Abstract
Over the past decade, the rate of drug attrition due to clinical trial failures has risen substantially. Unfortunately it is difficult to identify compounds that have unfavorable toxicity properties before conducting clinical trials. Inspired by the effective use of Sabermetrics in predicting successful baseball players, we sought to use a similar “moneyball” approach that analyzes overlooked features to predict clinical toxicity. We introduce a new data-driven approach (PrOCTOR) that directly predicts the likelihood of toxicity in clinical trials. PrOCTOR integrates properties of a compound’s targets and its structure to provide a new measure, the PrOCTOR score. Drug target network connectivity and expression levels, along with molecular weight, were identified as important indicators of adverse clinical events. Altogether, our method provides a data-driven broadly applicable strategy to identify drugs likely to possess manageable toxicity in clinical trials and will help drive the design of therapeutic agents with less toxicity.
Introduction
Failures in all phases of clinical trials have skyrocketed over the past three decades, with a substantial portion occurring for safety reasons (Hay et al., 2014; Ledford, 2011). This is occurring despite improvements in all stages of the drug development pipeline (Scannell et al., 2012). One of the key areas of improvement has been the screening for drugs likely to fail clinical trials.
Drug-likeness measures have been widely accepted as a useful guide for filtering out toxic molecules in the early stages of drug discovery. Lipinski first proposed this concept over a decade ago with his Rule of 5 (Ro5), a set of four physicochemical features associated with orally active drugs that were derived from analyzing clinical drugs that reached Phase II trials or beyond(Lipinski et al., 1997). This concept enhanced the drug discovery process by providing a set of practical filters that became widely adopted in drug development pipelines. However Lipinski noted that the Ro5 is a very conservative predictor and passing the rule does not guarantee drug-likeness (Lipinski, 2004). Modified rule sets have since been proposed, such as Veber’s Rule(Veber et al., 2002) and Ghose’s Rule(Ghose et al., 1999), to include more properties associated with bioavailability, such as Polar Surface Area, and to improve upon the concept proposed by Lipinski. More recently, the Quantitative Estimate for Drug-likeness (QED) was proposed as an alternative to rule-based methods (Bickerton et al., 2012).
The adoption of drug-likeness concepts early in the drug discovery process has been shown to reduce attrition rates (Leeson and Springthorpe, 2007). However despite these advances in identifying potentially toxic drugs, clinical trial attrition rates have continued to rise (Hay et al., 2014). While oral bioavailability is highly relevant to drug toxicity, there are other factors that also contribute to clinical trial toxicity events. To address this problem, we propose a new approach for predicting odds of clinical trial outcomes (PrOCTOR).
Results
Analysis of clinical trials data reveals limitations of structural-based approaches
Drug-likeness approaches have been important and informative in guiding the drug development process. However they cannot distinguish drugs with unmanageable toxicity profiles from safe ones (Bickerton et al., 2012; Leeson and Springthorpe, 2007). We verified this quantitatively by comparing drugs that have failed clinical trials with FDA approved drugs. To this end, we downloaded data from The Database for Aggregate Analysis of ClinicalTrials.gov (AACT) at ClinicalTrials.gov and extracted the names of the drugs associated with 108 clinical trials of any phase that were annotated as having failed for toxicity reasons. The comparative list was developed from the 1013 FDA approved drugs that were annotated as FDA approved in the DrugBank database(Law et al., 2014).
For the drugs in these lists, we tested existing methods for their ability to distinguish approved drugs from those that failed for toxicity in trials (FTT drugs). Most FDA approved drugs pass Lipinski’s Rule of Five(Lipinski et al., 1997) (80.6%) and Ghose’s(Ghose et al., 1999) (64.9%) rules, but so do most of the FTT drugs (73% Lipinski, 54% Ghose). In contrast, Veber’s rule(Veber et al., 2002) appears to be a far too conservative measure, with 75.2% of approved and 92% of FTT drugs being predicted to fail. Finally the QED approach, which calculates a continuous score(Bickerton et al., 2012), is also unable to significantly distinguish the two classes (p=0.1069, D=0.10703, Kolmogorov-Smirnov Test). This analysis further highlights the unmet need to develop strategies for predicting the likelihood of toxicity in clinical trials.
Computational approach accurately predicts likelihood of clinical trial failure
Because all of the drug-likeness methods consider only the chemical properties of a molecule, we reasoned that a new approach that includes overlooked features related to the results of a drugs performance could prove to be highly impactful, similar to the effect that adopting sabermetrics had on the baseball scouting process as described in Michael Lewis’s Moneyball(Lewis, 2003). A specific example is the consideration of target-related properties, such as tissue selectivity (an ideal target would be found only in diseased tissue and sparsely anywhere else). We suggest that such considerations could be useful in determining potential toxic effects.
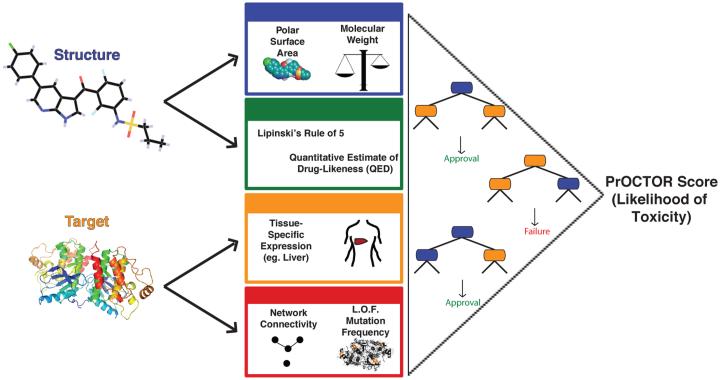
The inferences gained from the analysis of the various methods and the consideration of additional characteristics in the prediction of tolerable toxicity in clinical trials led to the development of our new approach for predicting odds of clinical trial outcomes using random-forest (PrOCTOR). PrOCTOR integrates established informative chemical features of the drugs with target-based features to produce a classifier that is able to distinguish FDA approved drugs from FTT drugs. Random forest(Breiman, 2001), a decision tree based machine learning model, is used to address the classification problem of clinical trial drug toxicity (Fig.1). The random forest model builds a set of 50 decision trees with a subset of features (see below) within each tree and assigns the predicted outcome to be the consensus of the trees.
Figure 1.
Method Schematic. Our approach integrates chemical properties, drug-likeness measures and target-based properties of a molecule into a random forest model to predict whether the drug is likely to be a member to fail clinical trials for toxicity reasons.
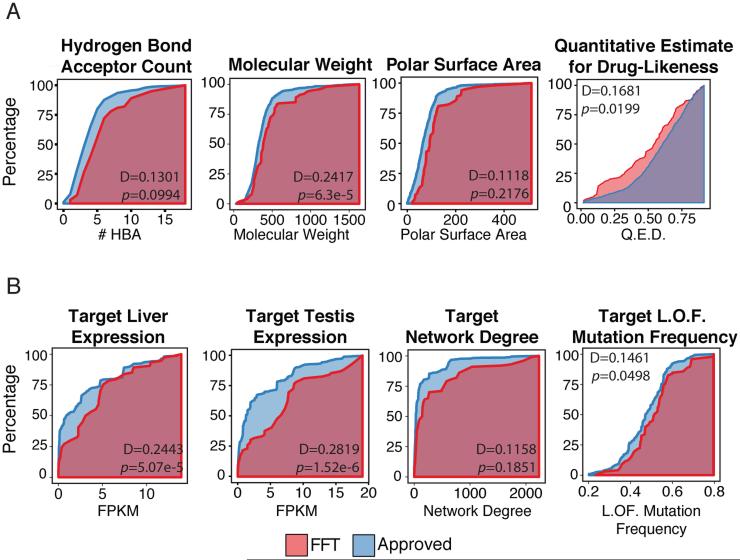
The set of 48 features describing each drug contains 10 molecular properties, 34 target-based properties and 4 drug-likeness rule features (see Supplementary Table S1). Given their established validity, we chose to include the molecular properties considered by the Lipinski, Veber and Ghose rules. We found that, individually, some of these properties had slight but significant power to discriminate between FDA approved drugs and FTT drugs when applied to our lists of drugs in the two categories (Fig.2a). Additional features represent the compatibility of the compounds with the drug-likeness approaches. Each drug’s known targets were annotated from the DrugBank dataset (Law et al., 2014) and used to derive an additional set of target-based properties. We considered the median expression of the gene targets in 30 different tissues, such as the liver and the brain, calculated from the Genotype-Tissue Expression (GTEx) project(Consortium, 2015). Other target-based features represent the network connectivity of the target, with gene degree and betweenness features, computed using an aggregated gene-gene interaction network(Aksoy et al., 2013; Das and Yu, 2012; Khurana et al., 2013), and a feature that represents the loss of function mutation frequency in the target gene, extracted from the Exome Aggregation Consortium (ExAC) database(Exome Aggregation Consortium (ExAC)). Like the chemical properties, we found that some of these target-based features also were able to weakly but significantly discriminate between FDA approved drugs and FTT drugs (Fig.2b). Not surprisingly, many of the features within the target-based or the chemical category were highly correlated with each other. Since we found the target expression values to be highly correlated (Fig.S1, see supplemental text), principle component analysis was applied to all target expression values in order to reduce the feature dimensionality. In place of the raw expression values, the first three principle components were instead used. However there was little correlation between the two classes of features (maximum Pearson correlation of r=0.1942). Thus the target-based features add information independent of the chemical features into the model. The full description of the features used in the model is described in Supplementary Table S1.
Figure 2.
Distributions of select (a) chemical features, and (b) target-based model features. The Kolmogorov-Smirnov D statistic and p-value are shown for the comparison of failed toxic clinical trial (FTT) drugs (red) and FDA approved drugs (blue).
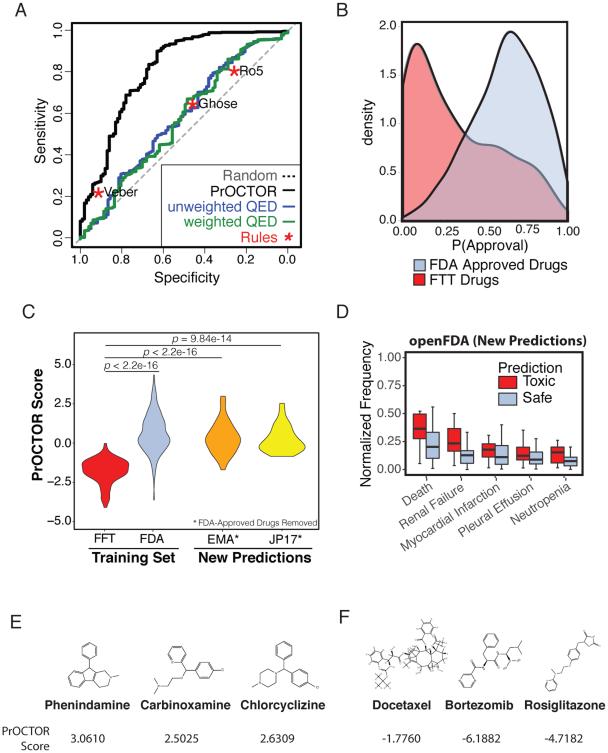
The approach was tested by performing 10-fold cross validation on a set of 784 FDA approved drugs with known targets and the drugs associated with 100 FTT that had at least one annotated target and known chemical structure. We found that PrOCTOR had significant predictive performance, with an area under the receiver operator curve (AUC) of 0.8263 (Fig.3a). At the optimal point of the curve the method achieved an accuracy (ACC) of 0.7529, with both high sensitivity (true positive rate (TPR) of 0.7544), and high specificity (true negative rate (TNR) of 0.7410). By comparison, on this same dataset the Ro5 and Ghose rules had a TPR of 0.8030 and 0.6468, respectively, and a TNR of 0.27 and 0.46 respectively. Application of the Veber method achieved a TPR of 0.2465, and a TNR of 0.92. (Fig.3a). The ROC curve of both the unweighted and weighted versions of the QED method fell significantly below that of PrOCTOR’s ROC curve (AUC=0.581, p<2.2e-16, Wilcoxon signed rank test), indicating that PrOCTOR is able to better distinguish the FTT and approved drug classes. Furthermore, PrOCTOR’s approval probability allows for the separation of the drugs of the FFT and FDA approved classes (D=0.5343, p< 2.2e-16, Kolmogorov-Smirnov test) (Fig.3b) on a continuous scale.
Figure 3.
Benchmarking Model performance. (a) Receiver operating characteristic (ROC) curves for PrOCTOR, three drug-likeness rules (Ro5, Veber, Ghose) and both the weighted and unweighted QED metrics. (b) PrOCTOR scores and the Q.E.D. metric for approved and failed toxic clinical trial (FTT) drugs. (c) PrOCTOR scores for the FDA approved and FTT drugs in the training set, as well as EMA-Approved and Japanese-Approved (JP17) drugs after removal of FDA approved drugs. Statistical significance was assessed for FDA, EMA, and JP17 vs FTT drugs using the Mann-Whitney U Test. (d) Reported frequencies, normalized to the most frequently reported adverse event, in the openFDA database for predicted toxic (red, score<−1) and predicted safe drugs from the DrugBank dataset. (e) The top three molecules predicted by PrOCTOR as most likely to be FDA approved are phenindamine, carbinoxamine, and chlorcyclizine. (f) The three molecules predicted by PrOCTOR as most likely to fail clinical trials for toxicity reasons are docetaxel, bortezomib, and rosiglitazone.
We further assessed the approach by applying PrOCTOR to drugs that are approved in Europe (EMA-Approved) or in Japan (JP17) but not annotated as being FDA approved in our dataset. When compared to the FTT drugs in our training set, we found that EMA-Approved (p<2.2e-16, Mann–Whitney UTest) and JP17 drugs (p=9.84e-14, Mann–Whitney U Test) were predicted to be significantly safer and had a similar distribution of PrOCTOR scores to the class of FDA Approved Drugs (Fig.3c).
Next, we applied PrOCTOR to 3,236 drugs that were in DrugBank and not in our training set (Supplementary Table 2). We found that the predicted toxic drugs had significantly more frequent reports of serious adverse events, such as death and renal failure, than predicted safe drugs in the openFDA resource of drug adverse events (https://open.fda.gov) (Fig.3d). Furthermore, we found that safe predictions were enriched for classes of drugs that are known to be relatively safe, such as antidepressants, stimulants, and serotonin-related drugs. In comparison, toxic predictions were enriched for known toxic classes of drugs, such as immunosuppressive agents and antineoplastic agents (Supplementary Table 3).
We also applied our approach to 137 drugs annotated as most-DILI-concern and 65 drugs of no-DILI-concern by the FDA. We found that the most-DILI-concern drugs had 1.5-fold higher odds of being classified as toxic by PrOCTOR than the no-DILI-concern drugs. More generally, the most-DILI-concern drugs had higher PrOCTOR scores than the no-DILI-concern drugs (p= 0.0005, Mann–Whitney UTest). This suggests that our model is able to generalize beyond the training set.
Identification of FDA drugs with increased likelihood of toxicity events
Next we looked to evaluate the predictions of our approach by analyzing PrOCTOR’s predictions for FDA approved drugs. A PrOCTOR score expressing the log2(odds of approval) was calculated taking the log2 of the ratio of the PrOCTOR-predicted probability of approval to the probability of failure.
The three molecules identified by PrOCTOR as most likely to receive FDA approval were phenindamine, carbinoxamine, and chlorcyclizine (Fig.3e). All three of these drugs are FDA approved antihistamines with highly tolerable side effects. Interestingly, all three of these drugs pass the Ro5 but have relatively low QED values (0.311, 0.242, and 0.499 respectively).
The three molecules with the worst PrOCTOR score and thus predicted as most likely to fail clinical trials for toxicity reasons were docetaxel, bortezomib, and rosiglitazone (Fig.3f). Of note, all are FDA approved drugs that have been associated with serious toxicity events. Docetaxel is a chemotherapy agent used to treat a number of cancers(Massacesi et al., 2004; Puisset et al., 2007). The most frequent adverse event associated with docetaxel is neutropenia, a potentially life threatening event that often results in delay of treatment(Puisset et al., 2007). It also fails the Ro5 and has an extremely low QED of 0.147, suggesting that this prediction is consistent with other drug screening methods. Bortezomib is a proteasome inhibitor used for treatment of relapse multiple myeloma that has a moderate QED value of 0.476 and passes the Ro5. While it was FDA approved due to its significant antitumor activity, it has been associated with frequent adverse events, such as peripheral neuropathy, that are thought to in part be due to nonproteasomal targets (Arastu-Kapur et al., 2011). Rosiglitazone is an antidiabetic drug that also passes the Ro5 and has a high QED value of 0.825. However it has been linked with an elevated risk of heart attack(Nissen and Wolski, 2007) and consequently was withdrawn from the market in Europe in 2010(Blind et al., 2011). This suggests that existing methods were not necessarily able to foresee the adverse events associated with these latter two compounds.
These compounds bring to attention the importance of context when considering toxicity events. In general, more frequent and serious side effects will be acceptable for drugs that are used to treat severe and otherwise untreatable conditions, such as cancer. This is an important consideration to keep in mind when determining acceptable score ranges in drug development. Additionally, it highlights the shortcomings of rule-based methods, which are unable to quantify the extent to which a drug may have undesirable characteristics since a molecule that just barely fails one requirement is equivalent to one that substantially fails all requirements.
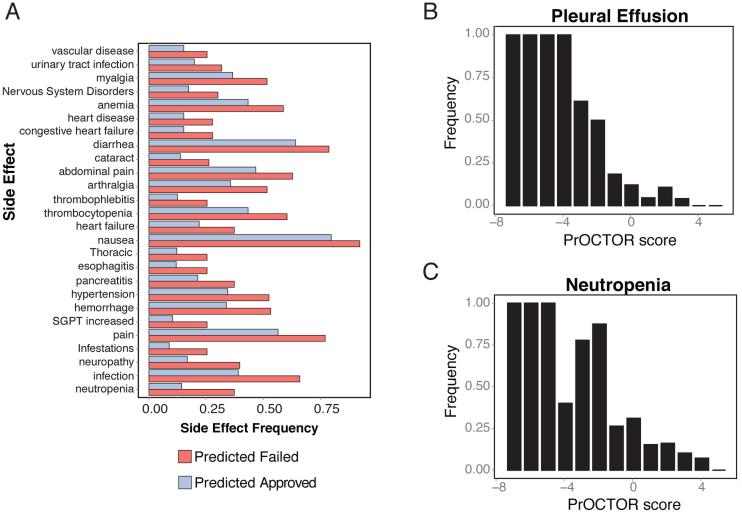
We further assessed what insights the predictions from PrOCTOR can offer regarding toxic effects using the SIDER side effect resource database (Kuhn et al., 2010). We hypothesized that drugs with better PrOCTOR scores would have less frequent severe side effects reported due to their more tolerable toxicity profiles. We first compared all drugs predicted to be approved by PrOCTOR (via cross-validation), including those misclassified, to those predicted to be of the FTT class. We found that the predicted FTT drugs had significantly more frequent severe side effects, such as neutropenia (37.3% vs 14.3%, p=1.78×10−7, Fisher-Exact test) (Fig.4A). When comparing the drugs with the top 10% best PrOCTOR scores to those within the bottom 10%, this distinction was even greater with severe toxic events, such as neutropenia (54.8% vs 13.4%, p=1.72×10−6, Fisher-Exact test) and pleural effusion (47.6% vs 5.2%, p=2.59×10−7, Fisher-Exact test), occurring far more frequently in the predicted FTT class (Fig.S2).
Figure 4.
Side Effects. (a) Adverse events that occur more frequently in predicted failed toxic clinical trial (FTT) drugs compared to predicted approved drugs. (b) Binned frequency of pleural effusion across PrOCTOR score bins. (c) Binned frequency of neutropenia across PrOCTOR score bins.
Furthermore, we found that these severe side effects were significantly negatively correlated with the PrOCTOR score. For example, the spearman’s correlation coefficient of the binned pleural effusion frequency against the PrOCTOR score was ρ=−0.9792 (Fig.4b) and for neutropenia was ρ=−0.9613 (Fig.4c). In comparison, the frequent side effect of dizziness still occurred more frequently in the predicted toxic drugs but had a much weaker correlation of ρ=−0.5070. Thus the predictions of PrOCTOR are consistent with reported adverse events, with the PrOCTOR score negatively correlating with the reported severe side effects that would ultimately contribute to a drug’s success in clinical trials.
Model reveals insights about how various properties can contribute to or help avert toxicity
We evaluated what insights PrOCTOR can offer about successful drugs. A feature importance analysis showed that both the chemical and target-based features contribute significantly to the performance of the PrOCTOR algorithm. The first expression principle component, QED metric, polar surface area, and the drug target’s network connectivity emerged as the four most important features (Fig.S3a), thus target-based features were identified as highly important features for predicting toxicity. Using target-based features alone, PrOCTOR achieved a significant predictive performance (ACC=0.7115). Our approach relies on existent annotation of drug targets to calculate these features. However this information is often not available during the drug development stage. We found that our method is robust to removal of targets (Fig.S3b) and additionally maintains a significant predictive performance (ACC= 0.6708) in absence of known target information. However PrOCTOR’s performance remains strongest when including both the chemical and target-based features (ACC=0.7529).
We next investigated the relationships between the features in the model. We found that certain combinations of uncorrelated features provided greater discriminative power. For example, Bickerton et al. (Bickerton et al., 2012) reported that the QED approach outperformed other drug-likeness methods when the threshold was set at 0.35. We found that 75% of drugs with QED<0.35 were approved. However when high testis expression (FPKM>10) was added into consideration, 88.5% of FTT drugs were accurately be classified (Fig.S4a). Additionally, tissue selectivity is a useful consideration in determining potential toxic effects. We hypothesize that this may be due to some tissue-specific toxicity events being associated with the drug target’s expression in normal tissue. We found that 84% (38/45) of drugs with high molecular weight (MW>500) but low general tissue expression (PC1< −2) were FDA approved. Thus if a gene appears to be a promising target for mechanistic reasons while appearing ill-suited due to high global expression profiles, it still may remain a viable candidate given that certain molecular properties are satisfied. To fully interpret how the model generated by PrOCTOR was able to capture these dependencies, we created a consensus decision tree which can be found in Figure S4b.
Discussion
Drug-likeness approaches, as first proposed by Lipinski almost two decades ago, have become a key tool for the pre-selection of compounds that are likely to have manageable toxicity in clinical studies. However all these methods consider only the molecular properties of the drug itself. We have proposed a data-driven approach (PrOCTOR) for predicting likelihood of toxic events in clinical trials that moves beyond existing drug likeness rules and measures by not only considering the chemical properties of a molecule, but also the properties of the drug’s target. When trained on failed clinical trials and FDA approved drugs, the PrOCTOR score performs at high accuracy, specificity and sensitivity. Furthermore, the PrOCTOR score strongly correlates with reported severe adverse events.
While phase I trials are designed to investigate safety, drugs can fail at any stage for toxicity reasons and additionally can fail phase I trails for non-safety reasons. Lipinski’s Ro5 was derived using the set drugs that had succeeded to phase II trials, under the assumption that undesirable drugs would have been eliminated in Phase I (Lipinski et al., 1997). However it has been observed that a substantial number of drugs fail in Phase II trials and beyond for safety reasons (Ledford, 2011). Additionally many of the drug-likeness measures were developed using larger representative datasets in place of clinical trial data(Bickerton et al., 2012). While these methods are important, they are focused on subtly different problems such as bioavailability. We have shown above that these approaches are not able to sufficiently capture clinical trial safety. There have been a number of other methods that have been developed to predict toxicity events as well. A recent DREAM Challenge focused on predicting cytotoxicity in lymphoblastoid cell lines, however primarily focused on environmental toxins(Eduati et al., 2015).
Similarly, the EPA’s extensive ToxCast dataset is covered predominantly by non-therapeutic chemicals(USEPA, 2016). Other toxicity prediction methods, such as those in AMBIT, have been developed to address other toxicity-based questions, including model organism and tissue-specific toxicities(Jeliazkova and Jeliazkov, 2011). QSAR models are also frequently used for toxicity prediction. However they have generally been applied to the prediction of specific toxicity endpoints, such as drug LD50 values, tissue-specific toxicity events or for the estimation of maximum tolerated dose levels (Patlewicz and Fitzpatrick, 2016). Finally, PK/PD models are highly valuable tools for identifying toxicological properties of drugs preclinically, but must be independently constructed for every drug and thus would benefit from more high-throughput methods for toxicity prediction(Sahota et al., 2016). Consequently, we selected the set of drugs that failed any phase of clinical trials for toxicity reasons to develop our approach.
We have also only addressed the issue of general clinical trial toxicity. However some indications, such as cancers, have more critical needs and consequently allow for higher toxicity levels. As a result, our model may predict some promising anti-cancer drugs to have unmanageable toxicity levels. Since PrOCTOR outputs a score, instead of just a prediction, a different threshold for allowable toxicity may be considered for different indications. A preliminary testing of this idea on cancer-only drugs with cancer type added as a feature demonstrated improved predictive power on this subset of drugs (ACC=0.74, AUC=0.80). However given the small sample size of this training set (n=89), this cancer-specific model is not optimal at this time. Additionally many new therapies are currently being developed to target specific isoforms and mutations. While our model is not currently accounting for these specific targets, it can straightforwardly be adapted using publicly available or user-provided target-based information. There are also areas in which PrOCTOR could be further improved such that leads to better predictive capacities. The use of 3D fingerprinting methods may allow for the structural features to be better represented. Co-expression networks from the GTEx data may also be useful features, as they may provide a stronger biological signal. Biological interaction networks are generally incomplete and also vary between cellular contexts and populations, which may limit the power of the network metrics. Finally, our method is largely dependent on existing target annotation for drugs, which is generally incomplete. Thus we will likely benefit from advancements in drug target identification.
Furthermore over two-thirds of clinical trials fail for other reasons, including efficacy, strategic and financial reasons (Ledford, 2011). The problem of efficacy is a highly complex issue, since each drug must demonstrate improvement over existing drugs in addition proving a context-specific efficacy. Thus while this problem remains important, it is not likely to be tractable using this style of approach.
Our approach has the potential to impact the preclinical drug development pipeline by quantifying how likely a given compound is to have manageable toxicity in clinical trials. In order to facilitate interaction with and application of our model, we have developed an interactive tool that we have made available on github (https://github.com/kgayvert/PrOCTOR). PrOCTOR may also help flag drugs for increased post-approval surveillance of adverse effects and toxicity. Perhaps even more importantly, the model will help design better drugs by providing insights about how various chemical and target-based properties can contribute to or help avert toxicity.
MATERIALS AND METHODS
Clinical Trials Training Set
We downloaded data from ClinicalTrials.gov from The Database for Aggregate Analysis of ClinicalTrials.gov (AACT) 10 10. To extract the names of the drugs associated with clinical trials that failed toxicity reasons, we identified any clinical trials that were annotated as “Terminated”, “Suspended” or “Withdrawn” and described as failing for toxicity reasons. The list of FDA approved drugs was obtained from the drug annotations within the DrugBank 4.0 database (Law et al., 2014).
Model Feature Derivation
Chemical Features
The structures (sdf format) were downloaded for all of the drugs in DrugBank. The molecular weight, polar surface area, hydrogen bond donor and acceptor counts, formal charge and number of rotatable bounds were extracted from the sdf file for each of these compounds. When that information was missing, it was filled in by querying PubChem or by computationally estimating these values using ChemmineR in R. The rule outcomes were then derived from these features. The QED values were computed using the author-released script.
Network features
We constructed the aggregated biological network by taking the union across multiple databases of gene-gene interactions. (Aksoy et al., 2013; Das and Yu, 2012; Khurana et al., 2013). The network degree of a gene was calculated as the number of gene neighbors that a particular gene has. For drug’s with multiple genes, the maximum value was take. The network betweenness for a particular gene (i.e. vertex) is defined as the number of shortest paths that travel through the vertex. This was calculated using the betweenness function in R’s igraph package(Csardi and Nepusz, 2006).
Tissue features
The Gene RPKM RNA-Seq data from the Genotype-Tissue Expression (GTEx) project(Consortium, 2015) was downloaded from http://www.gtexportal.org/home/. This dataset has 2921 samples spanning 30 tissues. For each tissue, the median RPKM was calculated for each gene. For drugs with more than one target gene, the maximum RPKM was used.
Target Loss Frequency
The Exome Aggregation Consortium (ExAC) database (Exome Aggregation Consortium (ExAC)) was downloaded from www.exac.broadinstitute.org. For each gene, we counted the deleterious and total number of mutations that was reported. We calculated the loss frequency to be percentage of mutations that were reported in the gene that were deleterious.
The PrOCTOR Model
We trained the PrOCTOR approach on the clinical trials dataset using the features described above. It was trained using the random forest model, an ensembl decision tree based approach, which constructs 50 bootstrapped decision trees. A sub-sampling approach was used to account for the imbalanced ratio of approved drugs to FTT drugs, by randomly sampling the FDA approved class of samples to the size of the FTT drugs. To reduce the odds of poor representatives being sampled, this was repeated 30 times.
The labels were assigned by taking the consensus across the set of bootstrapped trees and replicates. This approach also yields a probability for each test sample. This probability was used to calculate an odds . To better visualize the distribution of this score, the log2 of the odds score was used.
Independent Datasets
To further assess our approach, we applied PrOCTOR to European (EMA) and Japanese (JP17) approved drugs, as well as 3236 drugs in DrugBank (version 4.2) (Law et al., 2014). The list of EMA-approved drugs were downloaded from the EMA website (http://www.ema.europa.eu/ema) and the JP17 list was downloaded from KEGG(Anders et al., 2015). Drugs that were already annotated as FDA approved were removed from these lists and the trained PrOCTOR model was used to make predictions for the remaining drugs. The openFDA resource (https://open.fda.gov) was used to query adverse events of drugs in the DrugBank dataset but not in our training set. FDA annotated drug-induced liver toxicity (DILI). The DILI dataset was downloaded from the FDA website at http://www.fda.gov/ScienceResearch/BioinformaticsTools/LiverToxicityKnowledgeBase/ucm226811.htm. The SIDER side effect resource database (Kuhn et al., 2010) was used to annotate side effects of each drug in the clinical trials dataset. The meddra_adverse_effects.txt table was used to extract reported adverse events, using the MedDRA Preferred Term descriptor to group similar side effects.
Statistical Analyses
We used area under the receiver operating characteristic (ROC) curve and 10-fold cross validation to evaluate the predictive power of our approach. For the independent analysis of predictions in the DrugBank dataset, we tested for enrichment of drug classes using the binomial test. We tested for differences of serious adverse event frequency between predicted toxic (score<−1) and predicted safe (score>1) drugs in the DrugBank dataset and not in the training set using the unpaired Student’s t-test. For the EMA, JP17, and DILI datasets, we tested for differences in PrOCTOR scores between predictions using the Mann-Whitney U Test. For the side effects of drugs in the training set, we used the Fisher’s Exact Test to identify the side effects that occurred more frequently in predicted toxic drugs using a p-value cutoff of 0.01.
Supplementary Material
Highlights.
Computational approach predicts the likelihood of clinical trial toxicity
Identification of molecule and target properties associated with clinical toxicity
Development of a tool to facilitate interaction and interpretation of the model
Acknowledgements
The authors would like to thank H. Weinstein and the Elemento lab members for their feedback and discussions. This work was supported by the CAREER grant from National Science Foundation (DB1054964), NIH grant R01CA194547, the Starr Cancer Foundation, as well as by startup funds from the Institute for Computational Biomedicine. Support was also provided for K.G. and N.M. by the PhRMA Foundation Pre Doctoral Informatics Fellowship and by the Tri-Institutional Training Program in Computational Biology and Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.G. and O.E. conceived, designed, and developed methodology for this work. K.G, N.M., and O.E. analyzed and interpreted the data. O.E. supervised this study.
Supplementary Materials:
Supplementary Figures
Supplementary Table S1. PrOCTOR Features.
Supplementary Table S2. DrugBank predictions.
Supplementary Table S3. DrugBank enrichment.
Supplemental Tool, available at: https://github.com/kgayvert/PrOCTOR
eTOC Blurb
Gayvert et al. present a data-driven approach that accurately predicts the likelihood of clinical trial toxicity by integrating structural and target-based properties of a drug.
References
- AACT database Clinical Trials Transformation Initiative website.
- Aksoy BA, Gao J, Dresdner G, Wang W, Root A, Jing X, Cerami E, Sander C. PiHelper: an open source framework for drug-target and antibody-target data. Bioinformatics. 2013;29:2071–2072. doi: 10.1093/bioinformatics/btt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastu-Kapur S, Anderl JL, Kraus M, Parlati F, Shenk KD, Lee SJ, Muchamuel T, Bennett MK, Driessen C, Ball AJ, et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clin Cancer Res. 2011;17:2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nature chemistry. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind E, Dunder K, de Graeff PA, Abadie E. Rosiglitazone: a European regulatory perspective. Diabetologia. 2011;54:213–218. doi: 10.1007/s00125-010-1992-5. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Machine learning. 2001;45:5–32. [Google Scholar]
- Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006:1695. [Google Scholar]
- Das J, Yu H. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC systems biology. 2012;6:92. doi: 10.1186/1752-0509-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eduati F, Mangravite LM, Wang T, Tang H, Bare JC, Huang R, Norman T, Kellen M, Menden MP, Yang J, et al. Prediction of human population responses to toxic compounds by a collaborative competition. Nat Biotechnol. 2015;33:933–940. doi: 10.1038/nbt.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Aggregation Consortium (ExAC) (Cambridge, MA)
- Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. Journal of combinatorial chemistry. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Jeliazkova N, Jeliazkov V. AMBIT RESTful web services: an implementation of the OpenTox application programming interface. J Cheminform. 2011;3:18. doi: 10.1186/1758-2946-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS computational biology. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic acids research. 2011;39:D1035–1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Molecular systems biology. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic acids research. 2014;42:D1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Translational research: 4 ways to fix the clinical trial. Nature. 2011;477:526–528. doi: 10.1038/477526a. [DOI] [PubMed] [Google Scholar]
- Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- Lewis M. Moneyball : the art of winning an unfair game. 1st W.W. Norton; New York: 2003. [Google Scholar]
- Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews. 1997;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Massacesi C, Marcucci F, Rocchi MB, Mazzanti P, Pilone A, Bonsignori M. Factors predicting docetaxel-related toxicity: experience at a single institution. J Chemother. 2004;16:86–93. [PubMed] [Google Scholar]
- Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- Patlewicz G, Fitzpatrick JM. Current and Future Perspectives on the Development, Evaluation, and Application of in Silico Approaches for Predicting Toxicity. Chemical research in toxicology. 2016;29:438–451. doi: 10.1021/acs.chemrestox.5b00388. [DOI] [PubMed] [Google Scholar]
- Puisset F, Alexandre J, Treluyer JM, Raoul V, Roche H, Goldwasser F, Chatelut E. Clinical pharmacodynamic factors in docetaxel toxicity. Br J Cancer. 2007;97:290–296. doi: 10.1038/sj.bjc.6603872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahota T, Danhof M, Della Pasqua O. Pharmacology-based toxicity assessment: towards quantitative risk prediction in humans. Mutagenesis. 2016;31:359–374. doi: 10.1093/mutage/gev081. [DOI] [PubMed] [Google Scholar]
- Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- USEPA ToxCast & Tox21 Chemicals Distributed Structure-Searchable Toxicity Database from DSSTox_20151019. 2016 [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. Journal of medicinal chemistry. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.