D and E) and 0.5 µm (G). Arrowheads indicate bacteria targeted by IRGB10 (A and H), a layer of IRGB10 staining external to a layer of LPS staining (D), or a rupture on the LPS layer (G). Data are from two (A) or three (B–H) independent experiments.
(A) A phylogenetic tree depicting the sequence similarity of the full-length amino acid sequences from members of the mouse Immunity-related GTPases (IRG) family.
(B) Alignment of the predicted myristoylation motifs at the N-terminus of IRG proteins.
(C) Alignment of the amino acid region containing the putative transmembrane region 1 (top). The conservation of each amino acid residue in the putative transmembrane region 1 of the IRG family proteins is shown by a WebLogo representation (bottom). A major deletion was identified in the region spanning the proline residue within the amphipathic helix of IRGA2, IRGM1, IRGM2 and IRGM3.
(D) The standard helical wheel diagram showing the distribution of hydrophilic (green) versus hydrophobic (gold) residues (top) and the predicted anti-microbial potential of the putative transmembrane region 1 (bottom). Arrow in the helical wheel represents the direction of the hydrophobic moment. N, N-terminus of the sequence; C, C-terminus of the sequence.
(E) Alignment of the amino acid region containing the putative transmembrane region 2 (top). The conservation of each amino acid residue in the putative transmembrane region 3 of the IRG family proteins is shown by a WebLogo representation (bottom). IRGB5 carries a deletion in the transmembrane helix 2 region, which potentially render it unable to function as a transmembrane helix.
(F) The standard helical wheel diagram showing the distribution of hydrophilic (green) versus hydrophobic (gold) residues (top) and the predicted anti-microbial potential of the putative transmembrane region 2 (bottom). Arrow in the helical wheel represents the direction of the hydrophobic moment. N, N-terminus of the sequence; C, C-terminus of the sequence.
(A) Immunogold staining of IRGB10 in unprimed WT BMDMs 16 h after infection with E. coli. Images were acquired using transmission electron microscopy.
(B) Percentages of IRGB10+ bacteria with or without association with IRGB10-containing vesicular structures.
(C) Immunogold staining of IRGB10 in unprimed WT BMDMs 16 h after infection with F. novicida. Images were acquired as in (A).
(D) Immunofluorescence staining of GBP5 (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli (left). Quantification of the percentage of co-localization between GBP5 and IRGB10 staining in each field of uninfected (n = 10) and infected (= 20) samples (right).
(E) Immunofluorescence staining of DNA (blue), GBP5 (green) IRGB10 (red), and LPS (magenta) in unprimed WT BMDMs left untreated or 16 h after infection with E. coli. Images were taken using the Structured Illumination Microscopy (SIM) technique.
Scale bar, 500 nm (A [full-size image, left]), 100 nm (A [inset, right] and C), and 10 µm (D and E). Arrowheads indicate IRGB10 proteins within irregular-shaped E. coli cells (A), the interface between IRGB10 and F. novicida (C) or bacteria targeted by both IRGB10 and GBP5 (D). Asterisks indicate regular-shaped E. coli (A [inset, right]). ****P < 0.0001 (two-tailed t-test [D]). Each symbol represents an individual field (D). Data are from two independent experiments (A–E).
(A) Immunoblot analysis of iNOS and GAPDH (loading control) in unprimed WT and Irf1−/− BMDMs infected with F. novicida (MOI, 50).
(B) Immunofluorescence staining of iNOS (red) and DNA (blue) in unprimed WT, Nos2−/− and Irf1−/− BMDMs 16 h after infection with F. novicida.
(C) Immunoblot analysis of caspase-1 (top) and release of IL-1β and IL-18 and death (bottom) of unprimed WT and mutant BMDMs left untreated or assessed 20 h after infection with F. novicida (MOI, 100).
(D) Immunoblot analysis of iNOS, IRGB10, GBP2, GBP5 and GAPDH (loading control) of unprimed WT and Nos2−/− BMDMs infected with F. novicida (MOI, 50).
(E) Immunoblot analysis of iNOS and GAPDH (loading control) in unprimed WT and mutant BMDMs infected with F. novicida (MOI, 50).
(F) Immunoblot analysis as in (E).
(G) The cytosolic escape assay (top) and release of IL-1β (middle) and IL-18 (bottom) in WT and mutant BMDMs after 16 h of stimulation with Salmonella LPS alone, F. novicida (MOI, 100), F. novicida plus Salmonella LPS and F. novicida ΔmglA plus Salmonella LPS or after 10 h of transfection with Salmonella LPS.
(H) Real-time quantitative RT-PCR analysis of the gene encoding IL-1β (top) and IL-18 (bottom) in WT and mutant BMDMs 8 h after infection with F. novicida, presented relative to that of the gene encoding GAPDH.
Scale bar, 20 µm (B). NS, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (one-way ANOVA with Dunnett’s multiple-comparisons test [C and G]). Data are from two (H) three (A–G) independent experiments (mean and s.e.m. in C, G and H).
HIGHLIGHTS.
-
▪
IRGB10 is required for activation of multiple inflammasomes by bacteria
-
▪
Recruitment of IRGB10 to the bacteria is dependent on GBPs
-
▪
IRGB10 targeted bacterial cell membrane and mediated bacterial killing
-
▪
IRGB10 liberated ligands and served a conserved signaling hub for inflammasomes
Acknowledgments
We thank A. Burton, D. Horn, B. Sharma, F. Phillips, A. Hegab, V. Frohlich, L. Horner, R. Gursky, P.G. Thomas (St. Jude Children’s Research Hospital, SJCRH), V. M. Dixit, N. Kayagaki (Genentech), J.C. Howard, S. Könen-Waisman, T. Steinfeldt, C. Poschner (University of Cologne), W. Deng and B.B. Finlay (University of British Columbia) for reagents or technical support. Images were acquired at the SJCRH Cell & Tissue Imaging Center, which is supported by SJCRH and NCI P30 CA021765-35. Work from our laboratories is supported by the US National Institutes of Health (AI101935, AI124346, AR056296 and CA163507 to T.D.K.), the American Lebanese Syrian Associated Charities (to T.D.K.), JSPS KAKENHI (Grant Numbers 15H013770, 15H047450, 15H052750 and 15K151220 to M.Y.), and the R.G. Menzies Early Career Fellowship from the National Health and Medical Research Council of Australia (to S.M.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors Contributions
S.M.M., R.K., and T.D.K. conceptualized the study; S.M.M., R.K., S.K., and J.T. designed the methodology; S.M.M., R.K., M.S., D.E.P., S.K., J.T., S.F., Q.Z., R.K.S.M., T.K., J.L.P., and S.A.B. performed the experiments; S.M.M., R.K., D.E.P., S.K., J.T., S.F., J.L.P., and G.N. conducted the analysis; S.M.M. and T.D.K. wrote the manuscript. T.D.K. acquired the funding; J.C.H., M.Y., and T.D.K. provided resources; T.D.K. provided overall supervision.
REFERENCES
- Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, Glowalla E, Leptin M, Howard JC. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome biology. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein-Hanley I, Balsara ZR, Ulmer W, Coers J, Starnbach MN, Dietrich WF. Genetic analysis of susceptibility to Chlamydia trachomatis in mouse. Genes and immunity. 2006a;7:122–129. doi: 10.1038/sj.gene.6364285. [DOI] [PubMed] [Google Scholar]
- Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006b;103:14092–14097. doi: 10.1073/pnas.0603338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. The Journal of experimental medicine. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Coers J, Bernstein-Hanley I, Grotsky D, Parvanova I, Howard JC, Taylor GA, Dietrich WF, Starnbach MN. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. Journal of immunology. 2008;180:6237–6245. doi: 10.4049/jimmunol.180.9.6237. [DOI] [PubMed] [Google Scholar]
- Cordes FS, Bright JN, Sansom MS. Proline-induced distortions of transmembrane helices. Journal of molecular biology. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochimica et biophysica acta. 1999;1462:71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature immunology. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, Yamamoto M, Nagarajan UM, Miao EA, Coers J. Guanylate Binding Proteins Enable Rapid Activation of Canonical and Noncanonical Inflammasomes in Chlamydia-Infected Macrophages. Infection and immunity. 2015;83:4740–4749. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. The Journal of biological chemistry. 2012;287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, Frickel EM, Valdivia RH, Coers J. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS pathogens. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Peng L, Mirshahi T, Rohacs T, Chan KW, Sanchez R, Logothetis DE. The (beta)gamma subunits of G proteins gate a K(+) channel by pivoted bending of a transmembrane segment. Molecular cell. 2002;10:469–481. doi: 10.1016/s1097-2765(02)00659-7. [DOI] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Man SM, Malireddi RK, Gurung P, Vogel P, Lamkanfi M, Kanneganti TD. Concerted Activation of the AIM2 and NLRP3 Inflammasomes Orchestrates Host Protection against Aspergillus Infection. Cell host & microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signaling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong YC, Boothroyd JC, Reichmann G, et al. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cellular microbiology. 2010;12:939–961. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Chee JD, Bradfield CJ, Park ES, Kumar P, MacMicking JD. Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nature immunology. 2016;17:481–489. doi: 10.1038/ni.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nature reviews Immunology. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Hopkins LJ, Nugent E, Cox S, Gluck IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, Bryant CE. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunological reviews. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nature reviews Immunology. 2016;16:7–21. doi: 10.1038/nri.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nature immunology. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Place DE, Kuriakose T, Kanneganti TD. Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. Journal of leukocyte biology. 2016 doi: 10.1189/jlb.4MR0516-223R. pii: jlb.4MR0516-223R. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Sabel K, Lange R, Uthaiah R, Wolf E, Howard JC. Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: retargeting to plasma membrane induced by phagocytosis. Journal of immunology. 2004;173:2594–2606. doi: 10.4049/jimmunol.173.4.2594. [DOI] [PubMed] [Google Scholar]
- Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nature immunology. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyairi I, Tatireddigari VR, Mahdi OS, Rose LA, Belland RJ, Lu L, Williams RW, Byrne GI. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. Journal of immunology. 2007;179:1814–1824. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, Matsuura Y, Pann-Ghill S, Hayashi M, Ebisu S, et al. Role of mouse and human autophagy proteins in IFN-gamma-induced cell-autonomous responses against Toxoplasma gondii. Journal of immunology. 2014;192:3328–3335. doi: 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- Papic N, Hunn JP, Pawlowski N, Zerrahn J, Howard JC. Inactive and active states of the interferon-inducible resistance GTPase, Irga6, in vivo. The Journal of biological chemistry. 2008;283:32143–32151. doi: 10.1074/jbc.M804846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature immunology. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. TRIF Licenses Caspase-11-Dependent NLRP3 Inflammasome Activation by Gram-Negative Bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic acids research. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Steinfeldt T, Konen-Waisman S, Tong L, Pawlowski N, Lamkemeyer T, Sibley LD, Hunn JP, Howard JC. Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS biology. 2010;8:e1000576. doi: 10.1371/journal.pbio.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. Journal of immunology. 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman DP, Shrivastava IH, Ulmschneider MR, Sansom MS. Proline-induced hinges in transmembrane helices: possible roles in ion channel gating. Proteins. 2001;44:63–72. doi: 10.1002/prot.1073. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nature immunology. 2009;10:907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M, Di Tommaso P, Pulido D, Nogues MV, Notredame C, Boix E, Andreu D. AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics. 2012;28:130–131. doi: 10.1093/bioinformatics/btr604. [DOI] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam VA. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell. 2016;165:1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:959–963. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Microarray analysis of the expression of genes with higher or lower expression in unprimed Irf1−/−, Ifnar1−/− and Aim2−/− BMDMs 8 h after infection with F. novicida, relative to that of their WT counterparts. The 20 genes with the lowest levels of expression in Irf1−/− BMDMs compared with WT BMDMs are shown.
(B) Immunoblot analysis of IRGB10 and GAPDH (loading control) in WT BMDMs stimulated with IFN-β, IFN-γ, LPS and poly(I:C).
(C) Body weight of 3-week-old WT male mice (n = 8) and Irgb10−/− male mice (n = 9), 6–7 week-old WT male mice (n = 35) and Irgb10−/− male mice (n = 26), and 8–9 week-old WT male mice (n = 15) and Irgb10−/− male mice (n = 20).
(D) Flow cytometric analysis of side-scattered light (SSC), forward-scattered light (FSC) and expression of F4/80 and CD11c of bone marrow-derived macrophages generated from WT mice and Irgb10−/− mice.
NS, not statistically significant (one-way ANOVA with Tukey’s multiple-comparisons test [C]). Each symbol represents an individual mouse (C). Data are representative of one experiment with two biological replicates per genotype (A) or are one experiment representative of two (D) or three independent experiments (B).
(A) Total number of neutrophils, macrophages, DCs, CD4+ T cells, CD8+ T cells, and B cells in the spleen of WT mice (n = 5) and Irgb10−/− mice (n = 5).
(B) Total number of CD4+ T cells and CD8+ T cells in the thymus of WT mice (n = 5) and Irgb10−/− mice (n = 5).
(C) Total number of neutrophils, monocytes, CD4+ T cells, CD8+ T cells, and B cells in the bone marrow of WT mice (n = 5) and Irgb10−/− mice (n = 5).
NS, not statically significant (two-tailed t-test [A–C]). Each symbol represents an individual mouse (A–C). Data are one experiment representative of two independent experiments (A–C).
(A) Real-time quantitative RT-PCR analysis of the gene encoding IL-1β, IL-18, TNF and IFN-β in WT and mutant BMDMs 8 h after infection with F. novicida, presented relative to that of the gene encoding GAPDH.
(B) Release of TNF and IL-6 of unprimed WT and mutant BMDMs assessed 16 h after infection with F. novicida (MOI, 100).
(C) Real-time quantitative RT-PCR analysis of the gene encoding IRGB10, GBP2 and IL-1β in WT BMDMs treated with control or Irgb10-specific siRNA molecules, 8 h after infection with F. novicida, presented relative to that of the levels observed with control siRNA, set as 100%.
(D) Immunoblot analysis of caspase-1 in unprimed BMDMs treated with control or Irgb10-specific siRNA molecules and left untreated or assessed 20 h after infection with F. novicida.
(E) Release of IL-1β and TNF of unprimed BMDMs after treatment as in (D) or 20 h after infection with E. coli.
(F) Release of TNF and IL-6 of unprimed WT and mutant BMDMs assessed 16 h after infection with C. rodentium and E. coli (MOI, 20).
(G) Release of IL-1β and IL-18 of unprimed BMDMs assessed 16 h after infection with S. Typhimurium (STm) ΔfliCΔfljB (MOI, 20).
NS, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed t-test [A–C, E and F] and one-way ANOVA with Dunnett’s multiple-comparisons test [G]). Data are from two (A, C) or three (B, D–G) independent experiments (mean and s.e.m. in A–C and E–G).
(A) Immunofluorescence staining of the Citrobacter rodentium Tir protein (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with C. rodentium (MOI, 20).
(B) Immunoblot analysis of IRGB10, GBP2, GBP5 and GAPDH (loading control) in unprimed WT and mutant BMDMs infected with E. coli (MOI, 20).
(C) Immunoblot analysis of IRGB10, GBP2, GBP5 and GAPDH (loading control) in unprimed WT and Irgb10−/− BMDMs infected with F. novicida (MOI, 50).
(D) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with F. novicida. Three-dimensional images were taken using the Structured Illumination Microscopy (SIM) technique. Enlarged and orthogonal-view images of a F. novicida cell taken using SIM.
(E) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli. Three-dimensional images were taken using the Structured Illumination Microscopy (SIM) technique. A single z-plane of a 3D image is shown. White lines indicate a line scan analysis of the SIM image shown in (F).
(F) The line scan values for each signal were plotted and fitted with single and double Gaussian equations for analysis performed in (E).
(G) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli. Images were taken using SIM as in (E).
(H) Immunofluorescence staining of LPS (green), IRGB10 (red) and DNA (blue) in unprimed WT or mutant BMDMs 16 h after infection with E. coli. Images were taken using SIM as in (E).
Scale bar, 10 µm (A), 2 µm (H), 1 µm (D and E) and 0.5 µm (G). Arrowheads indicate bacteria targeted by IRGB10 (A and H), a layer of IRGB10 staining external to a layer of LPS staining (D), or a rupture on the LPS layer (G). Data are from two (A) or three (B–H) independent experiments.
(A) A phylogenetic tree depicting the sequence similarity of the full-length amino acid sequences from members of the mouse Immunity-related GTPases (IRG) family.
(B) Alignment of the predicted myristoylation motifs at the N-terminus of IRG proteins.
(C) Alignment of the amino acid region containing the putative transmembrane region 1 (top). The conservation of each amino acid residue in the putative transmembrane region 1 of the IRG family proteins is shown by a WebLogo representation (bottom). A major deletion was identified in the region spanning the proline residue within the amphipathic helix of IRGA2, IRGM1, IRGM2 and IRGM3.
(D) The standard helical wheel diagram showing the distribution of hydrophilic (green) versus hydrophobic (gold) residues (top) and the predicted anti-microbial potential of the putative transmembrane region 1 (bottom). Arrow in the helical wheel represents the direction of the hydrophobic moment. N, N-terminus of the sequence; C, C-terminus of the sequence.
(E) Alignment of the amino acid region containing the putative transmembrane region 2 (top). The conservation of each amino acid residue in the putative transmembrane region 3 of the IRG family proteins is shown by a WebLogo representation (bottom). IRGB5 carries a deletion in the transmembrane helix 2 region, which potentially render it unable to function as a transmembrane helix.
(F) The standard helical wheel diagram showing the distribution of hydrophilic (green) versus hydrophobic (gold) residues (top) and the predicted anti-microbial potential of the putative transmembrane region 2 (bottom). Arrow in the helical wheel represents the direction of the hydrophobic moment. N, N-terminus of the sequence; C, C-terminus of the sequence.
(A) Immunogold staining of IRGB10 in unprimed WT BMDMs 16 h after infection with E. coli. Images were acquired using transmission electron microscopy.
(B) Percentages of IRGB10+ bacteria with or without association with IRGB10-containing vesicular structures.
(C) Immunogold staining of IRGB10 in unprimed WT BMDMs 16 h after infection with F. novicida. Images were acquired as in (A).
(D) Immunofluorescence staining of GBP5 (green), IRGB10 (red) and DNA (blue) in unprimed WT BMDMs 16 h after infection with E. coli (left). Quantification of the percentage of co-localization between GBP5 and IRGB10 staining in each field of uninfected (n = 10) and infected (= 20) samples (right).
(E) Immunofluorescence staining of DNA (blue), GBP5 (green) IRGB10 (red), and LPS (magenta) in unprimed WT BMDMs left untreated or 16 h after infection with E. coli. Images were taken using the Structured Illumination Microscopy (SIM) technique.
Scale bar, 500 nm (A [full-size image, left]), 100 nm (A [inset, right] and C), and 10 µm (D and E). Arrowheads indicate IRGB10 proteins within irregular-shaped E. coli cells (A), the interface between IRGB10 and F. novicida (C) or bacteria targeted by both IRGB10 and GBP5 (D). Asterisks indicate regular-shaped E. coli (A [inset, right]). ****P < 0.0001 (two-tailed t-test [D]). Each symbol represents an individual field (D). Data are from two independent experiments (A–E).
(A) Immunoblot analysis of iNOS and GAPDH (loading control) in unprimed WT and Irf1−/− BMDMs infected with F. novicida (MOI, 50).
(B) Immunofluorescence staining of iNOS (red) and DNA (blue) in unprimed WT, Nos2−/− and Irf1−/− BMDMs 16 h after infection with F. novicida.
(C) Immunoblot analysis of caspase-1 (top) and release of IL-1β and IL-18 and death (bottom) of unprimed WT and mutant BMDMs left untreated or assessed 20 h after infection with F. novicida (MOI, 100).
(D) Immunoblot analysis of iNOS, IRGB10, GBP2, GBP5 and GAPDH (loading control) of unprimed WT and Nos2−/− BMDMs infected with F. novicida (MOI, 50).
(E) Immunoblot analysis of iNOS and GAPDH (loading control) in unprimed WT and mutant BMDMs infected with F. novicida (MOI, 50).
(F) Immunoblot analysis as in (E).
(G) The cytosolic escape assay (top) and release of IL-1β (middle) and IL-18 (bottom) in WT and mutant BMDMs after 16 h of stimulation with Salmonella LPS alone, F. novicida (MOI, 100), F. novicida plus Salmonella LPS and F. novicida ΔmglA plus Salmonella LPS or after 10 h of transfection with Salmonella LPS.
(H) Real-time quantitative RT-PCR analysis of the gene encoding IL-1β (top) and IL-18 (bottom) in WT and mutant BMDMs 8 h after infection with F. novicida, presented relative to that of the gene encoding GAPDH.
Scale bar, 20 µm (B). NS, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (one-way ANOVA with Dunnett’s multiple-comparisons test [C and G]). Data are from two (H) three (A–G) independent experiments (mean and s.e.m. in C, G and H).