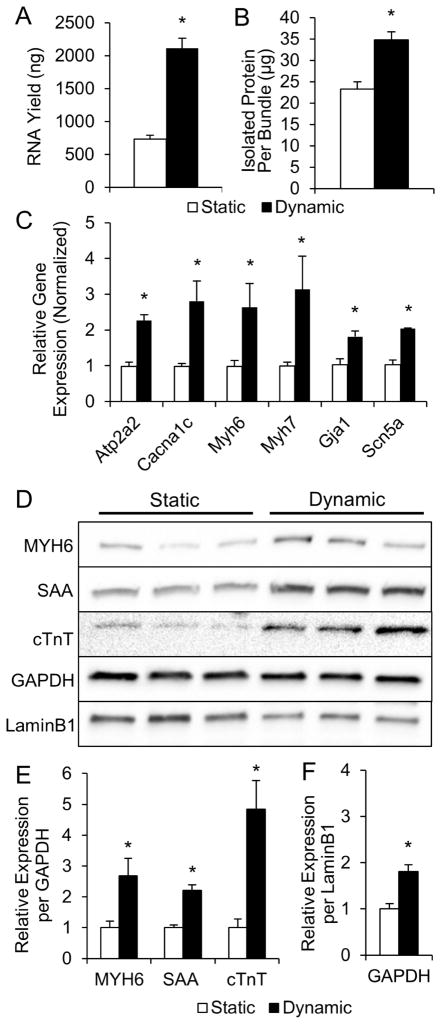
Figure 2. Molecular analysis of statically and dynamically cultured NRVM cardiobundles.
(A, B) RNA (A) and total protein (B) contents of cardiobundles cultured for 2 weeks under static or dynamic conditions (n = 9 samples per group from 3 cell isolations, each sample is 2–4 pooled cardiobundles). (C) Gene expression of cardiobundles (N = 3 cell isolations with 3 cardiobundles per group in each isolation). (D) Western blots of cardiobundles (N = 3 cell isolations with 3–4 cardiobundles per group in each isolation). (E) Quantification of protein expression from Western blots in (D) by densitometry normalized to GAPDH. (F) GAPDH normalized to LaminB1 estimates amount of total cellular protein per nucleus. *, p < 0.05 vs. static. Gene expression analyzed: SERCA (Atp2a2), L-type Ca2+ channel (Cacna1c), α- and β-myosin heavy chain (Myh6 and Myh7), connexin-43 (Gja1), sodium channel Nav1.5 (Scn5a). Protein expression analyzed: α-myosin heavy chain (MYH6), sarcomeric α-actinin (SAA), cardiac troponin-T (cTnT).