Abstract
C8-desaturated and C9-methylated glucosylceramide (GlcCer) is a fungal-specific sphingolipid that plays an important role in the growth and virulence of many species. In this work, we investigated the contribution of Aspergillus nidulans sphingolipid Δ8-desaturase (SdeA), sphingolipid C9-methyltransferases (SmtA/SmtB) and glucosylceramide synthase (GcsA) to fungal phenotypes, sensitivity to Psd1 defensin and Galleria mellonella virulence. We showed that ΔsdeA accumulated C8-saturated and unmethylated GlcCer, while gcsA deletion impaired GlcCer synthesis. Although increased levels of unmethylated GlcCer were observed in smtA and smtB mutants, ΔsmtA and wild-type cells showed a similar 9,Me-GlcCer content, reduced by 50 % in the smtB disruptant. The compromised 9,Me-GlcCer production in the ΔsmtB strain was not accompanied by reduced filamentation or defects in cell polarity. When combined with the smtA deletion, smtB repression significantly increased unmethylated GlcCer levels and compromised filamentous growth. Furthermore, sdeA and gcsA mutants displayed growth defects and raft mislocalization, which were accompanied by reduced neutral lipids levels and attenuated G. mellonella virulence in the ΔgcsA strain. Finally, ΔsdeA and ΔgcsA showed increased resistance to Psd1, suggesting that GlcCer synthesis and fungal sphingoid base structure specificities are relevant not only to differentiation but also to proper recognition by this antifungal defensin.
Keywords: glucosylceramide, Aspergillus nidulans, plant defensins, Pisum sativum defensin 1 (Psd1)
Graphical abstract
Introduction
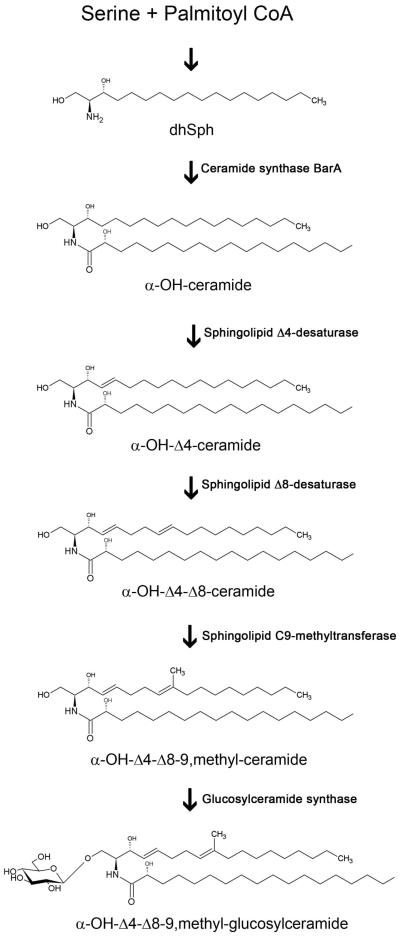
Glucosylceramide (GlcCer or CMH, ceramide monohexoside) is a neutral sphingolipid composed of a sphingoid base (or LCB, long chain base), a fatty acid chain and a glucose residue. GlcCer is found in most fungi, except (so far) in Saccharomyces cerevisiae and Candida glabrata (Saito et al., 2006), and is conserved in higher eukaryotes, such as plants and mammals. Although widely distributed among species, GlcCer structure and synthesis pathways vary in different organisms, generating distinct intermediates and products. In fungal cells, GlcCer synthesis is initiated by the condensation of palmitoyl-CoA and serine, producing 3-ketosphinganine, which is then reduced to dihydrosphingosine (dhSph, or sphinganine, Figure 1) (Barreto-Bergter et al., 2004). Ceramide synthase catalyzes the condensation of fatty acids to sphinganine, generating ceramide (Figure 1). Aspergillus nidulans ceramide synthase BarA has been previously characterized and shown to mediate the effects of a novel bacterial compound, HSAF (Li et al., 2006). Next, sphingolipid Δ4- and Δ8-desaturases promote the Δ4- and Δ8-desaturation of LCB in ceramide, and sphingolipid C9-methyltransferase catalyzes the introduction of a methyl group at C-9 of the LCB (Figure 1) (Ternes et al., 2002, Ternes et al., 2006). The product generated is a fungal GlcCer containing 9-methyl-4,8-sphingadienine as a sphingoid base, which differs from its mammalian counterpart 4-sphingenine. The last step of GlcCer synthesis is the transfer of a glucose residue from UDP-glucose to the ceramide moiety by glucosylceramide synthase (Figure 1) (Leipelt et al., 2001, Ternes et al., 2011). Once produced, GlcCer is exported from the Golgi to the plasma membrane or cell wall or is secreted to the extracellular space in vesicles (Rodrigues et al., 2000, Rodrigues et al., 2007, Rodrigues et al., 2008, Nimrichter et al., 2005). Both GlcCer synthesis and LCB structural modifications are necessary for fungal differentiation and pathogenesis. The disruption of Candida albicans sphingolipid Δ8-desaturase leads to reduced hyphal growth in solid medium, and this phenotype is attributed to the accumulation of 4-sphingenine (Oura & Kajiwara, 2008). Moreover, Cryptococcus neoformans cells lacking sphingolipid C9-methyltransferase (Δsmt1) and glucosylceramide synthase (Δgcs1), which do not produce methylated GlcCer (α-OH-Δ4-Δ8-9,methyl-GlcCer) or any GlcCer species, respectively, show a strong reduction of infectivity and tissue burden in a murine model of cryptococcosis (Rittershaus et al., 2006, Singh et al., 2012). The filamentous fungus Fusarium graminearum possesses two genes encoding functional methyltransferases: while ΔFgmt1 cells produce methylated GlcCer as the wild-type strain, FgMT2 disruption significantly reduces its levels, leading to the synthesis of up to 75 % non-methylated GlcCer (Ramamoorthy et al., 2009). The ΔFgmt2 strain also shows growth defects in solid medium, abnormal conidia formation and reduced pathogenicity in wheat and Arabidopsis thaliana, in contrast to the full virulence exhibited by the ΔFgmt1 strain (Ramamoorthy et al., 2009). Similarly, FgGCS1 deletion impairs mycelia growth and plant disease severity (Ramamoorthy et al., 2007). Taken together, these reports suggest that GlcCer plays a critical role in fungal virulence and that molecules that are capable of inhibiting enzymes involved in GlcCer biosynthesis or that specifically interact with fungal GlcCer structure may act as novel antifungal drugs. In fact, anti-GlcCer monoclonal antibody administration inhibits C. neoformans budding and growth and prevents cryptococcosis in mice (Rodrigues et al., 2000, Rodrigues et al., 2007). Moreover, GlcCer is the binding target of some plant defensins, cysteine-rich peptides that present antifungal activity (Thevissen et al., 2004, Ramamoorthy et al., 2007, Ramamoorthy et al., 2009), including Psd1, which was isolated and structurally characterized by our group (Almeida et al., 2001, Neves de Medeiros et al., 2014).
Figure 1.
The proposed fungal glucosylceramide pathway. dhSph, dihydrosphingosine.
Psd1 is a 54-amino-acid peptide that shows inhibitory activity against several fungal species, such as A. nidulans, A. niger, C. albicans and Fusarium solani (Almeida et al., 2001 and personal communication). A previous analysis of Psd1 chemical shift perturbations by NMR spectroscopy revealed that this peptide interacts with small unilamellar vesicles (SUV) containing F. solani GlcCer, specifically with the amino acids of loop 1 and turn 3 (de Medeiros et al., 2010). Furthermore, the Psd1 binding affinity to SUVs containing different sphingolipids was assessed through Surface Plasmon Resonance, and high association rates between Psd1 and pure GlcCerF. solani were observed, followed by weaker interactions with vesicles composed of phosphatidylcholine (PC):GlcCerF. solani 7:3 and PC alone (Neves de Medeiros et al., 2014). Interestingly, the Psd1 binding response to PC:GlcCersoybean was slightly higher than that to PC but still lower than that to PC:GlcCerF. solani. Soybean GlcCer is composed of 4,8-sphingadienine attached to a 2-hydroxypalmitic acid, while fungal GlcCer is usually constituted by 9-methyl-4,8-sphingadienine linked to 2-hydroxystearic acid. These results suggest that fatty acid structure and C9-methylation may play a role in fungal GlcCer recognition by Psd1. An interesting feature of Psd1 antifungal activity is that it does not solely rely on cell membrane anchoring because the peptide interacts with Neurospora crassa nuclear cyclin F, leading to cell cycle arrest (Lobo et al., 2007). Psd1 internalization was also observed in F. solani hyphae, which suggests a conserved mechanism among fungal species (Lobo et al., 2007). Furthermore, Psd1 was unable to internalize and efficiently inhibit the growth of C. albicans cells lacking the GlcCer synthase gene (HSX11), indicating that GlcCer production is also relevant for the Psd1 mechanism of action (Neves de Medeiros et al., 2014).
The present study used the model fungus A. nidulans to investigate the contribution of sphingolipid Δ8-desaturase (SdeA), sphingolipid C9-methyltransferases (SmtA and SmtB) and glucosylceramide synthase (GcsA) to fungal growth, lipid metabolism, virulence in Galleria mellonella larvae and Psd1 recognition. Lipid profiling revealed that the ΔgcsA mutant was unable to synthesize any GlcCer, while the ΔsdeA strain accumulated C8-saturated and unmethylated GlcCer. Surprisingly, smtA or smtB single deletion did not abolish methylated GlcCer production; however, an accumulation of unmethylated GlcCer was detected. Furthermore, smtA deletion along with smtB repression led to increased levels of unmethylated GlcCer and reduced filamentation. sdeA and gcsA deletion impaired filamentous growth, conidiophore formation and regular filipin staining. Intriguingly, all of the mutant strains exhibited increased resistance to cell wall (CW) stressing agents Calcofluor White and Congo Red, suggesting a compensatory mechanism in CW structure to retain viability. Finally, the ΔgcsA mutant had attenuated virulence in a G. mellonella larvae infection model, and as a ΔsdeA disruptant, showed increased resistance to Psd1, indicating that GlcCer synthesis and its proper structure are required for the peptide mechanism of action.
Results
Gene identification and structure
As an initial approach to identify A. nidulans putative sphingolipid Δ8-desaturase and glucosylceramide synthase-coding genes, sequence candidates were searched using C. albicans SLD1 and HSX11 genes as queries. The bidirectional BLAST analysis revealed AN4592 (here named sdeA; e-value = 1e-147; 57.9 % identity; 74.4 % similarity) and AN8806 (here named gcsA; e-value = 1e-58; 50 % identity; 68.2 % similarity) genes, respectively, whose predicted function was further confirmed in the Aspergillus genome databank (www.aspgd.org). The sdeA and gcsA gene models are 1,991 and 1,710 nucleotides long, with one intron each (sdeA: from 189 to 243; gcsA: from 147 to 197); their open reading frames (ORFs) contain 1,638 and 1,659 nucleotides encoding 545 and 552 amino acids or 63.2- and 62.0-kDa proteins.
Fungal sphingolipid C9-methyltransferases were first reported in a phylogenetic profile analysis that revealed the existence of two candidate genes in A. nidulans (Ternes et al., 2006). Here, AN5688 and AN7375 are referred to as smtA and smtB and seem to represent putative paralogues because they show high identity (e-value = 0.0; 60.7 % identity; 76.0 % similarity). The smtA and smtB gene models are 1,759 and 1,968 nucleotides in length and include three introns each (smtA: from 106 to 206, 694 to 746, and 1499 to 1549; and smtB: from 118 to 168, 1293 to 1339, and 1449 to 1506). Moreover, smtA and smtB ORFs possess 1,554 and 1,533 nucleotides that encode hypothetical proteins of 517 and 510 amino acids, or 58.2 and 57.7 kDa, respectively. All the gene models are supported by RNA-seq data (available at www.aspgd.org).
Protein domains were predicted using the SMART interface (http://smart.embl-heidelberg.de/) (Schultz et al., 1998, Letunic et al., 2015) and the Phyre2 protein homology tool (Kelley & Sternberg, 2009). Putative A. nidulans SmtA and SmtB were assumed to have two transmembrane domains each, and a Pfam domain was identified in the same CMAS methyltransferase region of both proteins (Supplementary Figure S1, PF02353, e-values = 3.3e-64 and 7.2e-61, respectively). The SdeA was predicted to contain five transmembrane domains (Supplementary Figure S1), two Pfam domains, a Cytochrome b5-like Heme/Steroid binding domain (PF00173, 1.35e-7) and a FA_desaturase domain (PF00487, 2.8e-43). Putative glucosylceramide synthase (GcsA) consisted of four transmembrane domains (Supplementary Figure S1), a Pfam domain and Glyco_transf_2_3 (PF13641, 6.9e-11).
A. nidulans ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and conditional double mutant ΔsmtA niiA::smtB analysis
To gain a deeper understanding of glucosylceramide pathway relevance in A. nidulans biology, we constructed sdeA, smtA, smtB and gcsA null mutants. A. nidulans wild-type protoplasts were transformed with deletion cassettes containing the pyrG sequence flanked by 5’ and 3’ UTR regions of target ORFs, and the transformant colonies were selected by their ability to grow in the absence of uracil and uridine (for more details, see Experimental Procedures). Homologous recombination was verified by digesting genomic DNA with EcoRV or BamHI restriction enzymes and exploring their differential recognition of pyrG and sdeA, smtA, smtB and gcsA sequences, followed by a Southern Blot analysis of the fragments using 1 kb of 5’ UTR as a hybridization probe (Supplementary Figure S2). This strategy led to the identification of differential DNA fragments in the wild-type and mutant strains, indicating that these genes were deleted and that a single integration event occurred in disrupted strains (Supplementary Figure S2). These results indicate that deletion cassettes were correctly inserted at the target loci, generating ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA strains deficient in three steps of the GlcCer synthetic pathway (see Figure 1).
We were able to generate at least five null mutants for each gene; a single null strain of each gene was selected for the majority of the analyses. To ensure that the deletion strains used in this manuscript (containing the pyrG+ deletion cassette) did not possess additional mutations, null mutants were sexually crossed with a strain containing a non-functional pyrG mutation (GR5 strain), which is auxotrophic for uracil and uridine. The subsequent null pyrG+ progeny, consisting of uracil and uridine prototrophs, was selected for further analysis. The phenotypic investigation of the progeny did not reveal any pyrG mutation cosegregating with either ΔsdeA, ΔsmtA, ΔsmtB or ΔgcsA, strongly indicating that the pyrG+ marker cosegregates with the deletion mutants and, ultimately, that deficient strains only present mutations in the sdeA, smtA, smtB and gcsA genes (Supplementary Table 1).
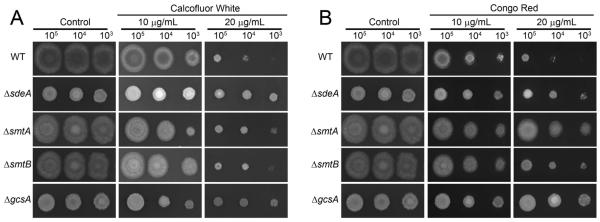
GlcCer presence has been previously reported in the fungal cell wall, plasma membrane and intracellular vesicles (Rodrigues et al., 2000, Rodrigues et al., 2007). Therefore, we initially analyzed GlcCer synthesis contribution to A. nidulans viability in the presence of cell-wall-damaging agents, such as Calcofluor White (CFW) and Congo Red (CR) (Figure 2). Surprisingly, all four mutants showed increased CFW and CR resistance at 10 and 20 μg.ml−1 (Figure 2).
Figure 2.
The glucosylceramide-defective mutants are more resistant to cell wall-damaging agents. In dropout experiments, 5 μl of ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and wild-type strains suspensions containing 2.107, 2.106 and 2.105 conidia.ml−1 was point inoculated in minimal media supplemented with different concentrations of CFW (A) or CR (B) and grown for 48 h at 37 °C.
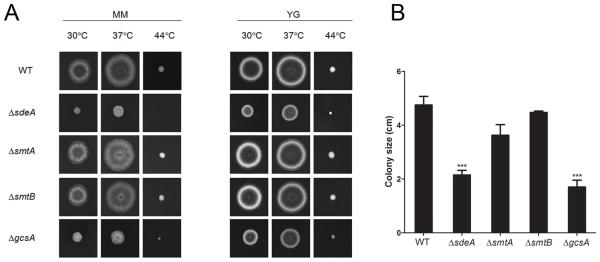
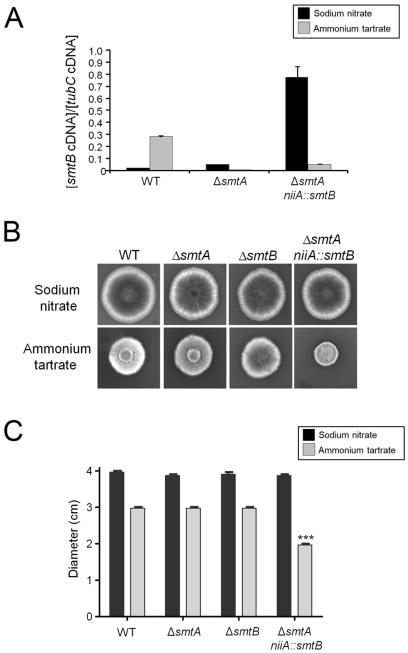
To investigate the relevance of LCB structural modifications and GlcCer production to A. nidulans growth and differentiation, we analyzed the ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA growth rates. Curiously, no significant difference was observed between the growth rates of cells lacking smtA or smtB and the parental strain after approximately 50 h of growth in liquid YUU (data not shown). However, sdeA and gcsA deletions severely compromised fungal growth in liquid medium (data not shown). Furthermore, ΔsdeA and ΔgcsA disruptants exhibited an approximately 55-65 % reduction in radial growth at 30 and 37 °C and a dramatic impairment of conidiation compared to the wild-type strain (Figures 3A and B). In contrast, the smtA null mutant showed a minor reduction in colony size (Figures 3B). To further elucidate the role of both smtA and smtB genes in fungal growth, we constructed the conditional double mutant ΔsmtA niiA::smtB. In this strain, smtB expression is under control of the nitrite reductase gene promoter (niiA), which is induced by sodium nitrate and repressed by ammonium tartrate (Punt et al., 1991). Indeed, ΔsmtA niiA::smtB cells grown in ammonium tartrate exhibited a 7-fold reduction in smtB mRNA levels compared to the culture maintained in sodium nitrate (Figure 4A). Additionally, smtB repression, along with smtA deletion, led to a 50 % reduction in A. nidulans growth (Figure 4B and 4C), indicating that sphingolipid C9-methyltransferases contributes to fungal differentiation and that the double mutant may not be viable.
Figure 3.
GlcCer formation and sphingoid base structural modifications contribute to A. nidulans hyphal growth and differentiation. The ΔsdeA and ΔgcsA mutants showed reduced growth compared to ΔsmtA, ΔsmtB and wild-type strains. (A) Conidial suspensions of WT, ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA strains were spotted in minimal (MM) or complete (YG) solid media supplemented with uracil, uridine and pyridoxine and grown at 30, 37 or 44 °C for 2 days. (B) The colony size was determined through growth diameter measurement at 37 °C. The results are the average of three repetitions ± SEM; ***p < 0.001.
Figure 4.
GlcCer C9-methylation is relevant for A. nidulans growth. The ΔsmtA niiA::smtB conditional double mutant exhibits impaired growth under niiA-repressing conditions. (A) Relative smtB mRNA levels in WT, ΔsmtA and ΔsmtA niiA::smtB strains grown for 16 h at 37 °C in the presence of niiA-inducer (10 mM sodium nitrate, black bars) or niiA-repressor agents (50 mM ammonium tartrate, grey bars). (B) Conidial suspensions of WT and ΔsmtA, ΔsmtB, ΔsmtA niiA::smtB mutants were point inoculated in MM containing nitrate or tartrate and uracil, uridine, pyridoxine. After 2 days of growth at 37 °C, images were taken, and (C) the colony diameter was measured. The results are the average of 3 replicates ± SEM; ***p < 0.001.
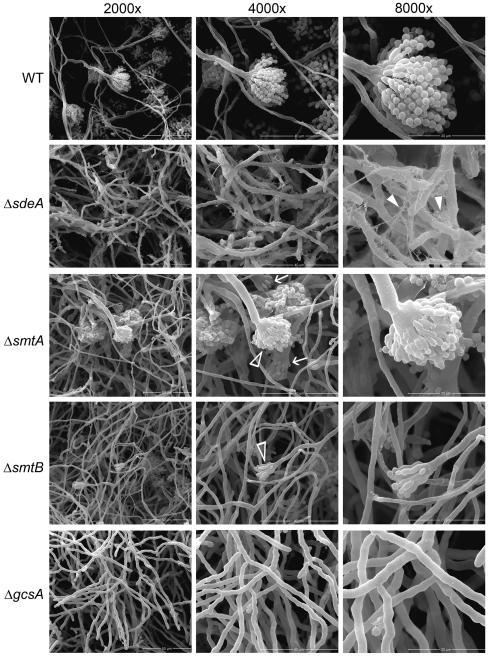
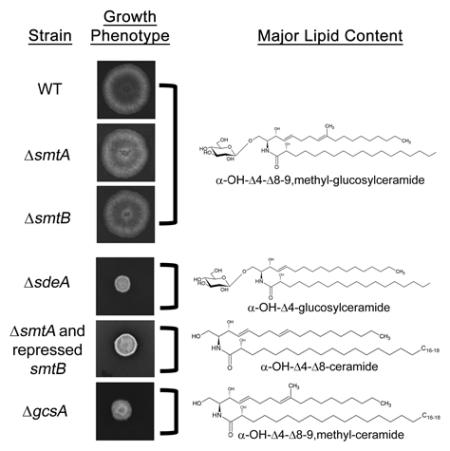
To determine whether GlcCer synthesis is also related to conidiophore formation, conidia were grown in supplemented liquid MM medium for 96 h, and conidiophore structure was assessed through Scanning Electron Microscopy (SEM). As expected, vegetative mycelia from the parental strain developed into mature conidiophores, with phialides linked to a chain of conidia (Figure 5). In contrast, ΔsdeA and ΔgcsA mutants were unable to form conidiophores, even after 120 h of growth (data not shown). Curiously, ΔsdeA mycelia exhibited a dense extracellular matrix (indicated by white filled arrowheads in Figure 5), which was less abundant in the parental strain. Despite some fields with regular conidiophores, ΔsmtA and ΔsmtB mutants generally showed a reduction in the number of conidiation structures (white arrowheads in Figure 5), along with abnormal conidiophore morphology in the smtA disruptant (white arrows in Figure 5). These data suggest that LCB structure and GlcCer synthesis are required for proper conidiophore development and, ultimately, for conidia production.
Figure 5.
The GlcCer synthesis pathway is required for conidiophore development. A total of 105 conidia of WT, ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA strains were grown in MM + UU + pyro media at 25 °C for 96 h, and mycelia topography was analyzed by SEM. The left, middle and right panels indicate 2,000X, 4,000X and 8,000X magnification, and the bars represent 50, 40 and 20 μm, respectively. Filled white arrowheads show the extracellular matrix, and white arrows and arrowheads highlight reduced and abnormal conidiophores.
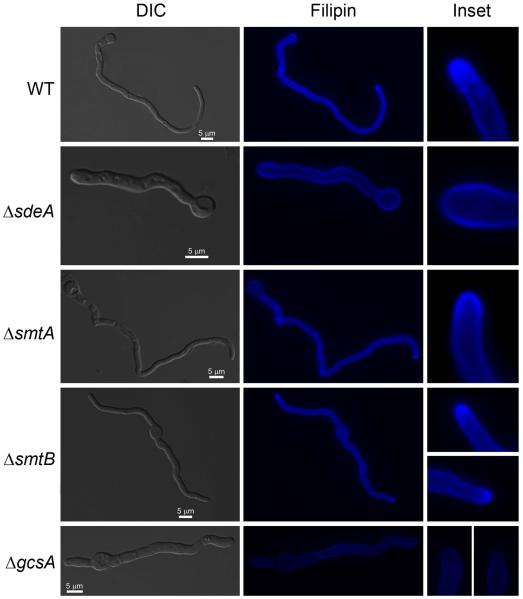
Lipid rafts are specialized membrane structures consisting of an aggregation of sphingolipids and ergosterol that mediates biosynthetic and endocytic processes by anchoring compounds to the plasma membrane (Alvarez et al., 2007). Lipid rafts serve as mounting and organizing centers for signaling molecules and are also relevant for cell polar organization (Alvarez et al., 2007). The highly polarized nature of fungal cells is a hallmark of their morphology as they grow through the insertion of a new membrane into the cell wall surface. For this purpose, vesicles loaded with components required for cell wall expansion are transported to active growth sites over a network of polarized microtubes (Gow, 1994). To investigate whether ΔsdeA and ΔgcsA reduced growth was accompanied by defects in cell polarity, we assessed lipid raft localization in germling cell membranes by staining with filipin, a fluorescent polyene antibiotic that binds sterols (Ghannoum and Rice, 1999). Filipin staining helps to determine whether membrane lipids are being delivered to the hyphal apex during polar growth. Intense filipin fluorescence was observed in 80 ± 17.5, 70 ± 12.7 and 71 ± 13.1 % of hyphal apexes in the wild-type, ΔsmtA and ΔsmtB strains, respectively (Figure 6). In contrast, a dispersed staining pattern was observed throughout the membrane of ΔsdeA and ΔgcsA germlings (Figure 6), which exhibited only 13.8 ± 4.5 and 10.8 ± 4.9 % of hyphal apexes with high binding affinity to filipin. Taken together, these findings strongly suggest that genes encoding enzymes involved in GlcCer synthesis contribute to cell wall structure, proper fungal growth and cell polarity.
Figure 6.
The polarized delivery of membrane lipids was confined to the hyphal apex in the wild-type, ΔsmtA and ΔsmtB strains but not in the ΔsdeA and ΔgcsA strains. Germlings were grown in MM + UU + pyro media for 16 hours at 25 °C and then stained with 25 μg.ml−1 filipin. The bars represent 5 μm. Insets show the hyphal apex of the wild-type and mutant strains.
Lipid content characterization in A. nidulans ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and ΔsmtA niiA::smtB strains
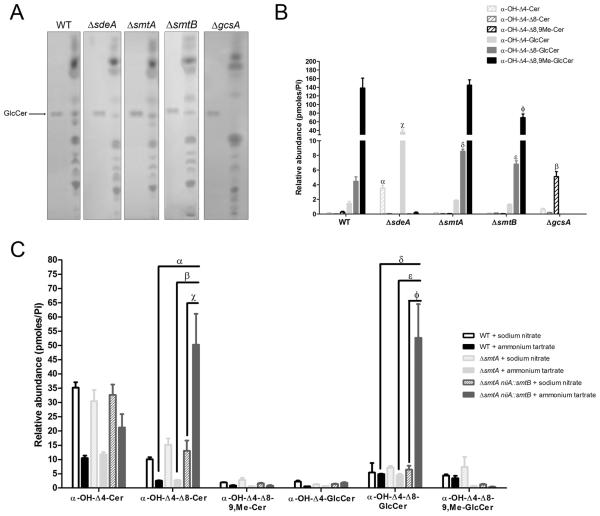
To further characterize the contribution of sdeA, smtA, smtB and gcsA to GlcCer synthesis, we initially investigated using thin layer chromatography (TLC) whether the mutant strains were able to synthesize this sphingolipid. A lipid band corresponding to GlcCer migration in chloroform: methanol: water 65:25:4 was observed in the wild-type strain and in the ΔsdeA, ΔsmtA and ΔsmtB extracts but not in ΔgcsA (Figure 7A), suggesting that deleted gcsA is truly an A. nidulans glucosylceramide synthase.
Figure 7.
(A) The ΔgcsA strain is unable to synthesize GlcCer. The ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and parental conidia were grown in YUU medium at 37 °C for 3-4 days. The total lipids were obtained according to Bligh & Dyer and submitted to TLC using chloroform: methanol: water 65:25:4 as the solvent mixture. Bands corresponding to distinct lipid classes were visualized after iodine staining, and 25 μg of soy GlcCer was used as the migration standard of the molecule (arrow). (B, C) The relative abundance of the intermediates involved in GlcCer synthesis after growth in YUU media (B) or under niiA-inducing/repressing conditions (C). (B) The relative abundance of α-OH-Δ4-ceramide (hatched light grey bars), α-OH-Δ4-Δ8-ceramide (hatched dark grey bars), α-OH-Δ4-Δ8-9,methyl-ceramide (hatched black bars) and their glycosylated products (α-OH-Δ4-GlcCer, filled light grey bars; α-OH-Δ4-Δ8-GlcCer, filled dark grey bars; and α-OH-Δ4-Δ8-9,methyl-GlcCer, filled black bars, respectively) in the ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and wild-type lipid extracts. All of the values represent the mean ± SEM of two independent experiments, as analyzed by a one-way ANOVA. αp < 0.001, ΔsdeA versus WT; βp < 0.001, ΔgcsA versus WT; χp < 0.001, ΔsdeA versus WT; δp < 0.01, ΔsmtA versus WT; εp < 0.05, ΔsmtB versus WT; φp < 0.05, ΔsmtB versus WT). (C) Relative abundance of GlcCer synthesis intermediates in the wild-type, ΔsmtA and ΔsmtA niiA::smtB strains grown in MMM medium supplemented with 10 mM sodium nitrate (black, hatched light grey and hatched dark grey bars, respectively) or 50 mM ammonium tartrate (filled black, filled light grey and filled dark grey bars, respectively) for niiA-induction/repression. The values represent the means ± SEM of three independent experiments, as analyzed by a one-way ANOVA. αp < 0.001, ΔsmtA niiA::smtB + ammonium tartrate versus wild-type + ammonium tartrate, βp < 0.001, ΔsmtA niiA::smtB + ammonium tartrate versus ΔsmtA + ammonium tartrate, χp < 0.01, ΔsmtA niiA::smtB + ammonium tartrate versus ΔsmtA niiA::smtB + sodium nitrate; δp < 0.001, ΔsmtA niiA::smtB + ammonium tartrate versus wild-type + ammonium tartrate, εp < 0.001, ΔsmtA niiA::smtB + ammonium tartrate versus ΔsmtA + ammonium tartrate, φp < 0.001, ΔsmtA niiA::smtB + ammonium tartrate versus ΔsmtA niiA::smtB + sodium nitrate.
Moreover, the relative abundance of the intermediates involved in GlcCer synthesis (α-OH-Δ4-ceramide, α-OH-Δ4-Δ8-ceramide, α-OH-Δ4-Δ8-9,methyl-ceramide) and its glycosylated forms (C8-saturated and unmethylated GlcCer: α-OH-Δ4-GlcCer, unmethylated GlcCer: α-OH-Δ4-Δ8-GlcCer, and the final product - desaturated and methylated GlcCer: α-OH-Δ4-Δ8-9,methyl-GlcCer) (see schematic pathway representation in Figure 1) was further quantified using mass spectrometry (Figure 7B). In the wild-type extract, high levels of α-OH-Δ4-Δ8-9,methyl-GlcCer and lower levels of other GlcCer species, such as α-OH-Δ4-GlcCer and α-OH-Δ4-Δ8-GlcCer, were observed (Figure 7B). Moreover, sdeA deletion impaired α-OH-Δ4-Δ8-9,methyl-GlcCer synthesis, leading to the accumulation of α-OH-Δ4-Cer and its conversion to α-OH-Δ4-GlcCer through gcsA activity. Intriguingly, smtA and smtB single deletions did not abolish methylated GlcCer production (Figure 7B). Nevertheless, smtB seemed to be more involved in adding a –CH3 group to the sphingoid base, as its depletion reduced by 50 % the abundance of α-OH-Δ4-Δ8-9,methyl-GlcCer, which was present in the ΔsmtA extract at levels comparable to those detected in the wild-type strain. Additionally, ΔsmtA and ΔsmtB extracts showed a significantly higher concentration of unmethylated GlcCer (α-OH-Δ4-Δ8-GlcCer) compared to that of the parental strain, indicating that, although single gene deletions did not impair α-OH-Δ4-Δ8-9,methyl-GlcCer synthesis, they still compromise ceramide methylation at some level.
The role of both sphingolipid C9-methyltransferases in GlcCer synthesis was analyzed in the wild-type, ΔsmtA and ΔsmtA niiA::smtB strains under niiA-inducing (10 mM sodium nitrate) and niiA-repressing (50 mM ammonium tartrate) conditions. Curiously, WT and ΔsmtA cells grown in MM + sodium nitrate or MM + ammonium tartrate (Figure 7C) exhibited an increased ceramide content and reduced methylated GlcCer abundance compared to those of the cultures maintained in rich (YUU) media (Figure 7B). Once GlcCer regulates fungal growth and differentiation, this distinct lipid profile may be correlated to the reduced growth observed in MM media (Figure 3). Additionally, in the wild-type extracts, similar contents of α-OH-Δ4-Δ8-GlcCer and α-OH-Δ4-Δ8-9,Me-GlcCer were observed, followed by a small amount of α-OH-Δ4-GlcCer (Figure 7C). Similar to those observed in YUU-grown cultures (Figure 7B), ΔsmtA cells grown in MM + sodium nitrate exhibited comparable levels of methylated GlcCer than did the wild-type strain (Figure 7C). Interestingly, smtB repression induced by ammonium tartrate along with smtA deletion caused a significant accumulation of α-OH-Δ4-Δ8-Cer and its conversion product (α-OH-Δ4-Δ8-GlcCer, Figure 7C). Taken together, these observations indicate that smtA and smtB play a crucial role in methylating the ceramide molecule and that a lack of these genes causes a remarkable production of unmethylated GlcCer.
Finally, gcsA deletion abolished the production of all GlcCer species, causing the accumulation of methylated ceramide (α-OH-Δ4-Δ8-9,methyl-ceramide). This result suggests that A. nidulans glucosylceramide synthase is the only enzyme involved in catalyzing glucose transfer to ceramide backbones.
The ΔgcsA null mutant shows reduced levels of neutral lipids
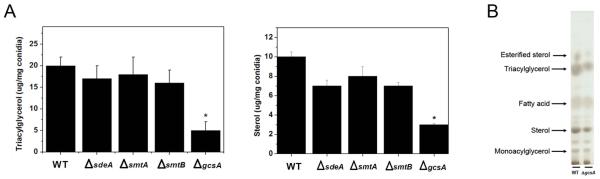
To investigate GlcCer synthesis contribution to neutral lipid homeostasis, triacylglycerol (TAG) and sterol abundance was analyzed in the wild-type strain and the ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA mutants. All of the deficient strains exhibited TAG and sterol levels similar to those in parental cells, except for the gcsA disruptant, in which significant reductions of these lipids were observed (Figure 8A). Indeed, a TLC analysis of the ΔgcsA lipid extract revealed a slight decrease in sterol and TAG and a major decrease in esterified sterol content (Figure 8B). These findings indicate that the ΔgcsA-altered growth phenotype may be correlated not only with GlcCer depletion but also with compromised neutral lipid production.
Figure 8.
GlcCer synthesis contributes to the maintenance of neutral lipid levels. A reduced abundance of triacylglycerols and sterols was observed in the gcsA disruptant. An analysis of the neutral lipid content: (A) Triacylglycerol and sterol levels were quantified in WT and ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA mutants. The results represent the mean value ± SEM, *p < 0.05, (B) For comparison of the TLC of ΔgcsA and WT extracts, lipid extraction was performed with equal weights of mycelia. The total lipids were suspended in chloroform: methanol 2:1, and identical volumes were applied to a silica plate. Neutral lipid separation was carried out using hexane: ethyl ether: acetic acid 6:4:1, and 25 μg of monoacylglycerol, triacylglycerol, sterol, fatty acids and esterified sterol standards was used as a control of the migration pattern. Lipid bands were visualized by Charring reagent staining.
GcsA is important for A. nidulans virulence in a Galleria mellonella infection model
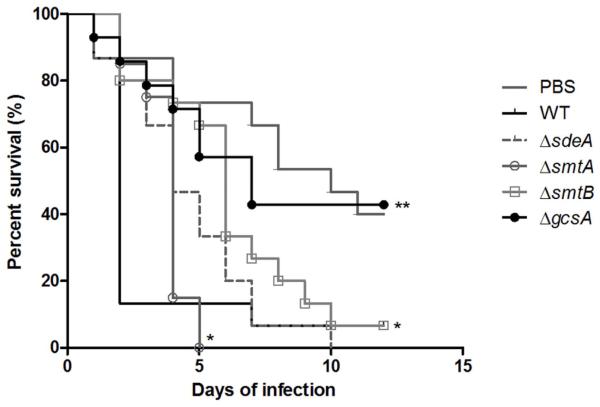
Both GlcCer production and LCB C9-methylation are essential for C. neoformans virulence in mice (Rittershaus et al., 2006, Singh et al., 2012). Curiously, GCS1 contribution to F. graminearum pathogenesis is host-dependent (Ramamoorthy et al., 2007). Moreover, FgMT2 deletion (which led to the production of unmethylated GlcCer) severely compromised fungal virulence in A. thaliana (Ramamoorthy et al., 2009). To better understand the role of GlcCer synthesis and structure in filamentous fungi pathogenesis, we analyzed the ability of A. nidulans to infect the alternative animal model Galleria mellonella, which has emerged as an infection model for Aspergillus (Jackson et al., 2009). Wild-type, ΔsdeA, ΔsmtA and ΔsmtB infections resulted in approximately 90 % mortality after 5 to 10 days of infection, while ΔgcsA showed a significantly reduced mortality rate (approximately 40 %) 12 days post-infection (Figure 9, p > 0.01 for the comparison between the wild-type and ΔsdeA, ΔsmtA and ΔsmtB using Log−rank - Mantel−Cox test and p < 0.01 for the comparison between the wild-type and ΔgcsA). These experiments were repeated twice with independent transformants for each deletion strain, and comparable results were obtained (Supplementary Figure S3).
Figure 9.
The A. nidulans gcsA mutant shows reduced virulence in G. mellonella. Larvae were treated with saline solution or infected with 1.106 conidia/caterpillar, and the survival rate was assessed for 10 days post inoculation. Each experimental group was composed of fifteen caterpillars, and the analyses were performed three times, presenting a similar pattern. No significant difference in the percentage viability was observed between ΔsdeA- and wild-type-infected insects, while ΔgcsA inoculation led to decreased virulence compared to the parental strain. The log-rank test was used to compare the survival rates; **p < 0.01 and *p < 0.05.
Psd1 antifungal activity depends on both GlcCer synthesis and proper structure
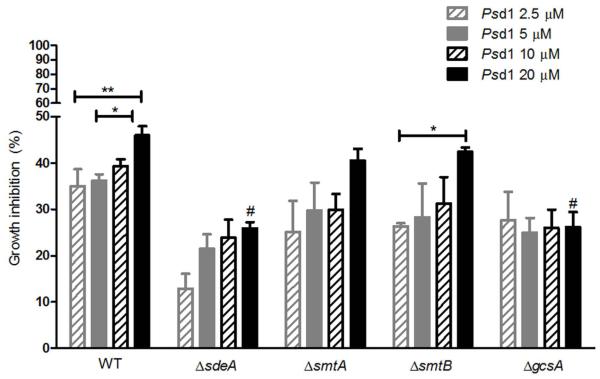
We previously reported that the deletion of the C. albicans glucosylceramide synthase gene confers resistance to Psd1 antifungal activity (Neves de Medeiros et al., 2014). Additionally, Psd1 does not show toxicity to some mammalian cell lineages (unpublished results), indicating that the peptide specifically recognizes fungal GlcCer structure. Indeed, Psd1 exhibited weaker binding affinity to vesicles containing soybean GlcCer, which lacks LCB C9-methylation (Sullards et al., 2000), than to those containing F. solani GlcCer (Neves de Medeiros et al., 2014). To determine whether, in addition to GlcCer synthesis, LCB structural alterations are also relevant for the Psd1 mechanism of action, we assessed the sensitivity of ΔsdeA, ΔsmtA and ΔsmtB strains (which accumulate α-OH-Δ4-GlcCer and α-OH-Δ4-Δ8-GlcCer, respectively) and the ΔgcsA mutant (which does not produce any GlcCer) to Psd1. For this purpose, germlings were incubated with 2.5-20 μM of the peptide, and the ability of A. nidulans to grow was measured at OD540, considering cell suspensions maintained in the absence of antifungal agents or 10 μM itraconazole as 0 and 100 % inhibition, respectively. Curiously, TNO2A3 germlings (here defined as wild-type) were intrinsically more resistant to Psd1 than were other A. nidulans strains, such as GR5 and A4 (data not shown). As expected, gcsA disruption diminished Psd1 antifungal activity, causing a 26 % growth inhibition in 20 μM-treated culture compared to 46 % observed in the wild-type strain (Figure 10). In addition, a dose-response pattern was not observed in the ΔgcsA mutant, indicating that this strain is resistant even to high concentrations of the peptide. Unmethylated GlcCer (α-OH-Δ4-Δ8-GlcCer) accumulation in ΔsmtA and ΔsmtB was not followed by reduced Psd1 activity, as 40 and 42 % growth inhibition was observed, respectively, in cultures grown in 20 μM of the peptide (Figure 10). The contribution of both smtA and smtB genes for Psd1 growth inhibition was also investigated; however, the high salt concentration (10 mM sodium nitrate and 50 mM ammonium tartrate) required for niiA-induction/repression impaired the peptide antifungal activity (data not shown). Furthermore, the peptide induced 29 % growth inhibition in the ΔsdeA null mutant, which was as resistant to 20 μM Psd1 as was the ΔgcsA strain (Figure 10). However, in contrast to ΔgcsA, ΔsdeA culture exhibited a dose-response profile to Psd1, suggesting that the fungal-specific LCB structure is as relevant as is the glucose moiety to higher concentrations of Psd1.
Figure 10.
Psd1 inhibitory activity depends partially on GlcCer synthesis and fungal sphingoid base structure. Wild-type, ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA cells were grown in YUU medium containing 2.5 (hatched grey bars), 5 (filled grey bars), 10 (hatched black bars) and 20 μM Psd1 (filled black bars) until OD540≥ 1.0. The growth inhibition in defensin-treated cultures was calculated using as parameters the suspensions maintained in the absence of antifungal drugs (0 % inhibition) or in 10 μM itraconazole (100 % inhibition). The values represent the means ± SEM, n = 3; *p < 0.05, 20 μM-treated WT versus 5 μM-treated WT and 20 μM-treated ΔsmtB versus 2.5 μM-treated ΔsmtB; **p < 0.01, 20 μM-treated WT versus 2.5 μM-treated WT; and #p < 0.001, 20 μM-treated ΔsdeA versus 20 μM-treated WT and 20 μM-treated ΔgcsA versus 20 μM-treated WT.
Discussion
GlcCer synthesis is relevant for fungal growth, differentiation and pathogenesis. Moreover, its distinctive sphingoid base structure, containing a Δ8-desaturation and a methyl group at C9, also seems to play a role in virulence, as C. albicans sld/sld cells showed reduced hyphal growth and the C. neoformans Δsmt1 strain became avirulent in a murine model of cryptococcosis (Oura & Kajiwara, 2008, Singh et al., 2012). Additionally, GlcCer is the binding target of plant defensins, such as Psd1, being crucial for peptide internalization and antifungal activity (Neves de Medeiros et al., 2014). In this work, we investigated the contribution of three enzymes involved in the GlcCer production pathway, sphingolipid Δ8-desaturase (SdeA), sphingolipid C9-methyltransferase (SmtA and SmtB) and glucosylceramide synthase (GcsA), to the A. nidulans phenotype, growth inhibition caused by Psd1 and G. mellonella virulence. To the best of our knowledge, this is the first report exploring the role of sphingolipid Δ8-desaturase in filamentous fungi growth and recognition by plant defensins.
It is interesting that sdeA, smtA, smtB and gcsA deletions promoted resistance to the cell-wall-damaging agents CFW and CR. Due to their hydrophobic properties, sphingolipids are thought to be membrane exclusive components. However, the presence of GlcCer also as a C. neoformans cell wall constituent was clearly demonstrated by the electron microscopy of yeasts labeled with immunogold antibody (Rodrigues et al., 2000). Thus, GlcCer synthesis impairment and, ultimately, the lack of its production or the accumulation of alternative compounds, such as α-OH-Δ4-GlcCer or α-OH-Δ4-Δ8-GlcCer, may have led to a more rigid CW architecture. Another hypothesis is the existence of a compensatory mechanism triggered by a thickened cell wall in null mutants to retain viability.
The determination of GlcCer abundance in null mutant strains provided new insight into the roles played by its synthesis pathway in fungal growth and differentiation. gcsA deletion completely impaired GlcCer synthesis, while sdeA, smtA and smtB disruptants were still able to produce the sphingolipid. The sdeA disruption led to the accumulation of saturated and unmethylated GlcCer (α-OH-Δ4-GlcCer) and a significant reduction of growth in solid media. These findings agree with the lipid analysis of C. albicans cells deficient in sphingolipid Δ8-desaturase, which contained 4-sphinganine as a GlcCer sphingoid base. Additionally, the sld/sld strain showed reduced filamentation on solid media, indicating that proper LCB structure is required for fungal growth (Oura & Kajiwara, 2008).
Surprisingly, smtA or smtB single deletion did not abolish 9,Me-GlcCer (α-OH-Δ4-Δ8-9,methyl-GlcCer) synthesis; however, the sphingolipid levels significantly decreased in the ΔsmtB lipid extract (50.4 %) compared to those in the ΔsmtA and wild-type cells. This suggests that smtA and smtB genes are redundant, both contributing to the addition of a –CH3 to the ceramide moiety, although smtB seems to code for a more functional methyltransferase. Despite the increased amount of unmethylated GlcCer (α-OH-Δ4-Δ8-GlcCer) observed in ΔsmtA and ΔsmtB lipid extracts, 9,Me-GlcCer still corresponded to the majority of the sphingolipid content.
These findings partially contrast with the lipid profile described for F. graminearum strains lacking the sphingolipid C9-methyltransferase-encoding genes FgMT1 and FgMT2. FgMT1 disruption did not compromise 9,Me-GlcCer synthesis, while the ΔFgMT2 strain showed reduced (25-35 %) levels of 9,Me-GlcCer and an accumulation (65-75 %) of unmethylated GlcCer (Ramamoorthy et al., 2009). Additionally, ΔFgMT2 cells exhibited growth defects compared to the ΔFgMT1 and wild type strains, suggesting that C9-methylated GlcCer is critical for hyphal differentiation. Indeed, it is interesting that in the ΔsmtB strain, the decreased 9,Me-GlcCer content was not followed by compromised filamentation.
The contribution of both sphingolipid C9-methyltransferases to fungal growth and GlcCer synthesis was further analyzed through ΔsmtA niiA::smtB conditional double mutant construction. In this strain, smtB expression is under the control of the niiA promoter in a smtA mutant background. Although niiA-repressing conditions did not abolish smtB expression, a striking reduction in mRNA levels was observed. Furthermore, smtA deletion and smtB repression led to the accumulation of unmethylated GlcCer, which was followed by a 50 % reduction in A. nidulans growth. These results indicate that sphingolipid C9-methyltransferases play a crucial role in fungal differentiation. Additionally, hyphal growth seems to be directly influenced by the [α-OH-Δ4,Δ8-GlcCer]/[α-OH-Δ4,Δ8-9,methyl-GlcCer] ratio. Indeed, ΔsmtA and ΔsmtB null mutants exhibited regular growth and higher levels of C9-methylated GlcCer than those of unmethylated GlcCer ([C9-methylated GlcCer] > [unmethylated GlcCer]). In contrast, ΔsmtA niiA::smtB strain produced more unmethylated GlcCer than C9-methylated GlcCer ([C9-methylated GlcCer] < [unmethylated GlcCer]) under smtB-repressing conditions, which was followed by compromised growth and differentiation. In fact, C. albicans mts1/mts1 cells showed impaired 9,Me-GlcCer synthesis, the accumulation of unmethylated GlcCer and a compromised yeast-to-hypha transition (Oura & Kajiwara, 2010).
An initial analysis of the role of A. nidulans GcsA in fungal growth was performed not through a gene deletion strategy but rather through the administration of EDO-P4 compound, a GCS inhibitor. Germlings maintained for 5 h in 20 μM EDO-P4 showed impaired growth, suggesting that GlcCer synthesis is required for hyphal extension (Levery et al., 2002). In fact, ΔgcsA strain, which was unable to produce any GlcCer, exhibited severe defects in radial growth and conidiation.
Lipid rafts are specialized microdomains enriched with ergosterol and sphingolipids that contribute to cell polarity establishment (Alvarez et al., 2007). Sterol-rich membranes have been identified at the tip of C. albicans growing hyphae but not in budding cells or pseudohyphae, strongly correlating these domains with active morphogenesis (Martin & Konopka, 2004). Moreover, the treatment of C. albicans hyphae with 100 μM myriocin, a selective serine palmitoyltransferase (SPT) inhibitor, impaired membrane polarization, which indicates that sphingolipid synthesis plays a crucial role in hyphal growth. Similarly, the deletion of A. nidulans barA, encoding a ceramide synthase, caused a severe reduction in sphingolipid content, followed by defects in polarized growth and lack of filipin staining at the hyphal tip (Li et al., 2006). Here, we investigated the effect of A. nidulans sdeA, smtA, smtB and gcsA disruption on lipid raft formation and hyphal morphology. Interestingly, ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA mutants exhibited regular nuclei distribution and septum formation/hyphal branching when compared to the wild-type strain (Supplementary Figure S4). Also, ΔsdeA and ΔgcsA cells did not exhibit sterol-rich domains at the hyphal apex followed by diminished ergosterol content in the gcsA disruptant, suggesting that reduced filamentation in these mutants can be attributed to impaired raft organization. Indeed, the contribution of GlcCer C9-methylation and glucose moiety to raft assembly has already been investigated in C. neoformans strains lacking smt1 and gcs1 genes. smt1 deletion led to an accumulation of unmethylated GlcCer and to a loss of the ability to form rafts in vitro (Singh et al., 2012). In contrast, the Δgcs1 mutant showed high levels of methylated ceramide and produced more rafts than did the wild-type strain, although they might not be functional (Del Poeta et al., 2014). In agreement with our findings in A. nidulans ΔsdeA and ΔgcsA mutants, it was then proposed that the C9-methyl group might contribute to the hydrophobic interaction of GlcCer with membrane lipids, while a glucose residue may be required for the interaction of GlcCer with sugar-coated sphingolipids, both promoting proper raft structure/formation.
In addition to impaired raft assembly, we investigated whether the disruption of GlcCer synthesis would also disturb the neutral lipid content. The triacylglycerol and sterol contents were quantified in ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA strains. Intriguingly, the lack of GlcCer synthesis in the gcsA disruptant was accompanied by a significant reduction in sterol abundance and a major depletion in triacylglycerol (TAG) levels, suggesting that the GlcCer biosynthesis pathway contributes to the maintenance of neutral lipid levels. Because TAG and steryl esters are stored in lipid droplets (LD), constituting their hydrophobic core, it remains to be elucidated whether GlcCer levels affect A. nidulans lipid trafficking.
Both GlcCer synthesis and C9-methylation are essential for C. neoformans pathogenesis in mice (Rittershaus et al., 2006, Singh et al., 2012). Additionally, when screening for regulators of C. albicans virulence, Noble et al., 2010 constructed approximately 3,000 homozygous deletion strains lacking 674 ORFs. Of these, 115 strains displayed reduced infectivity in mice, of which 89 showed growth rates resembling that of the wild-type strain (Noble et al., 2010). Interestingly, 4 mutants with an impaired ability to cause disease and normal proliferation were knocked out in GlcCer synthesis-related genes. These phenotypes were associated with Δ8-saturated and C9-unmethylated GlcCer synthesis in strains lacking orf19.260 (sphingolipid desaturase) and orf19.4831 (sphingolipid methyltransferase), respectively, while HSX11 disruption abolished GlcCer production. In filamentous fungi, C9-methylation and glucose moiety relevance to pathogenesis was initially explored in F. graminearum. The ability of ΔFggcs1 to cause disease was host-dependent, while the ΔFgmt2 strain showed reduced virulence in wheat heads even compared to ΔFggcs1, suggesting that C9-methylation is more relevant to F. graminearum pathogenesis than GlcCer production itself (Ramamoorthy et al., 2009). Here, the gcsA deletion impaired virulence in G. mellonella larvae, indicating that GlcCer synthesis is essential for A. nidulans pathogenesis in this host model. In fact, the lack of GlcCer production in the C. neoformans Δgcs1 strain was associated with compromised growth under a neutral/alkaline pH but not an acidic pH (Rittershaus et al., 2006). It is puzzling that the sdeA null mutation decreased growth in solid media but retained normal infectivity in G. mellonella larvae. Together, these observations suggest that ΔsdeA conidia spreading throughout the body cavity may be compromised, but pro-leg inoculum remains sufficient to induce lethality. The further quantification of fungal cells in fat bodies and other internal structures may elucidate the role of GlcCer C9-methylation and Δ8-desaturation in G. mellonella tissue colonization.
Plant defensins have emerged as antifungal molecules that specifically interact with fungal sphingolipids (Thevissen et al., 2004, Vriens et al., 2014). We recently showed that Psd1 binds more strongly to F. solani GlcCer than to soybean GlcCer, which has 4,8-sphingadienine as a sphingoid base (Neves de Medeiros et al., 2014). Moreover, the Psd1 antifungal activity partially depends on GlcCer synthesis, as C. albicans cells lacking GCS1 were approximately 30 % more resistant to this peptide. Because Δ8-desaturation and C9-methylation are distinguished features of fungal GlcCer, we investigated the relevance of these sphingoid base alterations to Psd1 inhibitory activity in A. nidulans. Cells lacking smtA or smtB genes exhibited similar Psd1 sensitivity to the wild-type strain; however, ΔsmtA and ΔsmtB null mutants still produced 9,Me-GlcCer. Therefore, to rule out –CH3 group contribution to Psd1 antifungal activity, it would be valuable to assess growth inhibition in P. pastoris or C. albicans strains deficient in sphingolipid C9-methyltransferases, which completely lack 9,Me-GlcCer production. Furthermore, sdeA disruption and, consequently, Δ4-GlcCer accumulation promoted resistance to Psd1 at levels comparable to those in ΔgcsA. Because C9-methylation and an additional Δ8-double bond are supposed to increase the physical distance between the hydrophobic core of GlcCer and other lipids (Del Poeta et al., 2014), it is possible that the lack of these modifications disrupts proper lipid organization throughout the plasma membrane, affecting Psd1 association with GlcCer/other cellular targets. Although plant defensins share high similarity in amino acid sequence, it is unlikely that a consensus mechanism of GlcCer recognition exists. In fact, LCB C9-methylation is required for AFP1 activity against C. albicans but not for the RsAFP2 inhibition of F. graminearum growth (Oguro et al., 2014, Ramamoorthy et al., 2009). Additionally, defensin entry into fungal cytoplasm, as described for Psd1 and NaD1, may also require interaction with distinct lipid motifs that are not involved in recognition by peptides as AFP1 and RsAFP2, whose mechanism of action solely relies on membrane permeabilization, as described so far. Therefore, further analyses are required to better understand C9-methylation and Δ8-desaturation contribution to plant defensin activity.
Experimental procedures
Strains, media and culture conditions
The A. nidulans strains that were used in this work are described in detail in Table 1. Aspergillus strains were grown in both minimal (high-nitrate salt solution, trace elements, 1 % glucose, 2 % agar, pH 6.5) and complete (YG (0.5 % yeast extract, 2 % glucose, trace elements) or YUU (YG supplemented with 5 mM uridine and 10 mM uracil)) media. smtB expression in the ΔsmtA niiA::smtB conditional double mutant was regulated by nitrate availability: for niiA induction, cells were grown in Modified Minimal Media (MMM: 1 % glucose, high nitrate salt solution without sodium nitrate, trace element solution without ammonium molybdate, 2 % agar, pH 6.5) supplemented with 10 mM sodium nitrate. Promoter repression was achieved by culturing cells in MMM plus 50 mM ammonium tartrate. Uracil, uridine and pyridoxine were added to both MMM media for qPCR and mass spectrometry analyses due to the auxotrophic requirements of the ΔsmtA and wild-type strains. The pRS426 plasmid (Christianson et al., 1992) and S. cerevisiae SC9721 strain (MATa his3-D200 URA 3-52 leu2D1 lys2D202 trp1D63) were used for yeast in vivo recombination. Initially, yeast cells were grown overnight in YPD medium (1 % yeast extract, 2 % peptone, 2 % glucose) at 30 °C, and the transformant colonies containing the deletion cassettes were selected on synthetic medium without uracil (SC, 0.7 % Difco yeast nitrogen base without amino acids, 2 % glucose, 0.1 g.L−1 leucine, 0.1 g.L−1 lysine, 0.1 g.L−1 tryptophan, 0.05 g.L−1 histidine, 2 % agar). The A. fumigatus pyrG sequence was amplified from the pcDA21 plasmid.
Table 1.
Genotype description of A. nidulans strains used in this work.
| Strains | Genotype | Reference |
|---|---|---|
| TNO2A3 (WT) |
pyroA4 pyrG89; chaA1; ΔnkuA::argB | (Nayak et al., 2006) |
| ΔsdeA |
pyroA4 pyrG89; chaA1; ΔnkuA::argB;
ΔsdeA::pyrG |
This work |
| ΔsmtA |
pyroA4 pyrG89; chaA1; ΔnkuA::argB;
ΔsmtA::pyrG |
This work |
| ΔsmtB |
pyroA4 pyrG89; chaA1; ΔnkuA::argB;
ΔsmtB::pyrG |
This work |
| ΔgcsA |
pyroA4 pyrG89; chaA1; ΔnkuA::argB;
ΔgcsA::pyrG |
This work |
| ΔsmtA
niiA::smtB |
pyroA4 pyrG89; chaA1; ΔnkuA::argB;
ΔsmtA::pyrG niiA::smtB::pyro |
This work |
For dropout experiments, 5 μl of ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and TNO2A3 (WT) suspensions containing 2.107, 2.106 and 2.105 conidia.ml−1 were point inoculated in minimal media supplemented with 5 mM uridine, 10 mM uracil and 50 ng.ml−1 of pyridoxine. Then, 10 and 20 μg.ml−1 of Calcofluor White (CFW) and Congo Red (CR) were used as cell wall stressing agents. Fungal cultures were grown for 48 h at 37 °C, after which the images were taken.
Construction of A. nidulans ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA mutant strains
A. nidulans gene disruption was performed as previously described through two different strategies. The sdeA (AN4592), smtA (AN5688) and gcsA (AN8806) deletion cassettes were obtained from the Fungal Genetics Stock Center and amplified in PCR reactions using TaKaRa Ex Taq DNA Polymerase (Clontech, USA) and P1-P2, P9-P10, and P11-P12 primer pairs, respectively (Supplementary Table S2). The smtB (AN7375) deletion cassette was constructed through the amplification of untranslated regions (UTR) and pyrG auxotrophic marker sequences, followed by the in vivo recombination of the fragments in S. cerevisiae (Malavazi & Goldman, 2012). Briefly, 1 kb of the 5’ and 3’ UTR regions, which flank smtB, and pyrG gene were PCR-amplified from the genomic DNA of wild-type cells and the pcDA21 vector (Chaveroche et al., 2000), respectively, using Phusion High-Fidelity DNA Polymerase (New England Biolabs, USA) and P3 to P8 primers, also listed in Supplemental Table 2. To allow for gene recombination in yeast, the P4-P7 and P5-P8 primer pairs were designed to contain 20 complementary base pairs. The P1-P3, P6, and P9-P12 primers included ends cohesive to the regions of pRS426 digested by EcoRI and BamHI (New England Biolabs, USA). Linearized pRS426, 5’ UTR, 3’ UTR and pyrG amplicons were inserted into S. cerevisiae SC9721 cells using the lithium acetate method, and once the plasmid contained the URA3 gene sequence, the transformant colonies were selected by their ability to grow in the absence of uracil. Then, yeast genomic DNA was extracted (Goldman et al., 2003), and the smtB deletion cassette was PCR-amplified using P3-P6 probes. Knockout constructions for sdeA, smtA and gcsA genes were also obtained in amplification reactions containing P1-P2 and P9 to P12 primers. Finally, the A. nidulans TNO2A3 (ΔnkuA) strain was transformed with the deletion cassettes described above. Because pyrG was used as auxotrophic marker in all of the deletion cassettes, the mutant strains were selected in agar media without uracil and uridine, and the integration of the deletion cassette and consequent gene disruption were confirmed by PCR and Southern Blot analyses.
Construction of A. nidulans ΔsmtA niiA::smtB strain
The A. nidulans ΔsmtA niiA::smtBstrain was constructed by replacing the endogenous promoter of the smtB gene with the niiA promoter (from the A. fumigatus nitrite reductase gene) in the ΔsmtA null mutant background. Initially, 1 kb of smtB 5’ UTR region and 2 kb of smtB ORF were PCR-amplified from A. nidulans wild-type genomic DNA using P23-P24 and P29-P30 primers (Supplementary Table S3). Then, 1.9 kb of the pyro gene and 1.2 kb of the niiA promoter were amplified from the Afpyro plasmid and A. fumigatus ΔakuB genomic DNA using P25-P28 primers, respectively. All of the fragments, along with linearized pRS426, were inserted into S. cerevisiae SC9721 cells, and yeast colonies containing the cassette 5’ UTR – pyro – niiA – smtB were selected by their ability to grow in the absence of uracil. Finally, the construct of interest was PCR-amplified from yeast genomic DNA using P23 and P29 probes, and transformation in A. nidulans ΔsmtA strain was performed as described above. Transformants that were able to grow in solid media lacking pyridoxine were selected by i) checking niiA::smtB integration by PCR or ii) quantifying smtB mRNA levels through qPCR in niiA-inducing (sodium nitrate) or niiA-repressing (ammonium tartrate) media (Punt et al., 1991).
Quantitative Real-Time PCR (qPCR)
A total of 107 conidia of wild-type, ΔsmtA and ΔsmtA niiA::smtB strains were inoculated into liquid MMM containing sodium nitrate or ammonium tartrate and cultured for 16 h at 37 °C. Mycelia were harvested by filtration, washed twice with water and ground in liquid nitrogen using a mortar and pestle. The total RNA was extracted using Trizol reagent (Invitrogen, USA), and the sample integrity was assessed using an Agilent 2100 Bioanalyzer system. Next, 20 μg of RNA was treated with RNAse-free DNase I and purified using an RNeasy kit (Qiagen, Germany). cDNA was generated using the SuperScript III First-Strand Synthesis system (Invitrogen, USA) with oligo(dT) primers according to the manufacturer’s protocol. Real-time PCR analysis was performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA) with SYBR Green detection and P31-P34 primers (Supplementary Table S3). The relative expression ratios were calculated by the ΔCt method, in which the tubulin C gene was used as a normalization reference.
Southern Blot analysis
To confirm the replacement of target genes for the pyrG sequence, genomic DNA of the parental and mutant strains was extracted and digested with 30 U of EcoRV or BamHI enzymes for 16 h at 37 °C. Then, the samples were fractionated in 1 % agarose gel and capillary transferred to a nylon membrane at an alkaline pH (Sambrook & Russell, 2001). The 5’ UTR regions that were used as probes were amplified using Phusion DNA Polymerase and P15 to P22 primers, listed in Table S2. Radiolabeling was performed with the Random Primer DNA Labelling System (Life Technologies, USA) according to the manufacturer’s instructions, and bands corresponding to digestion fragments were visualized by autoradiography (see Figure S2).
Scanning Electron Microscopy
The surface topography of the parental, ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA strains was assessed as previously reported (Vila et al., 2013) with some alterations. Initially, 105 conidia were grown on coverslips submerged on minimal media supplemented with uridine, uracil and pyridoxine for 96 h at 25 °C. Coverslips containing the adhered cells were fixed in a solution composed of 4 % formaldehyde and 2.5 % glutaraldehyde in 0.1 M sodium cacodylate buffer for 1 h at room temperature and further washed with 200 μl of 0.1 M cacodylate buffer. Then, cultures were post-fixed in 1.3 % potassium ferrocyanide and 1 % osmium tetroxide for 30 min and dehydrated for 15 min in increasing ethanol concentrations (30, 50, 70, 90 and 100 %, three times each). Finally, the samples were submitted to critical-point drying with CO2, covered with gold and observed in an FEI Quanta 250 Scanning Electron Microscope (FEI, Netherlands), where images were acquired in high-vacuum mode at 20 kV.
Lipid content assessment
To analyze the presence of GlcCer and to quantify the amount of each intermediate of its synthesis pathway in the ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA, ΔsmtA niiA::smtB and parental strains, the total lipids were extracted as previously described (Singh et al., 2012) with few modifications. Briefly, conidia were inoculated in 15 ml of YUU, MMM + sodium nitrate or MMM + ammonium tartrate medium and grown for 3-4 days at 37 °C and 200 rpm. Prior to lipid extraction, internal standards (C17-ceramide and C17-sphingosine) were added. The fungal suspensions were centrifuged to pellet down the mycelia, washed twice with 10 ml of sterile water, and submitted to Mandala Extraction (Mandala et al., 1995) and then to Bligh and Dyer Extraction (Bligh & Dyer, 1959). A quarter of each sample obtained from the Bligh and Dyer Extraction was reserved for inorganic phosphate (Pi) determination, while the remaining was submitted to thin layer chromatography (TLC) or alkaline hydrolysis and mass spectrometry analysis. For Pi measurement, each sample was initially incubated with 0.6 ml of ashing buffer (10 N sulfuric acid: 10 % perchloric acid: water 9:1:40), heated at 150-160 °C for 16 h and hydrated with 0.9 ml of water. Then, 0.5 ml of a 0.9 % ammonium molybdate solution and 0.2 ml of 9 % ascorbic acid solution were added to the tubes, which were heated to 45 °C for 30 min. The amount of Pi was determined by comparing the values of OD820nm for each sample to the standard curve, containing 5-80 nmol of a sodium dihydrogen phosphate solution. To analyze the presence of GlcCer in the lipid extracts through TLC, the samples were solubilized in chloroform: methanol 2:1 and spotted in a silica plate, which was dried and positioned in a tank containing chloroform: methanol: water 65:25:4 for 1 h. To visualize the bands corresponding to GlcCer, the silica plate was stained with iodine, and images were taken. Alkaline hydrolysis consisted of adding 0.5 ml of chloroform and 0.5 ml of 0.6 M potassium hydroxide in methanol to the samples, vortexing and incubating at room temperature for 1 h. Then, 0.325 ml of 1 M hydrochloride acid and 0.125 ml of water were added, and the tubes were vigorously agitated and centrifuged at 1700 g for 10 minutes. Finally, the organic phase was transferred to a new tube, dried and used for LC-MS analysis, which was performed in a Thermo Finnigan TSQ Quantum Ultra Mass Spectrometer (Thermo Fischer Scientific, USA). The relative abundance of α-OH-Δ4-ceramide (m/z 582.4), α-OH-Δ4-Δ8-ceramide (m/z 580.4), α-OH-Δ4-Δ8-9,methyl-ceramide (m/z 594.4), α-OH-Δ4-GlcCer (m/z 744.4), α-OH-Δ4-Δ8-GlcCer (m/z 742.4) and α-OH-Δ4-Δ8-9,methyl-GlcCer (m/z 756.4) was determined after the normalization of the respective product ions amount to the Pi value. To analyze the neutral lipid content in the ΔgcsA and parental strains, conidia were inoculated in 100 ml of YUU media and grown for 2 weeks at room temperature. Equal amounts (1 g) of dry mycelia were used for Bligh and Dyer Extraction, and lipid extracts were dried and suspended in the same volume of chloroform: methanol 2:1. The samples were spotted in a silica plate and submitted to chromatography in hexane: ethyl ether: acetic acid 60:40:1. Neutral lipid bands were observed after silica plate staining with Charring reagent (10 % copper sulfate, 80 % phosphoric acid) and heating at 200 °C.
Staining and microscopy
To investigate the sterol distribution profile in A. nidulans ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and parental strains, 105 conidia were grown in MM + UU + pyro media at 25 °C for 12 h and then stained with 25 μg.ml−1 filipin (Sigma Aldrich, USA) for 5 min. For nuclei and chitin staining, germlings were grown in the same conditions and incubated for 5 min with a 10 μg.ml−1 Hoechst (Life Technologies, USA) or 2 μg.ml−1 calcofluor white solution. Cell suspensions were washed in PBS buffer (140 mM sodium chloride, 2 mM potassium chloride, 10 mM disodium phosphate, 1.8 mM monopotassium phosphate, pH 7.4), coverslips were mounted and cultures were observed under a Zeiss Observer Z1 fluorescence (Zeiss, Germany) microscope with 100X magnification.
A. nidulans virulence analysis in Galleria mellonella
Larvae of the greater wax moth G. mellonella were obtained after the oviposition of the adult moths that were reared at the insectarium of Biochemistry Department, Chemistry Institute of the Federal University of Rio de Janeiro (UFRJ), Rio de Janeiro, Brazil. The stock culture was maintained in the dark at 28 °C inside plastic boxes with an artificial diet (120 g of honey, 120 g of glycerol, 200 g of milk, 60 g of yeast extract, 100 g of wheat germ, 100 g of wheat flour and 120 g of wheat bran) for the development of instars. G. mellonella larvae from the last instar used in this study were selected by the absence of grey pigmentation, with similar size (15-20 mm) and weight (approximately 200 mg). Prior to infection, A. nidulans TNO2A3, ΔsdeA, ΔsmtA, ΔsmtB and ΔgcsA strains were grown in minimal media supplemented with uracil, uridine and pyridoxine for 2 days at 37 °C. Then, the conidia were harvested and counted, and the suspensions were normalized to 1.108 cells.ml−1. Fifteen G. mellonella larvae were inoculated with 10 μl of fungal suspensions by injection into the hemocoel via the last left pro-leg, using a 25-μl Hamilton syringe. In addition, the same number of larvae were inoculated with 10 μl of PBS to monitor physical injury. After injection, the larvae were maintained in 90-mm glass dishes in the dark at 28 °C for up to 10 days. The insect viability was assessed daily; the animals were considered dead when no response was detected after touching them. Survival was expressed as the percentage of living larvae under A. nidulans infection, and statistical significance was plotted using Kaplan-Meier curves on GraphPad Prism 6.0 software.
Psd1 expression and purification
Psd1 was expressed and purified as previously described (Almeida et al., 2001). Briefly, a single colony of Pichia pastoris GS115 containing pPIC9-rPsd1 was grown in 10 ml of Buffered Minimal Glycerol media (BMG: 100 mM potassium phosphate buffer pH 6, 1.34 % Difco yeast nitrogen base without amino acids, 4.10−5 % biotin, 1 % glycerol) for 24 h at 28 °C and in 600 ml for 18 h at 30 °C. The culture was centrifuged at 4,000 g for 10 min, and the cell suspension was suspended in BBS media (100 mM potassium phosphate buffer pH 6, 4 g.L−1 ammonium chloride, 4.10−5 % biotin, 0.68 mM calcium chloride, 1.7 mM sodium chloride, 0.1 % magnesium solution, 0.01 % trace elements solution) supplemented with 0.7 % methanol and grown for 120 h at 28 °C under constant agitation. Recombinant Psd1 expression was induced by the daily addition of methanol to the growth culture. The culture was centrifuged, and the crude extract was applied to a Sephadex G50 Fine column (GE Healthcare, UK) with a constant flow of 0.2 ml.min−1 of 25 mM Tris-HCl pH 7.5. Then, the fractions containing low-molecular-weight peptides were grouped, dried and applied to a Luna C8 reverse phase column (Phenomenex, USA). Pure Psd1 elution was conducted in a linear gradient of 9-36 % acetonitrile for 30 min and under 30 % of the solvent. The fraction was dried and solubilized in water, and the peptide concentration was estimated using Lowry’s method (Lowry et al., 1951). Finally, the identity of Psd1 was confirmed after peptide digestion with trypsin and LC/MS-MS analysis, which included 90 % coverage of the defensin sequence (Centro de Espectrometria de Massas de Biomoléculas, CEMBIO-UFRJ).
A. nidulans antifungal assays
Antifungal assays were performed by microspectrophotometric analysis (UVM 340 ASYS, Biochrom, UK). For this analysis, 1.104 conidia.ml−1 of A. nidulans ΔsdeA, ΔsmtA, ΔsmtB, ΔgcsA and parental strains were inoculated in YUU medium in the presence of 2.5-20 μM Psd1, 10 μM itraconazole or water alone. Cell suspensions were grown for 100 h at 25 °C, and the OD540nm was measured during this time. The percentage of growth inhibition was calculated considering the culture absorbance in the absence of any antifungal compound or in the presence of itraconazole as 0 and 100 % inhibition, respectively. All of the experiments were conducted at least in triplicate.
Statistical analyses
Data were compared using a one-way ANOVA analysis of variance with Dunnett’s post-test. G. mellonella survival was analyzed by the Log-rank test, and statistical significance was considered when p values were < 0.05. All of the experiments were performed at least three times, except for lipid quantification by mass spectrometry, which was performed in duplicate.
Supplementary Material
Acknowledgements
Sources of funding for this work were from the Conselho Nacional deDesenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo a Pesquisa do Rio de Janeiro (FAPERJ), Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and from the National Institute of Health grants AI56168, AI100631 and by a Merit Review grant I01BX002624 from the Veterans Affairs Program in Biomedical Laboratory Research and Development to MDP. We thank Aline Sol Valle (MSc) for help in Psd1 expression and purification procedures and Mileane Busch for technical assistance. We also acknowledge Prof. Eliana Barreto-Bergter (IMPG-UFRJ) for initial GlcCer content analysis and Prof. André Luis Souza dos Santos (IMPG-UFRJ) for useful suggestions.
Footnotes
The authors declare that they have no conflicts of interest.
References
- Almeida MS, Cabral KS, de Medeiros LN, Valente AP, Almeida FC, Kurtenbach E. cDNA cloning and heterologous expression of functional cysteine-rich antifungal protein Psd1 in the yeast Pichia pastoris. Arch Biochem Biophys. 2001;395:199–207. doi: 10.1006/abbi.2001.2564. [DOI] [PubMed] [Google Scholar]
- Alvarez FJ, Douglas LM, Konopka JB. Sterol-rich plasma membrane domains in fungi. Eukaryot Cell. 2007;6:755–763. doi: 10.1128/EC.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-Bergter E, Pinto MR, Rodrigues ML. Structure and biological functions of fungal cerebrosides. An Acad Bras Cienc. 2004;76:67–84. doi: 10.1590/s0001-37652004000100007. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Chaveroche M-K, Ghigo J-M, d’Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic acids research. 2000;28:e97–e97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- de Medeiros LN, Angeli R, Sarzedas CG, Barreto-Bergter E, Valente AP, Kurtenbach E, Almeida FC. Backbone dynamics of the antifungal Psd1 pea defensin and its correlation with membrane interaction by NMR spectroscopy. Biochim Biophys Acta (BBA)-Biomembranes. 2010;1798:105–113. doi: 10.1016/j.bbamem.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Del Poeta M, Nimrichter L, Rodrigues ML, Luberto C. Synthesis and biological properties of fungal glucosylceramide. PLOS Pathog. 2014 doi: 10.1371/journal.ppat.1003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman GH, dos Reis Marques E, Ribeiro DCD, de Souza Bernardes L.A.n., Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell. 2003;2:34–48. doi: 10.1128/EC.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA. Growth and guidance of the fungal hypha. Microbiology. 1994;140:3193–3205. doi: 10.1099/13500872-140-12-3193. [DOI] [PubMed] [Google Scholar]
- Jackson JC, Higgins LA, Lin X. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PloS one. 2009;4:e4224. doi: 10.1371/journal.pone.0004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Leipelt M, Warnecke D, Zähringer U, Ott C, Müller F, Hube B, Heinz E. Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. Journal of Biological Chemistry. 2001;276:33621–33629. doi: 10.1074/jbc.M104952200. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levery SB, Momany M, Lindsey R, Toledo MS, Shayman JA, Fuller M, Brooks K, Doong RL, Straus AH, Takahashi HK. Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-Glc: ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 2002;525:59–64. doi: 10.1016/s0014-5793(02)03067-3. [DOI] [PubMed] [Google Scholar]
- Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo DS, Pereira IB, Fragel-Madeira L, Medeiros LN, Cabral LM, Faria J, Bellio M, Campos RC, Linden R, Kurtenbach E. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry. 2007;46:987–996. doi: 10.1021/bi061441j. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Malavazi I, Goldman GH. Gene disruption in Aspergillus fumigatus using a PCR-based strategy and in vivo recombination in yeast. Methods Mol Biol. 2012:99–118. doi: 10.1007/978-1-61779-539-8_7. [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton RA, Frommer BR, Curotto JE, Rozdilsky W, Kurtz MB, Giacobbe RA, Bills GF, Cabello MA, Martin I. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot (Tokyo) 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- Martin SW, Konopka JB. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell. 2004;3:675–684. doi: 10.1128/EC.3.3.675-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves de Medeiros L, Domitrovic T, Cavalcante de Andrade P, Faria J, Barreto Bergter E, Weissmüller G, Kurtenbach E. Psd1 binding affinity toward fungal membrane components as assessed by SPR: The role of glucosylceramide in fungal recognition and entry. Peptide Science. 2014;102:456–464. doi: 10.1002/bip.22570. [DOI] [PubMed] [Google Scholar]
- Nimrichter L, Rodrigues ML, Rodrigues EG, Travassos LR. The multitude of targets for the immune system and drug therapy in the fungal cell wall. Microbes Infect. 2005;7:789–798. doi: 10.1016/j.micinf.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro Y, Yamazaki H, Takagi M, Takaku H. Antifungal activity of plant defensin AFP1 in Brassica juncea involves the recognition of the methyl residue in glucosylceramide of target pathogen Candida albicans. Curr Genet. 2014;60:89–97. doi: 10.1007/s00294-013-0416-8. [DOI] [PubMed] [Google Scholar]
- Oura T, Kajiwara S. Disruption of the sphingolipid Δ8-desaturase gene causes a delay in morphological changes in Candida albicans. Microbiology. 2008;154:3795–3803. doi: 10.1099/mic.0.2008/018788-0. [DOI] [PubMed] [Google Scholar]
- Oura T, Kajiwara S. Candida albicans sphingolipid C9-methyltransferase is involved in hyphal elongation. Microbiology. 2010;156:1234–1243. doi: 10.1099/mic.0.033985-0. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Greaves PA, Kuyvenhoven A, van Deutekom JC, Kinghorn JR, Pouwels PH, van den Hondel CA. A twin-reporter vector for simultaneous analysis of expression signals of divergently transcribed, contiguous genes in filamentous fungi. Gene. 1991;104:119–122. doi: 10.1016/0378-1119(91)90476-r. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V, Cahoon EB, Li J, Thokala M, Minto RE, Shah DM. Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum. Mol Microbiol. 2007;66:771–786. doi: 10.1111/j.1365-2958.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V, Cahoon EB, Thokala M, Kaur J, Li J, Shah DM. Sphingolipid C-9 methyltransferases are important for growth and virulence but not for sensitivity to antifungal plant defensins in Fusarium graminearum. Eukaryot Cell. 2009;8:217–229. doi: 10.1128/EC.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Hennig M, Luberto C, Del Poeta M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest. 2006;116:1651. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Travassos LR, Miranda KR, Franzen AJ, Rozental S, de Souza W, Alviano CS, Barreto-Bergter E. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect Immun. 2000;68:7049–7060. doi: 10.1128/iai.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Takakuwa N, Ohnishi M, Oda Y. Presence of glucosylceramide in yeast and its relation to alkali tolerance of yeast. App Microbiol Biotechnol. 2006;71:515–521. doi: 10.1007/s00253-005-0187-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Wang H, Silva LC, Na C, Prieto M, Futerman AH, Luberto C, Del Poeta M. Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell Microbiol. 2012;14:500–516. doi: 10.1111/j.1462-5822.2011.01735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullards M, Lynch D, Merrill A, Adams J. Structure determination of soybean and wheat glucosylceramides by tandem mass spectrometry. J Mass Spectrom. 2000;35:347–353. doi: 10.1002/(SICI)1096-9888(200003)35:3<347::AID-JMS941>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Ternes P, Franke S, Zähringer U, Sperling P, Heinz E. Identification and characterization of a sphingolipid Δ4-desaturase family. J Biol Chem. 2002;277:25512–25518. doi: 10.1074/jbc.M202947200. [DOI] [PubMed] [Google Scholar]
- Ternes P, Sperling P, Albrecht S, Franke S, Cregg JM, Warnecke D, Heinz E. Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J Biol Chem. 2006;281:5582–5592. doi: 10.1074/jbc.M512864200. [DOI] [PubMed] [Google Scholar]
- Ternes P, Wobbe T, Schwarz M, Albrecht S, Feussner K, Riezman I, Cregg JM, Heinz E, Riezman H, Feussner I. Two pathways of sphingolipid biosynthesis are separated in the yeast Pichia pastoris. J Biol Chem. 2011;286:11401–11414. doi: 10.1074/jbc.M110.193094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K, Warnecke DC, François IE, Leipelt M, Heinz E, Ott C, Zähringer U, Thomma BP, Ferket KK, Cammue BP. Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem. 2004;279:3900–3905. doi: 10.1074/jbc.M311165200. [DOI] [PubMed] [Google Scholar]
- Vila TV, Ishida K, de Souza W, Prousis K, Calogeropoulou T, Rozental S. Effect of alkylphospholipids on Candida albicans biofilm formation and maturation. Journal of Antimicrobial Chemotherapy. 2013;68:113–125. doi: 10.1093/jac/dks353. [DOI] [PubMed] [Google Scholar]
- Vriens K, Cammue B, Thevissen K. Antifungal plant defensins: mechanisms of action and production. Molecules. 2014;19:12280–12303. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.