Pleomorphic xanthoastrocytoma (PXA, WHO grade II) and anaplastic PXA (WHO grade III) are astrocytic neoplasms that commonly harbor an activating mutation in BRAF (p.V600E, c.1799T>A) [3, 6], driving activation of the mitogen-activated protein kinase (MAPK) signaling pathway. Currently, PXA is diagnosed based solely on histopathologic features. Yet recent reports based on DNA methylation suggest anaplastic PXA-like tumors can masquerade as glioblastoma (GBM) [2]. Importantly these PXA-like tumors were associated with a more favorable prognosis than GBM and included both BRAF p.V600E mutant and non-mutant tumors in approximately equal proportions [2]. These findings suggest both histologic analysis and determination of BRAF p.V600E mutation status may not capture all tumors with the biologic behavior of anaplastic PXA. Here, we report on the identification of in-frame genomic rearrangements predicted to result in NRF1-BRAF and ATG7-RAF1, constitutively active kinase fusions, in two anaplastic PXA without BRAF p.V600E mutation.
Two BRAF p.V600 wild-type anaplastic PXA identified at the UCSF Brain Tumor Center were analyzed using targeted next-generation sequencing with the UCSF Clinical Cancer Genomics Laboratory. Patient clinical characteristics and tumor histopathology are summarized in Supplemental Table 1. Genomic profiling identified novel chromosomal rearrangements in both cases resulting in an in-frame fusion between the amino terminal portion of one gene encoding a homodimerization domain and the serine/threonine kinase domain of a RAF kinase family member. In PXA#1 the predicted fusion protein contains exons 1–5 of NRF1, lacks the RAS-binding domain of BRAF (exons 4–8), and retains the serine/threonine kinase domain of BRAF (Figure 1a). Nuclear Respiratory Factor 1 (NRF1) encodes a transcription factor that activates the expression of metabolic genes involved in cellular growth and development. Sanger sequencing for the NRF1-BRAF fusion breakpoint confirmed its presence in the most recent tumor and in tumor tissues from resections 2 and 4 years prior to the most recent resection, suggesting that the fusion arose early during tumor development (Supplemental Figure 1a).
Fig. 1. Novel RAF gene fusions identified in anaplastic PXA lacking BRAF p.V600E mutation.
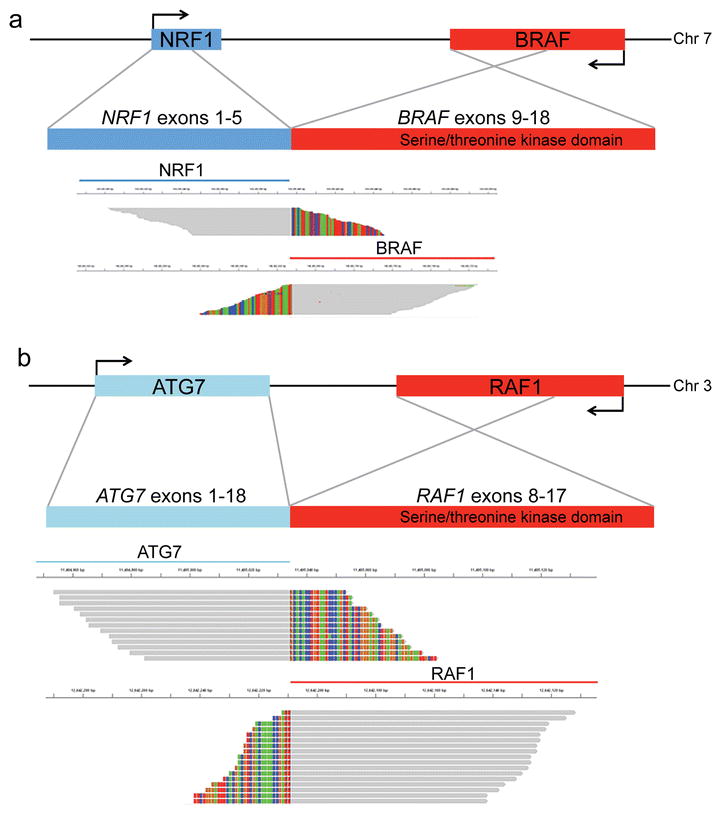
Sequencing reads across the fusion breakpoint in two BRAF p.V600 wild type anaplastic PXA. (a) PXA#1 contained an in-frame NRF1-BRAF fusion between NRF1 (left) and BRAF (right). (b) PXA#2 contained an in-frame ATG7-RAF1 fusion. Grey bar denotes sequence aligned to the reference human genome, and multicolor bases denote mismatched base pairs that instead align with the fusion partner.
PXA#2 harbored an ATG7-RAF1 fusion involving exons 1–18 of ATG7 and exons 8–17 of RAF1 (Figure 1b). Autophagy related 7 (ATG7) encodes an E1-like activating enzyme that is essential for autophagy and cytoplasmic to vacuole transport [5]. An ATG7-BRAF fusion has been reported previously in melanoma [4]. For PXA#2, molecular analysis of a subsequent recurrence confirmed the presence of the same ATG7-RAF1 fusion, consistent with persistence of this activating genetic driver (C. Kline et al. submitted). Immunohistochemistry for p-ERK confirmed robust MAPK signaling pathway activation in tumors with RAF-fusions and was similar to that observed in anaplastic PXA with BRAF p.V600E mutation (Figure 2). Therefore RAF kinase fusions represent an important alternative to BRAF p.V600E mutation to activate the MAP-kinase pathway in PXA.
Fig. 2. MAPK pathway activation in anaplastic PXA with BRAF p.V600E mutation or RAF kinase family gene fusions.
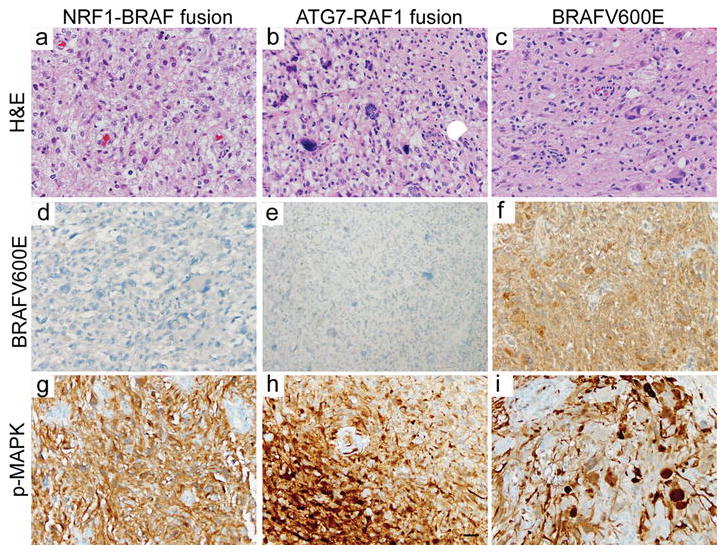
(a–c) PXA#1 and #2 and a representative BRAF p.V600E mutant tumor (PXA#3) have characteristic features of anaplastic PXA, WHO grade III, including pleomorphic, spindled, or epithelioid neoplastic cells with astrocytic morphology (H&E staining). (d–f) The BRAF p.V600E mutation can be detected by immunostaining using mutant-specific antibodies. Activation of the MAPK signaling pathway, as evidenced by phospho-MAPK immunostaining, in anaplastic PXA with NRF1-BRAF fusion (g), ATG7-RAF1 fusion (h), and BRAF p.V600E mutation (i). Scale bar, 30 μm.
Both tumors also had chromosomal deletion of the CDKN2A locus at 9p21. In addition, a hotspot mutation in TP53 (NM_000546: TP53 p.R273H) was identified in PXA#2 and two variants of unknown significance were identified, including a variant in KEAP1 (NM_012289, p.V79D) located in the BTB domain, in PXA #1 (Supplemental Table 2). Sanger sequencing for the KEAP1 variant confirmed its presence in the most recent tumor but was unable to detect the variant in tumor tissue from resections 2 and 4 years prior to the most recent resection (Supplemental Figure 1d–1f). KEAP1 encodes the protein Kelch Like ECH Associated Protein 1 and regulates NRF2 (nuclear factor erythroid 2-related factor 2) via ubiquitination. While the KEAP1 (p.V79D) variant has not previously been associated with cancer, this amino acid is highly conserved and other alterations in the KEAP1 BTB domain have been associated with constitutive activation of NRF2 in cancer [1]. Characteristic gene alterations of pediatric and adult infiltrating astrocytoma were not identified, including alterations in ATRX, SETD2, H3F3A, HIST1H3B, PTEN, EGFR, PDGFRA, PIK3CA, IDH1, and IDH2.
The diagnosis of anaplastic PXA can be challenging particularly in the absence of BRAF p.V600E mutation. Molecular characterization of RAF kinase family members beyond BRAF p.V600 status may improve the recognition of anaplastic PXA and help to identify patients most likely to benefit from inhibitors of the RAF pathway.
Supplementary Material
Acknowledgments
This study was funded in part by the UCSF Department of Pathology, UCSF Brain Tumor SPORE Tissue Core (NIH/NCI P50 CA097257), the Pediatric Brain Tumor Foundation (J.J.P and T.N.), and NIH/NINDS R01 (NS081117, J.J.P.). We thank and acknowledge the patients and families affected by PXA for their generous contributions to these studies.
References
- 1.Konstantinopoulos PA, Spentzos D, Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, Hecht JL, Cannistra SA. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 2011;71:5081–5089. doi: 10.1158/0008-5472.CAN-10-4668. [DOI] [PubMed] [Google Scholar]
- 2.Korshunov A, Ryzhova M, Hovestadt V, Bender S, Sturm D, Capper D, Meyer J, Schrimpf D, Kool M, Northcott PA, Zheludkova O, Milde T, Witt O, Kulozik AE, Reifenberger G, Jabado N, Perry A, Lichter P, von Deimling A, Pfister SM, Jones DTW. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol (Berl) 2015;129:669–678. doi: 10.1007/s00401-015-1405-4. [DOI] [PubMed] [Google Scholar]
- 3.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, Louis DN, Korshunov A, Pfister S, Hartmann C, Paulus W, Reifenberger G, von Deimling A. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol (Berl) 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 4.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, Boop FA, Lu C, Kandoth C, Ding L, Lee R, Huether R, Chen X, Hedlund E, Nagahawatte P, Rusch M, Boggs K, Cheng J, Becksfort J, Ma J, Song G, Li Y, Wei L, Wang J, Shurtleff S, Easton J, Zhao D, Fulton RS, Fulton LL, Dooling DJ, Vadodaria B, Mulder HL, Tang C, Ochoa K, Mullighan CG, Gajjar A, Kriwacki R, Sheer D, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Baker SJ, Ellison DW St Jude Children’s Research–Hospital Washington University Pediatric Cancer Genome Project. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


