Abstract
Purpose
To enable robust high spatio-temporal-resolution 3D Cartesian MRI using a scheme incorporating a novel variable density random k-space sampling trajectory allowing flexible and retrospective selection of the temporal footprint with compressed sensing (CS).
Methods
A “complementary Poisson-disc” k-space sampling trajectory was designed to allow view sharing and varying combinations of reduced view sharing with CS from the same prospective acquisition. These schemes were used for two-point-Dixon-based dynamic contrast-enhanced MRI (DCE-MRI) of the breast and abdomen. Results were validated in vivo with a novel approach using variable-flip-angle data, which was retrospectively accelerated using the same methods but offered a ground truth.
Results
In breast DCE-MRI, the temporal footprint could be reduced 2.3-fold retrospectively without introducing noticeable artifacts, improving depiction of rapidly enhancing lesions. Further, experiments with variable-flip-angle data showed that reducing view sharing improved accuracy in reconstruction and T1 mapping. In abdominal MRI, 2.3-fold and 3.6-fold reductions in temporal footprint allowed reduced motion artifacts.
Conclusion
The complementary-Poisson-disc k-space sampling trajectory allowed a retrospective spatiotemporal resolution tradeoff using CS and view sharing, imparting robustness to motion and contrast enhancement. The technique was also validated using a novel approach of fully acquired variable-flip-angle acquisition.
Keywords: Compressed sensing, view sharing, dynamic contrast-enhanced MRI
Introduction
Dynamic contrast-enhanced MRI (DCE-MRI) is commonly used for detection and characterization of primary and metastatic lesions, perfusion imaging, MR angiography, and imaging of renal function. In DCE-MRI, T1-weighted imaging is performed, typically with a 3D RF-spoiled gradient echo (SPGR, T1-fast field echo, or fast low-angle shot) sequence, before, during, and after intravenous injection of a gadolinium-based contrast agent. High spatial and temporal resolution is desirable, often forcing a tradeoff between these two conflicting aims. Characterization of contrast uptake requires high temporal resolution, while characterization of morphology requires high spatial resolution and sufficient anatomic coverage. To address the tradeoff between spatial and temporal resolution, schemes employing k-space undersampling have been proposed, including view sharing (VS) methods for 3D Cartesian imaging (1–5) and similar methods for radial imaging such as KWIC (6). View sharing methods undersample outer k-space for each time frame for high temporal resolution and fill in missing data using data from other time frames expanding the “temporal footprint.” Signal changes between time frames result in artifacts related to the point-spread-function of the undersampling scheme such as ringing for TRICKS, streaking for KWIC, and incoherent aliasing for pseudorandom patterns. Most methods additionally employ parallel imaging to improve the spatiotemporal resolution. Recently, methods employing pseudorandom trajectories for VS have gained considerable attention in DCE-MRI (7–10). The pseudorandom nature of the sampling trajectory imparts robustness to motion and contrast enhancement by dispersing aliasing artifacts. However, a drawback of all VS schemes is that they introduce temporal blurring of the high spatial frequencies, which makes them sensitive to both motion and signal changes due to contrast enhancement.
Compressed sensing (CS) MRI exploits the transform-domain sparsity of MR images using incoherent k-space sampling and nonlinear constrained reconstruction (11). While CS approaches have shown great promise, they are computationally expensive and not clinically available, which has limited their widespread use. The reconstruction is typically based on penalizing the l1 norm of a transform domain (e.g. wavelet) image representation, the sparsity of which determines the quality of the reconstruction. CS-MRI typically uses variable density random sampling to accelerate acquisition, and more sophisticated adaptive designs have been proposed (12–14). However, CS sampling schemes suffer from noise amplification when CS and parallel imaging are used jointly for acceleration. To balance requirements for uniformity and incoherence, one common approach that has empirically performed well is to generate 3D Cartesian sampling patterns according to gridded Poisson-disc distributions that are randomized but limit the size of “holes” between acquired samples (15–17). Some approaches have used CS with composite images to exploit subtraction sparsity (18,19) of high temporal resolution frames, which are similar to approaches using highly constrained backprojection (20). One drawback of these approaches is that the true temporal resolution and temporal footprint are elusive due to “leakage” from the composite or prior image, and current pseudorandom trajectories may not be optimal for CS reconstruction.
In addition to high spatiotemporal resolution, uniform fat suppression is challenging in DCE-MRI, especially at higher field strengths due to increased effects of B0 and B1 inhomogeneity. Conventional fat suppression techniques based on inversion or saturation pulses are not robust to B0 inhomogeneity, and 3-point or 2-point Dixon fat water separation (21–23) approaches offer superior fat suppression performance in adult body MRI applications and more flexible acquisition view-ordering.
A major challenge for validating accelerated imaging techniques in vivo is obtaining a ground truth image, since processes where accelerated imaging is needed prevent the acquisition of fully sampled data. Acquiring fully sampled images is infeasible, and performing multiple DCE-MRI acquisitions on the same patient is usually also impractical without the ability to inject contrast twice. Retrospectively undersampled DCE-MRI data has been used to assess the accuracy of quantitative DCE-MRI analysis with CS (24) and compare constrained reconstruction methods (25,26). Other methods have used computer-synthesized phantoms (27–29) or in vitro phantoms (30).
This work presents a novel k-space sampling trajectory that allows differing degrees of retrospective VS and a CS reconstruction for the resulting variable undersampling factors, thus enabling a retrospectively adjustable temporal footprint. Eliminating VS completely achieves a very short temporal footprint and low outer k-space sampling density, resulting in blurring. A combination of VS and CS can be used to trade off between temporal footprint and k-space sampling density. The approach is integrated in a clinical VS protocol (i.e. without CS acceleration) for breast and abdominal DCE-MRI, and spatial and temporal behaviors of various combinations of CS and VS reconstructions are compared. The different reconstruction schemes are validated with a novel approach that uses undersampled variable-flip-angle in vivo breast data, which provides a ground truth data set. The variable-flip-angle results are used to evaluate the image quality and accuracy of reduced VS schemes without the use of contrast injection.
Methods
Sampling Trajectory and Reconstruction
3D Cartesian variable density k-space VS schemes have been designed using a multi-level segmentation of k-space, first by segmenting into central (A) and peripheral annular regions (B, C, etc.), which are segmented into sub-regions (B1, B2, etc.) (4,5). One sub-region from each annular region is acquired in sequential order and all unique sub-regions are combined to “fill” each annular region. The number of sub-regions can be chosen to increase with the k-space radius of the annular region, and consequently, the temporal footprint expands with k-space radius. For example, using one annular region (B) segmented three-fold (B1, B2, B3), data acquired with the ordering AB1AB2AB3AB1 can be reconstructed with the underlined regions to generate the first phase. We refer to this VS scheme as AB1-B2-B3, with “-“ indicating an A region acquired but omitted from the reconstruction.
Conventional Poisson-disc sampling balances the requirements for joint CS parallel imaging reconstruction but does not attempt to satisfy temporal sampling requirements for VS and is hence incompatible with VS, while 3D Cartesian pseudorandom patterns such as differential subsampling with Cartesian ordering (DISCO) and time-resolved angiography with stochastic trajectories (4,5) allow VS. However, the latter approaches do not necessarily tolerate sample rejection, as their individual phases are not both uniform and incoherent. It is also desirable for sample distributions from arbitrary temporal combinations from VS and arbitrary temporal phases to have Poisson-disc-like properties. One previous method proposed for 3D Cartesian MRI aims to provide Poisson-disc sample distributions amenable to joint CS parallel imaging reconstruction within a variable data collection window for the purpose of acquiring a single image in an incomplete breathhold (31). In such a setting, motion occurs after data collection at a time that is unknown a priori, and k-space is segmented in nested regions. Inspired by this work, we consider a dynamic imaging scenario that requires a minimum frame rate and accurate reconstruction during the period of signal change, which can occur at any time. Without making any assumptions about the characteristics of motion and contrast enhancement, a novel two-level complementary Poisson-disc (CPD) k-space segmentation scheme consisting of disjoint regions and carefully chosen inter-sample spatiotemporal distance criteria was developed for DCE-MRI. CPD achieves sample distributions that closely approximate conventional Poisson-disc sampling both for individual time frames and for a range of temporal footprints.
CPD sampling patterns are generated by iterating random selection of a sample in k-t space from a set of candidates (code available online at https://github.com/evanlev/cpd). Candidate samples are determined based on allowed k-space locations and criteria for minimum distance to samples previously added to the pattern. Minimum distance constraints are relaxed when the set of sample candidates is empty (i.e. continuing becomes infeasible). Minimum distance criteria ensure that sub-regions and their combinations approximate Poisson-disc sampling distributions. To segment a region into NB sub-regions, a sample at k-space location k1 (in grid points) and acquired in sub-region t1 ∈ {1,2 … N} is chosen uniformly at random from the set of all candidates such that 1) k1 is not previously sampled in any sub-region, 2) (k1, t1) is at some distance d(k1,t1,k2,t2) at least dmin to any other sample (k2, t2), and 3) frame t1 contains less than the number of samples used to segment into regions of equal size. In this work, the distance metric used was
where C is a parameter that balances the spatial and temporal distances, and C=2 empirically provided a good balance and was used in this work. dmin is initialized to a value that is sufficiently large and reduced whenever new samples cannot otherwise be added to any frame. This sample is added to the pattern, and the procedure is repeated until all samples in the region have been assigned to a sub-region. The latter criterion limits the size of gaps in composite sampling patterns based on the temporal footprint by ensuring that sample distributions from an arbitrary temporal position and window approximate Poisson-disc sampling. A simple example illustrating the algorithm is shown in Figure 1. The time to generate a uniform density (256 × 256) CPD sampling trajectory is typically 100 msec.
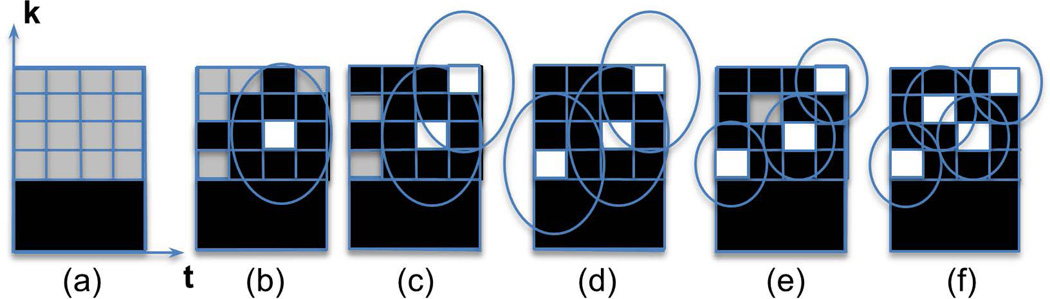
Fig. 1.
Illustration of uniform density 1D k-space segmentation of four samples into four time frames using the complementary Poisson-disc sampling method. An annular segment of k-space (a) is segmented into four frames from sample candidates shown in gray. As samples are added, shown in white (b–d), minimum distance criteria prohibit the addition of other samples, eventually leading to an overconstrained state (d), which requires minimum distance constraints to be relaxed (e). The final sampling pattern satisfies a relaxed minimum distance constraint (uniformity) and guarantees that k-space points are not acquired in multiple frames (efficiency for view sharing).
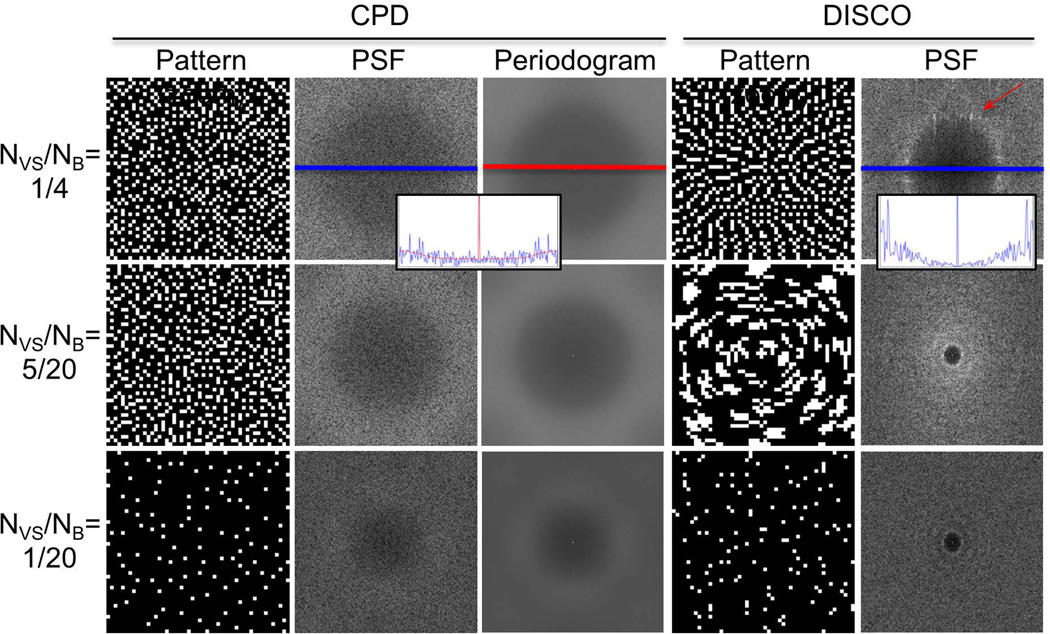
To demonstrate the properties of the proposed CPD sampling method, a square region was segmented into a number of subregions (denoted NB), a number (denoted NVS) of which were combined in composite sampling patterns. Figure 2 shows the resulting sampling patterns, with white and black pixels indicating acquired and non-acquired k-space samples respectively, comparing CPD with sampling patterns from a pseudorandom elliptical centric (DISCO) trajectory. Point spread functions (PSFs) were tone-mapped using a square root scale and scaling the central peak, and periodograms were computed as means of 300 randomly generated CPD PSFs and shown in red along with sample PSFs. The DISCO trajectory is generated by first sorting samples (ky, kz) with polar coordinates (kr, θ) in the annular region according to the value FOVykr + θ/2π to achieve a smooth trajectory, then assigning each to a sub-region in sequential order. Both sets of CPD and DISCO sampling patterns are complementary and somewhat incoherent. However, CPD sampling patterns achieve more uniform sample distributions than DISCO sampling patterns. Because of the second CPD distance criterion, composite sampling patterns shown in the middle row of Figure 2 are also Poisson-disc like and are comparable to those shown in the top row, which have the same reduction factor in a single frame. One advantage of CPD is that the PSF has lower power at lower frequencies, also a property of Poisson-disc sample distributions called the blue noise property (32). It is reasonable to design the PSF with the blue noise property to distribute aliased energy outside of the object, and the radius of the transition region can be modulated using dmin (33). A second advantage is that the combination of uniformity and randomness is known to achieve very low g-factor in parallel imaging (15). A third advantage is that the PSF is radially symmetric with energy distributed evenly between frames. DISCO PSFs are not radially symmetric and can have peaks in the PSF side lobes, as indicated by the red arrow in Figure 2.
Fig. 2.
Comparison of pseudorandom (DISCO) and CPD sampling patterns after segmenting a square region of k-space into NB disjoint sub-regions and view sharing with NVS of these sub-regions, denoted NVS /NB. CPD provides uniform and random sampling patterns with arbitrary choices of segmentation and view sharing parameters as seen from three representative examples. CPD point spread functions are radially symmetric and have low power at low frequencies, also seen in periodograms from 300 averaged PSFs. DISCO point spread functions can have peaks at side lobes (arrow). The CPD point spread functions for NVS/NB = 5/20 and 1/4 are approximately equal, demonstrating the “complementary” nature, which allows an arbitrary, retrospectively selectable NVS to maintain properties of Poisson-disc sampling. For comparison, the case of NVS/NB = 1/20 shows a lower effective FOV.
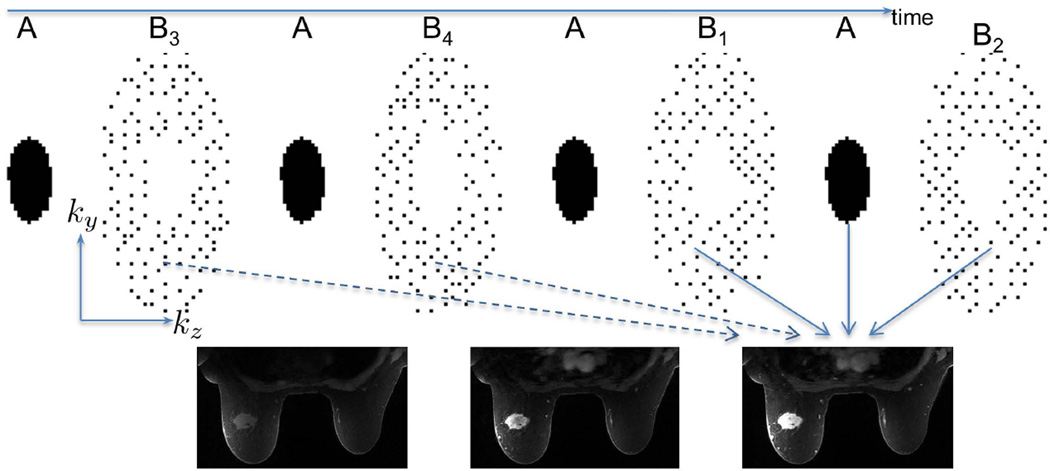
A two-level variable density k-space segmentation scheme was designed using uniform density CPD sampling to segment an annular region of k-space (Figure 3), providing both VS images and prospective CS undersampling. Fully VS data (solid and dashed arrows in Figure 3) were reconstructed online with a phase-preserving non-iterative acPI method using the vendor supplied reconstruction engine. After selecting the VS temporal footprint retrospectively (example shown with solid arrows in Figure 3 using two B regions), undersampled data were reconstructed with eigenvector-based iterative self-consistent parallel imaging reconstruction from arbitrary k-space (ESPIRiT) (34) with l1-regularization applied to each echo along the phase encoding directions (y and z), which uses parallel imaging and CS jointly to remove undersampling artifacts. Coil sensitivity maps were estimated from the fully VS data for each temporal phase from the sum of in- and opposed-phase echoes of the fully VS data using ESPIRiT. The open-source Berkeley Advanced Reconstruction Toolbox (35) was used, enabling CS reconstruction with 6 minutes of additional processing time for a matrix of size 180×100. Fat and water images were obtained using a two-point Dixon processing algorithm (22).
Fig. 3.
Schematic showing data sampling and reconstruction in DCE acquisition. A central region “A” was acquired with each temporal phase or flip angle and complementary Poisson-disc sampling patterns B1–B4 segmented from an outer region, 5× accelerated for acPI, were interleaved. The footprint of the B3B4B1AB2 view sharing scheme (solid and dashed arrows) was reduced (e.g. by using only B1AB2 data, solid arrows) and data was reconstructed with ESPIRiT.
Variable-flip-angle Acquisition
To validate CS and VS schemes in vivo, we introduce a novel approach using a fully acquired variable-flip-angle acquisition. Because the signal in different tissues varies with flip angle, this acquisition is a surrogate for DCE-MRI signal enhancement, in that it has both spatial and temporal signal variation. However, variable-flip-angle data offers a fully sampled ground truth, which is not available in a DCE-MRI experiment due to the spatiotemporal tradeoffs that novel methods try to address. A variable-flip-angle acquisition is performed under similar in vivo conditions and with the same imaging parameters as the DCE-MRI acquisition, but by retrospectively applying the same k-t sampling trajectory and CS/VS schemes used in DCE-MRI to fully sampled variable-flip-angle data in k-α space (α=flip angle), images and signal intensity-flip angle curves can be compared to a ground truth dataset in vivo.
A fully sampled variable-flip-angle Dixon bilateral breast acquisition was acquired on a healthy subject with 9 flip angles (α) from 2° to 18° in steps of 2° on a GE 3.0T MRI scanner (MR750, GE Healthcare) with a 3D SPGR sequence without contrast injection. Scan parameters were TE1/TE2/TR = 2.2/3.3/6ms, 512×386×192 matrix, 0.8 mm slice thickness. Variable-flip-angle data were retrospectively undersampled in k-α space with the same trajectory later used in breast DCE-MRI, which acquires the central part of k-space (A) for every flip angle and one of four regions (B1–B4) segmented from the inner annulus of peripheral k-space (B) using the CPD method described above. A 2.5×2 autocalibrating parallel imaging (acPI) factor was also used, resulting in B region data with accelerations of 6.7, 10, and 20 for reduced VS (B4-B1AB2, B1AB2, AB2) and 5 for VS (B3-B4-B1AB2) respectively.
An initial experiment applied VS schemes directly to the original variable-flip-angle data, which provided tissue-dependent signal intensity-flip angle curves. A second experiment was performed by synthesizing a new dataset with four initial phases from an image that is zero everywhere followed by four phases, each with data from the 4° flip angle i.e. mimicking a step response with replicated data. The four VS schemes described above and original fully acquired images were compared using coronal reformatted images. T1 maps were computed from reconstructed images from the first experiment and used to assess the quantitative accuracy of CS and VS schemes. Signal intensity curves measured from a small ROI were compared in the second “step response” experiment.
Prospective Breast DCE-MRI Acquisition
Bilateral breast DCE-MRI acquisitions were performed on three subjects with suspected breast cancer on a GE 3.0T MRI scanner (MR750, GE Healthcare) with a 3D RF-spoiled gradient echo (SPGR) 2-point Dixon sequence (7). Scan parameters: TE1/TE2/TR = 2.2/3.3/6 ms, 512×386×192 matrix, 0.8 mm slice thickness. The central 16% of k-space area (A) was acquired with 12.7s temporal resolution, and the inner annulus of peripheral k-space (B) was segmented into 4 non-overlapping sampling patterns (B1–B4) using the CPD method described above with a 2.5×2 autocalibrating parallel imaging (acPI) factor. An outermost annular region of k-space (C) was acquired only pre- and post-contrast, and C was not used in reconstruction of dynamic phases. Following contrast injection, regions A and Bi were repeatedly acquired, allowing reconstruction of VS images and alternatively, reconstruction with a prospectively undersampled B region. Images from reduced VS schemes (B1AB2, 19.9 second temporal footprint, R=10 and AB2, 12.7 second temporal footprint, R=20) were compared to VS images (B3-B4-B1AB2, 45.3 second temporal footprint, R=5) using reformatted coronal images, maps of initial slope, signal intensity-time curves measured from lesion ROIs. Although the original axial images are higher resolution, coronal images are best for comparison because they are aligned with the phase encode plane.
The impact of reducing VS on the derived semiquantitative and pharmacokinetic parameters was also assessed. CS reconstructions have been shown to achieve predictable performance and accuracy when applied retrospectively to fully-sampled breast DCE-MRI acquisitions in pharmacokinetic modeling (24,36). For an acquisition using prospective acceleration, bias in pharmacokinetic parameters was instead assessed. Maps of Ktrans, kep, and maximum slope were derived from B1AB2 and B3-B4-B1AB2 images for four lesions. For quantitative DCE-MRI analysis, a Tofts model (37) was used to relate the contrast agent concentration Ct(t) to the arterial input function (AIF) Cp(t) and values of Ktrans, kep by a discretization of the relation
A precontrast T1 of 1500 ms and a population-based AIF (38) were used. Differences between measurements derived with each method were compared by using a paired t-test. A 95% confidence interval was used and p < 0.05 was considered significant.
Motion-tolerant Abdominal DCE-MRI
To demonstrate the benefit of reduced VS schemes for coping with motion artifacts in a clinical scenario, the technique was used for abdominal DCE-MRI imaging. Abdominal images were acquired using a CPD trajectory with contrast injection in patients referred for liver evaluation. Patients were scanned using a CPD acquisition with 4 B regions, 0.16 of acquired data in region A, 2×2.5 acPI acceleration. Scan parameters: TE1/TE2/TR = 1.2/2.2/3.9ms, 260×202×72 matrix, 3 mm slice thickness. Temporal phases were acquired with 4.8 second temporal resolution, and VS images were reconstructed from B1AB2-B3-B4 data (17.1 second temporal footprint). Reduced VS schemes using l1-ESPIRiT and B1AB2 (7.1 second temporal footprint) and AB2 (4.8 second temporal footprint) data were compared to fully VS images.
Results
Breast DCE and Variable-flip-angle Experiments
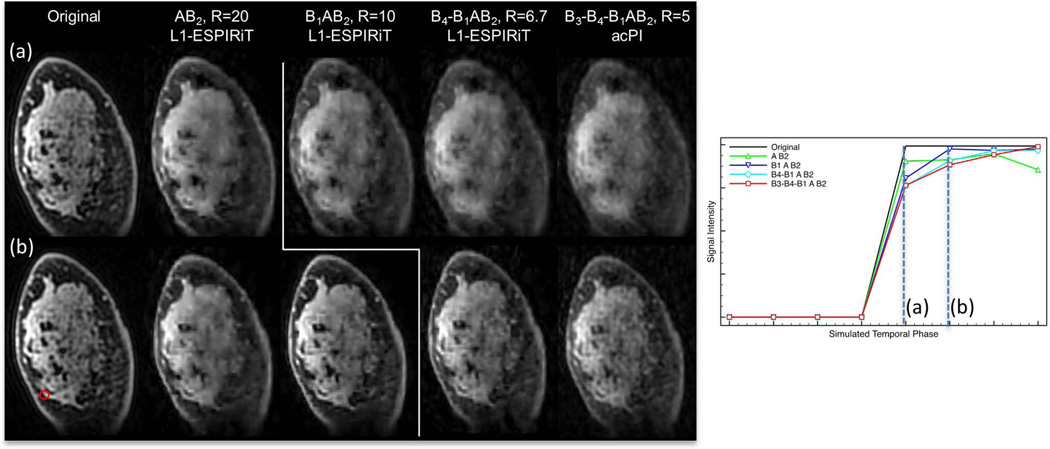
In the first experiment with variable-flip-angle data using all the acquired flip angles, B1AB2 data was sufficient to allow accurate CS reconstruction at each flip angle, while CS reconstruction from AB2 data showed noticeable blurring (Figure 4a). Derived T1 maps (Figure 4b) show that B1AB2 images achieve the lowest error of 12.8% RMSE, compared to 16.6%, 17.6%, and 22.7% RMSE for AB2, B4-B1-AB2, and B3-B4-B1AB2 images respectively.
Fig. 4.
Fully acquired variable-flip-angle images were retrospectively under-sampled and reconstructed with four CS and view sharing schemes. Coronal reformats of images and derived T1 maps were compared to original fully sampled images.
In the second experiment where variable-flip-angle data was used to mimic a step contrast uptake curve, AB2 images showed the fastest signal change initially but underestimation and spatial blurring in later phases (Figure 5a). Images from VS schemes showed blurring across the simulated temporal phases. The variable-flip-angle experiment using a step function sweeps the range of temporal footprints and signal changes, which together illustrate the CPD PSF for any temporal footprint (similar to Figure 2) and the effect of using CS to estimate missing data rather than zero-filling it. Although B1AB2 images showed blurring in Figure 5a, they showed consistent and accurate reconstruction in the later phases and appeared much sharper than AB2 images (Figure 5b).
Fig. 5.
To mimic instantaneous contrast enhancement (a step function), a dataset consisting of an image of zero replicated over four phases followed by variable-flip-angle data from flip angle 4° replicated over four phases was created. The same CS and view sharing schemes were used to undersample and reconstruct data. Images show the buildup in image quality over the first two temporal phases following the change from an image of all zeroes to the 4° flip angle for view sharing and CS schemes. View sharing zero-fills missing k-space data in images to the right of the solid line, while CS improves image quality by reconstructing it.
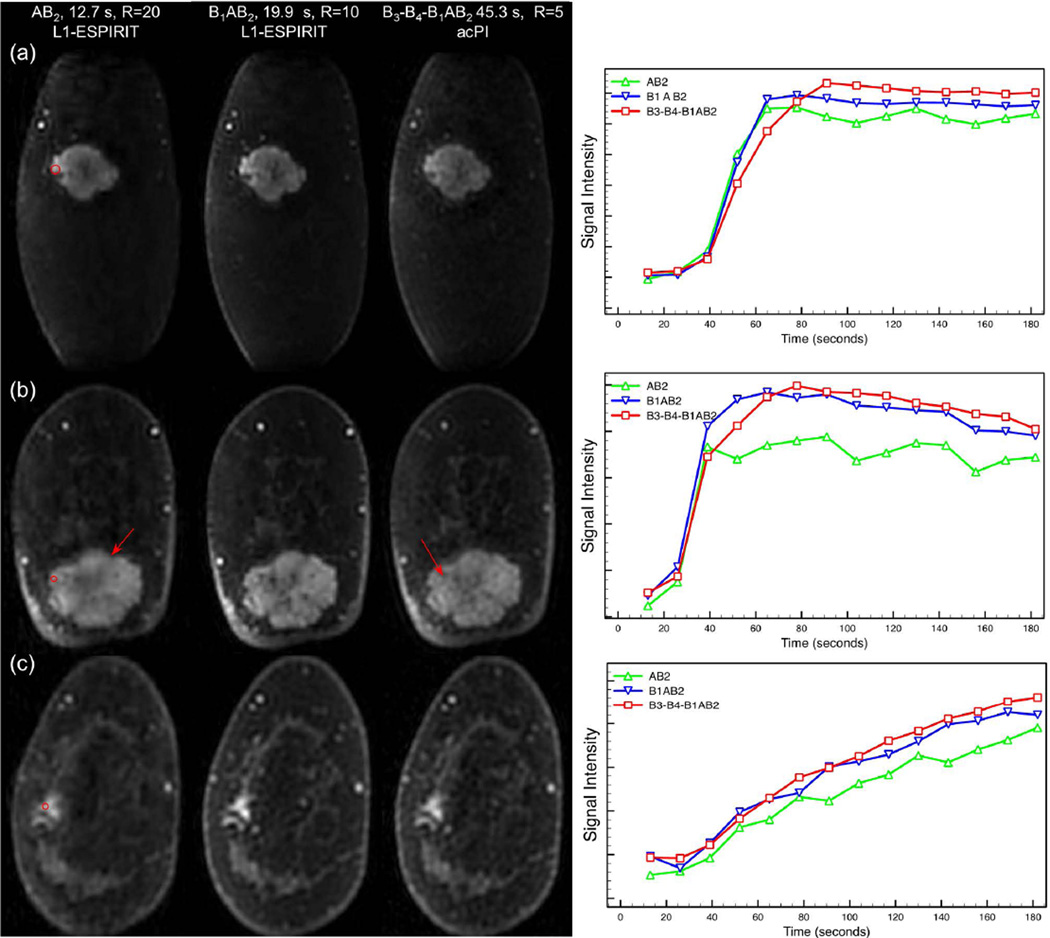
Figure 6 shows reformatted coronal breast DCE water images with different temporal footprints from six patients. All reconstructions showed acceptable fat suppression performance without fat-water swaps. The B1AB2 images show image quality comparable to that of fully VS images, but with a 2.3-fold reduction in temporal footprint. The AB2 images show blurring due to high undersampling in the B region. Signal intensity vs. time curves from ROIs show faster uptake in rapidly enhancing lesions (Figure 6a,b) with shorter temporal footprints, with 29% and 25% increases in slope of enhancement in AB2 and B1AB2 images from Figure 6a and b respectively. Figure 6c shows uptake curves from slowly enhancing tissues. Pharmacokinetic parameter maps and initial slope maps derived from B1AB2 images showed higher Ktrans and initial slope in lesions with rapid enhancement, particularly near boundaries (Figure 7). Bland-Altman plots (Figure 8) also showed higher Ktrans and initial slope, which was confirmed by a paired t-test (Table 1).
Fig. 6.
Coronal reformatted images reconstructed from view sharing and reduced view sharing schemes from four patients with suspected breast cancer. B1AB2 images show reduced spatial blurring inside the lesion and at the lesion boundary (red arrows) immediately following contrast injection compared to AB2 and fully view shared images. ROIs shown were used to compare signal intensity-time courses, shown in the right column. Signal intensity-time curves and maps of initial slope suggest better ability to discriminate rapidly enhancing lesions (a, b) with reduced view sharing temporal footprint and similar depiction of slowly-enhancing tissues (c).
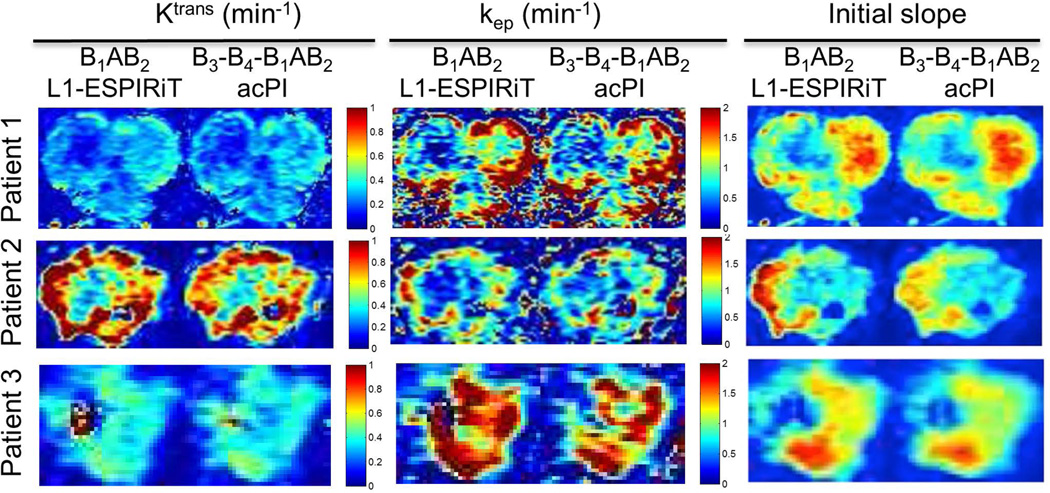
Fig. 7.
Maps of initial slope, Ktrans, kep derived from view shared images (B3-B4-B1AB2 data) and images from reduced view sharing and CS (B1AB2 data) for 4 lesions. Higher Ktrans and initial slope were derived from B1AB2 images in lesions with rapid enhancement.
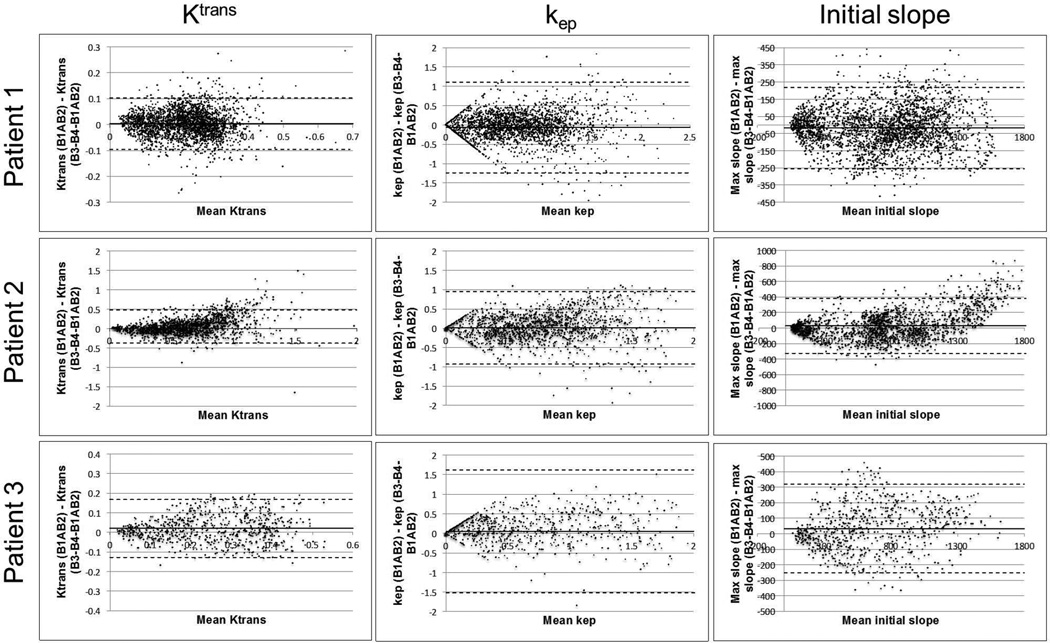
Fig. 8.
Bland Altman plots comparing Ktrans, kep, and maximum slope measurements between view shared images (B3-B4-B1AB2 data) and images from reduced view sharing and CS (B1AB2 data) from ROIs covering the 4 lesions in Figure 7. Solid and dashed lines correspond to means and limits of agreement respectively.
Table 1.
Paired differences between DCE parameter measurements from view shared images (B3-B4-B1AB2 data) and images from reduced view sharing and CS (B1AB2 data) show a bias in DCE parameters, including higher Ktrans and initial slope in lesions with rapid wash-in consistent with the expected temporal blurring effect.
| B3-B4-B1AB2 | B1AB2 | Difference | P-value | ||
|---|---|---|---|---|---|
| Patient 1 | Ktrans (min−1) | 0.215±0.004 | 0.218±0.004 | 0.003±0.002 | 0.002 |
| kep (min−1) | 0.76±0.03 | 0.70±0.02 | −0.067±0.02 | <0.001 | |
| Initial slope (au) | 742±14 | 724±14 | −19±4 | <0.001 | |
| Patient 2 | Ktrans (min−1) | 0.595±0.014 | 0.648±0.018 | 0.052±0.011 | <0.001 |
| kep (min−1) | 0.74±0.03 | 0.75±0.02 | 0.006±0.02 | 0.600 | |
| Initial slope (au) | 731±18 | 759±22 | 28±8 | <0.001 | |
| Patient 3 | Ktrans (min−1) | 0.238±0.009 | 0.258±0.010 | 0.020±0.006 | <0.001 |
| kep (min−1) | 0.66±0.06 | 0.71±0.04 | 0.05±0.06 | 0.11 | |
| Initial slope (au) | 640±25 | 672±27 | 32±10 | <0.001 |
Motion-tolerant Abdominal DCE-MRI
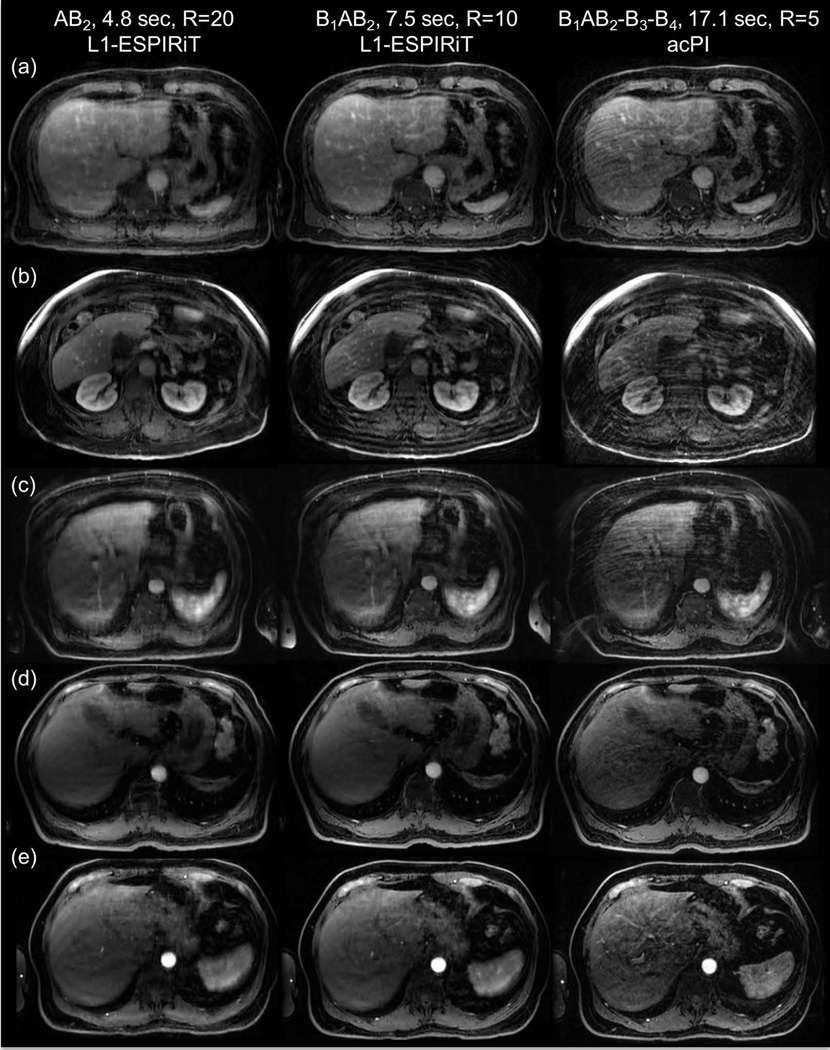
Figure 9 shows images from five patients with temporal blurring artifacts in B1AB2-B3-B4 reconstructions reduced in B1AB2 or AB2 images. (a–d) show motion artifacts, and (e) shows vessels enhancing early in VS images. Note that (b) shows motion artifacts even in B1AB2 images, but artifacts are further reduced in AB2 images at the cost of spatial blurring.
Fig. 9.
Liver images from five patients with temporal blurring artifacts in B1AB2-B3-B4 data not seen in images from B1AB2 or AB2 data. (a–d) show motion artifacts, and (e) shows vessels enhancing early in VS images. Images from one patient (b) show motion artifact even from B1AB2 data but not in images from AB2 data due to the shorter temporal footprint.
Discussion
This work introduces a novel complementary Poisson-disc sampling trajectory that remains random and uniform after retrospective selection of the VS temporal footprint, allowing variable VS with CS reconstruction. The proposed scheme can retrospectively enhance the temporal resolution of DCE-MRI. As DCE-MRI precludes comparison with a ground truth, a novel variable-flip-angle acquisition was used to demonstrate the impact of the spatiotemporal resolution tradeoff.
The CPD data sampling and reconstruction strategy parallels non-Cartesian imaging methods such as golden angle radial sampling (39) and KWIC (6,40) that use complementary sampling and a retrospective spatial and temporal resolution tradeoff. Golden angle radial sampling provides sample distributions from any time frame and temporal footprint with fairly uniform k-space coverage, allowing the temporal footprint to be selected retrospectively. KWIC also uses a variable density k-space segmentation scheme to increase the temporal footprint with k-space radius but allows the temporal footprint to be reduced retrospectively. In 3D Cartesian dynamic MRI, samples can be distributed in the phase encode plane to achieve similar goals. To our knowledge, there is no prior Cartesian approach that is analogous to Golden-angle radial scanning in allowing a truly retrospective tradeoff between spatial and temporal resolution.
The parameters of the CPD trajectory and reconstruction scheme investigated in this work are the number of B regions used in VS (NVS) of NB sub-regions and acceleration factor from acPI (RacPI), which relate to the k-space sampling density according to
For the same temporal footprint and sample density, a larger number of sub-regions and a lower acPI factor could be used without changing the spatiotemporal resolution. In this work, parameters were chosen to integrate CS reconstruction in the VS methodology (DISCO) used at our institution. NB = 4 and RacPI =4 or 5 were used so that fully VS data had uniform acPI sampling, allowing non-iterative reconstruction of the fully VS data for efficient clinical workflow. However, choosing RacPI = 1 (no acPI) and NB to achieve the same sample density could better approximate true Poisson-disc sample distributions similar to those shown in Figure 2, potentially improving both CS and VS images.
In experiments with the original variable-flip-angle data, the quantitative accuracy of T1 mapping suggests that B1AB2 data could optimally balance blurring from CS acceleration and blurring across flip angles. While the first experiment with variable-flip-angle data illustrates a case where VS works well even in the first flip angle, the second experiment mimics rapid contrast enhancement, demonstrating a case where VS fails by effectively zero-filling data due to the rapid signal change. Results mirror those from DCE-MRI experiments, which show rapid enhancement, but provide a ground truth for comparison. Variable-flip-angle experiments did not simulate changes within the acquisition window of ABi data, which could be included by interpolating data between flip angles or continuously varying the flip angle during acquisition. However, it is reasonable to expect similar results when the original signal does not change much faster than the spacing between A regions, which is a basic assumption of these methods. Variable-flip-angle experiments using the original data also suggest that a combination of VS and CS could be used to accelerate T1 mapping, but further investigation is required.
From in vivo DCE data, signal intensity vs. time curves for ROIs suggest better ability to discriminate rapidly enhancing lesions with reduced VS schemes (Figure 6a,b) and consistency for slowly enhancing tissues (Figure 6c). Images from AB2 data show spatial blurring artifacts due to high undersampling in the B region, while images from B1AB2 data appear no less sharp than VS images, suggesting that the B1AB2 scheme offers a better tradeoff of spatial and temporal resolution. Signal curves from AB2 data are noisy due to residual artifacts of the random PSF. Introducing moderate VS helps to smooth these artifacts by 1) providing more data, which improves the reconstruction accuracy and 2) introducing correlations in the PSFs for consecutive frames. AB2 images also underestimate the signal because at high reduction factors, incomplete recovery of the B region causes blurring, which results in underestimation of hyperintense features.
In rapidly enhancing lesions (Figure 6a,b), B1AB2 images appear sharper than full VS images. VS images use B3 and/or B4 data from the precontrast period, which results in zero-filling. The effect is exactly simulated with variable-flip-angle data, which shows similar artifacts in Figure 5. In the variable-flip-angle experiment, image quality is always improved by considering pre-contrast data missing and reconstructing it with CS rather than effectively zero-filling it with VS (Figure 5). By providing the appropriate sample distribution after retrospective selection of the temporal footprint, CPD renders artifacts of contrast enhancement noise-like and thus correctable with CS.
Our study investigates the use of prospective undersampling, which precludes comparison with a ground truth for semi-quantitative and quantitative DCE parameters. Nevertheless, the ability to measure a higher Ktrans and initial slope is not expected to be an artifact, since previous work using fully-sampled acquisitions did not indicate a bias in CS reconstruction and the current work also omits temporal constraints that could bias the derived pharmacokinetic parameters (24). The sharpness of parameter maps derived in B1AB2 images indicates that remaining bias due to spatial regularization in CS is minor.
Results from the motion-tolerant abdominal imaging application shown in Figure 9 demonstrate the ability to trade off between temporal footprint and motion artifacts. Although pseudorandom VS schemes often work well, they can fail due to motion, which creates coherent ghosting artifacts. Retrospectively reducing the temporal footprint with CS imparts robustness to motion but results in spatial blurring at very low temporal footprints. In Figure 9, an optimal temporal footprint is achieved in B1AB2 images without noticeable blurring, whereas in some cases with severe motion artifacts, AB2 data must be used to salvage motion-free images, but with noticeable blurring. In abdominal imaging applications, either AB2 or B1AB2 images could be used when the breathhold is lost very early or initiated late depending on the characteristics of motion, obviating the need for an additional low-resolution sequence for poor breath holders. In time frames where the underlying signal dynamics have a significant spatial variation, multiple temporal footprints would provide useful information. For example, simultaneously measuring an arterial input function from the aorta and enhancement at a lesion boundary would require both short and long temporal footprints for each respective task, whereas accounting for breathhold loss may require only one.
Other constrained reconstruction methods could be used with CPD and reduced VS to improve the reconstruction quality. Composite images can be used to estimate slowly varying quantities such as field maps and channel sensitivities. For example, a field map can be used to integrate a two-point Dixon model in the reconstruction, allowing constraints to be applied to fat and water images. In some cases, we have found that it is beneficial to use VS for both the field map and fat image, which can be subtracted because fat does not enhance and thus is static in the absence of motion. In most cases, we have found that using a field map from VS images with an integrated two-point Dixon model and regularizing the water and fat images does not improve accuracy compared to sequential CS and Dixon processing. Other VS schemes using a different number of B sub-regions can be designed for different acceleration factors compared to fully VS data. For example, 3 B sub-regions could be used to achieve a 1.5-fold reduction factor with B1AB2 data compared to VS data. In this work, temporal sparsity was purposefully not exploited to maintain a clear notion of spatiotemporal resolution, but temporal image models have been applied to DCE-MRI (41–44) and could be used with VS to balance spatiotemporal resolution. In combination with temporal models, reconstruction from variable-flip-angle data could be included, as the same models have been used to exploit sparsity along the parametric dimension for T1 mapping (45,46). Partial Fourier acceleration was also not used because is nontrivial to incorporate in two-point Dixon-based fat water separation.
Our study had some limitations. The clinical VS protocol used acPI acceleration and uniform density sampling, which may not be ideal for CS reconstruction and Poisson-disc sampling. The method does not propose an intelligent way to choose the optimal temporal footprint. In future work, the A region data or respiratory bellows could be used to detect breath hold loss. Future studies will be directed toward evaluation in a larger number of patients in applications demanding robustness to motion and contrast enhancement. While variable-flip-angle is a good surrogate for DCE-MRI to evaluate image reconstruction accuracy under similar in vivo conditions, more complex models could be generated from replicated, reordered, and/or interpolated variable-flip-angle data. While the approach to quantitative DCE analysis is intended to show biases in the derived parameters, future work is necessary to determine an appropriate pharmacokinetic model for lesions with rapid wash-in and k-space trajectory parameters that meet its spatiotemporal resolution requirements.
Conclusion
A judicious combination of CS and VS can achieve robust high spatiotemporal resolution dynamic MRI in a number of applications by reducing the temporal footprint retrospectively, which does not modify and compromise the original acquisition and reconstruction without CS and does not require computationally intensive reconstruction. Without a priori knowledge of the exact severity and characteristics of signal intensity changes, the optimal tradeoff of spatial and temporal resolution that should be imposed by the reconstruction must be determined retrospectively. To address this tradeoff, a CPD trajectory that distributes samples so that data from any time frame and temporal footprint approximates Poisson-disc sampling is developed. Spatial-domain CS is used in the standard VS methodology for reconstruction with a reduced temporal footprint. Examples from variable-flip-angle acquisition, DCE-MRI, and abdominal imaging indicate that spatial-domain CS can be used to reduce temporal footprint without introducing noticeable artifacts. The proposed approach using variable-flip-angle acquisition can offer a ground truth and a method to validate different CS/VS schemes in vivo without using multiple contrast injections.
Acknowledgments
This work was supported by NIH grant P41EB015891, R01-EB009055, R01-EB009690, R01-EB017739
References
- 1.Van Vaals JJ, Brummer ME, Thomas Dixon W, Tuithof HH, Engels H, Nelson RC, Gerety BM, Chezmar JL, Den Boer JA. “Keyhole” method for accelerating imaging of contrast agent uptake. J. Magn. Reson. Imaging. 1993;3:671–675. doi: 10.1002/jmri.1880030419. [DOI] [PubMed] [Google Scholar]
- 2.Korosec FR, Frayne R, Grist TM, Mistretta CA. Time-resolved contrast-enhanced 3D MR angiography. Magn. Reson. Med. 1996;36:345–351. doi: 10.1002/mrm.1910360304. [DOI] [PubMed] [Google Scholar]
- 3.Madhuranthakam AJ, Hu HH, Barger AV, Haider CR, Kruger DG, Glockner JF, Huston J, Riederer SJ. Undersampled elliptical centric view-order for improved spatial resolution in contrast-enhanced MR angiography. Magn. Reson. Med. 2006;55:50–58. doi: 10.1002/mrm.20726. [DOI] [PubMed] [Google Scholar]
- 4.Song T, Laine AF, Chen Q, Rusinek H, Bokacheva L, Lim RP, Laub G, Kroeker R, Lee VS. Optimal k-space sampling for dynamic contrast-enhanced MRI with an application to MR renography. Magn. Reson. Med. 2009;61:1242–1248. doi: 10.1002/mrm.21901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saranathan M, Rettmann DW, Hargreaves BA, Clarke SE, Vasanawala SS. DIfferential subsampling with cartesian ordering (DISCO): A high spatio-temporal resolution dixon imaging sequence for multiphasic contrast enhanced abdominal imaging. J Magn. Reson. Imaging. 2012;35:1484–1492. doi: 10.1002/jmri.23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song HK, Dougherty L. k-Space weighted image contrast (KWIC) for contrast manipulation in projection reconstruction MRI. Magn. Reson. Med. 2000;44:825–832. doi: 10.1002/1522-2594(200012)44:6<825::aid-mrm2>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 7.Saranathan M, Rettmann DW, Hargreaves BA, Lipson JA, Daniel BL. Variable spatiotemporal resolution three-dimensional dixon sequence for rapid dynamic contrast-enhanced breast MRI. J. Magn. Reson. Imaging. 2014;40:1392–1399. doi: 10.1002/jmri.24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Y, Kipfer H, Majidi S, Holz S, Dale B, Geppert C, Kroeker R, Lin C. Application of time-resolved angiography with stochastic trajectories (TWIST)-Dixon in dynamic contrast-enhanced (DCE) breast MRI. J. Magn. Reson. Imaging. 2013;38:1033–1042. doi: 10.1002/jmri.24062. [DOI] [PubMed] [Google Scholar]
- 9.Herrmann K-H, Baltzer PA, Dietzel M, Krumbein I, Geppert C, Kaiser WA, Reichenbach JR. Resolving arterial phase and temporal enhancement characteristics in DCE MRM at high spatial resolution with TWIST acquisition. J. Magn. Reson. Imaging. 2011;34:973–982. doi: 10.1002/jmri.22689. [DOI] [PubMed] [Google Scholar]
- 10.Le Y, Kroeker R, Kipfer HD, Lin C. Development and evaluation of TWIST Dixon for dynamic contrast-enhanced (DCE) MRI with improved acquisition efficiency and fat suppression. J. Magn. Reson. Imaging. 2012;36:483–491. doi: 10.1002/jmri.23663. [DOI] [PubMed] [Google Scholar]
- 11.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 12.Seeger M, Nickisch H, Pohmann R, Schölkopf B. Optimization of k-space trajectories for compressed sensing by Bayesian experimental design. Magn. Reson. Med. 2010;63:116–126. doi: 10.1002/mrm.22180. [DOI] [PubMed] [Google Scholar]
- 13.Ravishankar S, Bresler Y. Adaptive sampling design for compressed sensing MRI. 2011; Annu. Int. Conf. IEEE Eng. Med. Biol. Soc; 2011. pp. 3751–3755. [DOI] [PubMed] [Google Scholar]
- 14.Liu D-D, Liang Dcd, Zhang Ncd, Liu Xcd, Zhang Y-Tce. Undersampling trajectory design for compressed sensing based dynamic contrast-enhanced magnetic resonance imaging. J. Electron. Imaging. 2015;24 [Google Scholar]
- 15.Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn. Reson. Med. 2010;64:457–471. doi: 10.1002/mrm.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Athalye V, Lustig M, Uecker M. Parallel magnetic resonance imaging as approximation in a reproducing kernel Hilbert space. Inverse Probl. 2015;31:45008. doi: 10.1088/0266-5611/31/4/045008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasanawala SS, Murphy MJ, Lai P, Keutzer K, Alley MT, Pauly JM, Lustig M. Practical Parallel Imaging Compressed Sensing MRI: Summary of Two Years of Experience in Accelerating Body MRI of Pediatric Patients. 2011:1–5. doi: 10.1109/ISBI.2011.5872579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapacchi S, Natsuaki Y, Plotnik A, Gabriel S, Laub G, Finn JP, Hu P. Reducing view sharing using compressed sensing in time-resolved contrast-enhanced magnetic resonance angiography. Magn. Reson. Med. 2014;74:474–481. doi: 10.1002/mrm.25414. [DOI] [PubMed] [Google Scholar]
- 19.Rapacchi S, Han F, Natsuaki Y, Kroeker R, Plotnik A, Lehrman E, Sayre J, Laub G, Finn JP, Hu P. High spatial and temporal resolution dynamic contrast-enhanced magnetic resonance angiography using compressed sensing with magnitude image subtraction. Magn. Reson. Med. 2014;71:1771–1783. doi: 10.1002/mrm.24842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mistretta CA, Wieben O, Velikina J, Block W, Perry J, Wu Y, Johnson K. Highly constrained backprojection for time-resolved MRI. Magn. Reson. Med. 2006;55:30–40. doi: 10.1002/mrm.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon WT. Simple proton spectroscopic imaging. Radiology. 1984;153:189–194. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- 22.Ma J. Breath-hold water and fat imaging using a dual-echo two-point dixon technique with an efficient and robust phase-correction algorithm. Magn. Reson. Med. 2004;52:415–419. doi: 10.1002/mrm.20146. [DOI] [PubMed] [Google Scholar]
- 23.Reeder SB, Markl M, Yu H, Hellinger JC, Herfkens RJ, Pelc NJ. Cardiac CINE imaging with IDEAL water-fat separation and steady-state free precession. J. Magn. Reson. Imaging. 2005;22:44–52. doi: 10.1002/jmri.20327. [DOI] [PubMed] [Google Scholar]
- 24.Smith DS, Welch EB, Li X, Arlinghaus LR, Loveless ME, Koyama T, Gore JC, Yankeelov TE. Quantitative effects of using compressed sensing in dynamic contrast enhanced MRI. Phys. Med. Biol. 2011;56:4933–4946. doi: 10.1088/0031-9155/56/15/018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Schabel MC, DiBella EVR. Reconstruction of dynamic contrast enhanced magnetic resonance imaging of the breast with temporal constraints. Mag Reson Imag. 2010;28:637–645. doi: 10.1016/j.mri.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adluru G, Tasdizen T, Schabel MC, DiBella EVR. Reconstruction of 3D dynamic contrast-enhanced magnetic resonance imaging using nonlocal means. JMRI. 2010;32:1217–1227. doi: 10.1002/jmri.22358. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Block WF, Turski Pa, Mistretta Ca, Rusinak DJ, Wu Y, Johnson KM. Noncontrast dynamic 3D intracranial MR angiography using pseudo-continuous arterial spin labeling (PCASL) and accelerated 3D radial acquisition. J. Magn. Reson. Imaging. 2014;39:1320–1326. doi: 10.1002/jmri.24279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Usman M, Prieto C, Schaeffter T, Batchetlor PG. k-t group sparse: A method for accelerating dynamic MRI. Magn. Reson. Med. 2011 doi: 10.1002/mrm.22883. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen H, Kozerke S, Ringgaard S, Nehrke K, Kim WY. k-t PCA: Temporally constrained k-t BLAST reconstruction using principal component analysis. J. Magn. Reson. Imaging. 2009;62:706–716. doi: 10.1002/mrm.22052. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Miao Y, Zhou K, et al. Feasibility of high temporal resolution breast DCE-MRI using compressed sensing theory. Med. Phys. 2010;37:4971. doi: 10.1118/1.3483094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gdaniec N, Eggers H, Börnert P, Doneva M, Mertins A. Robust abdominal imaging with incomplete breath-holds. Magn. Reson. Med. 2014;71:1733–1742. doi: 10.1002/mrm.24829. [DOI] [PubMed] [Google Scholar]
- 32.Talmor D. Well-spaced points for numerical methods. University of Minnesota; 1997. [Google Scholar]
- 33.Lagae A, Dutré P. A comparison of methods for generating poisson disk distributions. Comput. Graph. Forum. 2008;27:114–129. [Google Scholar]
- 34.Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn. Reson. Med. 2014;71:990–1001. doi: 10.1002/mrm.24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uecker M, Ong F, Tamir J, Bahri D, Virtue P, Cheng J, Zhang T, Lustig M. Berkeley Advanced Reconstruction Toolbox. Annu. Meet. ISMRM, Toronto 2015, Proc. Intl. Soc. Mag. Reson. Med. 232486. 2015;23:2486. [Google Scholar]
- 36.Smith DS, Li X, Gambrell JV, Arlinghaus LR, Quarles CC, Yankeelov TE, Welch EB. Robustness of quantitative compressive sensing MRI: the effect of random undersampling patterns on derived parameters for DCE- and DSC-MRI. IEEE Trans. Med. Imaging. 2012;31:504–511. doi: 10.1109/TMI.2011.2172216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tofts PS, Brix G, Buckley DL, et al. Estimating Kinetic Parameters From Dynamic Contrast-Enhanced T1-Weighted MRI of a Diffusable Tracer: Standardized Quantities and Symbols. J Magn Reson Imag. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 38.Walker-Samuel S, Leach MO, Collins DJ. Evaluation of response to treatment using DCE-MRI: the relationship between initial area under the gadolinium curve (IAUGC) and quantitative pharmacokinetic analysis. Phys. Med. Biol. 2006;51:3593–3602. doi: 10.1088/0031-9155/51/14/021. [DOI] [PubMed] [Google Scholar]
- 39.Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An Optimal Radial Profile Order Based on the Golden Ratio for Time-Resolved MRI. Med. Imaging, IEEE Trans. 2007;26:68–76. doi: 10.1109/TMI.2006.885337. [DOI] [PubMed] [Google Scholar]
- 40.Song HK, Dougherty L. Dynamic MRI with projection reconstruction and KWIC processing for simultaneous high spatial and temporal resolution. Magn. Reson. Med. 2004;52:815–824. doi: 10.1002/mrm.20237. [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Cheng JY, Potnick AG, Barth RA, Alley MT, Uecker M, Lustig M, Pauly JM, Vasanawala SS. Fast pediatric 3D free-breathing abdominal dynamic contrast enhanced MRI with high spatiotemporal resolution. J. Magn. Reson. Imaging. 2015;41:460–473. doi: 10.1002/jmri.24551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otazo R, Candès E, Sodickson DK. Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components. Magn. Reson. Med. 2015;73:1125–1136. doi: 10.1002/mrm.25240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebel RM, Jones J, Ferre J-C, Law M, Nayak KS. Highly accelerated dynamic contrast enhanced imaging. Magn. Reson. Med. 2014;71:635–644. doi: 10.1002/mrm.24710. [DOI] [PubMed] [Google Scholar]
- 44.Veliknia J, Morrison C, Bancroft L, Strigel R, Holmes J, Wang K, Samsonov AA, Korosec FR. An application of compressed sensing for improved temporal fidelity in DCE breast MRI; ISMRM Workshop on MRI in the Management of Breast Disease; 2015. [Google Scholar]
- 45.Velikina JV, Alexander AL, Samsonov A. Accelerating MR parameter mapping using sparsity-promoting regularization in parametric dimension. Magn. Reson. Med. 2013;70:1263–1273. doi: 10.1002/mrm.24577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T, Pauly JM, Levesque IR. Accelerating parameter mapping with a locally low rank constraint. Magn. Reson. Med. 2014;661:655–661. doi: 10.1002/mrm.25161. [DOI] [PMC free article] [PubMed] [Google Scholar]