Abstract
Background
Cutaneous squamous cell carcinoma (cSCC) is the second most common type of non-melanoma skin cancer (NMSC) globally. The aims of this study were to further systematically clarify the potential association of rs833061 (-460 C>T) and rs1570360 (-1154 G>A), two SNPs of VEGF, with the risk of cSCC and the prognostic impacts on cSCC patients.
Material/Methods
This hospital-based case-control study analyzed peripheral venous blood collected from 100 cSCC patients and 124 healthy controls, and gathered personal information on patients. Genotypes of the VEGF gene -460C>T and -1154G>A polymorphism were detected using polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) method. Different distributions of allele frequencies and genotype in the case and control group were measured, comparing different genotype differences in the survival of patients with cSCC.
Results
Distributions of allele frequencies and genotype of -460 C>T in the case and control group were statistically different; the TT + CT genotype was significantly correlated with a decrease risk of cSCC (OR=0.36, 95% CI=0.21–0.63, P<0.001). There was no difference in the distribution of allele frequencies and genotype of -1154 G>A between control and case groups. For -1154460C>T, the CC genotype was an adverse factor, associated with a significant decrease in the survival status of cSCC patients (P<0.001). For VEGF-1154 G>A, the AA genotype was significantly correlated with the reduced overall survival in cSCC patients, with the mean survival time of 23.88 months (P=0.009).
Conclusions
The VEGF gene -460 C>T polymorphism and -1154 G>A polymorphism may serve as potential genetic markers for the risk and prognosis of cSCC.
MeSH Keywords: Polymorphism, Single-Stranded Conformational; Prognosis; Vascular Endothelial Growth Factor, Endocrine-Gland-Derived
Background
Cutaneous squamous cell carcinoma (cSCC) is the second most common type of non-melanoma skin cancer (NMSC) globally, with a constantly increasing incidence [1,2]. The known risk factors for cSCC include aging, fair complexion, homosexual intercourse, chronic skin ulcers, burn scars, immune suppression, and exposure to ultraviolet light and chemical carcinogens [3]. The vast majority of cSCCs are curable with simple surgical resection, and prognosis is usually favorable [4]. However, approximately 4% of the patients develop metastatic disease and 1.5% eventually die from the disease [5]. Metastatic cSCCs present serious therapeutic problems due to lack of consistently effective treatments [6]. There are few treatments for patients with advanced cSCCs, primarily due to the lack of knowledge regarding genomic alternations that induce metastatic cSCC. Furthermore, there are no specific molecular biomarkers predict treatment response or disease behavior [4]. Therefore, it is important to identify specific molecules involved in metastatic cSCC that can potentially become targets for diagnosis and enable the development of new therapy strategies.
Angiogenesis, the formation of new blood vessels from pre-existing vasculature, plays a key role in tumor development, invasion, and metastases formation [7,8]. Angiogenesis is a complicated process stimulated by a variety of factors, among which vascular endothelial growth factor (VEGF) is crucial [9]. VEGF is one of the most potent endothelial cell growth factors; it promotes formation of new blood vessels and stimulates vascular permeability of endothelial cells [10]. VEGF initiates intracellular signal transduction pathways by specifically binding 2 transmembrane VEGF receptors on endothelial cells [11]. While initiating intracellular signal transduction pathways, VEGF supplies the newly formed blood vessels with nutrients and oxygen. This induces aggressive local growth and metastases formation, resulting in poor prognosis and low survival rates of cancer patients [12].
The VEGF gene is located at chromosome 6p21.3 and consists of 8 exons and 7 introns. It is very polymorphic, with at least 30 functional single-nucleotide polymorphisms (SNPs) in the 5′-untranslated region, 3′-untranslated region (UTR) and promoter [13]. Single-nucleotide polymorphisms (SNPs) of VEGF that can be found in the promoter or other regulatory regions may regulate VEGF expression or activity [14]. The involvement of VEGF polymorphisms in cancer progression has been demonstrated in several types of tumors [15–18]. VEGF polymorphisms have previously been described in solid carcinomas, containing lung cancer [7,19], non-Hodgkin’s lymphoma [20], cervical cancer [21], colorectal cancer [22], esophageal squamous cell carcinoma (ESCC) [10, 23], and oral squamous cell carcinoma (OSCC) [9,24–26]. The impact of the VEGF SNPs on clinical outcomes of patients with cancer has been previously described [7,12,26,27]. However, the impact of VEGF polymorphism during follow-up has been much less studied. The -1154460C>T SNP (rs833061) and -1154G>A SNP (rs1570360) are located at the promoter region and may regulate promoter activity. The association of these 2 functional SNPs with OSCC risk has been previously studied [9,12]. However, the effect of VEGF gene -460C>T SNP (rs833061) and -1154G>A SNP (rs1570360) on cSCC risk and prognosis remains unclear [28,29].
In the current study, the -460C>T SNP (rs833061) and -1154G>A SNP (rs1570360) within the VEGF gene were detected in cSCC patients; a regular follow-up was conducted for all the patients. This study aimed to further clarify the potential association of these 2 SNPs with the cSCC risk and to evaluate the prognostic impact of VEGF-related SNPs on cSCC patients.
Material and Methods
Study population
This study consisted of 100 patients with cSCC. They were treated in the department of Surgery at Shandong Provincial Hospital Affiliated to Shandong University between 2010 and 2014; the group included 55 men and 45 women. The ages of patients ranged from 34 to 76 years old, with the average age of 56.04±10.41 years old. No patients underwent radiotherapy, chemotherapy, or hormone therapy before surgery. The control group contained 124 healthy subjects who came from our hospital’s physical checkup center during the same period of time. All control subjects had no history of blood system disease or cancer. Written permission was obtained from all the participants and the study was approved by the Research Ethics Committee of Shandong Provincial Hospital Affiliated to Shandong University.
DNA extraction
Peripheral venous blood samples (5 mL) were acquired from all the participants and stored at −20°C until the extraction of genomic DNA. Genomic DNA was isolated with the QIAamp DNA mini kit (Qiagen, Hamburg Germany) according to the manufacturer’s protocol. The extracted DNA was stored at −80°C for further use.
Detection of VEGF gene polymorphism
Genotyping of the VEGF gene -460C>T and -1154G>A polymorphisms were detected using PCR – RFLP method. The primers of -1154460C>T were 5′-CGAGAGTGAGGACGTGTGTG-3′(forward) and 5′-ATTGGAATCCTGGAGTGACC-3′(reverse); the primers for -11541154G>A were 5′-TCCTGCTCCCTCCTCGCCAATG-3′(forward) and 5′-GGCGGGGACAGGCGAGCATC-3′(reverse), purchased from Sangon Biotech (Shanghai, China). The PCR reaction was performed in a 20-μl reaction mixture containing 40–160 ng DNA template, 0.25 units Taq DNA polymerase (Sangon Biotech), 250 μM each deoxyribonucleotide triphosphate (dNTP) (Sangon Biotech), 0.5 μM forward primer, 0.5 μM reverse primer, and 1×PCR buffer with 1.2 mM MgCl2. A negative control without DNA template was used to evaluate contamination. The PCR reaction was carried out in a thermocycler (MJ Research, USA) and reaction conditions consisted of 94 °C pre-denaturizing for 5 min, 94°C denaturizing for 45 s, 61°C annealing for 45 s, 72°C extension for 45 s, and 72°C continuous extension for 7 min after 35 cycles. The amplified products were digested with Bshl236I and MnlI restriction endonuclease (Takara, Japan) incubated at 37°C for 16 h. The PCR and digestion products were analyzed with 3% agarose gels.
Follow-up
Follow-up files were created for all the cSCC patients upon admission, including complete pathological, clinical, and follow-up data. A regular follow-up of 2~5 years for 100 patients was performed by hospital visit, treatment, and telephone interview. Follow-up start time was defined as the discharge time of patients after system treatment and the follow-up deadline was January 2015. Follow-up time was counted monthly until death or the end of the follow-up time.
Statistical analysis
All statistical analyses in this study were performed by SPSS 19.0 software packages. Differences between ages were analyzed with unpaired t-tests. Counted data were analyzed with the chi-square test. Hardy-Weinberg equilibrium (HWE) was calculated with Hardy-Weinberg equilibrium theory (p=allele frequency, q=1–p, p2+q2=1). The different distributions of allele frequencies and genotypes in the case and control group were analyzed using the chi-square test. Logistic regression analysis was used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs). Kaplan-Meier method was used to plot survival curves and the statistical differences were analyzed with the log-rank test. P value <0.05 was defined as statistically significant.
Results
General research characteristics
The patient group consisted of 100 cSCC cases. There were 55 men and 45 women in the patient group; 46 patients were under 60 years old and 54 patients were 60 years old or older (average age: 56.04±10.41 years old; range: 34–76 years old). There were 40 poorly-differentiated cases (the percentage of differentiated cells <50%), 34 moderately-differentiated cases (the differentiated cells between 50–75%), and 26 well-differentiated cases (the differentiated cells >75%), according to Broders classification [30]. Thirty-five cases with lymph node involvement and 65 cases without lymph node involvement were identified. The control group contained 124 healthy subjects attending the hospital physical checkup center during the same period. There were 66 males and 58 females in the control group; 59 subjects were under 60 years old and 65 patients were 60 years old or older (average age: 53.79±11.89 years; range: 30–78 years). Age and sex in the two groups showed no significant differences (P>0.05) (Table 1).
Table 1.
Characteristics of patients with CSCC and control group.
| Characteristics | CSCC (n=100) | Controls (n=124) | P-value |
|---|---|---|---|
| Age (years old), n (%) | |||
| Mean ±SD | 56.04±10.41 | 53.79±11.89 | 0.138 |
| ≥60 | 54 (54.00%) | 65 (52.42%) | |
| <60 | 46 (46.00%) | 59 (47.58%) | |
| Sex, n (%) | |||
| Male | 55 (55.00%) | 66 (53.23%) | 0.836 |
| Female | 45 (45.00%) | 58 (46.77%) | |
| Histopathological grade, n (%) | |||
| Well-differentiated | 26 (26.00%) | ||
| Moderately-differentiated | 34 (34.00%) | ||
| Poorly-differentiated | 40 (40.00%) | ||
| Lymph node involvement, n (%) | |||
| Yes | 35 (35.00%) | ||
| No | 65 (65.00%) | ||
CSCC – cutaneous squamous cell carcinoma; SD – standard deviation.
Genotyping of the VEGF gene -460C>T and -1154G>A polymorphisms
Genotypes of the VEGF-460C>T and -1154G>A were successfully detected in all the DNA samples using the PCR-RFLP method. The 175-bp PCR product of VEGF-460C>T was digested with Bshl236I restriction endonuclease. The homozygous T/T genotype remained uncut at 175bp; the homozygous C/C genotype was completely digested into 155 bp and 20 bp fragments; 175 bp, 155 bp, and 20 bp fragments corresponded to the heterozygous C>T genotype (Figure 1). The 206-bp PCR product of VEGF-1154G>A was digested with MnlI restriction endonuclease and the G allele was replaced by the A allele with a loss of digestion site. The homozygous A/A genotype was digested into 184bp and 22bp fragments; the homozygous G/G genotype was completely digested into 150 bp, 34 bp and 22 bp fragments; all 4 fragments (184 bp, 150 bp, 34bp, and 22 bp) corresponded to the heterozygous G>A genotype (Figure 2).
Figure 1.
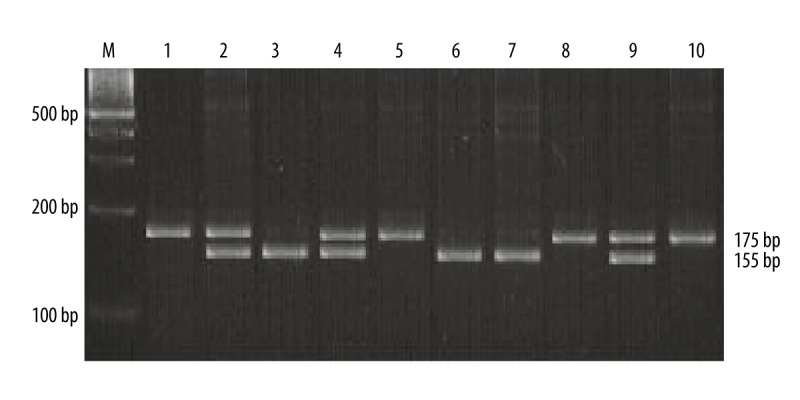
VEGF-460C>T genotyping by PCR-RFLP analysis followed by separation on 3% agarose gel as described in text. M – 100 bp DNA marker; lanes 1, 5 and 8 – T/T homozygous genotype; lanes 2, 4 and 9 – C>T heterozygous genotype; lanes 3, 6 and 7 – C/C homozygous genotype; lane 10 – PCR product.
Figure 2.
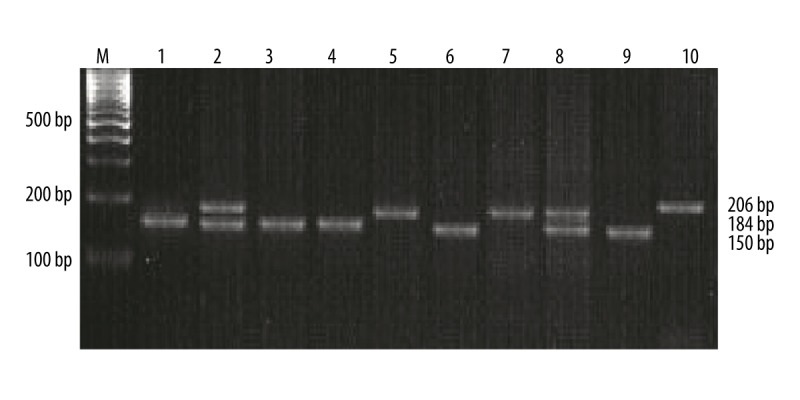
VEGF-1154G>A genotyping by PCR-RFLP analysis followed by separation on 3% agarose gel as described in text. M – 100 bp DNA marker; lanes 1, 3, 4, 6 and 9 – G/G homozygous genotype; lanes 2 and 8 – G>A heterozygous genotype; lanes 5 and 7 – A/A homozygous genotype; lane 10: PCR product.
The association between clinicopathological characteristics and VEGF polymorphism
The distribution of the VEGF-460C>T SNP genotype in patients is presented in Table 2. The genotype frequencies of CC, CT, and TT in the well-differentiated group was 46.15%, 38.46%, and 15.38%, respectively; in the moderately-differentiated group, the genotype frequencies of CC and CT were 44.11% and 55.89%, respectively; the genotype frequencies of CC and CT were 95.00% and 5.00%, respectively, in the poorly-differentiated group. There were hardly any carriers of homozygote TT in either moderately- or poorly-differentiated grades. Differences between the 3 groups were significant (P<0.001). The genotype distribution of VEGF-1154 G>A was also associated with histological differentiation of cSCC (P<0.001). The genotype frequencies of GG in the well-differentiated group, the moderately-differentiated group, and the poorly-differentiated group were 96.15%, 47.06%, and 25.00%, respectively; the genotype frequencies of GA in the well-differentiated group, the moderately-differentiated group, and the poorly-differentiated group were 4.00%, 47.06%, and 60.00%, respectively. The genotype distribution of VEGF-460C>T and VEGF-1154 G>A showed no significant correlation with age, sex, or lymph node metastasis (all P>0.05) (Table 2).
Table 2.
Correlations between clinicopathological characteristics and VEGF polymorphisms.
| Characteristics | n | -460C>T | P-value | -1154G>A | P-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | GG | GA | AA | ||||
| Age (years) | 0.858 | 0.128 | |||||||
| ≥60 | 54 | 34 | 18 | 2 | 24 | 25 | 2 | ||
| <60 | 46 | 31 | 13 | 2 | 27 | 16 | 6 | ||
| Sex | 0.908 | 0.124 | |||||||
| Male | 55 | 35 | 18 | 2 | 33 | 19 | 3 | ||
| Female | 45 | 30 | 13 | 2 | 18 | 22 | 5 | ||
| Histopathological grade | <0.001 | <0.001 | |||||||
| Well-differentiated | 26 | 12 | 10 | 4 | 25 | 1 | 0 | ||
| Moderately-differentiated | 34 | 15 | 19 | 0 | 16 | 16 | 2 | ||
| Poorly-differentiated | 40 | 38 | 2 | 0 | 10 | 24 | 6 | ||
| Lymph node involvement | 0.606 | 0.141 | |||||||
| Yes | 35 | 25 | 9 | 1 | 22 | 11 | 2 | ||
| No | 65 | 40 | 22 | 3 | 29 | 30 | 6 | ||
The genotype and allele distribution of VEGF-460C>T and -1154G>A SNP
The VEGF-460C>T and -1154G>A genotype and allele distribution of the cSCC patients and of the control group are presented in Table 3. The genotype distribution of the VEGF-460C>T and -1154G>A polymorphism did not deviate from Hardy-Weinberg equilibrium (P>0.05). The CT and CC genotype frequencies of VEGF-460C>T in the control group were 50.81% and 41.13%, which were significantly different from those in the patient group (31.00% and 65.00%, respectively) (CT vs. CC, OR=0.39, 95%CI: 0.22–0.68, P=0.001). Compared to the CC genotype, the TT+CT genotype significantly decreased the risk of cSCC (OR=0.36, 95% CI: 0.21–0.63, P<0.001). The C allele frequencies in the control group and patient group were 66.53% and 80.50%, respectively; while the T allele frequencies in the control group and patient group were 33.47% and 19.50%, respectively; the differences in the 2 groups were statistically significant (T vs. C, OR=0.48, 95%CI: 0.31–0.75, P=001). Comparing the genotype distributions of the VEGF gene -1154 G>A polymorphism in cSCC patients to that in the controls revealed no significant differences (P>0.05) and the differences in allele frequencies were also not statistically significant.
Table 3.
VEGF genotype and allele frequency in patients with cSCC (n=100) and control (n=124).
| Groups | Controls (n=124) | Patients (n=100) | P-value* | OR (95% CI) | P-value# |
|---|---|---|---|---|---|
| -1154460 C>T | |||||
| CC | 51 (41.13%) | 65 (65.00%) | – | 1.00 (reference) | – |
| CT | 63 (50.81%) | 31 (31.00%) | 0.001 | 0.39 (0.22–0.68) | 0.043 |
| TT | 10 (8.06%) | 4 (4.00%) | 0.052 | 0.31 (0.09–1.06) | 0.715 |
| TT+CT | 73 (58.87%) | 35 (35.00%) | <0.001 | 0.36 (0.21–0.63) | <0.001 |
| Allele frequency | |||||
| C | 165 (66.53%) | 161 (80.50%) | – | 1.00 (reference) | – |
| T | 83 (33.47%) | 39 (19.50%) | 0.001 | 0.48 (0.31–0.75) | – |
| -11541154 G>A | |||||
| GG | 54 (43.55%) | 51 (51.00%) | – | 1.00 (reference) | – |
| GA | 58 (46.77%) | 41 (41.00%) | 0.305 | 0.75 (0.43–1.30) | 0.247 |
| AA | 12 (9.68%) | 8 (8.00%) | 0.482 | 0.71 (0.27–1.87) | 0.654 |
| GA+AA | 70 (56.45%) | 49 (49.00%) | 0.267 | 0.74 (0.44–1.26) | 0.155 |
| Allele frequency | |||||
| G | 166 (66.94%) | 143 (71.50%) | – | 1.00 (reference) | – |
| A | 82 (33.06%) | 57 (28.50%) | 0.299 | 0.81 (0.54–1.21) | – |
VEGF – vascular endothelial growth factor; OR – odds ratio; CI – confidence interval.
Two-sided χ2 test for distribution of genotypic and allelic frequencies.
Compared by binary logistic multivariate regression model with adjustment of sex and age for cases and controls.
VEGF gene -460C>T and -1154G>A polymorphism and the prognosis of cSCC
A systematic follow-up of 2~5 years for 100 cSCC patients was performed. The follow-up deadline was January 2015; the longest follow-up time was 60 months and the shortest follow-up time was 24 months. Eleven patients were lost during the follow-up period; the follow-up rate was 89%. For VEGF-460C>T SNP, the mean survival time of cSCC patients with CC, CT, and TT genotype were 28.92, 53.54, and 58.75 months, respectively. The CC genotype was an adverse factor that negatively affected the survival time of cSCC patients when compared with heterozygote CT and homozygote TT (P<0.001, Figure 3). For VEGF-1154 G>A SNP, the mean survival time of cSCC patients with GG, GA, and AA genotypes were 41.19, 36.41, and 23.88 months, respectively; compared to the GG and GA genotypes, the AA genotype had a significant correlation with decreased overall survival time (P=0.009, Figure 4).
Figure 3.
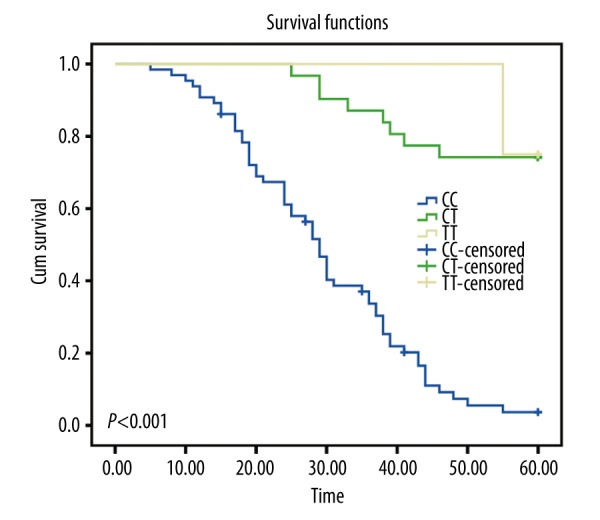
Kaplan-Meier curves of VEGF-460 C>T, compared by the log-rank test.
Figure 4.
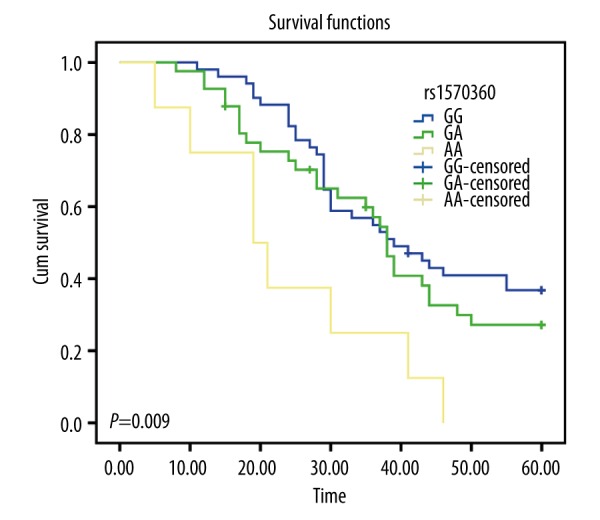
Kaplan-Meier curves of VEGF-1154 G>A, compared by the log-rank test.
Discussion
Angiogenesis is an important process for tumor development, invasion, and metastasis; it is stimulated by a variety of factors. VEGF is considered to be one of the key factors involved [7,8]. Recently, several VEGF SNPs have been identified and their important effects have attracted considerable attention. Correlation between VEGF SNPs and susceptibility or prognosis in various forms of cancers has been reported in many studies [7,10,25,31]. In the present study, the -460 C>T and -1154 G>A polymorphisms of VEGF in 100 cSCC patients and 124 healthy subjects were observed. In order to analyze the relation of VEGF gene polymorphisms with cSCC risk, the patients were stratified by age, sex, histopathological grade, and lymph node involvement. The results demonstrate that the genotype distribution of VEGF-460 C>T shows no correlation with age, sex, or lymph node metastasis (all P>0.05), but there was association with histological differentiation of cSCC (P<0.001). A significant difference was found between the genotype distribution and allele frequency of the VEGF gene -460 C>T polymorphism of the control group and that of the patients with cSCC. Furthermore, a higher percentage of the T allele and the TT homozygote were distributed in cSCC patients compared to the controls. Therefore, we can reason that there was a much lower cSCC risk for individuals with the CT heterozygote genotype in their VEGF-460 C>T region, indicating that the VEGF-460 C>T polymorphism could act as a candidate genetic biomarker in cSCC patients. The association of the VEGF-460 C>T polymorphism with clinical outcomes in cSCC patients has not been evaluated. However, there were several studies concerning VEGF-460 C>T polymorphism in other cancers. For example, Ku et al. indicated that the T allele and TT genotype of VEGF-460 C>T are significantly associated with OSCC [32]. Borase et al. demonstrated that the T allele and TT genotype of VEGF-460 C>T can greatly increase the risk for OSCC patients [26]. In the analysis of a correlation between clinical characteristics and the VEGF gene -1154 G>A polymorphism, the results demonstrate that the genotype distribution of VEGF-1154 G>A shows no relation with age, sex, or lymph node metastasis (all P>0.05), but there was an association with histological differentiation of cSCC (P<0.001). The decreased frequency of the VEGF-1154 A allele (AA and GA genotypes) in cSCC patients did not differ significantly from that of the control. In a previous study, Kämmerer et al. demonstrated that the genotype distribution of VEGF-1154 G>A in OSCC patients and controls had no significant difference [9]. Unal et al. examined 89 healthy subjects and 57 patients with laryngeal squamous cell carcinoma; they found that the -1154 GG genotype of VEGF appears to be a higher risk for laryngeal SCC patients [33].
Prognostic factors play an essential role when come to dividing patients into different risk groups in order to predict outcomes and provide adequate therapy strategies [34]. In the current study, the relationship between VEGF gene -460 C>T polymorphism, -1154 G>A polymorphism, and the survival of patients with cSCC was examined for the first time. The results demonstrate that the TT genotype of -460 C>T is a protective factor in comparison to genotypes of CT and CC, positively affecting the survival time of cSCC patients (P<0.001). The AA genotype of -1154 G>A has a significant correlation with the decreased overall survival time in cSCC patients when compared with homozygote GG and heterozygote GA (P=0.009).
The association of VEGF gene polymorphism with survival was controversial in previous studies. Kämmerer et al. observed that the VEGF-460 C>T SNP had no significant impact on survival in advanced OSCCs, while the -1154 AA genotype was associated with significantly worse survival in the univariate analysis [12]. In contrast, Supic et al. found that the -1154 GG genotype may be a prognostic marker associated with significantly worse survival in advanced-stage OSCC patients with a similar sample size [8]. Other studies revealed the -1154 GG genotype to be an adverse survival factor in colorectal cancer [35], but it was associated with a higher overall survival of the 1154 A allele in advanced breast cancer [36]. To sum up, results of clinical studies are inconsistent, possibly due to differences in ethnic groups studied, tumor locations, and sample sizes.
In this study, only 2 SNPs in the promoter were analyzed, which might be a limiting factor. The results of 1 specific SNP may be influenced by the other SNPs in the angiogenesis pathway. In addition, the sample sizes of the present study are somewhat small, even though similar studies were previously performed [26,33]. Hence, more studies, including larger sample sizes and more SNPs, distributed not only in the promoter but also in the 5′ and 3′ un-translated region, should be conducted.
Conclusions
Our data suggest that the VEGF gene -460 C>T polymorphism is related to the risk of cSCC, and both VEGF gene -460 C>T polymorphism and -1154 G>A polymorphism are correlated with the prognosis of cSCC patients. Therefore, the VEGF gene -460 C>T polymorphism and -1154 G>A polymorphism may serve as a potential genetic marker for the risk and prognosis of cSCC.
Footnotes
Disclosure of conflict of interest
None of the authors have conflicts of interest to report with regard to this manuscript.
Source of support: This study was funded by the Natural Science Foundation of China (81171492/H1102)
References
- 1.Guy GP, Jr, Machlin SR, Ekwueme DU, Yabroff KR. Prevalence and costs of skin cancer treatment in the U.S., 2002–2006 and 2007–2011. Am J Prev Med. 2015;48:183–87. doi: 10.1016/j.amepre.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu GA, Lynn Su Chang A. Overall and progression-free survival of stage 4 cutaneous squamous cell carcinoma at a single large referral center. J Am Acad Dermatol. 2015;73:165–66. doi: 10.1016/j.jaad.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 3.Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;44:975–83. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 4.Li YY, Hanna GJ, Laga AC, et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin Cancer Res. 2015;21:1447–56. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantsch KD, Meisner C, Schonfisch B, et al. Analysis of risk factors determining prognosis of cutaneous squamous-cell carcinoma: A prospective study. Lancet Oncol. 2008;9:713–20. doi: 10.1016/S1470-2045(08)70178-5. [DOI] [PubMed] [Google Scholar]
- 6.Wells JL, III, Shirai K. Systemic therapy for squamous cell carcinoma of the skin in organ transplant recipients. Am J Clin Oncol. 2012;35:498–503. doi: 10.1097/COC.0b013e318201a3ef. [DOI] [PubMed] [Google Scholar]
- 7.Maeda A, Nakata M, Yasuda K, et al. Influence of vascular endothelial growth factor single nucleotide polymorphisms on non-small cell lung cancer tumor angiogenesis. Oncol Rep. 2013;29:39–44. doi: 10.3892/or.2012.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supic G, Jovic N, Zeljic K, et al. Association of VEGF-A genetic polymorphisms with cancer risk and survival in advanced-stage oral squamous cell carcinoma patients. Oral Oncol. 2012;48:1171–77. doi: 10.1016/j.oraloncology.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 9.Kammerer PW, Toyoshima T, Eletr S, et al. Single nucleotide polymorphisms of the vascular endothelial growth factor gene associated with incidence of oral squamous cell carcinoma. J Oral Pathol Med. 2010;39:786–92. doi: 10.1111/j.1600-0714.2010.00904.x. [DOI] [PubMed] [Google Scholar]
- 10.Yang PW, Hsieh MS, Huang YC, et al. Genetic variants of EGF and VEGF predict prognosis of patients with advanced esophageal squamous cell carcinoma. PLoS One. 2014;9:e100326. doi: 10.1371/journal.pone.0100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz MT, Biselli PM, Maniglia JV, et al. Genetic variability of vascular endothelial growth factor and prognosis of head and neck cancer in a Brazilian population. Braz J Med Biol Res. 2010;43:127–33. doi: 10.1590/s0100-879x2009007500036. [DOI] [PubMed] [Google Scholar]
- 12.Kammerer PW, Koch FP, Schiegnitz E, et al. Associations between single-nucleotide polymorphisms of the VEGF gene and long-term prognosis of oral squamous cell carcinoma. J Oral Pathol Med. 2013;42:374–81. doi: 10.1111/jop.12026. [DOI] [PubMed] [Google Scholar]
- 13.Jain L, Vargo CA, Danesi R, et al. The role of vascular endothelial growth factor SNPs as predictive and prognostic markers for major solid tumors. Mol Cancer Ther. 2009;8:2496–508. doi: 10.1158/1535-7163.MCT-09-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien MH, Liu YF, Hsin CH, et al. Impact of VEGF-C gene polymorphisms and environmental factors on oral cancer susceptibility in Taiwan. PLoS One. 2013;8:e60283. doi: 10.1371/journal.pone.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Wang Y, Yu X, et al. Expression of TP53, BCL-2, and VEGFA genes in esophagus carcinoma and its biological significance. Med Sci Monit. 2015;21:3016–22. doi: 10.12659/MSM.894640. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Tu J, Wang S, Zhao J, et al. rs833061 and rs699947 on promoter gene of vascular endothelial growth factor (VEGF) and associated lung cancer susceptibility and survival: A meta-analysis. Med Sci Monit. 2014;20:2520–6. doi: 10.12659/MSM.891394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao-Yun X, Zhao-Hui M, Ke C, et al. Glucagon-like peptide-1 improves proliferation and differentiation of endothelial progenitor cells via upregulating VEGF generation. Med Sci Monit. 2011;17(2):BR35–41. doi: 10.12659/MSM.881383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauss L, Volland D, Kunkel M, et al. Dual role of VEGF family members in the pathogenesis of head and neck cancer (HNSCC): Possible link between angiogenesis and immune tolerance. Med Sci Monit. 2005;11:BR280–92. [PubMed] [Google Scholar]
- 19.Naik NA, Bhat IA, Afroze D, et al. Vascular endothelial growth factor A gene (VEGFA) polymorphisms and expression of VEGFA gene in lung cancer patients of Kashmir Valley (India) Tumour Biol. 2012;33:833–39. doi: 10.1007/s13277-011-0306-y. [DOI] [PubMed] [Google Scholar]
- 20.Diao LP, Yu XM, Gao YH, et al. Association of VEGF genetic polymorphisms with the clinical characteristics of non-Hodgkin’s lymphoma. J Cancer Res Clin Oncol. 2009;135:1473–81. doi: 10.1007/s00432-009-0650-0. [DOI] [PubMed] [Google Scholar]
- 21.Kim YH, Kim MA, Park IA, et al. VEGF polymorphisms in early cervical cancer susceptibility, angiogenesis, and survival. Gynecol Oncol. 2010;119:232–36. doi: 10.1016/j.ygyno.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Kim JG, Chae YS, Sohn SK, et al. Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with colorectal cancer. Clin Cancer Res. 2008;14:62–66. doi: 10.1158/1078-0432.CCR-07-1537. [DOI] [PubMed] [Google Scholar]
- 23.Golozar A, Beaty TH, Gravitt PE, et al. Oesophageal squamous cell carcinoma in high-risk Chinese populations: Possible role for vascular epithelial growth factor A. Eur J Cancer. 2014;50:2855–65. doi: 10.1016/j.ejca.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seki S, Fujiwara M, Matsuura M, et al. Prediction of outcome of patients with oral squamous cell carcinoma using vascular invasion and the strongly positive expression of vascular endothelial growth factors. Oral Oncol. 2011;47:588–93. doi: 10.1016/j.oraloncology.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Kammerer PW, Al-Nawas B, Kalkan S, et al. Angiogenesis-related prognosis in patients with oral squamous cell carcinoma-role of the VEGF +936 C/T polymorphism. J Oral Pathol Med. 2015;44:429–36. doi: 10.1111/jop.12254. [DOI] [PubMed] [Google Scholar]
- 26.Borase AP, Ganvir SM, Hazarey VK, et al. Estimation of vascular endothelial growth factor gene -460 C/T polymorphism as a biomarker in oral squamous cell carcinoma patients from the Indian subcontinent. J Investig Clin Dent. 2015;6:267–72. doi: 10.1111/jicd.12103. [DOI] [PubMed] [Google Scholar]
- 27.Savas S. A curated database of genetic markers from the angiogenesis/VEGF pathway and their relation to clinical outcome in human cancers. Acta Oncol. 2012;51:243–46. doi: 10.3109/0284186X.2011.636758. [DOI] [PubMed] [Google Scholar]
- 28.Moussai D, Mitsui H, Pettersen JS, et al. The human cutaneous squamous cell carcinoma microenvironment is characterized by increased lymphatic density and enhanced expression of macrophage-derived VEGF-C. J Invest Dermatol. 2011;131:229–36. doi: 10.1038/jid.2010.266. [DOI] [PubMed] [Google Scholar]
- 29.Kluger MS, Colegio OR. Lymphangiogenesis linked to VEGF-C from tumor-associated macrophages: Accomplices to metastasis by cutaneous squamous cell carcinoma? J Invest Dermatol. 2011;131:17–19. doi: 10.1038/jid.2010.347. [DOI] [PubMed] [Google Scholar]
- 30.Broders AC. Squamous-Cell Epithelioma of the Skin: A Study of 256 Cases. Ann Surg. 1921;73:141–60. doi: 10.1097/00000658-192102000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masago K, Fujita S, Kim YH, et al. Effect of vascular endothelial growth factor polymorphisms on survival in advanced-stage non-small-cell lung cancer. Cancer Sci. 2009;100:1917–22. doi: 10.1111/j.1349-7006.2009.01253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku KT, Wan L, Peng HC, et al. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for oral cancer. Oral Oncol. 2005;41:497–502. doi: 10.1016/j.oraloncology.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Unal ZN, Unal M, Bagdatoglu OT, et al. Genetic polymorphism of VEGF-1154 (A/G) in laryngeal squamous cell carcinoma. Arch Med Res. 2008;39:209–11. doi: 10.1016/j.arcmed.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Schiegnitz E, Kammerer PW, Koch FP, et al. GDF 15 as an anti-apoptotic, diagnostic and prognostic marker in oral squamous cell carcinoma. Oral Oncol. 2012;48:608–14. doi: 10.1016/j.oraloncology.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Koutras AK, Antonacopoulou AG, Eleftheraki AG, et al. Vascular endothelial growth factor polymorphisms and clinical outcome in colorectal cancer patients treated with irinotecan-based chemotherapy and bevacizumab. Pharmacogenomics J. 2012;12:468–75. doi: 10.1038/tpj.2011.37. [DOI] [PubMed] [Google Scholar]
- 36.Schneider BP, Wang M, Radovich M, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26:4672–78. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]


