Abstract
The study of complex genetics in autoimmune diseases has progressed at a tremendous pace over the last 4 years, as a direct result of the enormous gains made by genome wide association studies (GWAS). Novel genetic findings are continuously being reported alongside the rapid development of genetic technologies, sophisticated statistical analysis, and larger sample collections. It is now becoming clear that multiple genes contribute to disease risk in many complex genetic disorders including rheumatoid arthritis (RA) and that there are common genetic risk factors that underlie a spectrum of autoimmune diseases. This review details the current genetic landscape of RA, and describes what GWAS has taught us in terms of missing heritability, subsets of disease, existence of genetic heterogeneity, and shared autoimmune risk loci. Finally, this review addresses the initial challenges faced in translating the wealth of genetic findings into determining the biological mechanisms that contribute to the relationship between genotype and phenotype. Unraveling the mechanism of how genes directly influence the cause of RA will lead to a better understanding of the disease and will ultimately have a direct clinical impact, informing the development of new therapies that can be utilized in the treatment of RA.
Keywords: rheumatoid arthritis, GWAS, fine mapping, immunochip, bioinformatics
Introduction
Rheumatoid arthritis (RA) is a complex, chronic autoimmune disease (AID) that affects approximately 0.5%–1% of the population worldwide. It is characterized by inflammation and progressive destruction of the synovial joints leading to pain, long-term disability, and reduced quality of life in many patients.1 Although RA primarily affects joints it can also result in extra-articular disease manifestations, which tend to occur in patients with more severe disease.2 According to most epidemiological studies, RA is more prevalent in women and the peak age of onset is typically around the fifth decade, although there is evidence to suggest this is rising.3 There are significant variations of disease incidence and prevalence amongst different populations, suggesting an association of RA with ethnicity.4 RA continues to impose a substantial economic burden on society resulting from the significant morbidity and premature mortality associated with the disease, with the life expectancy of RA patients shortened by 3–10 years.5
The etiology of RA remains largely unknown, although it is established as a multifactorial disease resulting from a complex interplay between genetic and environmental factors. A genetic link to RA was first established through the observation of familial clustering in cases,6 siblings of affected patients having an increased relative risk of RA compared to the general population (λs), ranging between 5 and 10.7 Twin studies have also provided compelling evidence to support this genetic component, with disease concordance between monozygotic twins (15%) being considerably higher compared to dizygotic twins (3.6%).8,9 From such studies, the overall heritability of RA has been estimated to be between 50% and 60%.10 It is documented that genetic risk factors contribute to both occurrence and severity of disease.11 Low concordance rates between monozygotic twins emphasize the importance of environmental factors in RA susceptibility. Long-term smoking remains the only validated environmental factor that contributes to an increased risk of developing seropositive RA,12 although studies have implicated other potential environmental risk factors including hormones, pollution, diet, and infectious agents.13–15 Ongoing work will continue to evaluate the role of environmental factors and place importance on dissecting gene–environment (G–E) interactions.
Genetics of RA
The need to uncover the full genetic risk component of RA is imperative for improving understanding of the disease and may inform the development of new therapies, improved diagnosis, prevention, and potentially prediction of disease risk and severity.
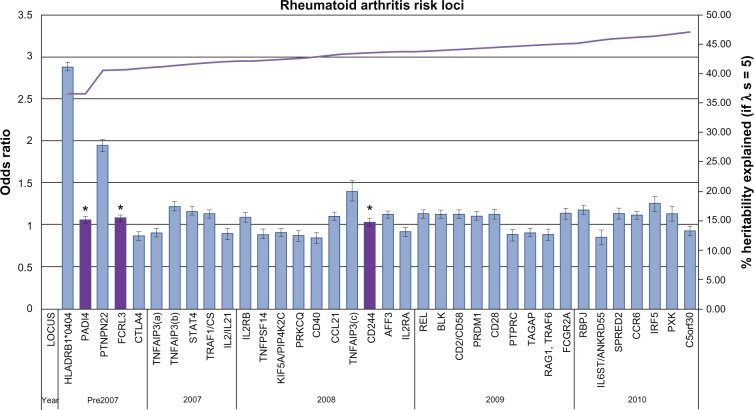
The last 60 years have seen mounting evidence to support the genetic basis of RA through the identification of genetic susceptibility variants. Three main approaches have been used to identify these susceptibility loci: candidate gene, linkage, and genome wide association studies (GWAS), the third being the most successful. The latest GWAS meta-analysis association study has brought the total number of confirmed RA risk loci to 34.16 A comprehensive discussion of all the identified genetic loci is beyond the scope of this review; however the major landmarks in the search of susceptibility loci will be covered. Figure 1 illustrates all confirmed RA loci to date in approximate order of discovery complete with heritability estimates.
Figure 1.
Current rheumatoid arthritis (RA) genetic risk loci. Each confirmed RA risk locus has been plotted in order of approximate discovery from left to right on the bottom axis. For each locus the odds ratio and confidence interval of the most significant allele has been plotted against the y-axis (left). On the y-axis (right) the increase in heritability explained by confirmed loci has been plotted (λs = 5). * validated in east-Asian populations only.
Prior to the development of GWAS, RA susceptibility loci were discovered through candidate gene and linkage studies. Although these approaches led to the discovery of only a small number of loci, they identified the two most significant RA risk loci that remain to date: human leukocyte antigen (HLA-DRB1) and protein tyrosine phosphatase, nonreceptor type 22 (PTPN22). Together HLA-DRB1 and PTPN22 are estimated to account for approximately 40% of the total genetic risk for RA.17 More recently these approaches have also led to the identification of the signal transducer and activator of transcription 4 (STAT4) locus.
HLA-DRB1
The HLA region remains the most statistically significant and important genetic association for RA. The knowledge of a marked involvement of HLA-DRB1 with the immune response identified it as a good candidate gene for RA. Stastny first demonstrated an association of HLA-DRw4 with RA in the 1970s using serological techniques in population studies.18 This initial finding evolved over time to include association of several distinct alleles of the class II gene, HLA-DRB1. Molecular typing techniques were developed to investigate the complexity of the locus and work by Gregersen et al led to the observation that the associated alleles encode for a conserved amino acid sequence in the third hypervariable region of the DRB1 chain.19 Subsequently this region was termed the shared epitope (SE). The SE hypothesis is based on the theory that these class II alleles are directly involved in the pathogenesis of RA. The molecular basis of this association remains elusive but present models suggest that structural differences between alleles could be involved at the level of antigen presentation or in the alteration of the T cell repertoire.20
The alleles in the HLA-DRB1 locus have been consistently associated with RA in many populations through linkage and association studies despite the fact that SE allele frequencies differ significantly between ethnic groups. More recently, studies have shown evidence of association for additional RA risk alleles within the major histocompatibility complex (MHC) region, adding further complexity,21 although the extended linkage disequilibrium (LD) across this region makes interpretation of these associations very difficult.22 Ongoing work has led to suggestions that MHC associations with RA cannot wholly be accounted for by the SE hypothesis and there have been several attempts to modify and redefine this concept.23,24 Although the MHC locus confers the largest risk for RA susceptibility, it is neither necessary nor sufficient to cause disease.
PTPN22
PTPN22 was the first locus outside the MHC to demonstrate consistently validated association with RA. PTPN22 is recognized as the second most important risk loci to the MHC in populations of European descent. The minor allele of a nonsynonymous single nucleotide polymorphism (SNP), (rs2476601, 1858 C > T, R620W) in PTPN22 was first associated with type 1 diabetes (T1D) in a candidate gene study.25 This variant was subsequently confirmed at a similar time in RA through assaying 87 putative functional SNPs in RA candidate genes and/or linkage regions.26 Since then, the association with PTPN22 has been replicated in several other populations,27–34 and the same variant is also associated with a variety of other autoimmune and inflammatory diseases.35–39 Interestingly no variants within this gene have been confirmed to be associated with RA in Asian populations where the second largest RA genetic risk factor is recognized as PADI4.40 The PTPN22 gene is a compelling biological candidate being a key component in the regulation of T cell receptor signaling.25 The associated risk allele encodes an arginine to tryptophan substitution at residue 620 in the polypeptide chain disrupting the binding to Csk. There is evidence that this change confers increased function to the PTPN22 protein, with the 620W variant enhancing the inhibitory effect on T cell receptor signaling.41
STAT4
The association of STAT4 with RA was discovered through a combination of linkage and candidate gene studies. In contrast with HLA-DRB1 and PTPN22, the association of STAT4 with RA is more modest. This locus was identified using a fine mapping approach to examine 13 candidate genes previously found under a peak of linkage identified on chromosome 2q.42 STAT4 is an example of a loci that confers susceptibility to RA across all populations tested to date as well as being associated with a variety of other AIDs.43–46 STAT4 itself is part of a family of seven transcription factors involved in cytokine receptor signaling, initially activated by IL-12 signaling in T cells, and then phosphorylated by janus kinases. Subsequently, STAT4 is translocated to the cell nucleus where it initiates transcription of its target genes and results in the production of interferon-γ and Th1 responses. Transgenic knockout mice for STAT4 have proved that it is critical to the development and function of T helper cells. Defective mice are resistant to proteoglycan-induced arthritis, do not respond to IL-12, and lack Th1 responses.47 Although rodent models of arthritis do not perfectly duplicate the clinical pathology of RA they have demonstrated good predictability and efficacy exemplified by the successful development of therapeutics for clinical use.48
The use of traditional linkage and candidate gene studies to uncover genetic susceptibility has not led to definitive answers regarding the etiology of RA. The limited success in the application of linkage analysis to complex diseases is predominately due to the inadequate power and resolution to detect variants of modest effect.49 Candidate gene studies have in the past resulted in numerous associations that are difficult to replicate, and in some cases, to overestimated genetic effects.50 This approach is also limited by its reliance on prior knowledge of the biology of the disease and by the highly subjective selection of potential genes. The alternative approach to these methods are GWAS which entail a systematic search of the entire genome for susceptibility variants. Over the past 4 years, the implementation of GWAS has brought tremendous gains to knowledge in many complex diseases.
GWAS
GWAS have become the most powerful and extensively employed approach in discovering susceptibility variants for complex disease traits. Advances in technology, reduced costs, and the ascertainment of large well-characterized case cohorts and panels of controls have made GWA scans widely available. GWAS have also been made possible through several other major advances in the last decade. The completion of the Human Genome Project in 200151 and the initial release of the International Hap Map project in 200352 paved the way forward for this approach, allowing the design of genotyping arrays to capture variation across the entire genome. These studies are based on genetic association, determined if genetic variants are seen more or less frequently in disease affected cohorts compared to non affected cohorts in the same population. The genetic markers typically used in these studies are SNPs, as they are abundant and easy to genotype. The SNP genotyping arrays, however, have very limited coverage and power to detect rare variation associated to disease, so the underlying rationale for GWAS is the common disease common variant hypothesis (CDCV).53 This proposes that common disease alleles (SNPs) with a minor allele frequency (MAF) >1% underlie most common diseases.54
The success and popularity of this strategy has been marked, evidenced by an online catalog which contained 812 published GWAS for 476 different complex traits as of March 2011, and is still growing.55 As with other complex diseases, GWAS has expanded the number of confirmed RA susceptibility loci dramatically with 34 genetic loci now confidently associated (Figure 1). A summary of these findings gives an insight to the current genetic landscape of RA.
In the same year that STAT4 was identified as a risk locus for RA the first major GWAS was conducted by the Wellcome Trust Case-Control Consortium (WTCCC). The WTCCC is an assembly of 50 research groups across the UK established in 2005. Its primary purpose is to accelerate efforts in deciphering the human genome sequence variation and to identify variants responsible for the major causes of morbidity and mortality.
WTCCC – GWAS
In 2007, a revolutionary GWAS was undertaken by the WTCCC in 14,000 cases of seven common diseases, including 2000 United Kingdom RA cases collected by the Arthritis Research United Kingdom Epidemiology Unit and ∼3000 shared controls. This was the largest single GWAS of its time. Cases and controls were genotyped using the Affymetrix Gene Chip (Santa Clara, CA) 500 k array.56 The success of this study is demonstrated by the identification of 24 independently associated regions across all seven diseases. Association with RA at genome-wide significance level (P < 5 × 10−8) was validated at the robustly associated susceptibility loci: HLA-DRB1 and PTPN22. Nine other loci were associated in the next tier of significance (Tier 2, P = 1 × 10−5 − 5 × 10−7) none of which had been previously associated with RA. In Tier 3, 49 SNPs were identified with modest association with RA (P = 1 × 10−4 − 1 × 10−5). Many of these suggestive associations have subsequently been validated as RA susceptibility loci in replication studies.
Since the first WTCCC GWAS, many different strategies have been used in attempting to identify additional RA risk loci. These include validation studies, further GWAS, meta-analysis studies, candidate gene studies, and bioinformatics approaches.
GWAS validation studies
With the extensive number of significant associations being reported through GWAS, it is imperative that the findings are validated through replication in large independent cohorts and potentially confirmed in other populations to ensure that they are real findings. GWAS validation studies have been employed extensively since the first WTCCC study. Many follow-up studies were initiated for the Tier 2 and Tier 3 WTCCC SNPs, and as a result, confirmed numerous novel associations. The 6q23 locus provides a good example of the use of this strategy. Following the WTCCC GWAS, Thomson et al followed up nine RA Tier 2 SNPs in a large independent UK cohort of 5063 cases and 3849 controls.57 One of these SNPs, rs6920220, was replicated reaching a level of genome wide significance (P = 1.1 × 10−8). Simultaneously, this SNP and a second independent polymorphism was detected in the 6q23 region in a GWAS of a US population which confirmed its importance as a risk loci for RA.58 Fine mapping of this region has revealed the complexity of this locus which has at least three independent effects. Two of these alleles, rs6920220 and rs5029937, confer risk and one, rs13207033, confers protection to RA. Also, two of the variants map to an intergenic region between OLIG3 and TNFAIP3 and one (rs5029937) maps to the second intron of TNFAIP3i.59 Since OLIG3 is involved in the development and differentiation of neuronal cells, TNFAIP3 has been selected as an attractive candidate gene for the 6q23 region. The product of TNFAIP3, A20, is a potent anti-inflammatory protein that negatively regulates the transcription factor nuclear factor kappa-beta (NF)-κB responses to tumor necrosis factor (TNF) α toll-like receptor and NOD2 signaling.60 Inappropriate NF-κB activity has been linked with many AIDs and inflammatory diseases.61,62 This hypothesis is further supported by TNFAIP3 knockout mice, which develop severe multi-organ inflammation including inflammation of the joints.63 Interestingly, a recent study has shown TNFAIP3 protein expression in human synovium and in vitro evidence of altered TNFAIP3 transcription by 6q23 intergenic SNPs associated with RA, further supporting its role in the pathogenesis of RA.64
Further GWAS
Since the initial WTCCC GWAS, many additional GWAS have been carried out in RA.55,57,65–68 Table 1 is a summary of the major RA GWAS from 2007 to present, their study designs, and the significant SNP associations reported in each study.
Table 1.
Major independent RA GWAS from 2007 to present
| Author/journal/study | Number SNPs passing QC | Study sample size | Replication sample | Region | Reported gene(s) | Associated SNP | Largest reported P-value | OR | References |
|---|---|---|---|---|---|---|---|---|---|
| WTCCC | 469,557 | 1860 cases, | – | 6p21.32 | MHC | rs6457617 | 5 × 10−75 | 2.36 | 56 |
| Nature | 2938 controls | 6p21.32 | HLA-DRB1 | rs615672 | 8 × 10−27 | NR | |||
| Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls (June 2007) | 1p13.2 | PTPN22 | rs6679677 | 6 × 10−25 | 1.98 | ||||
| Plenge | 297,086 | 1522 cases, | 997 cases | 6p21.32 | HLA-DRB1 | rs660895 | 1 × 10−100 | 3.62 | 68 |
| New England Journal of Medicine | 1850 controls | 1780 controls | 9q34 | TRAF1-C5 | rs3761847 | 4 × 10−14 | 1.32 | ||
| TRAF1-C5 as a risk locus for rheumatoid arthritis-a genome wide study (Sep 2007) | 1p13.2 | PTPN22 | rs2476601 | 2 × 10−11 | 1.72 | ||||
| Plenge | 87,962 | 393 cases, | 2283 cases, | 6q23.3 | TNFAIP3, OLIG3 | rs10499194 | 1 × 10−9 | 1.33 | 58 |
| Nature Genetics | 1211 related | 3258 controls | 6q23.3 | TNFAIP3, OLIG3 | rs6920220 | 1 × 10−7 | 1.22 | ||
| Two independent alleles at 6q23 associated with risk of rheumatoid arthritis (Dec 2007) | Framingham individuals as controls | ||||||||
| Julia | 299,918 | 400 cases, | 410 cases, | 6p21.32 | HLA-DQA1, | rs6457617 | 1 × 10−9 | NR | 66 |
| Arthritis and Rheumatism | 400 controls | 394 controls | HLA-DQA2 | ||||||
| Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility (Aug 2008) | |||||||||
| Gregersen | 278,502 | 2418 cases, | 2604 cases, | 1p13.2 | PTPN22 | rs2476601 | 2 × 10−21 | NR | 67 |
| Nature Genetics | 4504 controls | 2882 controls | 2p16.1 | REL | rs13017599 | 2.60 × 10−12 | 1.21 | ||
| REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis (July 2009) | 2q33.2 | CTLA4 | rs231735 | 6.25 × 10−9 | 1.17 | ||||
| 8p23.1 | BLK | rs2736340 | 5.69 × 10−9 | 1.19 | |||||
| 9q33.2 | TRAF1, C5 | rs881375 | 4.09 × 10−8 | NR | |||||
| Kochi | 393,217 | 2303 Japanese | 7069 cases | 6p21.32 | HLA-DRB1 | rs13192471 | 1.9 × 10−58 | 1.97 | 65 |
| Nature Genetics | cases, 3380 | 20727 controls | 6q27 | CCR6 | rs3093024 | 7.7 × 10−19 | 1.19 | ||
| A regulatory variant in CCR6 is associated with rheumatoid arthritis (June 2010) | Japanese controls |
Notes: Only genes with SNPs associated at genome-wide significance (P < 5 × 10−8) have been shown; P-values and odds ratios from the largest sample size have been recorded where they are reported.
Abbreviations: NR, not reported; QC, quality control; SNP, single nucleotide polymorphism.
Independent RA GWAS have led to the identification and validation of many novel RA susceptibility loci. The major GWAS detailed in Table 1 evidence novel associations with TRAF1/C5, TNFAIP3, REL, and CCR6. REL represents a good example of an RA locus identified in independent GWAS not previously implicated by association studies.66 Gregersen et al initially genotyped 278,502 SNPs in 2418 cases and 4504 controls from North America to identify the association of the marker SNP rs13031237 with REL. Subsequently this association was confirmed in an independent replication cohort of 2064 cases and 2882 controls and provided convincing evidence in the combined data set (P = 3.08 × 10−14). The association with rs1031237 which maps to an intron in REL67 has since been confirmed in a UK population of 3962 cases and 3531 controls.69 REL is a strong candidate gene for RA as it encodes c-Rel, which is a member of the NF-κB family of transcription factors that are involved in immune system regulation. c-Rel-deficient mice are resistant to induction of collagen-induced arthritis and have also shown deficiency in Th1 type immune responses. c-Rel has also been implicated in the involvement of CD40 signaling pathways with effects on B cell proliferation and survival.66
As well as identifying novel RA loci, independent GWAS studies have been a useful strategy for the validation of previously reported loci and the implication of other highly suggestive associations, some of which have subsequently been confirmed as RA susceptibility loci, eg, BLK. It is well- documented that larger sample sizes increase the power to maximize the probability of being able to detect novel associations with the small effect sizes expected of complex diseases.70 This has since led to the establishment of larger sample collections and the implementation of larger meta-analysis studies.
Meta-analysis studies
Many novel RA loci have been discovered more recently through meta-analysis studies. Meta-analyses combine data from several studies and increase the power of a study to detect novel associations of modest effect. The association with the CD40 susceptibility locus was discovered through a meta-analysis of WTCCC and US GWAS. This association was validated in a replication study of 3929 cases and 5807 matched controls from eight case-control collections.71 Subsequently, it has also been confirmed in a large UK population.72 Association with CD40 has since been replicated in Graves disease73–75 and multiple sclerosis (MS).76 The RA susceptibility variant rs4810485 is located in the second intron of the CD40 gene, with the gene itself encoding a protein that is a member of the TNF receptor super-family (TNFSR5). The protein expressed by CD40 is found on the surface of antigen presenting cells including B cells, dendritic cells, and macrophages.70 It is essential in stimulating a broad range of immune and inflammatory responses in several autoimmune conditions which implicates it as a good candidate gene for RA.
The most recent RA meta-analysis included a total of 12,307 patients and 20,169 controls of European descent. Seven novel RA risk alleles were identified at genome-wide significance following replication in an independent data set of 6768 RA cases and 8806 controls.16 The novel SNPs are located in genes related to immune function including CCR6, IL6ST, IRF5, PXK, RBPJ, and SPRED2. Additionally the study confirmed 24 previously established RA loci and further findings of borderline significance were reported that will require further validation.
Candidate gene studies in the GWAS era
Identification of RA loci previously associated with other AIDs
Another strategy that has proved useful in finding new associations with RA is candidate gene studies based on GWAS findings from RA and other AIDs. A good example of this is the 4q27 region which contains four genes: KIAA1109, Tenr, IL2, and IL21. Originally, this region was first associated with T1D and celiac disease in two GWAS.77,78 Based on these findings, Zhernakova et al went on to investigate this disease locus in RA in a case-control study population and found evidence for association with rs6822844.79 Subsequently, this association has been validated in independent Dutch and UK populations. The 4q27 region has since shown association with ulcerative colitis,80 juvenile idiopathic arthritis,81 psoriatic arthritis,82 and Crohn’s disease,83 further implying that this locus is a general risk factor for multiple AIDs. The IL2 and IL21 genes represent strong candidate genes for this region because the proteins encoded by these genes are cytokines that play a significant role in the development and control of inflammation in RA. Animal models of RA have demonstrated a pathogenic role for both IL-2 and IL-21. Mice deficient in IL-2 develop severe autoimmunity because of defective regulatory T-cell production.84 In DBA/1 mice with collagen-induced arthritis, the administration of IL-21 receptor Fc fusion proteins has led to an improvement in clinical symptoms and histological parameters.85 Despite the potential of both these genes as therapeutic targets in RA susceptibility, it will be difficult to identify the causal polymorphisms in this region due to the high degree of LD.
Pathway-based bioinformatics approaches
Gene relationships across implicated loci (GRAIL) developed by Raychaudhuri et al is a method of prioritizing SNPs to follow up in a candidate gene validation study in order to identify novel associations.86 Raychaudhuri et al selected 370 SNPs from 179 loci that reached P < 0.001 in a previous independent GWAS meta-analysis for investigation by GRAIL. GRAIL initially defines a genomic region in LD with each candidate SNP and then selects all genes within the region. It then uses statistical text mining of the available literature in PubMed to assess and score the relatedness of the implicated loci with genomic regions already known to be previously associated with disease. High GRAIL scores implicated 22 loci with functional connectivity. SNPs representing these candidate loci were then genotyped in an independent study of 8096 cases and 11,822 matched controls.87 Three of these loci, CD28, CD2/CD58, and PRDM1, were convincingly replicated. Limitations of the GRAIL method that should be considered include its use of established knowledge bases, which may bias towards more characterized genes, and its assumption that each region only contains one pathogenic gene. It has however proved to be a productive method of analysis to identify novel susceptibility genes.
The wealth of novel susceptibility loci identified in recent GWAS and meta-analysis studies demonstrates that the GWAS approach is still proving to be hugely successful. The use of a more informed, focused candidate–gene approach alongside bioinformatics in the GWAS era has also proved fruitful. The use of these alternative strategies has in turn led to the discovery, replication, and validation of all 34 RA susceptibility loci to date. The wealth of genetic findings for RA has led to challenges and opportunities to consider that are likely to take focus in the post-GWAS era.
What else has GWAS taught us in RA?
GWAS has been greatly successful in identifying numerous RA risk alleles. Many themes have emerged during the search for these loci including: the well-debated concept of ‘missing heritability’, the possibility of genetically distinct clinical subsets of disease, the existence of ethnic heterogeneity, and the evidence of shared risk loci across multiple AIDs. Each of these topics poses a challenge to the complete understanding of the pathogenesis of RA. Ongoing research in this field is vital to help realize the ultimate goal of establishing a relationship between genotype and phenotype.
Missing heritability
Although GWAS have provided valuable insights into the genetic basis of human disease, the identified associations for RA (with some exceptions) have a modest effect size with odds ratios (OR) <1.5. Figure 1 illustrates the heritability estimates for established RA loci. If we consider that the total sibling recurrence risk ratio (λs) is estimated to be between 5 and 10 for RA then based on λs, the established susceptibility loci at present contribute approximately 33%–47% of total heritability for RA.88 This suggests that it is likely that >50% of the genetic risk to RA remains unknown. The concept of ‘missing heritability’ is one of conflicting opinion. Questions remain as to why so much heritability has been left unexplained by GWAS findings when they capture up to 90% of common genetic variation and what it is therefore likely to be. Many explanations of ‘missing heritability’ have been suggested, including; larger numbers of common SNPs of smaller effect sizes (CDCV), rarer and structural variants which are poorly covered in current genotyping arrays, and the concept that the ‘missing’ heritability can be explained by reevaluating the proportion of genetic variance explained by current associations.
Structural variants including copy number variations (CNVs), insertions and deletions, and copy neutral variation such as translocations and inversions are an abundant source of genetic variation. To date, discovery of structural variation has been limited due to the direct focus on SNPs as mapping tools and probable causal variants. CNVs have gained particular attention as methods have improved to detect them but there has been limited work in determining their potential impact on disease risk. Several associations have been made between CNVs and autoimmunity in humans such as psoriasis,89 systemic lupus erythematosus,90 RA,91 Crohn’s disease,92 and T1D.93 A recent GWAS of CNVs in 16,000 cases of eight common diseases and 3000 common controls was carried out by the WTCCC. The findings from this study suggested that common CNVs that can be typed on current genotyping platforms seemed unlikely to account for a large proportion of ‘missing heritability’.94 Other types of structural variation including inversions and translocations have been implicated in rare Mendelian conditions, but remain largely unexplored in complex traits.
The fraction of genetic risk to common diseases that is attributable to rare variants and structural variants is debatable. The detection of rare variation presents a challenge in current genotyping arrays as they have little power to detect SNPs with MAF < 1%, and structural variation is generally under represented in current databases to study effectively. Until the full extent of this variation can be ascertained, it would be premature to conclude what impact it will have on ‘missing heritability’. The pilot phase of the 1000 genomes project has now been completed which has brought the total number of known SNPs to over 15 million, and structural variants to over 20,000.95 The next 5 years will also see a move towards the availability of affordable whole genome sequencing which will help facilitate detailed analysis of rare and structural variants. This will undoubtedly lead to the further identification and refinement of causal variations and help resolve some of the controversy surrounding the concept of ‘missing heritability’.
Other challenges faced in studying complex disease genetics include the role of gene–gene, G–E, and epigenetic modification of the genome. Established G–E associations exist between smoking and HLA-DRB1 SE alleles, PTPN22, and antibodies to cyclic citrullinated peptides;96 however replication of other findings has been poor. These studies are primarily lacking power to detect effects and face controversy and uncertainty in the best ways to model interaction. G–E interactions specifically lack well-characterized environmental exposure data and would benefit from established consortia with uniform data collection and better assessment of exposure variables.97 In essence, better study design, larger sample sizes, and development of analysis techniques will prove essential before substantial progress is made in tackling these issues.
Clinical subsets of disease
RA is currently defined by a broad range of criteria that has been revised recently by the American College of Rheumatology and European League Against Rheumatism.98 RA exhibits extensive phenotypic heterogeneity especially in the early stages of disease and as such is not recognized as a discrete clinical entity. It has been accepted for a long time that there are likely to be several distinct subtypes of RA that will correlate to different genotypes.99
RA is already classified into two subsets of disease by the presence or absence of anti-citrullinated protein antibodies (ACPA), with two-thirds of patients with early RA being ACPA-positive. ACPA are autoantibodies detected in RA patients and are highly predictive of the future development of RA,100 ACPA-positive disease being associated with a more destructive phenotype with higher rates of joint destruction compared to ACPA-negative disease.101 This supports the notion that different mechanisms may be involved in the development of ACPA-positive and ACPA-negative RA. Indeed, evidence is emerging that ACPA-positive and ACPA-negative disease may well vary in associations to different genetic and environmental risk factors.102
The majority of genetic risk factors identified for RA to date have been found in ACPA-positive cohorts. HLA-DRB1 SE alleles predispose mainly to ACPA-positive RA with a smaller effect on ACPA-negative disease,90 contributing 18% to heritability of ACPA-positive disease compared to only 2.4% in ACPA-negative disease.103 Far fewer genetic risk factors have been associated with ACPA-negative RA, but include IRF5, C-type lectin genes, and a suggestive, nonreplicated association with HLA-DRB1*03.102 The lack of risk factors associated with ACPA-negative RA could be due to increased heterogeneity within this disease subset or due to a lack of studies with large numbers of ACPA-negative patients. The heritability of ACPA-negative RA is similar to that of ACPA-positive RA, indicating that many genetic risk factors remain to be identified for ACPA-negative RA. It should be recognized that there are limitations in using an antibody-based biomarker like ACPA. There is still ambiguity in whether ACPA presentation is a cause or effect of disease and whether differences result from interindividual variation in immune response rather than from the underlying disease process.
One goal arising from GWAS is the possibility to stratify patients further than the simple ACPA classification based on the predominant genetic background or biological pathway that is driving their disease. It has been proposed that treating RA as a quantitative trait would be a better approach to dissect its genetic heterogeneity under the assumption that it is a common disorder affected by multiple underlying genes.104 This idea assumes that phenotypes vary along a continuous gradient and that a disorder such as RA can be considered as the extreme of a quantitative trait. Essentially this implies that using the present case-control design will confound genetic analysis, as controls are diluted with individuals who nearly meet the classification criteria. Refinement of statistical analyses will be important in pursuing the identification of sets of variants that could potentially relate to different underlying disease mechanisms and to the verification of different subtypes of disease. Undoubtedly, this will help refine diagnosis, predictions of outcome and treatment response, and will provide the opportunity to develop selective therapies that are specific for an individual’s phenotype.
Ethnic heterogeneity in RA risk loci
Through GWAS, clear differences have emerged amongst the major ethnic groups with regards to the genes underlying RA. Some genetic polymorphisms are restricted to specific ethnic populations, indicating a presence of genetic heterogeneity in RA, whilst some are common across multiple ethnic groups, eg, TNFAIP3 and STAT4.40 One of the most striking differences seen between different populations is the nonsynonymous coding SNP of the PTPN22 gene (1858C > T). Although widely replicated across European populations, this polymorphism is rarely found in Asian populations,105 indicating that genetic heterogeneity might be explained by the presence of exclusive susceptibility alleles in specific populations. There are also clear differences in the association of the PADI4 gene. PADI4 was the first non-HLA genetic risk factor for RA initially demonstrated in a Japanese population.106 It has since been confirmed through replication in multiple East-Asian populations,107,108 but has exhibited reports of conflicting associations in European populations30,109–111 despite a comparable allele frequency between the two populations. Risk alleles of the HLA-DRB1 locus have also been shown to vary significantly between ethnic groups. Genetic variants restricted to specific ethnic groups could reflect the result of migration, natural selection, or mutation and could explain the significant differences that have been observed between prevalence rates of disease. Studies have shown a much lower prevalence of RA in developing countries compared to European and American populations.112 Differences in prevalence data could however result from heterogeneity in the distribution of environmental exposures, or age-distribution differences between populations. There are many methodological errors such as different classification criteria for case assessment, reporter bias and access to medical care that could also result in differences between prevalence that are not specific to underlying genetic heterogeneity.40 Populations of European ancestry have dominated the majority of GWAS, which could explain the limited replication of genetic associations in other ethnic populations. Some studies that have reported negative findings are based on relatively small sample sizes, which could indicate inadequate statistical power. Challenges lie in collecting large cohorts of homogenous samples from a variety of different populations. As well as conducting GWAS in a variety of larger well-characterized populations, further dissection of genetic and environmental determinants and their interactions will help clarify genetic heterogeneity between ethnic populations with regards to AIDs.
Shared autoimmune risk loci
Another theme that has emerged in the search for RA susceptibility genes is the concept that some genetic susceptibility variants predispose to multiple AIDs which gives an insight to the possibility of shared genetic pathways. Table 2 shows a selection of the RA susceptibility risk factors that have shown to be associated in multiple AIDs.37–39,44,113–121,122–147
Table 2.
Selection of RA susceptibility loci associated with other AID
| Association with Other diseases | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | AS | ASA | BD | CD | CeD | GD | HT | JIA | MG | MS | PsA | PV | SLE | SS | SSc | T1D | UC | WG | |||
| Locus | Location | References | |||||||||||||||||||
| PTPN22 | 1p13 | 37–39, 113–115 | |||||||||||||||||||
| STAT 4 | 2q32 | 42, 44–46, 116–121 | |||||||||||||||||||
| CTLA4 | 2q33 | 113,116,122–127 | |||||||||||||||||||
| HLA | 6p21 | 113,128–136 | |||||||||||||||||||
| TNFAIP3 | 6q23 | 120,137–140 | |||||||||||||||||||
| TRAF1/C5 | 9q33.1 | 68,140–144 | |||||||||||||||||||
| IL2RA | 10p15 | 140–143,145,146 | |||||||||||||||||||
| CD40 | 20q13 | 73,147 | |||||||||||||||||||
Abbreviations: AD, Addison’s disease; AS, ankylosing spondylitis; ASA, asthma; BD, Bechet’s disease; CeD, celiac disease; CD, Crohn’s disease; GD, Graves’ disease; HT, Hashimoto’s thyroiditis; JIA, juvenile idiopathic arthritis; MG, myasthenia gravis; MS, multiple sclerosis; PsA, psoriatic arthritis; PV, pemphigus vulgaris; SLE, systemic lupus erythematosus; SS, Sjögren syndrome; SSC, systemic sclerosis; T1D, Type 1 diabetes; UC, ulcerative colitis; WG, Wegener’s granulomatosis.
Since the application of GWAS the number of loci predisposing to multiple AIDs has increased rapidly. It is evident that many loci predispose to more than one AID. A review of recent genetic studies by Zhernakova et al revealed that shared genetic factors between AIDs indicate the possibility of common etiological pathways.140 This notion is supported by the clustering of AIDs within families, the overlap in clinical manifestations, and observation that the HLA locus is associated with most common AIDs. In most cases the same allele is associated with risk across different diseases, which highlights the likelihood of common underlying mechanisms, eg, STAT4. In some cases, the same allele predisposes to risk in one AID and is protective in another.148 A good example of this would be the IL2RA gene region where allelic heterogeneity is known to exist between MS and T1D. At rs11594656 the minor allele A is associated with susceptibility to MS (OR 1.17) and protection from T1D (OR 0.87).145 Opposing risk effects at a locus suggest that predisposition to related diseases could be controlled by over- or underexpression of genes and genomic elements in biological pathways. Alternatively, there are variants that show association between multiple AIDs, but at a different position within the same locus. Multiple genetic variants are associated differently in TNFAIP3 between SLE and RA. The causal polymorphism for genetic loci post-GWAS is generally poorly understood and it is possible if different SNPs are associated with different diseases that the likely role of the gene product in pathogenesis for each of the diseases will vary.
It appears that although there is much sharing of loci between similar diseases, not all loci overlap with all AIDs. This could prove useful in determining subsets of biological pathways perturbed in different diseases, giving an insight into how far common diseases share underlying susceptibility, and locate genes or pathways that uniquely affect single diseases, perhaps determining which one of the related diseases someone is likely to develop.
Challenges in the post-GWAS era: a long way to go
The ultimate goal of genetic research in any disease is to unravel the connection between genotype and phenotype. With a wealth of established RA genetic loci and many emerging themes, we are in a greater position to understand the pathogenesis of RA but there is still a long way to go. Most GWAS studies have concentrated on finding SNP associations that are associated with disease susceptibility but have rarely invested resources in the functional characterization of the identified risk loci. Whilst there are 34 statistically valid genetic loci associated with RA, the majority of causal variants for these associations are not known, with the current exception of PTPN22. This is because GWAS are based on the principle of LD. In regions of strong LD it becomes difficult to distinguish between the causal variant and the neighboring markers in high LD with the causal variant. There are other challenges that lie in the way of assigning functionality to susceptibility SNPs. Although GWAS are uncovering more risk variants, it is apparent that the majority of associations have modest effect sizes (OR: 1.1–1.5) which implies that the functional effect of these SNPs could be subtle and hard to establish. In addition, risk alleles found in GWAS are often assigned to the most compelling gene candidate within the region based on either its proximity to the marker or the biological plausibility of its likely involvement in the pathogenesis of disease, making disease pathway analysis imprecise.
With such a wealth of genetic associations, it seems sensible to invest greater effort in the functional annotation of the GWAS findings to confidently assign association to genes. Initial progress in the post-GWAS era will be first made from identifying the causal variant/s for each RA locus, and then localizing the genetic signal to identify which target gene/s it may affect. The second stage will be to investigate the biological mechanism of the risk allele on causality of disease. There is no established gold standard approach to resolve these aims. This review is going to focus on the initial stage of functional characterization in refinement and localization of the causal genetic variants, the stage currently occupied by many of the 34 confirmed RA loci. Strategies including fine-mapping and bioinformatics tools are logical starting methods that can be applied to associated regions.
Fine mapping/resequencing
The fine mapping approach aims to narrow a region of association and pinpoint the causal variant/s responsible. It involves genotyping all known SNPs within a region to resolve information about which genes in a region are likely to be responsible. The confirmed RA susceptibility loci already show different trends in their associations. For some loci, it appears that there are distinct single variant effects (PTPN22, REL, CTLA4) and others multiple independent effects or haplotypes that confer risk (TNFAIP3 and STAT4, respectively). Ultimately the aim of fine mapping these established susceptibility regions is to identity the causal SNPs and to refine the associations at these loci further. Park et al concluded from their findings that fine mapping studies of reported loci have so far failed to find novel common variants with larger effect sizes than their tagging SNPs.149 Despite this, TNFAIP3 is a good example of how the fine-mapping approach resolved the existence of three independent effects at the RA susceptibility locus, two conferring susceptibility (rs6920220 and rs5029937) and one conferring protection (rs13207033).58 Although the success of fine mapping appears to be minimal, it is in its infancy and the Wellcome Trust has recently completed a large-scale fine-mapping study called Immunochip. Immunochip is a customized Illumina chip with 195,806 SNPs designed for fine mapping and replication of established loci across a multitude of AIDs including ∼4500 cases from UK RA patients. This is the first systematic and comprehensive application of fine mapping to date and analysis of the data should help refine all current RA genetic associations, identify causal variants, clarify the existence of haplotypes, and confirm single or multiple independent effects at each locus. There are several limitations to the Immunochip project, which must be considered. This custom chip has been designed for use in white European populations and therefore will not help to refine or replicate associations in other ethnic groups. The chip contains all known SNPs to date from the dbSNP database and from the pilot data release of the 1000 genomes project in June 2010 but will miss unknown SNPs and rare variants that have not yet been discovered. It is also thought that the Immunochip will not be 100% accurate in genotyping rare and structural variants as it is a difficult process and it is envisaged that the availability of whole genome sequencing will prove to be more informative for these types of variation.150
The above mentioned limitations of fine mapping can be overcome by resequencing susceptibility loci. Resequencing has not been extensively used to identify causal variants at complex diseases risk regions, because it is much more expensive than fine mapping. However, sequencing costs are currently decreasing, which will make possible the utilization of this approach to establish comprehensive maps of genomic variation at disease loci in the near future.
Following the application of fine mapping and resequencing, regions with a number of putative associated SNPs that cannot be separated any further by strength of genetic association will still exist. After defining the target region with identification of candidate SNP variant/s the next step is to use computational methods for SNP prioritization and to build up evidence for their likely functional impact.
Bioinformatics tools to prioritize SNPs
Bioinformatics tools are a further refinement step for the prioritization of causal SNPs. Many tools exist to enable identification of a candidate for the causal variant by utilizing prediction of functional effects to prioritize SNPs for downstream analysis. One such tool is the ENCyclopedia Of DNA Elements (ENCODE) which is hosted by the University of California Santa Cruz (UCSC). The aim of ENCODE is to find and document all the functional elements that exist in the genome in both coding and noncoding regions. This database essentially gathers its data from wet lab experiments. It includes data from a range of experiments in a variety of tissues and cell types including transcription factor binding sites, chromatin profiling, and histone modification. Data generated from wet lab experiments potentially offer greater evidence of putative function compared with the current predictive algorithms. Such a wealth of data is available through UCSC and although incredibly useful is difficult and time consuming to interrogate. Martin et al have developed a bioinformatics program (ASSIMILATOR) that quickly assimilates a concise summary of experimental data for inputted SNPs from UCSC making it much easier to compare and assess their biological relevance.151 This is a very promising method of building functional evidence to support prioritization of SNPs.
Another step towards functional characterization of loci is to explore associations between candidate SNPs and gene expression. Treating gene expression as a quantitative trait makes it possible to correlate gene transcript levels with SNPs (expression quantitative trait loci [eQTLs]). Gene expression levels have been shown to have a strong heritable component and variation can be due to polymorphisms close to the gene locus (cis) or in a different chromosome (trans). Many genetic variants resulting in phenotypic differences are mediated through changes in gene expression, and correlation between gene expression and DNA polymorphisms can be used to aid the interpretation of genetic association, indicating which particular transcript is most likely to be influenced by the associated SNP. Indeed, several studies have already demonstrated that trait-associated SNPs are more likely to be eQTLs.152,153 Whole genome mRNA expression data are becoming publicly available for a growing number of cell and tissue types enabling resources, such as the Gene Regulators in Disease project, to aid the performance of in silicoeQTL analysis and the correlation of genotype to different transcript levels.154 This is a strategy that can potentially help connect risk variants to their target genes. These are just two of the bioinformatic approaches available to refine the location of causal variants and implicate the leading positional candidate SNPs based on functional evidence. Several molecular methods have also proved useful to further investigate genetic localization of association signals, including Allele Specific Expression and 3C, but are beyond the scope of this review.
The secondary challenge of moving from an associated variant to a mechanism of action that explains disease causality is more substantial and will require many additional resources including validated functional assays, animal models, or in vitro models in which the causal variants can be assessed. This will require a lot more time, patience, and expertise to develop. It is anticipated that in the future the functional underpinning of many GWA tag SNPs can be elucidated. Closing the gap between genotype and phenotype is complicated especially since most known loci are outside protein coding regions and many have modest effects. It is important to remember that the attempt to functionally characterize all disease-associated SNPs is just at the beginning and is very far from proving causality of all disease-associated risk loci.
Conclusion
The GWAS approach has been used extensively to find the common genetic risk factors associated with RA. The results of these scans have been remarkable and strong associations with both known and novel genomic regions have been uncovered, expanding the number of robustly associated RA loci to 34. Larger meta-analysis studies have more recently been employed to increase the power to detect novel associations of smaller effect sizes and to find common associations between related AIDs. Ongoing GWA studies alongside statistical and epidemiological refinement will undoubtedly deliver novel associations and potentially implicate further pathways that could be important in the pathogenesis of RA.
GWAS has uncovered several themes that will undoubtedly contribute valuable knowledge to the complete understanding of the pathogenesis of RA. Heritability estimates have suggested that more than 50% of the genetic risk to RA remains unknown. The concept of ‘missing heritability’ is likely to become much clearer once whole genome sequencing can be implemented on a larger scale and the detection of all common, rare and, structural variants is possible. Larger, well-characterized cohorts of patients from different ethnic backgrounds are necessary to fully resolve the existence of genetic heterogeneity between populations.
Whilst the current genetic discoveries have implicated many important pathways, the largest knowledge gap lies in how the established DNA variations contribute to the pathogenesis of disease. It is apparent that the need to establish causal variants and elucidate their biological role in disease is imperative. It is anticipated that the use of fine-mapping data from the Immunochip project will help refine all the current regions of association with RA to identify the causal variants as well as giving more insight into the concept of shared AID loci. Bioinformatics tools can then be used to prioritize SNPs that can be taken forward into functional studies for validation of biological function. The integration of both genomic and functional data will be necessary to prove the role that implicated pathways and genes play in complex diseases. Identification of novel biological pathways susceptible to pharmacological intervention is possible, which in turn will enable the development of effective preventative and therapeutic agents. In addition the likely stratification of RA patients into more genetically homogenous subgroups could well facilitate prediction of disease progression and response to treatment.
Acknowledgments
The authors thank Jane Worthington for critical reading of the manuscript. KM is funded by Arthritis Research United Kingdom. GO is supported by the European Union (Marie Curie IEF Fellowship PIEF-GA-2009-235662).
Footnotes
Disclosure
The authors report no conflict of interest in this work.
References
- 1.Worthington J. Investigating the genetic basis of susceptibility to rheumatoid arthritis. J Autoimmun. 2005;25(Suppl):16–20. doi: 10.1016/j.jaut.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Young A, Koduri G. Extra-articular manifestations and complications of rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2007;21(5):907–927. doi: 10.1016/j.berh.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376(9746):1094–1108. doi: 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- 4.Alamanos Y, Drosos AA. Epidemiology of adult rheumatoid arthritis. Autoimmun Rev. 2005;4(3):130–136. doi: 10.1016/j.autrev.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Reginster JY. The prevalence and burden of arthritis. Rheumatology (Oxford) 2002;41(Suppl 1):3–6. [PubMed] [Google Scholar]
- 6.Deighton CM, Walker DJ. The familial nature of rheumatoid arthritis. Ann Rheum Dis. 1991;50(1):62–65. doi: 10.1136/ard.50.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wordsworth P, Bell J. Polygenic susceptibility in rheumatoid arthritis. Ann Rheum Dis. 1991;50(6):343–346. doi: 10.1136/ard.50.6.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarvinen P, Aho K. Twin studies in rheumatic diseases. Semin Arthritis Rheum. 1994;24(1):19–28. doi: 10.1016/0049-0172(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 9.Silman AJ, MacGregor AJ, Thomson W, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br J Rheumatol. 1993;32(10):903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- 10.MacGregor AJ, Snieder H, Rigby AS, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43(1):30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 11.Van der Linden MP, Feitsma AL, le Cessie S, et al. Association of a single-nucleotide polymorphism in CD40 with the rate of joint destruction in rheumatoid arthritis. Arthritis Rheum. 2009;60(8):2242–2247. doi: 10.1002/art.24721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolt P, Bengtsson C, Nordmark B, et al. Quantification of the influence of cigarette smoking on rheumatoid arthritis: results from a population based case-control study, using incident cases. Ann Rheum Dis. 2003;62(9):835–841. doi: 10.1136/ard.62.9.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobon GJ, Youinou P, Saraux A. The environment, geo-epidemiology, and autoimmune disease: rheumatoid arthritis. Autoimmun Rev. 2010;9(5):A288–A292. doi: 10.1016/j.autrev.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Symmons DP. Environmental factors and the outcome of rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2003;17(5):717–727. doi: 10.1016/s1521-6942(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 15.Oliver JE, Silman AJ. Risk factors for the development of rheumatoid arthritis. Scand J Rheumatol. 2006;35(3):169–174. doi: 10.1080/03009740600718080. [DOI] [PubMed] [Google Scholar]
- 16.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan AW, Robinson JI, Conaghan PG, et al. Evaluation of the rheumatoid arthritis susceptibility loci HLA-DRB1, PTPN22, OLIG3/TNFAIP3, STAT4 and TRAF1/C5 in an inception cohort. Arthritis Res Ther. 2010;12(2):R57. doi: 10.1186/ar2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298(16):869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 19.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 20.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4(4):e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HS, Lee AT, Criswell LA, et al. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Mol Med. 2008;14(5–6):293–300. doi: 10.2119/2007-00123.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22(2):109–118. doi: 10.1097/BOR.0b013e328336474d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding B, Padyukov L, Lundstrom E, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60(1):30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michou L, Croiseau P, Petit-Teixeira E, et al. Validation of the reshaped shared epitope HLA-DRB1 classification in rheumatoid arthritis. Arthritis Res Ther. 2006;8(3):R79. doi: 10.1186/ar1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36(4):337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 26.Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farago B, Talian GC, Komlosi K, et al. Protein tyrosine phosphatase gene C1858T allele confers risk for rheumatoid arthritis in Hungarian subjects. Rheumatol Int. 2009;29(7):793–796. doi: 10.1007/s00296-008-0771-9. [DOI] [PubMed] [Google Scholar]
- 28.Kokkonen H, Johansson M, Innala L, Jidell E, Rantapaa-Dahlqvist S. The PTPN22 1858C/T polymorphism is associated with anti-cyclic citrullinated peptide antibody-positive early rheumatoid arthritis in northern Sweden. Arthritis Res Ther. 2007;9(3):R56. doi: 10.1186/ar2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierer M, Kaltenhauser S, Arnold S, et al. Association of PTPN22 1858 single-nucleotide polymorphism with rheumatoid arthritis in a German cohort: higher frequency of the risk allele in male compared to female patients. Arthritis Res Ther. 2006;8(3):R75. doi: 10.1186/ar1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plenge RM, Padyukov L, Remmers EF, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77(6):1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simkins HM, Merriman ME, Highton J, et al. Association of the PTPN22 locus with rheumatoid arthritis in a New Zealand Caucasian cohort. Arthritis Rheum. 2005;52(7):2222–2225. doi: 10.1002/art.21126. [DOI] [PubMed] [Google Scholar]
- 32.Viken MK, Olsson M, Flam ST, et al. The PTPN22 promoter polymorphism −1123G > C association cannot be distinguished from the 1858C > T association in a Norwegian rheumatoid arthritis material. Tissue Antigens. 2007;70(3):190–197. doi: 10.1111/j.1399-0039.2007.00871.x. [DOI] [PubMed] [Google Scholar]
- 33.Wesoly J, Hu X, Thabet MM, et al. The 620W allele is the PTPN22 genetic variant conferring susceptibility to RA in a Dutch population. Rheumatology (Oxford) 2007;46(4):617–621. doi: 10.1093/rheumatology/kel381. [DOI] [PubMed] [Google Scholar]
- 34.Zhernakova A, Eerligh P, Wijmenga C, Barrera P, Roep BO, Koeleman BP. Differential association of the PTPN22 coding variant with autoimmune diseases in a Dutch population. Genes Immun. 2005;6(6):459–461. doi: 10.1038/sj.gene.6364220. [DOI] [PubMed] [Google Scholar]
- 35.Chelala C, Duchatelet S, Joffret ML, et al. PTPN22 R620W functional variant in type 1 diabetes and autoimmunity related traits. Diabetes. 2007;56(2):522–526. doi: 10.2337/db06-0942. [DOI] [PubMed] [Google Scholar]
- 36.Chung SA, Criswell LA. PTPN22: its role in SLE and autoimmunity. Autoimmunity. 2007;40(8):582–590. doi: 10.1080/08916930701510848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Criswell LA, Pfeiffer KA, Lum RF, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76(4):561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinks A, Worthington J, Thomson W. The association of PTPN22 with rheumatoid arthritis and juvenile idiopathic arthritis. Rheumatology (Oxford) 2006;45(4):365–368. doi: 10.1093/rheumatology/kel005. [DOI] [PubMed] [Google Scholar]
- 39.Orozco G, Sanchez E, Gonzalez-Gay MA, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52(1):219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 40.Kochi Y, Suzuki A, Yamada R, Yamamoto K. Genetics of rheumatoid arthritis: underlying evidence of ethnic differences. J Autoimmun. 2009;32(3–4):158–162. doi: 10.1016/j.jaut.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 41.Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37(12):1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 42.Korman BD, Kastner DL, Gregersen PK, Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008;8(5):398–403. doi: 10.1007/s11882-008-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HS, Remmers EF, Le JM, Kastner DL, Bae SC, Gregersen PK. Association of STAT4 with rheumatoid arthritis in the Korean population. Mol Med. 2007;13(9–10):455–460. doi: 10.2119/2007-00072.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi S, Ikari K, Kaneko H, et al. Association of STAT4 with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Japanese population. Arthritis Rheum. 2008;58(7):1940–1946. doi: 10.1002/art.23494. [DOI] [PubMed] [Google Scholar]
- 45.Martinez A, Varade J, Marquez A, et al. Association of the STAT4 gene with increased susceptibility for some immune-mediated diseases. Arthritis Rheum. 2008;58(9):2598–2602. doi: 10.1002/art.23792. [DOI] [PubMed] [Google Scholar]
- 46.Zervou MI, Mamoulakis D, Panierakis C, Boumpas DT, Goulielmos GN. STAT4: a risk factor for type 1 diabetes? Hum Immunol. 2008;69(10):647–650. doi: 10.1016/j.humimm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19(21):2577–2584. doi: 10.1038/sj.onc.1203485. [DOI] [PubMed] [Google Scholar]
- 48.Kannan K, Ortmann RA, Kimpel D. Animal models of rheumatoid arthritis and their relevance to human disease. Pathophysiology. 2005;12(3):167–181. doi: 10.1016/j.pathophys.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 50.Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. Int J Biol Sci. 2007;3(7):420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 52.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 54.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant … or not? Hum Mol Genet. 2002;11(20):2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 55.Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A catalog of published genome-wide association studies. Dec 12, 2010. [Accessed April 21, 2011]. Available at: http://www.genome.gov/gwastudies/
- 56.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orozco G, Hinks A, Eyre S, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23. Hum Mol Genet. 2009;18(14):2693–2699. doi: 10.1093/hmg/ddp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coornaert B, Carpentier I, Beyaert R. A20: central gatekeeper in inflammation and immunity. J Biol Chem. 2009;284(13):8217–8221. doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vereecke L, Beyaert R, van LG. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30(8):383–391. doi: 10.1016/j.it.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 62.Brown KD, Claudio E, Siebenlist U. The roles of the classical and alternative nuclear factor-kappaB pathways: potential implications for autoimmunity and rheumatoid arthritis. Arthritis Res Ther. 2008;10(4):212. doi: 10.1186/ar2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee EG, Boone DL, Chai S, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289(5488):2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elsby LM, Orozco G, Denton J, Worthington J, Ray DW, Donn RP. Functional evaluation of TNFAIP3 (A20) in rheumatoid arthritis. Clin Exp Rheumatol. 2010;28(5):708–714. [PMC free article] [PubMed] [Google Scholar]
- 65.Kochi Y, Okada Y, Suzuki A, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet. 2010;42(6):515–519. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 66.Julia A, Ballina J, Canete JD, et al. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 2008;58(8):2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- 67.Gregersen PK, Amos CI, Lee AT, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41(7):820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis – a genomewide study. N Engl J Med. 2007;357(12):1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eyre S, Hinks A, Flynn E, et al. Confirmation of association of the REL locus with rheumatoid arthritis susceptibility in the UK population. Ann Rheum Dis. 2010;69(8):1572–1573. doi: 10.1136/ard.2009.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5(5):e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orozco G, Eyre S, Hinks A, et al. Association of CD40 with rheumatoid arthritis confirmed in a large UK case-control study. Ann Rheum Dis. 2010;69(5):813–816. doi: 10.1136/ard.2009.109579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomer Y, Concepcion E, Greenberg DA. A C/T single-nucleotide polymorphism in the region of the CD40 gene is associated with Graves’ disease. Thyroid. 2002;12(12):1129–1135. doi: 10.1089/105072502321085234. [DOI] [PubMed] [Google Scholar]
- 74.Kim TY, Park YJ, Hwang JK, et al. A C/T polymorphism in the 5′-untranslated region of the CD40 gene is associated with Graves’ disease in Koreans. Thyroid. 2003;13(10):919–925. doi: 10.1089/105072503322511319. [DOI] [PubMed] [Google Scholar]
- 75.Mukai T, Hiromatsu Y, Fukutani T, et al. A C/T polymorphism in the 5′ untranslated region of the CD40 gene is associated with later onset of Graves’ disease in Japanese. Endocr J. 2005;52(4):471–477. doi: 10.1507/endocrj.52.471. [DOI] [PubMed] [Google Scholar]
- 76.Australia and New Zealand Multiple Sclerosis Genetics Consortium Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 77.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39(7):827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhernakova A, Alizadeh BZ, Bevova M, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81(6):1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Festen EA, Goyette P, Scott R, et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009;58(6):799–804. doi: 10.1136/gut.2008.166918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Albers HM, Kurreeman FA, Stoeken-Rijsbergen G, et al. Association of the autoimmunity locus 4q27 with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60(3):901–904. doi: 10.1002/art.24296. [DOI] [PubMed] [Google Scholar]
- 82.Liu Y, Helms C, Liao W, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4(3):e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marquez A, Orozco G, Martinez A, et al. Novel association of the interleukin 2-interleukin 21 region with inflammatory bowel disease. Am J Gastroenterol. 2009;104(8):1968–1975. doi: 10.1038/ajg.2009.224. [DOI] [PubMed] [Google Scholar]
- 84.Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 85.Young DA, Hegen M, Ma HL, et al. Blockade of the interleukin-21/interleukin-21 receptor pathway ameliorates disease in animal models of rheumatoid arthritis. Arthritis Rheum. 2007;56(4):1152–1163. doi: 10.1002/art.22452. [DOI] [PubMed] [Google Scholar]
- 86.Raychaudhuri S, Plenge RM, Rossin EJ, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5(6):e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raychaudhuri S, Thomson BP, Remmers EF, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41(12):1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orozco G, Barrett JC, Zeggini E. Synthetic associations in the context of genome-wide association scan signals. Hum Mol Genet. 2010;19(R2):R137–R144. doi: 10.1093/hmg/ddq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huffmeier U, Bergboer JG, Becker T, et al. Replication of LCE3C-LCE3B CNV as a risk factor for psoriasis and analysis of interaction with other genetic risk factors. J Invest Dermatol. 2010;130(4):979–984. doi: 10.1038/jid.2009.385. [DOI] [PubMed] [Google Scholar]
- 90.Mamtani M, Rovin B, Brey R, et al. CCL3L1 gene-containing segmental duplications and polymorphisms in CCR5 affect risk of systemic lupus erythaematosus. Ann Rheum Dis. 2008;67(8):1076–1083. doi: 10.1136/ard.2007.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McKinney C, Merriman ME, Chapman PT, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67(3):409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 92.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40(9):1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grayson BL, Smith ME, Thomas JW, et al. Genome-wide analysis of copy number variation in type 1 diabetes. PLoS One. 2010;5(11):e15393. doi: 10.1371/journal.pone.0015393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464(7289):713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mahdi H, Fisher BA, Kallberg H, et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat Genet. 2009;41(12):1319–1324. doi: 10.1038/ng.480. [DOI] [PubMed] [Google Scholar]
- 97.Thomas D. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu Rev Public Health. 2010;31:21–36. doi: 10.1146/annurev.publhealth.012809.103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 99.Weyand CM, Klimiuk PA, Goronzy JJ. Heterogeneity of rheumatoid arthritis: from phenotypes to genotypes. Springer Semin Immunopathol. 1998;20(1–2):5–22. doi: 10.1007/BF00831996. [DOI] [PubMed] [Google Scholar]
- 100.Berglin E, Padyukov L, Sundin U, et al. A combination of autoantibodies to cyclic citrullinated peptide (CCP) and HLA-DRB1 locus antigens is strongly associated with future onset of rheumatoid arthritis. Arthritis Res Ther. 2004;6(4):R303–R308. doi: 10.1186/ar1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van der WD, Huizinga TW. Every shared epitope allele for itself? Nat Rev Rheumatol. 2009;5(9):477–478. doi: 10.1038/nrrheum.2009.174. [DOI] [PubMed] [Google Scholar]
- 102.Padyukov L, Seielstad M, Ong RT, et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann Rheum Dis. 2010;70(2):259–265. doi: 10.1136/ard.2009.126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van der WD, Houwing-Duistermaat JJ, Toes RE, et al. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60(4):916–923. doi: 10.1002/art.24385. [DOI] [PubMed] [Google Scholar]
- 104.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10(12):872–828. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 105.Kochi Y, Suzuki A, Yamada R, Yamamoto K. Ethnogenetic heterogeneity of rheumatoid arthritis-implications for pathogenesis. Nat Rev Rheumatol. 2010;6(5):290–295. doi: 10.1038/nrrheum.2010.23. [DOI] [PubMed] [Google Scholar]
- 106.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylargininedeiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 107.Ikari K, Kuwahara M, Nakamura T, et al. Association between PADI4 and rheumatoid arthritis: a replication study. Arthritis Rheum. 2005;52(10):3054–3057. doi: 10.1002/art.21309. [DOI] [PubMed] [Google Scholar]
- 108.Kang CP, Lee HS, Ju H, Cho H, Kang C, Bae SC. A functional haplotype of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Koreans. Arthritis Rheum. 2006;54(1):90–96. doi: 10.1002/art.21536. [DOI] [PubMed] [Google Scholar]
- 109.Burr ML, Naseem H, Hinks A, et al. PADI4 genotype is not associated with rheumatoid arthritis in a large UK Caucasian population. Ann Rheum Dis. 2010;69(4):666–670. doi: 10.1136/ard.2009.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barton A, Bowes J, Eyre S, et al. A functional haplotype of the PADI4 gene associated with rheumatoid arthritis in a Japanese population is not associated in a United Kingdom population. Arthritis Rheum. 2004;50(4):1117–1121. doi: 10.1002/art.20169. [DOI] [PubMed] [Google Scholar]
- 111.Iwamoto T, Ikari K, Nakamura T, et al. Association between PADI4 and rheumatoid arthritis: a meta-analysis. Rheumatology (Oxford) 2006;45(7):804–807. doi: 10.1093/rheumatology/kel023. [DOI] [PubMed] [Google Scholar]
- 112.Alamanos Y, Voulgari PV, Drosos AA. Incidence and prevalence of rheumatoid arthritis, based on the 1987 American College of Rheumatology criteria: a systematic review. Semin Arthritis Rheum. 2006;36(3):182–188. doi: 10.1016/j.semarthrit.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Brand O, Gough S, Heward J. HLA, CTLA-4 and PTPN22: the shared genetic master-key to autoimmunity? Expert Rev Mol Med. 2005;7(23):1–15. doi: 10.1017/S1462399405009981. [DOI] [PubMed] [Google Scholar]
- 114.Dieude P, Guedj M, Wipff J, et al. The PTPN22 620W allele confers susceptibility to systemic sclerosis: findings of a large case-control study of European Caucasians and a meta-analysis. Arthritis Rheum. 2008;58(7):2183–2188. doi: 10.1002/art.23601. [DOI] [PubMed] [Google Scholar]
- 115.Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity. 2007;40(6):453–461. doi: 10.1080/08916930701464897. [DOI] [PubMed] [Google Scholar]
- 116.Daha NA, Kurreeman FA, Marques RB, et al. Confirmation of STAT4, IL2/IL21, and CTLA4 polymorphisms in rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1255–1260. doi: 10.1002/art.24503. [DOI] [PubMed] [Google Scholar]
- 117.Glas J, Seiderer J, Nagy M, et al. Evidence for STAT4 as a common autoimmune gene: rs7574865 is associated with colonic Crohn’s disease and early disease onset. PLoS One. 2010;5(4):e10373. doi: 10.1371/journal.pone.0010373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG. Association between the rs7574865 polymorphism of STAT4 and rheumatoid arthritis: a meta-analysis. Rheumatol Int. 2010;30(5):661–666. doi: 10.1007/s00296-009-1051-z. [DOI] [PubMed] [Google Scholar]
- 119.Moon CM, Cheon JH, Kim SW, et al. Association of signal transducer and activator of transcription 4 genetic variants with extra-intestinal manifestations in inflammatory bowel disease. Life Sci. 2010;86(17–18):661–667. doi: 10.1016/j.lfs.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 120.Prahalad S, Hansen S, Whiting A, et al. Variants in TNFAIP3, STAT4, and C12 or f30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60(7):2124–2130. doi: 10.1002/art.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhernakova A, Eerligh P, Barrera P, et al. CTLA4 is differentially associated with autoimmune diseases in the Dutch population. Hum Genet. 2005;118(1):58–66. doi: 10.1007/s00439-005-0006-z. [DOI] [PubMed] [Google Scholar]
- 123.Zhao SX, Pan CM, Cao HM, et al. Association of the CTLA4 gene with Graves’ disease in the Chinese Han population. PLoS One. 2010;5(3):e9821. doi: 10.1371/journal.pone.0009821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lester S, Downie-Doyle S, Rischmueller M. CTLA4 polymorphism and primary Sjogren’s syndrome. Arthritis Res Ther. 2007;9(3):401. doi: 10.1186/ar2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Einarsdottir E, Soderstrom I, Lofgren-Burstrom A, et al. The CTLA4 region as a general autoimmunity factor: an extended pedigree provides evidence for synergy with the HLA locus in the etiology of type 1 diabetes mellitus, Hashimoto’s thyroiditis and Graves’ disease. Eur J Hum Genet. 2003;11(1):81–84. doi: 10.1038/sj.ejhg.5200903. [DOI] [PubMed] [Google Scholar]
- 126.Chistiakov DA, Turakulov RI. CTLA-4 and its role in autoimmune thyroid disease. J Mol Endocrinol. 2003;31(1):21–36. doi: 10.1677/jme.0.0310021. [DOI] [PubMed] [Google Scholar]
- 127.Ayadi H, Hadj KH, Rebai A, Farid NR. The genetics of autoimmune thyroid disease. Trends Endocrinol Metab. 2004;15(5):234–239. doi: 10.1016/j.tem.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 128.Ban Y, Tomer Y. Genetic susceptibility in thyroid autoimmunity. Pediatr Endocrinol Rev. 2005;3(1):20–32. [PubMed] [Google Scholar]
- 129.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6(12):683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Holmes S, Friese MA, Siebold C, Jones EY, Bell J, Fugger L. Multiple sclerosis: MHC associations and therapeutic implications. Expert Rev Mol Med. 2005;7(3):1–17. doi: 10.1017/S1462399405008914. [DOI] [PubMed] [Google Scholar]
- 131.Iwasa K, Kato-Motozaki Y, Furukawa Y, et al. Up-regulation of MHC class I and class II in the skeletal muscles of myasthenia gravis. J Neuroimmunol. 2010;225(1–2):171–174. doi: 10.1016/j.jneuroim.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 132.Loiseau P, Lepage V, Djelal F, et al. HLA class I and class II are both associated with the genetic predisposition to primary Sjogren syndrome. Hum Immunol. 2001;62(7):725–731. doi: 10.1016/s0198-8859(01)00253-1. [DOI] [PubMed] [Google Scholar]
- 133.Pender MP, Greer JM. Immunology of multiple sclerosis. Curr Allergy Asthma Rep. 2007;7(4):285–292. doi: 10.1007/s11882-007-0043-x. [DOI] [PubMed] [Google Scholar]
- 134.Rioux JD, Goyette P, Vyse TJ, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106(44):18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thomas GP, Brown MA. Genetics and genomics of ankylosing spondylitis. Immunol Rev. 2010;233(1):162–180. doi: 10.1111/j.0105-2896.2009.00852.x. [DOI] [PubMed] [Google Scholar]
- 136.Waterman M, Xu W, Stempak JM, et al. Distinct and overlapping genetic loci in crohn’s disease and ulcerative colitis: correlations with pathogenesis. Inflamm Bowel Dis. 2010 Dec 10; doi: 10.1002/ibd.21579. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dieude P, Guedj M, Wipff J, et al. Association of the TNFAIP3 rs5029939 variant with systemic sclerosis in the European Caucasian population. Ann Rheum Dis. 2010;69(11):1958–1964. doi: 10.1136/ard.2009.127928. [DOI] [PubMed] [Google Scholar]
- 138.Eyre S, Hinks A, Bowes J, et al. Overlapping genetic susceptibility variants between three autoimmune disorders: rheumatoid arthritis, type 1 diabetes and coeliac disease. Arthritis Res Ther. 2010;12(5):R175. doi: 10.1186/ar3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fung EY, Smyth DJ, Howson JM, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10(2):188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 140.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10(1):43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 141.Nishimoto K, Kochi Y, Ikari K, et al. Association study of TRAF1-C5 polymorphisms with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese. Ann Rheum Dis. 2010;69(2):368–373. doi: 10.1136/ard.2008.104315. [DOI] [PubMed] [Google Scholar]
- 142.Kurreeman FA, Goulielmos GN, Alizadeh BZ, et al. The TRAF1-C5 region on chromosome 9q33 is associated with multiple autoimmune diseases. Ann Rheum Dis. 2010;69(4):696–699. doi: 10.1136/ard.2008.106567. [DOI] [PubMed] [Google Scholar]
- 143.Hinks A, Eyre S, Ke X, et al. Overlap of disease susceptibility loci for rheumatoid arthritis and juvenile idiopathic arthritis. Ann Rheum Dis. 2010;69(6):1049–1053. doi: 10.1136/ard.2009.110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Behrens EM, Finkel TH, Bradfield JP, et al. Association of the TRAF1-C5 locus on chromosome 9 with juvenile idiopathic arthritis. Arthritis Rheum. 2008;58(7):2206–2207. doi: 10.1002/art.23603. [DOI] [PubMed] [Google Scholar]
- 145.Maier LM, Lowe CE, Cooper J, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5(1):e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brand OJ, Lowe CE, Heward JM, et al. Association of the interleukin-2 receptor alpha (IL-2R alpha)/CD25 gene region with Graves’ disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf) 2007;66(4):508–512. doi: 10.1111/j.1365-2265.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- 147.Blanco-Kelly F, Matesanz F, Alcina A, et al. CD40: novel association with Crohn’s disease and replication in multiple sclerosis susceptibility. PLoS One. 2010;5(7):e11520. doi: 10.1371/journal.pone.0011520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang K, Baldassano R, Zhang H, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19(10):2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Park JH, Wacholder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Martin P, Barton A, Eyre S. ASSIMILATOR: a new tool to inform selection of associated genetic variants for functional studies. Bioinformatics. 2011;27(1):144–146. doi: 10.1093/bioinformatics/btq611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 154.Ge B, Pokholok DK, Kwan T, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41(11):1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]