Abstract
Engineered cardiac tissues hold promise for cell therapy and drug development, but exhibit inadequate function and maturity. In this study, we sought to significantly improve the function and maturation of rat and human engineered cardiac tissues. We developed dynamic, free-floating culture conditions for engineering “cardiobundles”, 3-dimensional cylindrical tissues made from neonatal rat cardiomyocytes or human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) embedded in fibrin-based hydrogel. Compared to static culture, 2-week dynamic culture of neonatal rat cardiobundles significantly increased expression of sarcomeric proteins, cardiomyocyte size (~2.1-fold), contractile force (~3.5-fold), and conduction velocity of action potentials (~1.4-fold). The average contractile force per cross-sectional area (59.7 mN/mm2) and conduction velocity (52.5 cm/sec) matched or approached those of adult rat myocardium, respectively. The inferior function of statically cultured cardiobundles was rescued by transfer to dynamic conditions, which was accompanied by an increase in mTORC1 activity and decline in AMPK phosphorylation and was blocked by rapamycin. Furthermore, dynamic culture effects did not stimulate ERK1/2 pathway and were insensitive to blockers of mechanosensitive channels, suggesting increased nutrient availability rather than mechanical stimulation as the upstream activator of mTORC1. Direct comparison with phenylephrine treatment confirmed that dynamic culture promoted physiological cardiomyocyte growth rather than pathological hypertrophy. Optimized dynamic culture conditions also augmented function of human cardiobundles made reproducibly from cardiomyocytes derived from multiple hPSC lines, resulting in significantly increased contraction force (~2.5-fold) and conduction velocity (~1.4-fold). The average specific force of 23.2 mN/mm2 and conduction velocity of 25.8 cm/sec approached the functional metrics of adult human myocardium. In conclusion, we have developed a versatile methodology for engineering cardiac tissues with a near-adult functional output without the need for exogenous electrical or mechanical stimulation, and have identified mTOR signaling as an important mechanism for advancing tissue maturation and function in vitro.
Keywords: Cardiac tissue engineering, Conduction velocity, Contractile function, mTOR, Human pluripotent stem cells
INTRODUCTION
Engineered cardiac tissues can be used for in vitro studies of heart development [1, 2], modeling of cardiovascular disease [3–5], and high-throughput screening of drug effects on cardiomyocyte (CM) survival and function [6, 7]. However, fidelity of these assays is currently limited by poor functional properties of cultured CMs compared to adult CMs [8, 9]. Specifically, engineered 3D cardiac tissue constructs generate contractile stresses that are an order of magnitude lower than those of adult ventricular myocardium and conduct action potentials at a significantly slower speed, likely due to CM immaturity and a low volume fraction of CMs within the tissue [10]. Engineering tissues that accurately mimic the structure and function of the native myocardium would enable more predictive in vitro studies and promote cell therapies for myocardial infarction and heart failure [2, 11]. In the latter case, achieving highest velocity of action potential conduction and contractile stress would be two fundamental requirements to ensure electrical safety and functional efficacy of the therapy, respectively.
The adult heart consists of densely packed myocytes with high metabolic demand, which necessitates at least one capillary per CM to supply nutrients [12]. Local depletion of oxygen has been observed in the vicinity of cultured CMs [13–15], highlighting the need to enhance nutrient and oxygen transport in 3D engineered cardiac tissues. Previous studies have utilized tissue perfusion [15–17] or bioreactor culture [18–20] to improve mass transport in engineered myocardium, with low-shear conditions being preferable due to CM susceptibility to shear-induced damage [17, 20, 21]. Many of these pioneering reports involved the use of porous polymer scaffolds; however, natural hydrogels (fibrin, collagen) have proven superior for the generation of highly functional cardiac tissues [22–24] because they support uniform cell density, macroscopic contractions, and application of tension to support myocyte spreading and alignment [25, 26].
We therefore sought to develop a versatile approach to culture hydrogel-based cardiac tissue constructs under low-shear dynamic conditions that would support high tissue cellularity while maintaining mechanical tension requisite for cell survival and functional cardiogenesis. Specifically, we designed cylindrically shaped engineered cardiac tissues (“cardiobundles”) anchored within porous flexible frames that supported both a chronic auxotonic loading and free-floating culture of CM tissues. Dynamic culture was applied using standardized tissue culture plates and platform rockers, eliminating the need for custom-designed bioreactors or perfusion chambers. The effects of dynamic culture on tissue density, CM size, and electrical and mechanical function were first established for cardiobundles made of neonatal rat CMs, followed by the application of optimized conditions to hPSC-CMs. For both rat and human tissues, we observed significant improvements in myocardial mass, CM maturation, and tissue function with dynamic culture, and identified mTOR signaling as a critical mechanistic component of this response. The resulting cardiac tissue model is simple for implementation and represents promising platform for future applications in cardiac disease modeling, drug screening, and regenerative therapy.
METHODS
Additional details and assessment methodologies are provided in Supplemental Materials.
Neonatal rat CM isolation and fabrication of engineered cardiobundles
Neonatal rat ventricular myocytes (NRVMs) were isolated from the hearts of 2-day-old Sprague-Dawley rats using previously described methods [19, 27] and embedded in a fibrin-based hydrogel cast into 7 mm × 2 mm troughs of pre-fabricated polydimethylsiloxane (PDMS) molds (Fig. S1). Obtained cardiobundles, each constructed from 3.75 × 105 cells, were anchored at each end by a porous nylon frame (Cerex®) and removed from troughs to allow free movement in culture media. Cardiobundles were cultured on a static (non-moving) or dynamic (rocking, ± 30° tilt, 0.4 Hz [28], Supplemental Video 1) platform for a period of 14 days. For some experiments, tissues were switched between static and dynamic conditions on day 7.
hPSC-CM differentiation and cardiobundle fabrication
Human cardiobundles were constructed using CMs differentiated from 2 hESC lines (H9, RUES2) and one iPSC line (derived at the Duke University Core Facility). Cardiac differentiation was carried out using small molecules CHIR99021 and IWP-2 to upregulate and inhibit Wnt signaling, respectively, as previously described [29]. In some studies, hPSC-CMs were purified using glucose-free chemically defined medium (CDM3) applied at differentiation days 10–12 [30, 31]. After 12–22 days of differentiation, derived hPSC-CMs were utilized to fabricate cardiobundles as described for NRVMs.
Immunohistology
Intact cardiobundles and 10 μm thick cross-sections were fixed and immunostained using standard techniques.
Western blotting and qPCR
Expression of cardiac genes and proteins was analyzed using standard qPCR protocols and Western blot.
Optical mapping of action potential propagation
Optical mapping of action potentials was performed as previously described [32, 33]. Cardiobundles were stained with a transmembrane voltage-sensitive dye (di-4-ANEPPS) and paced at different rates by suprathreshold point stimulus to map propagation of action potentials. Calcium transients were imaged using lentiviral expression of a genetically encoded calcium indicator (GCaMP6) driven by a myocyte-specific promoter (MHCK7) [34], or using Rhod-2 AM.
Measurement of isometric contractile force generation
Isometric contractile forces were measured as previously described [33, 35]. Cardiobundles were immersed in Tyrode’s solution containing 1.8 mM CaCl2 and connected to a force transducer. Contractions were elicited by electric field stimulus from parallel platinum electrodes.
Statistics
Data are expressed as mean ± SEM. Statistically significant differences were determined by one-way ANOVA with Tukey’s post hoc test and p < 0.05 was considered significantly different.
RESULTS
Dynamic culture of cardiobundles stimulates growth of neonatal rat cardiomyocytes
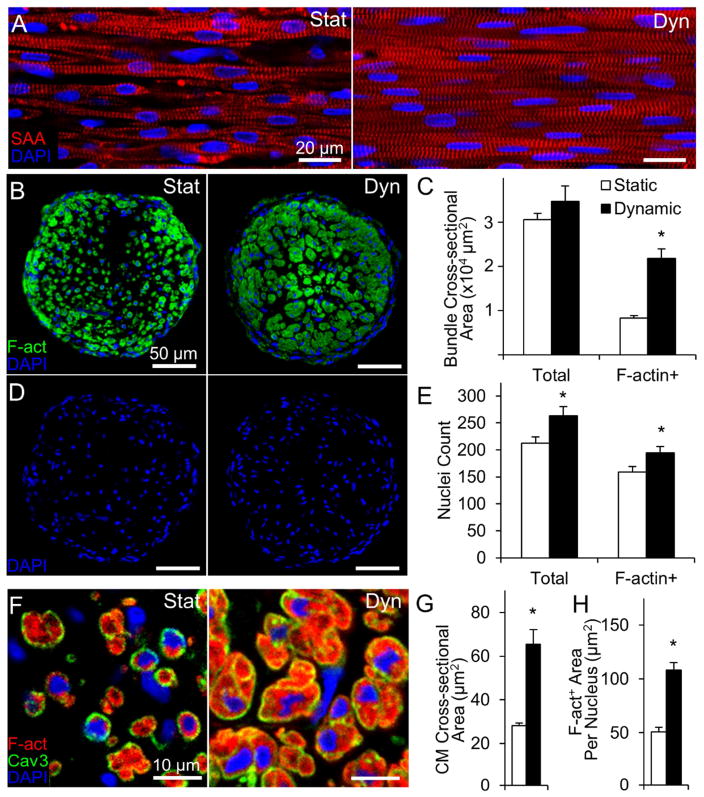
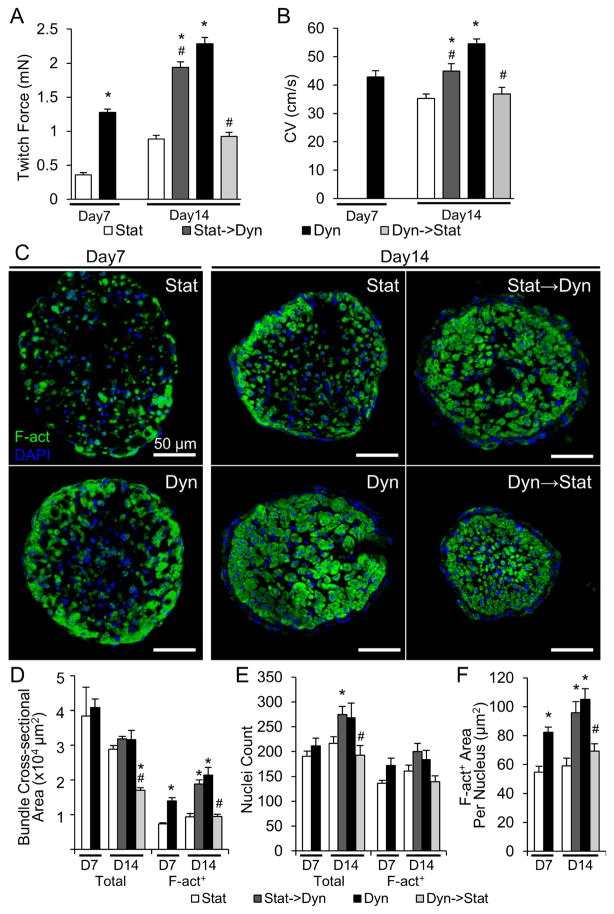
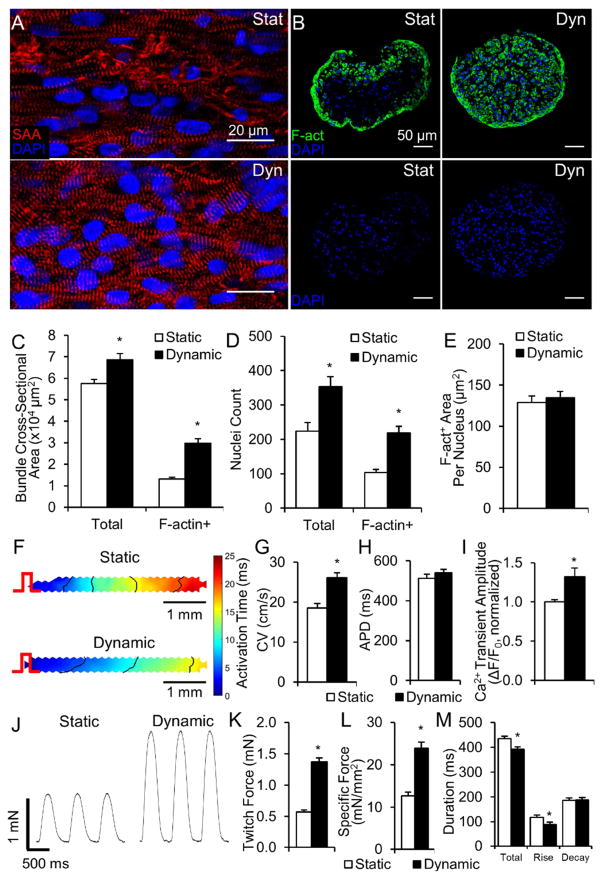
After 2 weeks of culture, cardiobundles consisted of uniformly aligned, cross-striated cardiomyocytes that robustly expressed electrical and mechanical junctions (Fig. 1A, Fig. S2). In both static and dynamic conditions, NRVM cardiobundles contracted synchronously and spontaneously at rates that initially increased and then decreased to completely cease after culture day 10 (Fig. S3, Supplemental Video 2). In longitudinal sections, CMs cultured under dynamic conditions appeared larger and more densely packed than CMs in static culture (Fig. 1A). In transverse cross-sections (Fig. 1B), dynamically cultured cardiobundles exhibited a 2.5-fold larger muscle area (Fig. 1C, right), as quantified from F-actin stainings (Fig. S4), and only 1.2-fold higher nuclei number (Fig. 1D–E) compared to statically cultured cardiobundles. Similar cellularities in static and dynamic culture were confirmed by two additional assays (Fig. S5). Consistent with comparable cell numbers, static and dynamic cardiobundles had comparable expressions of hypoxia marker PHD2 (Fig. S6), while PHD3 expression was 2-fold higher in static condition (Fig. S6). Regarding previous reports of ~5-fold increased PHD2 and ~40-fold increased PHD3 expression in 0.5–1% oxygen vs. normoxia [36, 37], these results suggested that static culture exhibited only a mild hypoxia relative to dynamic culture. From transverse cross-sections, we estimated a 2.1-fold larger CM size in dynamically cultured cardiobundles by dividing the total muscle (F-actin+) area with number of muscle nuclei (Fig. 1H). The larger CM size was further confirmed by quantifying area of individual cells from caveolin-3 staining (Fig. 1F–G). Fibroblasts and collagen 1 were located preferentially on the cardiobundle exterior and their abundance was increased in dynamic vs. static culture (Fig. S7).
Figure 1. Effects of static and dynamic culture on structure of engineered NRVM cardiobundles.
(A) Representative images of cardiobundles cultured for 2 weeks under static (Stat) or dynamic (Dyn) conditions, stained for sarcomeric α-actinin (SAA, red) and nuclei (DAPI, blue). (B) Representative cardiobundle cross-sections stained for filamentous actin (F-act, green) and nuclei (blue). (C) Total and F-actin+ area of cardiobundle cross-sections (n = 8–10 cardiobundles per group from 4 cell isolations). (D) Cross-section stains from (B) without F-act shown. (E) Count of total and F-actin+ nuclei per cross-section (n = 8–10 cardiobundles per group from 4 cell isolations). (F) Higher magnification images of cross-sections stained for F-actin (red), Caveolin-3 (Cav3, green), and nuclei (blue). (G) Cross-sectional area of CMs (n = 4 cardiobundles per group from 2 cell isolations, with >25 CMs analyzed per cardiobundle). (H) F-actin+ area per nucleus (n = 8–10 cardiobundles per group from 4 cell isolations) *, p < 0.05 vs. static.
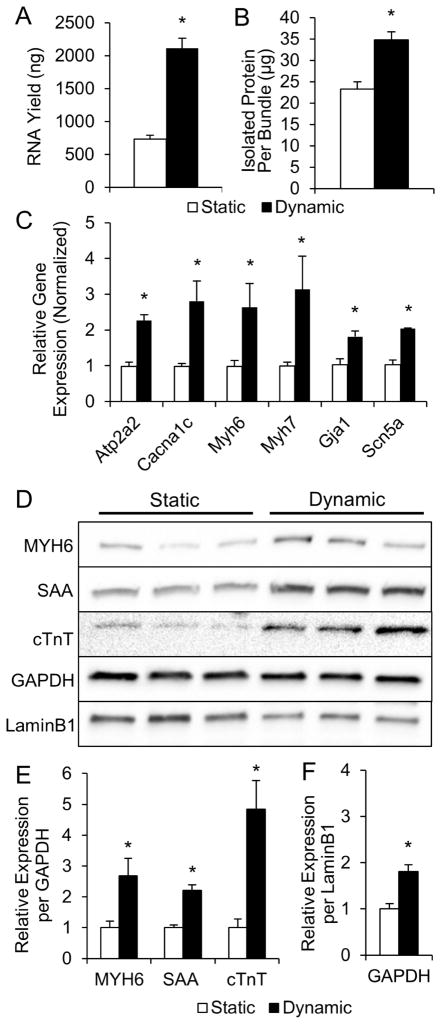
Consistent with immunostaining results, we found significant upregulation of total RNA (Fig. 2A) and protein (Fig. 2B) contents in dynamic cultures. This 3-fold higher RNA expression indicated higher transcriptional activity, a marker of cellular hypertrophy [38–40]. Furthermore, qPCR revealed upregulated expression of several genes coding for proteins governing cardiac electrical and contractile function, including SERCA pump, L-type Ca2+ channel, α- and β-MHC, gap junctional protein connexin-43, and fast sodium channel Nav1.5 (Fig. 2C, Supplementary Table S4). From Western blot analyses, the amount of several sarcomeric proteins (myosin heavy chain, sarcomeric α-actinin, cardiac troponin-T) per total cellular protein (GAPDH) was also higher in dynamic vs. static cultures (Fig. 2D–E). Consistent with the 2-fold larger immunostained CM area (Fig. 1E–G), the amount of total cellular protein per nucleus (GAPDH per Lamin B1 ratio) was ~2-fold higher in dynamically cultured cardiobundles (Fig. 2F), further indicating enhanced CM hypertrophy. Overall, these results indicated that dynamic culture significantly increased the myocardial mass within engineered NRVM tissues, primarily by increasing CM size and protein expression, and to a lesser extent by enhancing the survival, binucleation, or proliferation of CMs.
Figure 2. Molecular analysis of statically and dynamically cultured NRVM cardiobundles.
(A, B) RNA (A) and total protein (B) contents of cardiobundles cultured for 2 weeks under static or dynamic conditions (n = 9 samples per group from 3 cell isolations, each sample is 2–4 pooled cardiobundles). (C) Gene expression of cardiobundles (N = 3 cell isolations with 3 cardiobundles per group in each isolation). (D) Western blots of cardiobundles (N = 3 cell isolations with 3–4 cardiobundles per group in each isolation). (E) Quantification of protein expression from Western blots in (D) by densitometry normalized to GAPDH. (F) GAPDH normalized to LaminB1 estimates amount of total cellular protein per nucleus. *, p < 0.05 vs. static. Gene expression analyzed: SERCA (Atp2a2), L-type Ca2+ channel (Cacna1c), α- and β-myosin heavy chain (Myh6 and Myh7), connexin-43 (Gja1), sodium channel Nav1.5 (Scn5a). Protein expression analyzed: α-myosin heavy chain (MYH6), sarcomeric α-actinin (SAA), cardiac troponin-T (cTnT).
Dynamic culture improves electrophysiological function of NRVM cardiobundles
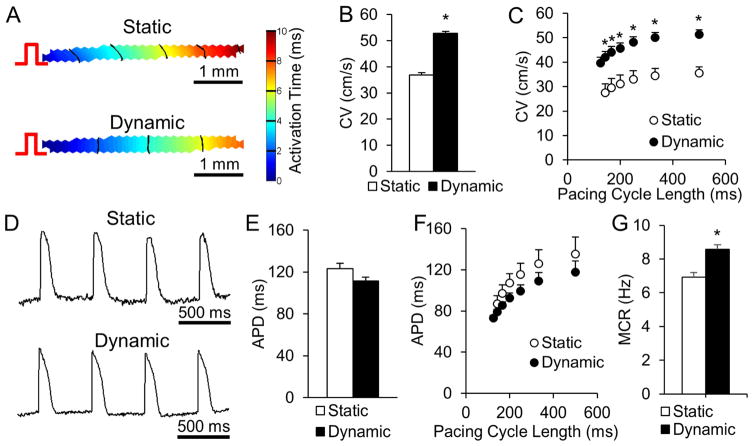
For assessment of electrical function, we paced cardiobundles at one end and applied a voltage-sensitive dye (Di-4-ANEPPS) and optical mapping to generate movies of action potential propagation (Supplemental Video 3). Both static and dynamic cardiobundles showed highly uniform propagation (Fig. 3A) allowing the precise measurement of conduction velocity (CV) along the bundle axis (Fig. S8). Consistent with qPCR results (Fig. 2C), dynamically cultured cardiobundles exhibited a 41% faster CV than statically cultured cardiobundles (52.5 ± 0.9 cm/s compared to 37.1 ± 1.8 cm/s, Fig. 3B). Both static and dynamic cardiobundles showed expected physiological slowing (restitution) of CV at higher pacing rates (Fig. 3C). Action potentials in static and dynamic cultures (Fig. 3D) had similar durations at basal pacing rate of 2 Hz (123 ± 10 ms vs. 115 ± 6 ms, Fig. 3E) and at faster pacing rates (Fig. 3F). Dynamic culture supported higher maximum capture rate (8.5 Hz) than static culture (6.9 Hz, Fig. 3G), approaching the heart rate in adult rats during exercise (9 Hz) [41].
Figure 3. Effects of static and dynamic culture on electrical function of NRVM cardiobundles.
(A) Representative isochrone maps of action potential (AP) propagation in response to 2 Hz point pacing (pulse sign) in cardiobundles cultured for 2 weeks under static or dynamic conditions. (B) Cardiobundle conduction velocity (CV) at a pacing rate of 2 Hz. (C) Dependence of CV on pacing period (cycle length) from 500 ms (2 Hz) to 125 ms (8Hz). (D) Representative optical action potentials recorded in cardiobundles during 2 Hz pacing. (E) AP duration (APD) measured from activation time to 80% repolarization during pacing at 2 Hz (for B and E, n = 43 cardiobundles per group from 9 cell isolations). (F) APD dependence on pacing rate (2–8 Hz). (G) Maximum capture rate (MCR) in cardiobundles (for C, F and G, n = 12 cardiobundles per group from 3 cell isolations). *, p < 0.05 vs. static.
Dynamic culture improves contractile function of NRVM cardiobundles
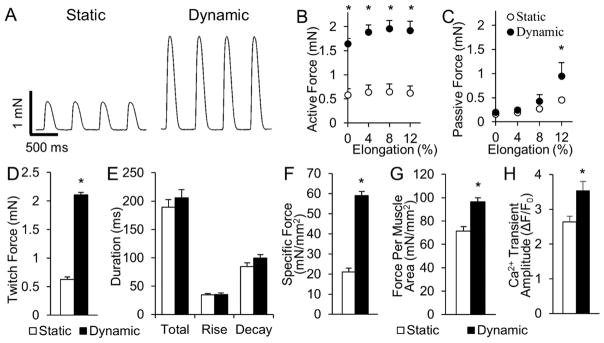
In addition to improved electrical function, dynamically cultured cardiobundles generated 3.5-fold higher isometric contractile force than statically cultured cardiobundles (2.10 ± 0.10 mN vs. 0.59 ± 0.09 mN, Fig. 4A,D), without a change in the duration or kinetics of twitch contractions (Fig. 4E). Both groups exhibited physiological force-length relationships and maximum contractile force when elongated 8% beyond culture length (Fig. 4B, Fig. S9). In agreement with histological findings (Fig. S7), dynamically cultured cardiobundles generated more passive force at 12% elongation (0.95 ± 0.28 mN vs. 0.45 ± 0.11 mN, Fig. 4C), consistent with increased tissue stiffness. Maximum specific force (contractile force normalized by total cardiobundle cross-sectional area) was increased from 21.8 ± 4.6 mN/mm2 in statically cultured to 59.7 ± 4.3 mN/mm2 in dynamically cultured cardiobundles (Fig. 4F). Contractile force normalized by muscle (F-actin+) cross-sectional area was less prominently increased (Fig. 4G), suggesting that the enhanced contractile function in dynamic culture resulted partially from the increase in myocardial mass alone. Moreover, we conducted fluorescence-based calcium imaging and found that, consistent with expression of calcium handling genes (Fig. 2C), calcium transient amplitude was higher in dynamically cultured cardiobundles (Fig. 4H), which additionally contributed to their increased contractile force.
Figure 4. Effects of static and dynamic culture on mechanical function of NRVM cardiobundles.
(A) Representative twitch force traces recorded from cardiobundles cultured for 2 weeks under static or dynamic conditions and paced at 2 Hz. (B–C) Active force (twitch amplitude, B) and passive tension (C) curves as a function of cardiobundle elongation (n = 6 cardiobundles per group from 3 cell isolations). (D) Maximum twitch forces recorded during 2 Hz pacing at optimal tissue elongation of 8% (n = 55–60 cardiobundles from 13 cell isolations). (E) Twitch kinetics, measured by total duration, rise time, and decay time of the twitch (n = 55–60 cardiobundles from 13 cell isolations). (F–G) Specific force (max twitch force normalized by total cardiobundle cross-section area, F) and muscle-only specific force (max twitch force normalized by muscle-only, F-actin+ area, G) (n = 17–18 bundles per group from 4 cell isolations). (H) Calcium transient amplitude measured as a relative change in GCaMP6 fluorescence signal (ΔF/F0) during 1 Hz pacing. (n = 11 cardiobundles per group from 3 cell isolations). *, p < 0.05 vs. static.
Switching from static to dynamic culture rescues function of NRVM cardiobundles
Since inferior function of 14-day static cardiobundles was primarily a result of smaller cell size rather than cell death, we investigated if the effects of static vs. dynamic culture were reversible and could be manipulated to study underlying mechanisms (experimental flow shown in Fig. S10). We began with cardiobundles cultured for 7 days that had inferior properties to their 14-day counterparts (Fig. 5). The day 7 cardiobundles cultured in dynamic conditions showed significantly higher forces (Fig. 5A), faster CVs (Fig. 5B) and more cross-sectional myocardial area (Fig. 5C–D) compared to age-matched static cardiobundles. When the day 7 static cardiobundles were switched to dynamic culture for the next 7 days (denoted as stat→dyn), they exhibited a remarkable 5.4-fold increase in contractile force, nearing the force of cardiobundles cultured dynamically (dyn) for the entire 14-day period (1.98 ± 0.09 mN for stat→dyn vs. 2.29 ± 0.10 mN for dyn group, Fig. 5A). A similar trend was observed for the conduction velocity, although stat→dyn cardiobundles displayed intermediate CVs (stat: 35.3 ± 1.8 cm/s; stat→dyn: 44.9 ± 3.2 cm/s; dyn: 54.5 ± 2.2 cm/s, Fig. 5B) instead of a near-full recovery as seen for contractile function. Conversely, day 7 dynamic cardiobundles that were switched to static culture for the next 7 days (denoted as dyn→stat) exhibited a decrease in contractile force and CV to levels comparable with those of cardiobundles cultured statically the entire 14 days (Fig. 5A,B). Cross-sectional stainings of cardiobundles revealed increase in myocardial area when static cultures were switched to dynamic, and decrease in myocardial area in dyn→stat group (Fig. 5C–D). These changes in myocardial area were mostly due to differences in cell size rather than CM number (Fig. 5E–F). Interestingly, dyn→stat cardiobundles had a similar cross-sectional myocardial area as stat cardiobundles, but CMs were packed much more tightly (Fig. 5C–D). This finding suggests that CM hypertrophy in dynamic culture was associated with irreversible remodeling or loss of ECM, such that the switch to static conditions caused compaction of atrophied CMs and decrease of the cardiobundle size.
Figure 5. Effects of switch between static and dynamic culture on structure and function of NRVM cardiobundles.
(A–B) Max twitch force (A) and CV (B) at 2 Hz pacing in static (Stat) and dynamic (Dyn) cardiobundles on culture day 7 and 14, cardiobundles cultured static for 7 days then dynamic for 7 days (Stat→Dyn), and cardiobundles cultured dynamic for 7 days then static for 7 days (Dyn→Stat) (n = 12–14 cardiobundles per group from 3 cell isolations). (C) Representative cross-sections of cardiobundles stained for F-actin (green) and nuclei (DAPI, blue). (D–F) Quantification of D) total and F-actin+ area, E) total and F-actin+ nuclei, and F) F-actin+ area per F-actin+ nucleus in cardiobundle cross-sections (n = 5–7 cardiobundles per group from 2 cell isolations). *, p < 0.05 vs. static at same culture point; #, p < 0.05 vs. dynamic at the same culture point.
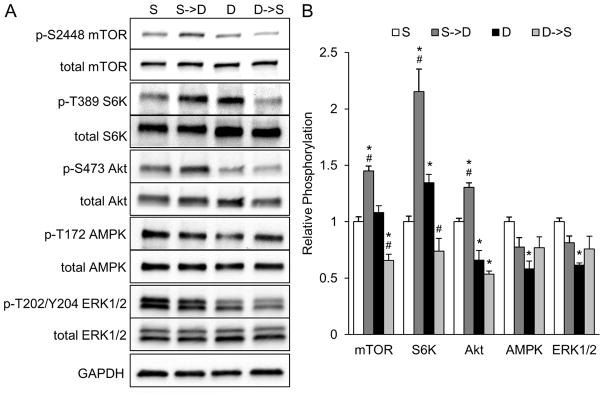
mTOR signaling mediates response of CMs to dynamic culture
Mammalian target of rapamycin (mTOR) is a protein kinase that serves as a master regulator of protein synthesis and cell growth based on availability of nutrients [42, 43]. Thus, we studied the role of mTOR signaling in the structural and functional response of cardiobundles to dynamic culture. Specifically, we analyzed activity of multiple mTOR-related kinases in stat, stat→dyn, dyn, and dyn→stat cardiobundles on day 8 (Fig. S10). This experimental strategy enabled analysis of both transient signaling effects immediately (24 hours) after switching culture conditions, and sustained effects of chronic dynamic culture (> 7 days after initiation). Phosphorylation of mTOR and ribosomal S6 kinase (S6K), which is the downstream effector of mTOR complex 1 (mTORC1), were both increased after switching from static to dynamic culture, and conversely decreased after switching from dynamic to static culture (Fig. 6A–B). mTORC1 activity, measured by S6K phosphorylation, was elevated in chronic dynamic culture relative to chronic static culture. Akt phosphorylation, a positive regulator of mTORC1, was increased upon switching to dynamic culture but, interestingly, decreased in chronic dynamic culture. AMPK, a negative regulator of mTORC1, exhibited reduced activity in chronic dynamic culture relative to chronic static culture. We also assessed if differences in mechanical environment between dynamic and static culture were responsible for the observed effects in cardiobundles. However, ERK1/2 phosphorylation, which is upregulated in CMs in response to mechanical stress (e.g. cyclic stretch [44, 45], pulsatile interstitial flow [46]), was not increased by either transient or chronic dynamic culture, and was rather decreased by chronic dynamic culture (Fig. 6A–B).
Figure 6. Studies of nutrient and mechanical stress related mediators of mTORC1 signaling in NRVM cardiobundles.
(A) Representative Western blots for phosphorylated (Ser2448) and total mTOR, phosphorylated (Thr389) and total S6K, phosphorylated (Ser473) and total Akt, phosphorylated (Thr172) and total AMPK, phosphorylated (Thr202/Tyr204) and total ERK1/2, and GAPDH (loading control) at 8 days of static (S) or dynamic (D) culture or 24 hours after switching from static to dynamic culture (S→D) or from dynamic to static culture (D→S) at culture day 7. (B) Quantified ratio of phosphorylated/total protein for each kinase in (A), normalized to static (S) group (n = 4 samples per group from 2 cell isolations, each sample is lysate of 4 cardiobundles pooled together). *, p < 0.05 vs. static; #, p < 0.05 vs. dynamic.
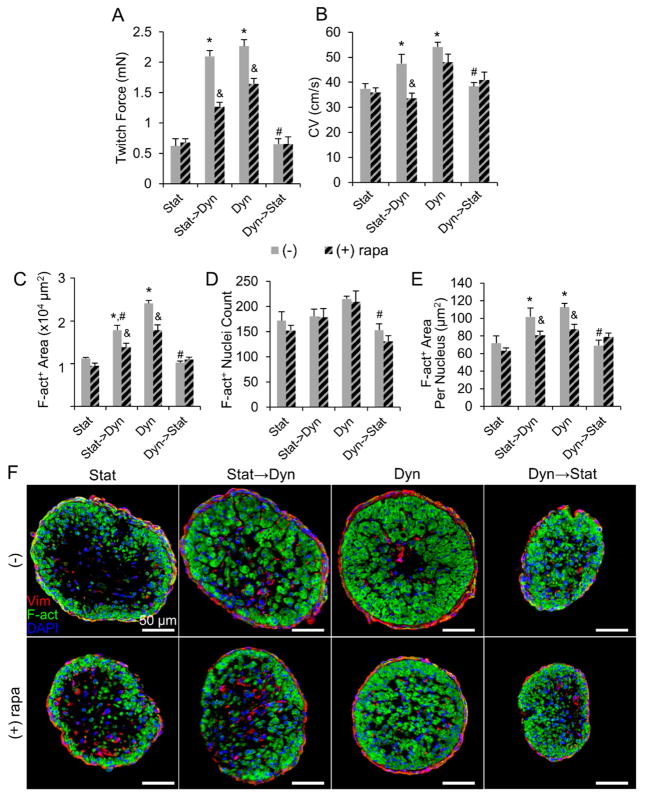
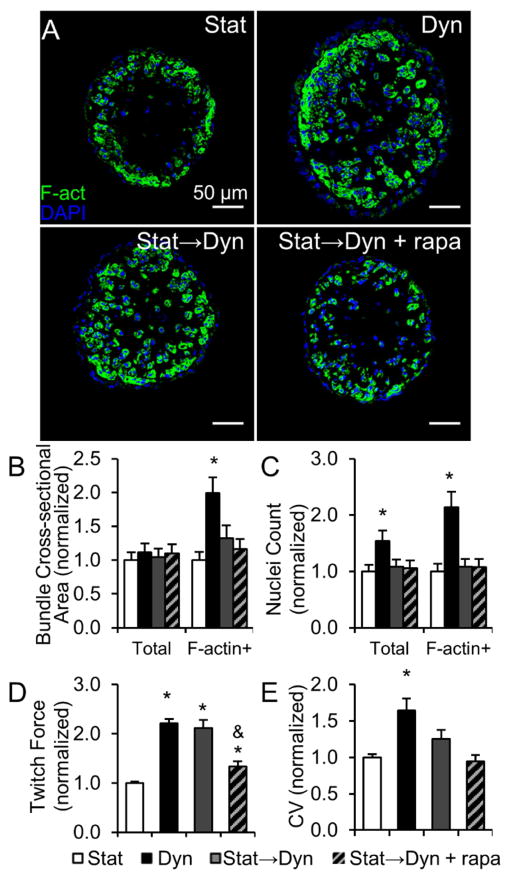
Inhibition of mTORC1 with rapamycin significantly reduced the contractile force increase between 7 and 14 days of dynamic culture in both stat→dyn group (2.10 ± 0.10 mN without rapamycin, 1.27 ±0.07 mN with rapamycin) and dyn group (2.27 ± 0.11 mN without rapamycin, 1.64 ± 0.09 mN with rapamycin, Fig. 7A). Rapamycin decreased the CV of stat→dyn cardiobundles (from 47.4 ± 3.7 cm/s to 33.6 ± 2.0 cm/s, Fig. 7B), but had no significant effect on cardiobundles dynamically cultured for 14 days. These functional changes exerted by rapamycin on dynamically cultured cardiobundles were associated with a decrease in cross-sectional muscle area (Fig. 7C) and no effect on muscle nuclei count (Fig. 7D) compared to drug-free controls. Quantification of muscle area per nucleus (Fig. 7E) and higher magnification cross-sectional images (Fig. S12) further suggested that rapamycin in dynamic cultures acted by preventing CM hypertrophy. In contrast to dynamic cultures, rapamycin had no effect on contractile force, CV, cross-sectional muscle area, or nuclei number in dyn→stat or static cultures (Fig. 7A–F). Furthermore, inhibition of mechanosensitive channels with effective doses of streptomycin or gadolinium [47] did not attenuate the functional improvements by dynamic culture (Fig. S11). Along with no increase in ERK1/2 phosphorylation (Fig. 6A–B), this result suggested increased nutrient availability rather than mechanical stimulation as the main trigger for mTORC1-mediated effects of dynamic culture.
Figure 7. Effects of mTORC1 inhibition on structure and function of NRVM cardiobundles.
(A–B) Max twitch forces (A) and CV (B) in cardiobundles switched from static to dynamic culture (Stat→Dyn) or from dynamic to static culture (Dyn→Stat) at culture day 7. (Stat) and (Dyn) denote static- and dynamic-only culture for 14 days. Cardiobundles were cultured with or without the mTORC1 inhibitor rapamycin (500 nM, rapa, diagonally patterned bars) (n = 10–13 cardiobundles per group from 3 cell isolations). (C–E) F-actin+ area (C), nuclei count (D) and F-actin+ area/nuclei (E) in cardiobundle cross-sections (n = 6–8 cardiobundles per group from 2 cell isolations). (F) Representative cross-sections stained for vimentin (Vim, red), F-actin (F-act, green), and nuclei (DAPI, blue). *, p < 0.05 vs. static at the same culture point; #, p < 0.05 vs. dynamic at the same culture point; &, p < 0.05 vs. corresponding non-rapamycin control.
Effects of dynamic culture on cardiobundles differ from pathological cardiac hypertrophy
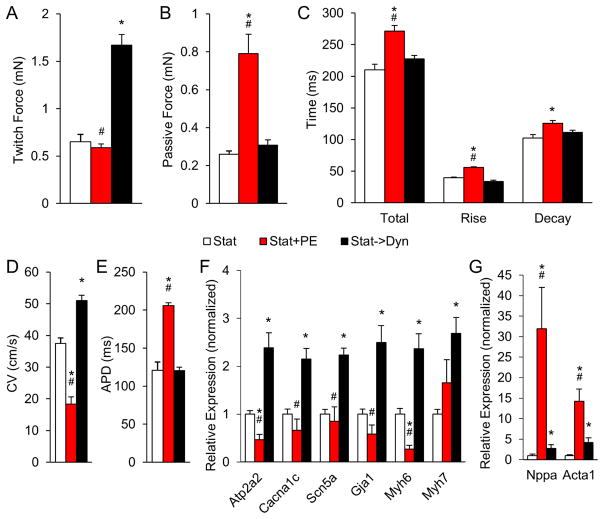
We also investigated if dynamically cultured cardiobundles underwent changes characteristic of the pathological cardiac hypertrophy. Specifically, molecular and functional effects of dynamic culture were compared with traditional model of pathological hypertrophy, treatment of CMs with phenylephrine (PE) [48, 49]. Day 7 statically cultured cardiobundles were exposed for additional 7 days to dynamic culture or 20 μM PE. In contrast to dynamic culture, PE selectively increased passive force rather than active force, prolonged both APD and twitch kinetics, and significantly slowed CV (Fig. 8A–E). Furthermore, PE had no effect or decreased the expression of cardiac functional genes improved by dynamic culture, instead inducing a disproportionate increase in expression of pathological hypertrophy markers atrial natriuretic peptide (ANP) and α-skeletal actin (Fig. 8F, Table S4). Collectively, these results suggested that dynamic culture induced physiological growth and functional maturation of CMs akin to postnatal cardiac development rather than adult pathological hypertrophy.
Figure 8. Comparison of effects of dynamic culture and phenylephrine treatment on function and gene expression of NRVM cardiobundles.
(A–B) Twitch force (A) and passive force (B) at 8% elongation of NRVM cardiobundles cultured statically for the entire 14 days (Stat) or treated after first 7 days of static culture with either 20 μM phenylephrine (Stat+PE) or dynamic culture (Stat→Dyn) for the following 7 days. (C) Twitch kinetics, measured by total duration (Total), rise time (Rise), and decay time (Decay) of the twitch. (D–E) Conduction velocity (CV, D) and action potential duration (APD, E) of cardiobundles during 2 Hz point stimulation. (F–G) Fold change in expression of indicated genes (Table S4) relative to static (Stat) group (n = 7–10 cardiobundles per group from 2 cell isolations for each figure panel). *, p < 0.05 vs. Stat, #, p < 0.05 vs. Stat→Dyn.
Dynamic culture induces mTOR-dependent functional improvement of hPSC-CM cardiobundles
We further studied cardiobundles made of hPSC-CMs and found consistent responses to dynamic versus static culture with those observed in NRVM tissues. Specifically, under optimized conditions, hPSC-CMs in 14-day dynamically cultured cardiobundles were aligned, cross-striated, and robustly interconnected by electrical and mechanical junctions (Fig. S13A–C). They exhibited better structural organization compared to statically cultured cardiobundles (Fig. 9A), but still inferior cell elongation and alignment compared to NRVMs (Fig. 1A). From transverse cross-sections (Fig. 9B), dynamically cultured hPSC-CM cardiobundles exhibited a 1.2-fold and 2.3-fold higher total and myocardial (F-actin+) area (Fig. 9C), respectively, and a 1.6-fold and 2.1-fold higher total and myocardial nuclei numbers (Fig. 9D), compared to static cardiobundles. However, in contrast to NRVMs, hPSC-CM size in static and dynamic cultures was not different (Fig. 9E).
Figure 9. Effects of static and dynamic culture on structure and function of hPSC-CM cardiobundles.
(A–B) Representative longitudinal (A) and cross-sectional (B) images of human cardiobundles cultured for 2 weeks under static (Stat) or dynamic (Dyn) conditions, stained for sarcomeric α-actinin (SAA, red), F-actin (green), and nuclei (DAPI, blue). (C–E) Quantification of the total and F-actin+ cross-sectional area (C) and nuclei counts (D), and cross-sectional area per F-actin+ nuclei (E) in human cardiobundles (n = 24–26 cardiobundles per group from 7 differentiations). (F) Representative isochrone maps of action potential propagation in human cardiobundles in response to 1 Hz point pacing (pulse sign). (G–H) Quantification of conduction velocity (CV, G) and AP duration (APD, H) (n = 31–38 cardiobundles per group from 10 differentiations). (I) Ca2+ transient amplitude measured as a relative change in GCaMP6 fluorescence signal (ΔF/F0) during 1 Hz pacing (n = 10–14 cardiobundles per group from 3 differentiations). (J) Representative twitch force traces recorded from human cardiobundles during 1.5 Hz stimulation. (K–L) Quantification of max twitch force (K), specific force (per total cardiobundle cross-sectional area, L), and force kinetics (M) in human cardiobundles. (n = 23–33 cardiobundles per group from 6–9 differentiations). *, p < 0.05 vs. static culture.
Furthermore, while both statically and dynamically cultured hPSC-CM cardiobundles supported continuous and uniform action potential propagation (Fig. 9F, Supplemental Video 4), dynamic culture yielded a 1.4-fold increase in CV compared to static culture (25.8±1.2 cm/s vs. 18.2±1.2 cm/s, Fig. 9G) without significant change in APD (Fig. 9H). Dynamically cultured cardiobundles also demonstrated a 1.6-fold increase in Ca2+ transient amplitude relative to static group suggesting improved CM maturation (Fig. 9I). In response to electrical stimulation, hPSC-CM cardiobundles exhibited forceful contractions (Fig. 9J) and demonstrated a physiological force-length relationship (Fig. S13D–E). Similar to findings in NRVM cardiobundles, compared to static culture, dynamic culture of hPSC-CM cardiobundles yielded a 2.5-fold higher active force (1.3 ± 0.051 mN vs. 0.52 ± 0.027 mN, Fig. 9K) and a 1.9-fold increase in specific force (23.2 ± 1.6 mN/mm2 vs. 12.2 ± 1.0 mN/mm2, Fig. 9L), further suggesting enhanced CM maturation. The total twitch time and rise time both slightly but significantly decreased, while twitch decay time remained unchanged (Fig. 9M). We then applied the same dynamic culture method to produce human cardiobundles from hPSC-CMs that were differentiated using 2 distinct protocols and 3 different hPSC lines, and obtained similarly advanced cardiobundle function with maximum achieved forces > 2 mN, specific forces of > 30 mN/mm2 and CVs of > 40 cm/sec (Fig. S13F–I).
Finally, hPSC-CM cardiobundles cultured under static conditions for 7 days were switched to dynamic culture, with or without addition of rapamycin, for an additional week prior to histological and functional assessment. From the cross-sectional images (Fig. 10A), we found that switching from static to dynamic conditions had no significant effects on the total and myocardial (F-actin+) areas or nuclei counts (Fig. 10B); however, it yielded a 2.3-fold higher contractile force and a trend towards higher CV compared to static group (Fig. 10D–E). Addition of the mTORC1 inhibitor rapamycin after switching to dynamic culture reduced force production to levels not statistically different from those measured in static cultures (Fig. 10D), suggesting that similar to NRVMs, dynamic cardiobundle culture promoted functional maturation of human CMs and that this effect was at least in part mediated by mTOR signaling.
Figure 10. Effects of switch between static and dynamic culture on structure and function of hPSC-CM cardiobundles.
(A) Representative cross-section images of human cardiobundles cultured for 2 weeks in static or dynamic conditions stained with F-actin (green) and nuclei (DAPI, blue). (B–C) Quantification of areas (B) and nuclei counts (C) from cross-sections of human cardiobundles shown relative to static group (n = 14–17 cardiobundles per group from 4 differentiations). (D–E) Quantification of twitch force amplitude (D) and CV (E) in human cardiobundles relative to static group (n > 25 cardiobundles per group from 6–7 differentiations). *, p < 0.05 vs. static; &, p < 0.05 vs. corresponding non-rapamycin group.
DISCUSSION
In this study, we described a versatile approach for engineering highly cellular rat and human cardiac tissues (“cardiobundles”) capable of rapid action potential conduction and strong force generation. In contrast to traditional static culture of hydrogel-based engineered heart tissues [25, 50–56], this method relies on the application of free-floating dynamic culture conditions that lead to upregulation of mTORC1 signaling and robust structural and functional improvements associated with increased cardiomyocyte density, hypertrophy, and maturation. Reproducibility of the increased functional output (across different species, hPSC lines and methods of hPSC-CM differentiation) and the simplified, cost-effective nature of the system (reusable PDMS molds, no specialized bioreactors or exogenous electromechanical stimulation) offer an easy-to-implement platform for production of highly functional engineered cardiac tissues for various in vitro and, potentially, in vivo applications.
One of the unique features of this system is the use of laser-cut, porous polymer frames to provide anchorage and mechanical tension to fibrin-based tissue, while simultaneously enabling free-floating, dynamic conditions to improve mass transport around metabolically demanding cardiomyocytes. The final cardiobundle diameter (Fig. 1B, Fig. 9B) is comparable to that of trabeculae from adult rat (40 – 360 μm) [57–59] and human (< 1mm) [60] hearts, which are the standard cardiac tissue preparations for ex vivo studies of myocardial function. Optimized cardiobundles require relatively low cell number (3.75 × 105), have dense and uniformly aligned cardiomyocytes throughout the tissue volume, and allow accurate quantification of cellular composition and architecture in transverse tissue cross-sections. Importantly, the use of anchoring frames facilitates transfer and mounting of cardiobundles to allow rapid assessments of both electrical (optical mapping) and mechanical (isometric force testing) function.
Consistent with previous studies that applied other forms of dynamic culture [15–18, 20, 61, 62], free-floating dynamic culture of cardiobundles yielded significantly increased myocardial mass and expression of contractile proteins and functional genes compared to static culture (Fig. 2). For neonatal rat CMs, the larger myocardial mass of cardiobundles was contributed by a slightly (1.2-fold) higher CM nuclei number and substantially (2.1-fold) increased CM cross-sectional area (Fig. 1C,E–H). Conversely, dynamically cultured human cardiobundles exhibited a 2.1-fold higher CM nuclei number (via enhanced cell survival, binucleation, or proliferation) compared to static cultures, without significant change in CM size (Fig. 9D–E). Furthermore, compared to static culture, dynamic culture of NRVM cardiobundles had proportionally increased number of both CMs and non-CMs without altering the CM to non-CM ratio (~3:1) over 14-day period (Fig. 5E). Moreover, the total NRVM number in either static or dynamic fibrin-based cardiobundles remained relatively constant with time of culture (Fig. 5E). The observed stability in cell number and cell population is in contrast to a previous report where NRVMs underwent continuous apoptotic death over 12 days of collagen gel culture, yielding a ~8 fold decrease in the CM number and the final CM to non-CM ratio of ~1:4 [1].
Unlike previous studies, we also compared statically and dynamically cultured engineered cardiac tissues for their action potential conduction and isometric force generation. The average CV of 52.5 cm/s (range of 41.9 – 61.1 cm/s) achieved in dynamically cultured NRVM cardiobundles is the highest reported in the field, significantly higher than the CV of neonatal rat myocardium (22 – 27 cm/s) [19, 63], but still lower than the longitudinal CV of adult rat myocardium (66 – 69 cm/s)[64, 65]. Faster conduction in dynamic vs. static culture could be contributed to increase in CM size [66] and gene expression of Cx43 (Gja1) and fast Na+ current (SCN5A) (Fig. 2C). Similarly, the average CV in dynamically cultured hPSC-CM cardiobundles (25.8 cm/s, range of 15.0 – 42.1 cm/s) is among the highest reported in the field [23, 51, 67]. Dynamic culture also robustly increased contractile force generation of rat and human cardiac tissues by 3.5-fold and 2.5-fold respectively, resulting in average specific forces of 59.7 mN/mm2 (range 43.8 – 70.1 mN/mm2) and 23.2 mN/mm2 (range 16.2 – 32.1 mN/mm2), i.e., the highest reported in the field, and in the case of 3D NRVM tissues, the first time matching those of adult rat ventricular myocardium (40 – 71 mN/mm2) [59, 68, 69]. However, achieving adult-level specific force in thicker patches suitable for cardiac repair will be significantly more challenging. The higher contractile force per CM area (Fig. 4G), Ca2+ transient amplitude (Fig. 4H, 9I), maximum capture rate (Fig. 3G), and cellular expression of functional genes and sarcomeric proteins (Fig. 2C–E), all suggested improved CM maturation in dynamically cultured cardiobundles, although CMs still exhibited certain immature features including relatively small size and diffuse expression of Cx43 (Fig. S2B).
In the heart, mTOR signaling is required for postnatal cardiomyocyte growth and is generally associated with physiological hypertrophy [70, 71]. Active mTOR complex 1 (mTORC1) increases protein synthesis from mRNA by phosphorylation of S6 kinase and inhibition of 4E binding protein (4E-BP1), while also activating transcription factors such as STAT3 to increase mRNA synthesis [72, 73]. Despite the known role of mTOR in coupling nutrient availability to cellular growth, it has not been studied in the context of improving nutrient delivery to engineered 3D tissues via enhanced mass transport. In the present study, dynamic culture increased mTORC1 activity in cardiobundles via a transient activation of Akt and a sustained suppression of AMPK activity (Fig. 6), and inhibition of mTORC1 with rapamycin attenuated the effects of dynamic culture (Fig. 7). Akt is activated by insulin or IGF1 signaling and is known to promote physiological CM hypertrophy [74–77], suggesting that dynamic culture improves delivery of serum-derived nutrients and growth factors to cells in engineered tissues. The reduced Akt activation in chronic dynamic culture, which is likely due to negative feedback of S6K on mTORC2, PI3K, and insulin receptors [78–80], could mirror previously observed biphasic responses of Akt to mTORC1 modulation [78, 81] and prevent pathological consequences of constitutively active Akt in the heart [76]. Since PI3K also stimulates ERK1/2 signaling through several mechanisms [82], the negative feedback of S6K on PI3K likely explains the suppression of ERK1/2 in dynamic culture (Fig. 6). Another mTOR signaling mediator, AMPK, is activated by accumulation of intercellular AMP, typically when the cell is deprived of metabolic substrates, and inhibits mTORC1 activity to conserve energy. The suppression of AMPK by dynamic culture could be a result of improved availability of oxygen (Fig. S6) and glucose to cardiomyocytes. Interestingly, chronic static and chronic dynamic cultures exhibited equal Ser2448 phosphorylation of mTOR, which is present in both mTORC1 and mTORC2 [83]. The elevated mTORC1 activity (S6K phospho-Thr389) in dynamic culture and elevated mTORC2 activity (Akt phospho-Ser473) [84] in static culture generally suggest that dynamic culture facilitates growth signaling, as opposed to survival signaling in static culture [85, 86]. The most important changes in mTORC1-related signaling by dynamic culture are summarized in Fig. S14.
It is well known that CM hypertrophy is the main contributor to postnatal heart growth [87, 88]. The average diameter of day 14 neonatal rat CMs in this study was increased from approximately 6.0 μm in static culture to 9.1 μm in dynamic culture (estimated from CM areas in Fig. 1G), which was comparable to CM diameter of P12 rats (9–10 μm [1, 89]) yet significantly smaller than adult CM diameter (18–24 μm [1, 89–91]). This suggests that the increased size of neonatal CMs in response to dynamic culture mirrors normal developmental growth rather than a pathological response with abnormally large CMs. To further assess physiological vs. pathological hypertrophy, we directly compared effects of dynamic culture to treatment with the α-adrenergic agonist phenylephrine, a known in vitro and in vivo inducer of pathological hypertrophy [48, 49, 92, 93]. Unlike dynamic culture, PE failed to improve contractile force and expression of several cardiac genes, instead causing increased passive tension, decreased CV, prolonged APD, significant upregulation of ANP and α-skeletal actin, and selective decrease in α-MHC with trend of higher β-MHC (Fig. 8). Collectively, these results indicate that dynamic culture promotes normal growth and maturation rather than causing functional decline and re-activation of fetal genes classically associated with adult pathology [76, 77]. The increased activity of mTORC1, but not ERK1/2, further supported that CM growth induced by dynamic culture was physiological rather than pathological [77]. Finally, we substantiate this conclusion by comparing the molecular and functional changes caused by dynamic culture, PE treatment, postnatal cardiac development, and pathological cardiac hypertrophy in Table S6.
In summary, we have developed a method to engineer trabeculae-sized cardiac tissues that consistently generate near-adult levels of contractile force and conduction velocity without requiring application of electrical stimulation or mechanical stretch. A critical aspect of the high cellularity and function of cardiobundles is dynamic culture, which was also a stimulus for enhanced maturation of CMs from both neonatal rats and human pluripotent stem cells. The involvement of mTOR signaling as a mediator of the dynamic culture effects could have broader implications for tissue engineering field. In addition to yielding a realistic in vitro model of miniature cardiac muscle, this approach may provide a framework to scale up size of engineered tissues for use in myocardial repair in vivo.
Supplementary Material
Acknowledgments
A. Krol, L. Li, and Y. Li for assistance with NRVM isolation and culture; I. Shadrin for assistance with hPSC-CM differentiation and culture; S. Earp for use of laser cutter; C. Gersbach and P. Thakore for use of Western blot imaging system; J. Merritt of Cerex Advanced Fabrics for material samples. The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Funding: Grants HL104326 and HL132389 from National Heart, Lung, and Blood Institute; Grant UH3TR000505 from the NIH Common Fund for the Microphysiological Systems Initiative; Grant from the Foundation Leducq; NSF Graduate Research Fellowship to C. Jackman.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tiburcy M, Didié M, Boy O, Christalla P, Döker S, Naito H, et al. Terminal differentiation, advanced organotypic maturation, and modeling of hypertrophic growth in engineered heart tissue. Circulation Research. 2011;109:1105–14. doi: 10.1161/CIRCRESAHA.111.251843. [DOI] [PubMed] [Google Scholar]
- 2.Liau B, Zhang D, Bursac N. Functional cardiac tissue engineering. Regenerative Medicine. 2012;7:187–206. doi: 10.2217/rme.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirt MN, Werner T, Indenbirken D, Alawi M, Demin P, Kunze A-C, et al. Deciphering the microrna signature of pathological cardiac hypertrophy by engineered heart tissue- and sequencing-technology. Journal of Molecular and Cellular Cardiology. 2015;81:1–9. doi: 10.1016/j.yjmcc.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of barth syndrome with ipsc and heart-on-chip technologies. Nature medicine. 2014;20:616–23. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson SA, Copeland CR, Reich DH, Tung L. Mechanical coupling between myofibroblasts and cardiomyocytes slows electric conduction in fibrotic cell monolayers. Circulation. 2011;123:2083–U71. doi: 10.1161/CIRCULATIONAHA.110.015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eder A, Hansen A, Uebeler J, Schulze T, Neuber C, Schaaf S, et al. Effects of proarrhythmic drugs on relaxation time and beating pattern in rat engineered heart tissue. Basic Res Cardiol. 2014;109:1–15. doi: 10.1007/s00395-014-0436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, et al. Human ipsc-based cardiac microphysiological system for drug screening applications. Scientific Reports. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eschenhagen T, Eder A, Vollert I, Hansen A. Physiological aspects of cardiac tissue engineering. American Journal of Physiology - Heart and Circulatory Physiology. 2012;303:H133–H43. doi: 10.1152/ajpheart.00007.2012. [DOI] [PubMed] [Google Scholar]
- 9.Jackman CP, Shadrin IY, Carlson AL, Bursac N. Human cardiac tissue engineering: From pluripotent stem cells to heart repair. Current Opinion in Chemical Engineering. 2015;7:57–64. doi: 10.1016/j.coche.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirt MN, Hansen A, Eschenhagen T. Cardiac tissue engineering: State of the art. Circulation Research. 2014;114:354–67. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Pabon L, Murry CE. Engineering adolescence: Maturation of human pluripotent stem cell–derived cardiomyocytes. Circulation Research. 2014;114:511–23. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh PCH, Davis ME, Lisowski LK, Lee RT. Endothelial-cardiomyocyte interactions in cardiac development and repair. Annual review of physiology. 2006;68:51–66. doi: 10.1146/annurev.physiol.68.040104.124629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine K, Kagawa Y, Maeyama E, Ota H, Haraguchi Y, Matsuura K, et al. Oxygen consumption of human heart cells in monolayer culture. Biochemical and Biophysical Research Communications. 2014;452:834–9. doi: 10.1016/j.bbrc.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Kagawa Y, Matsuura K, Shimizu T, Tsuneda S. Direct measurement of local dissolved oxygen concentration spatial profiles in a cell culture environment. Biotechnology and Bioengineering. 2015;112:1263–74. doi: 10.1002/bit.25531. [DOI] [PubMed] [Google Scholar]
- 15.Vollert I, Holnthoner W, Redl H, Reichenspurner H, Hansen A, Eschenhagen T, et al. In vitro perfusion of engineered heart tissue through endothelialized channels. Tissue engineering Part A. 2014;20:854–63. doi: 10.1089/ten.TEA.2013.0214. [DOI] [PubMed] [Google Scholar]
- 16.Radisic M, Yang L, Boublik J, Cohen RJ, Langer R, Freed LE, et al. Medium perfusion enables engineering of compact and contractile cardiac tissue. American Journal of Physiology-Heart and Circulatory Physiology. 2004;286:H507–H16. doi: 10.1152/ajpheart.00171.2003. [DOI] [PubMed] [Google Scholar]
- 17.Dvir T, Levy O, Shachar M, Granot Y, Cohen S. Activation of the erk1/2 cascade via pulsatile interstitial fluid flow promotes cardiac tissue assembly. Tissue Engineering. 2007;13:2185–93. doi: 10.1089/ten.2006.0364. [DOI] [PubMed] [Google Scholar]
- 18.Carrier RL, Papadaki M, Rupnick M, Schoen FJ, Bursac N, Langer R, et al. Cardiac tissue engineering: Cell seeding, cultivation parameters, and tissue construct characterization. Biotechnology and Bioengineering. 1999;64:580–9. doi: 10.1002/(sici)1097-0290(19990905)64:5<580::aid-bit8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 19.Bursac N, Papadaki M, Cohen RJ, Schoen FJ, Eisenberg SR, Carrier R, et al. Cardiac muscle tissue engineering: Toward an in vitro model for electrophysiological studies. American Journal of Physiology - Heart and Circulatory Physiology. 1999;277:H433–H44. doi: 10.1152/ajpheart.1999.277.2.H433. [DOI] [PubMed] [Google Scholar]
- 20.Papadaki M, Bursac N, Langer R, Merok J, Vunjak-Novakovic G, Freed LE. Tissue engineering of functional cardiac muscle: Molecular, structural, and electrophysiological studies. American journal of physiology Heart and circulatory physiology. 2001;280:H168–78. doi: 10.1152/ajpheart.2001.280.1.H168. [DOI] [PubMed] [Google Scholar]
- 21.Vunjak-Novakovic G, Tandon N, Godier A, Maidhof R, Marsano A, Martens TP, et al. Challenges in cardiac tissue engineering. Tissue Engineering Part B: Reviews. 2009;16:169–87. doi: 10.1089/ten.teb.2009.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang BY, et al. Biowire: A platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781–7. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D, Shadrin IY, Lam J, Xian H-Q, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human esc-derived cardiomyocytes. Biomaterials. 2013;34:5813–20. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godier-Furnémont AFG, Tiburcy M, Wagner E, Dewenter M, Lämmle S, El-Armouche A, et al. Physiologic force-frequency response in engineered heart muscle by electromechanical stimulation. Biomaterials. 2015;60:82–91. doi: 10.1016/j.biomaterials.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circulation research. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bian W, Liau B, Badie N, Bursac N. Mesoscopic hydrogel molding to control the 3d geometry of bioartificial muscle tissues. Nature protocols. 2009;4:1522–34. doi: 10.1038/nprot.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito H, Melnychenko I, Didié M, Schneiderbanger K, Schubert P, Rosenkranz S, et al. Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation. 2006;114:I72–I8. doi: 10.1161/CIRCULATIONAHA.105.001560. [DOI] [PubMed] [Google Scholar]
- 28.Juhas M, Bursac N. Roles of adherent myogenic cells and dynamic culture in engineered muscle function and maintenance of satellite cells. Biomaterials. 2014;35:9438–46. doi: 10.1016/j.biomaterials.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical wnt signaling. Proc Natl Acad Sci U S A. 2012;109:1848–57. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–37. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Meth. 2014 doi: 10.1038/nmeth.2999. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McSpadden LC, Kirkton RD, Bursac N. Electrotonic loading of anisotropic cardiac monolayers by unexcitable cells depends on connexin type and expression level. Am J Physiol-Cell Physiol. 2009;297:C339–C51. doi: 10.1152/ajpcell.00024.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bian W, Jackman CP, Bursac N. Controlling the structural and functional anisotropy of engineered cardiac tissues. Biofabrication. 2014;6:024109. doi: 10.1088/1758-5082/6/2/024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. eLife. 2015;4:e04885. doi: 10.7554/eLife.04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liau B, Christoforou N, Leong KW, Bursac N. Pluripotent stem cell-derived cardiac tissue patch with advanced structure and function. Biomaterials. 2011;32:9180–7. doi: 10.1016/j.biomaterials.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schödel J, Klanke B, Weidemann A, Buchholz B, Bernhardt W, Bertog M, et al. Hif-prolyl hydroxylases in the rat kidney: Physiologic expression patterns and regulation in acute kidney injury. The American Journal of Pathology. 2009;174:1663–74. doi: 10.2353/ajpath.2009.080687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Appelhoff RJ, Tian Y-M, Raval RR, Turley H, Harris AL, Pugh CW, et al. Differential function of the prolyl hydroxylases phd1, phd2, and phd3 in the regulation of hypoxia-inducible factor. Journal of Biological Chemistry. 2004;279:38458–65. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 38.Schäfer M, Pönicke K, Heinroth-Hoffmann I, Brodde O-E, Piper HM, Schlüter K-D. Beta-adrenoceptor stimulation attenuates the hypertrophic effect of alpha-adrenoceptor stimulation in adult rat ventricular cardiomyocytes. Journal of the American College of Cardiology. 2001;37:300–7. doi: 10.1016/s0735-1097(00)01065-2. [DOI] [PubMed] [Google Scholar]
- 39.Zimmer H-G, Irlbeck M, Kolbeck-Rühmkorff C. Response of the rat heart to catecholamines and thyroid hormones. Mol Cell Biochem. 1995;147:105–14. doi: 10.1007/BF00944790. [DOI] [PubMed] [Google Scholar]
- 40.Millar BC, Schluter KD, Zhou XJ, McDermott BJ, Piper HM. Neuropeptide y stimulates hypertrophy of adult ventricular cardiomyocytes. The American journal of physiology. 1994;266:C1271–7. doi: 10.1152/ajpcell.1994.266.5.C1271. [DOI] [PubMed] [Google Scholar]
- 41.Bolter CP, Atkinson KJ. Maximum heart rate responses to exercise and isoproterenol in the trained rat. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 1988;254:R834. doi: 10.1152/ajpregu.1988.254.5.R834. [DOI] [PubMed] [Google Scholar]
- 42.Howell JJ, Manning BD. Mtor couples cellular nutrient sensing to organismal metabolic homeostasis. Trends in Endocrinology & Metabolism. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D-H, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. Mtor interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 44.Salameh A, Wustmann A, Karl S, Blanke K, Apel D, Rojas-Gomez D, et al. Cyclic mechanical stretch induces cardiomyocyte orientation and polarization of the gap junction protein connexin43. Circ Res. 2010;106:1592–602. doi: 10.1161/CIRCRESAHA.109.214429. [DOI] [PubMed] [Google Scholar]
- 45.Kudoh S, Komuro I, Hiroi Y, Zou Y, Harada K, Sugaya T, et al. Mechanical stretch induces hypertrophic responses in cardiac myocytes of angiotensin ii type 1a receptor knockout mice. Journal of Biological Chemistry. 1998;273:24037–43. doi: 10.1074/jbc.273.37.24037. [DOI] [PubMed] [Google Scholar]
- 46.Dvir T, Levy O, Shachar M, Granot Y, Cohen S. Activation of the erk1/2 cascade via pulsatile interstitial fluid flow promotes cardiac tissue assembly. Tissue Eng. 2007;13:2185. doi: 10.1089/ten.2006.0364. [DOI] [PubMed] [Google Scholar]
- 47.Thompson SA, Copeland CR, Reich DH, Tung L. Mechanical coupling between myofibroblasts and cardiomyocytes slows electric conduction in fibrotic cell monolayers. Circulation. 2011;123:2083–93. doi: 10.1161/CIRCULATIONAHA.110.015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirt MN, Sörensen NA, Bartholdt LM, Boeddinghaus J, Schaaf S, Eder A, et al. Increased afterload induces pathological cardiac hypertrophy: A new in vitro model. Basic Res Cardiol. 2012;107:1–16. doi: 10.1007/s00395-012-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saadane N, Alpert L, Chalifour LE. Expression of immediate early genes, gata-4, and nkx-2.5 in adrenergic-induced cardiac hypertrophy and during regression in adult mice. British Journal of Pharmacology. 1999;127:1165–76. doi: 10.1038/sj.bjp.0702676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kensah G, Roa Lara A, Dahlmann J, Zweigerdt R, Schwanke K, Hegermann J, et al. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. European Heart Journal. 2013;34:1134–46. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- 51.Thavandiran N, Dubois N, Mikryukov A, Massé S, Beca B, Simmons CA, et al. Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proceedings of the National Academy of Sciences. 2013;110:E4698–E707. doi: 10.1073/pnas.1311120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaaf S, Shibamiya A, Mewe M, Eder A, Stöhr A, Hirt MN, et al. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS ONE. 2011;6:e26397. doi: 10.1371/journal.pone.0026397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turnbull IC, Akar FG, Hajjar RJ, Hulot J-S, Costa KD, Karakikes I, et al. Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2014;28:644–54. doi: 10.1096/fj.13-228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Black LD, 3rd, Meyers JD, Weinbaum JS, Shvelidze YA, Tranquillo RT. Cell-induced alignment augments twitch force in fibrin gel-based engineered myocardium via gap junction modification. Tissue engineering Part A. 2009;15:3099–108. doi: 10.1089/ten.tea.2008.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boudou T, Legant WR, Mu AB, Borochin MA, Thavandiran N, Radisic M, et al. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A. 2012;18:910–9. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sondergaard CS, Mathews G, Wang L, Jeffreys A, Sahota A, Wood M, et al. Contractile and electrophysiologic characterization of optimized self-organizing engineered heart tissue. The Annals of thoracic surgery. 2012;94:1241–9. doi: 10.1016/j.athoracsur.2012.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Layland J, Kentish JC. Positive force- and [ca2+]i-frequency relationships in rat ventricular trabeculae at physiological frequencies. American Journal of Physiology - Heart and Circulatory Physiology. 1999;276:H9–H18. doi: 10.1152/ajpheart.1999.276.1.H9. [DOI] [PubMed] [Google Scholar]
- 58.Hiranandani N, Varian KD, Monasky MM, Janssen PML. Frequency-dependent contractile response of isolated cardiac trabeculae under hypo-, normo-, and hyperthermic conditions. Journal of Applied Physiology. 2006;100:1727–32. doi: 10.1152/japplphysiol.01244.2005. [DOI] [PubMed] [Google Scholar]
- 59.Raman S, Kelley M, Janssen P. Effect of muscle dimensions on trabecular contractile performance under physiological conditions. Pflugers Arch - Eur J Physiol. 2006;451:625–30. doi: 10.1007/s00424-005-1500-9. [DOI] [PubMed] [Google Scholar]
- 60.Bartel S, Stein B, Eschenhagen T, Mende U, Neumann J, Schmitz W, et al. Protein phosphorylation in isolated trabeculae from nonfailing and failing human hearts. Mol Cell Biochem. 1996;157:171–9. doi: 10.1007/BF00227896. [DOI] [PubMed] [Google Scholar]
- 61.Cheng M, Moretti M, Engelmayr GC, Freed LE. Insulin-like growth factor-i and slow, bidirectional perfusion enhance the formation of tissue-engineered cardiac grafts. Tissue Eng Part A. 2008;15:645–53. doi: 10.1089/ten.tea.2008.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown MA, Iyer RK, Radisic M. Pulsatile perfusion bioreactor for cardiac tissue engineering. Biotechnology Progress. 2008;24:907–20. doi: 10.1002/btpr.11. [DOI] [PubMed] [Google Scholar]
- 63.Sun LS, Legato MJ, Rosen TS, Steinberg SF, Rosen MR. Sympathetic innervation modulates ventricular impulse propagation and repolarization in the immature rat-heart. Cardiovascular Research. 1993;27:459–63. doi: 10.1093/cvr/27.3.459. [DOI] [PubMed] [Google Scholar]
- 64.Nygren A, Kondo C, Clark RB, Giles WR. Voltage-sensitive dye mapping in langendorff-perfused rat hearts. American Journal of Physiology - Heart and Circulatory Physiology. 2003;284:H892–H902. doi: 10.1152/ajpheart.00648.2002. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann W-H, Melnychenko I, Wasmeier G, Didié M, Naito H, Nixdorff U, et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nature Medicine. 2006;12:452–8. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- 66.Spach MS, Heidlage JF, Dolber PC, Barr RC. Changes in anisotropic conduction caused by remodeling cell size and the cellular distribution of gap junctions and na(+) channels. Journal of Electrocardiology. 2001;34(Suppl):69–76. doi: 10.1054/jelc.2001.28833. [DOI] [PubMed] [Google Scholar]
- 67.Guyette JP, Charest JM, Mills RW, Jank BJ, Moser PT, Gilpin SE, et al. Bioengineering human myocardium on native extracellular matrix. Circulation Research. 2016;118:56–72. doi: 10.1161/CIRCRESAHA.115.306874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solaro RJ, Lee JA, Kentish JC, Allen DG. Effects of acidosis on ventricular muscle from adult and neonatal rats. Circulation Research. 1988;63:779–87. doi: 10.1161/01.res.63.4.779. [DOI] [PubMed] [Google Scholar]
- 69.Hasenfuss G, Mulieri L, Blanchard E, Holubarsch C, Leavitt B, Ittleman F, et al. Energetics of isometric force development in control and volume- overload human myocardium. Comparison with animal species. Circulation research. 1991;68:836–46. doi: 10.1161/01.res.68.3.836. [DOI] [PubMed] [Google Scholar]
- 70.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circulation Research. 2014;114:549–64. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kemi OJ, Ceci M, Wisloff U, Grimaldi S, Gallo P, Smith GL, et al. Activation or inactivation of cardiac akt/mtor signaling diverges physiological from pathological hypertrophy. Journal of Cellular Physiology. 2008;214:316–21. doi: 10.1002/jcp.21197. [DOI] [PubMed] [Google Scholar]
- 72.Laplante M, Sabatini DM. Regulation of mtorc1 and its impact on gene expression at a glance. Journal of Cell Science. 2013;126:1713–9. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of stat3 during cntf signaling is mediated by the rapamycin target mtor. Current Biology. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 74.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase b by insulin and igf-1. The EMBO Journal. 1996;15:6541–51. [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng W-H, Quirion R. Insulin-like growth factor-1 (igf-1) induces the activation/phosphorylation of akt kinase and camp response element-binding protein (creb) by activating different signaling pathways in pc12 cells. BMC Neuroscience. 2006;7:1–10. doi: 10.1186/1471-2202-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maillet M, van Berlo JH, Molkentin JD. Molecular basis of physiological heart growth: Fundamental concepts and new players. Nat Rev Mol Cell Biol. 2013;14:38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McMullen JR, Jennings GL. Differences between pathological and physiological cardiac hypertrophy: Novel therapeutic strategies to treat heart failure. Clinical and Experimental Pharmacology and Physiology. 2007;34:255–62. doi: 10.1111/j.1440-1681.2007.04585.x. [DOI] [PubMed] [Google Scholar]
- 78.Dibble CC, Asara JM, Manning BD. Characterization of rictor phosphorylation sites reveals direct regulation of mtor complex 2 by s6k1. Molecular and Cellular Biology. 2009;29:5657–70. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harrington LS, Findlay GM, Lamb RF. Restraining pi3k: Mtor signalling goes back to the membrane. Trends in Biochemical Sciences. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 80.Manning BD. Balancing akt with s6k: Implications for both metabolic diseases and tumorigenesis. The Journal of Cell Biology. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, et al. Mtor kinase inhibition causes feedback-dependent biphasic regulation of akt signaling. Cancer Discovery. 2011;1:248–59. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aksamitiene E, Kiyatkin A, Kholodenko Boris N. Cross-talk between mitogenic ras/mapk and survival pi3k/akt pathways: A fine balance. Biochemical Society Transactions. 2012;40:139. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- 83.Rosner M, Siegel N, Valli A, Fuchs C, Hengstschläger M. Mtor phosphorylated at s2448 binds to raptor and rictor. Amino Acids. 2010;38:223–8. doi: 10.1007/s00726-008-0230-7. [DOI] [PubMed] [Google Scholar]
- 84.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of akt/pkb by the rictor-mtor complex. Science. 2005;307:1098. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 85.Laplante M, Sabatini DM. Mtor signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bracho-Valdés I, Moreno-Alvarez P, Valencia-Martínez I, Robles-Molina E, Chávez-Vargas L, Vázquez-Prado J. Mtorc1- and mtorc2-interacting proteins keep their multifunctional partners focused. IUBMB Life. 2011;63:896–914. doi: 10.1002/iub.558. [DOI] [PubMed] [Google Scholar]
- 87.Anversa P, Olivetti G, Loud AV. Morphometric study of early postnatal development in the left and right ventricular myocardium of the rat. I. Hypertrophy, hyperplasia, and binucleation of myocytes. Circulation Research. 1980;46:495–502. doi: 10.1161/01.res.46.4.495. [DOI] [PubMed] [Google Scholar]
- 88.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–35. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lasher RA, Pahnke AQ, Johnson JM, Sachse FB, Hitchcock RW. Electrical stimulation directs engineered cardiac tissue to an age-matched native phenotype. Journal of tissue engineering. 2012;3:2041731412455354–15. doi: 10.1177/2041731412455354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin D, Zhang Z-H, Caref EB, Boutjdir M, Jain P, El-Sherif N. Cellular and ionic basis of arrhythmias in postinfarction remodeled ventricular myocardium. Circulation Research. 1996;79:461–73. doi: 10.1161/01.res.79.3.461. [DOI] [PubMed] [Google Scholar]
- 91.Gerdes AM, Onodera T, Wang X, McCune SA. Myocyte remodeling during the progression to failure in rats with hypertension. Hypertension. 1996;28:609–14. doi: 10.1161/01.hyp.28.4.609. [DOI] [PubMed] [Google Scholar]
- 92.Taylor JM, Rovin JD, Parsons JT. A role for focal adhesion kinase in phenylephrine-induced hypertrophy of rat ventricular cardiomyocytes. Journal of Biological Chemistry. 2000;275:19250–7. doi: 10.1074/jbc.M909099199. [DOI] [PubMed] [Google Scholar]
- 93.Tigchelaar W, de Jong AM, Bloks VW, van Gilst WH, de Boer RA, Silljé HHW. Hypertrophy induced kif5b controls mitochondrial localization and function in neonatal rat cardiomyocytes. Journal of Molecular and Cellular Cardiology. 2016;97:70–81. doi: 10.1016/j.yjmcc.2016.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.