Abstract
Introduction
In the United States, respiratory syncytial virus (RSV) is responsible for the majority of infant hospitalizations resulting from viral infections, as well as a leading source of pneumonia and bronchiolitis in young children and the elderly. In the absence of vaccine prophylaxis or an effective antiviral for improved disease management, the development of novel anti-RSV therapeutics is critical.
Areas Covered
Several advanced drug development campaigns of the past decade have focused on blocking viral infection. These efforts have returned a chemically distinct panel of small-molecule RSV entry inhibitors, but binding sites and molecular mechanism of action appeared to share a common mechanism, resulting in comprehensive cross-resistance and calling for alternative druggable targets such as viral RNA-dependent RNA-polymerase complex.
Expert Opinion
In this review, we discuss the current status of the mechanism of action of RSV entry inhibitors, recent structural insight into the organization of the polymerase complex that have revealed novel drug targets sites, and outline a path towards the discovery of next-generation RSV therapeutics.
Keywords: respiratory syncytial virus, small molecule, antiviral, fusion, viral replication, paramyxovirus, RNA-dependent RNA-polymerase, therapeutics, virus entry
1.1 Introduction
Respiratory syncytial virus (RSV) belongs to the paramyxovirus family of the order mononegavirales. Paramyxovirus particles are composed of a non-segmented RNA genome encapsidated by the viral nucleocapsid protein, viral RNA-dependent RNA-polymerase (RdRp) components, a lipid envelope, and matrix and envelope proteins that organize virion assembly and mediate host cell entry, respectively. In addition to RSV, the family includes major animal and human pathogens such as Newcastle disease virus (NDV), measles virus (MeV), and mumps virus (MuV) amongst others. The paramyxoviruses are further divided into two subfamilies, the Pneumovirinae and the Paramyxovirinae, with RSV belonging to the Pneumovirinae subfamily. All members of the paramyxovirus family spread through the respiratory route and most cause acute disease. In addition, most paramyxoviruses, RSV included, are extremely contagious. While vaccines are available for several of the paramyxovirinae, very few treatment options exist causing an urgent need for the development of effective antiviral therapeutics.
RSV infects through the respiratory route [1], and initial infection of airway epithelia cells is followed by rapid spread from the nasopharynx to the lower airways that can affect respiratory function through excessive mucus, necrotic epithelial debris, and inflammatory cells obstructing the airways. Despite heightened research efforts, no safe and effective RSV vaccine has been approved to date. In the United States, RSV causes over 120,000 infant hospitalizations and is the foremost cause of infant mortality from viral respiratory disease [2]. Mortality from RSV disease is highest in the very young – the virus is responsible for 6.7% of all deaths in children between the ages of 1 month to 1 year [3]. Globally, RSV disease is estimated to cause the deaths of 66,000–199,000 children below the age of five per year [4]. Those especially at risk of severe RSV disease are premature infants, children with bronchopulmonary dysplasia or congenital heart defects, the elderly, and the immunosuppressed [2, 4–6].
Several clinical studies have challenged the paradigm that RSV pathogenesis is the result of host immunopathology alone. Although the relationship between viral load and severe RSV disease involving lower respiratory infection (LRI) is not fully defined, a correlation between upper respiratory tract illness and viral load was demonstrated in adults [7]. Furthermore, higher viral load served as a predictor of RSV LRI in naturally infected infants [8]. Among hospitalized RSV-infected children less than two years of age, viral load on day three of hospitalization was associated with a requirement for intensive care and respiratory failure, highlighting a potential window of opportunity for therapeutic intervention [9].
1.2 Current Therapy and Developmental Status of Anti-RSV Therapeutics
While RSV elicits both innate and adaptive immune responses, the virus is poorly immunogenic and neutralizing antibody titers wane quickly after infection [1]. Poor immunogenicity allows for reinfections throughout life and compromises vaccine development [1]. Presently, approved therapeutic options are limited to the monoclonal antibody Palivizumab which targets the RSV fusion protein, and the small molecule therapeutic Ribavirin. Palivizumab immunoprophylaxis, however, remains reserved for high-risk individuals due to prohibitive treatment costs [10, 11], while Ribavirin is compromised by toxicity and teratogenicity issues and limited antiviral efficacy [12, 13]. Current RSV disease management remains therefore frequently restricted to supportive care, such as oxygen therapy, creating an urgent clinical need for new, efficacious and cost effective therapies [14].
The development of next-generation therapeutics against RSV has largely focused on two major target areas, the viral entry machinery and the viral RdRp complex (Figure 1). Of a multitude of RSV inhibitors that have been described in recent years, only two orally available drug candidates have advanced to phase 2 clinical trials, GS-5806 and ALS-8176. The former functions as an allosteric inhibitor of the RSV fusion (F) protein, blocking viral entry [15]. By contrast, ALS-8176 is a nucleoside analog that competitively inhibits the RSV RdRp complex [16]. Both candidates decreased viral load and disease symptoms in challenge studies [17]. In this review, we will discuss the current state and future of RSV drug development.
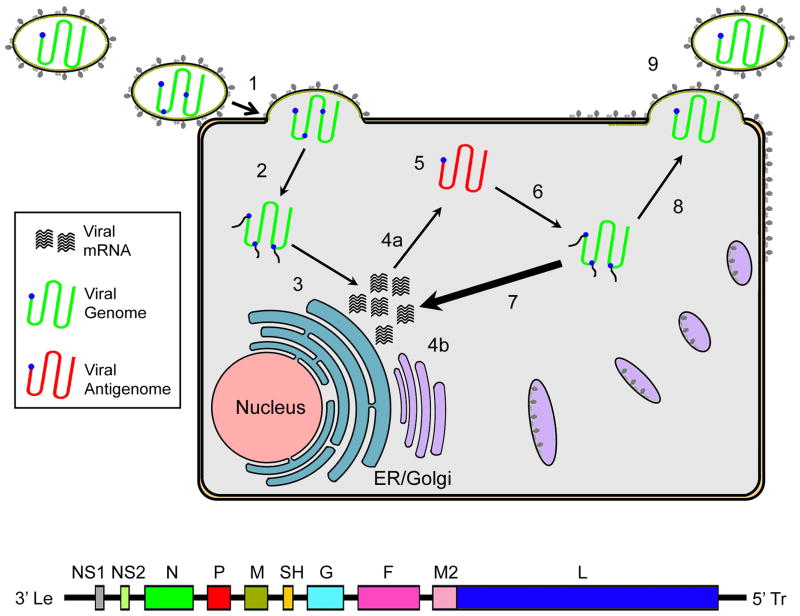
Figure 1.
Schematic of the RSV life cycle. (1) Infection commences with viral attachment, fusion of the viral envelope with target membranes, and entry. (2) The viral nucleocapsid genome and RdRp complex are released into the cytoplasm. (3) Together with host factors, the RdRp complex begins primary transcription to synthesize viral mRNAs. (4) Viral proteins are synthesized, glycoproteins are transported to the plasma membrane through the host cell secretory system. (5) Switch of RdRp to replicase mode results in the production of full-length antigenomes of positive polarity. (6) Antigenomes then serve as templates for the creation of progeny genomic RNA. (7) Newly synthesized genomes template the majority of viral mRNAs through secondary transcription. (8) For particle assembly, progeny genomes with associated RdRp components are shuttled to budding sites at the plasma membrane. (9) Virion assembly. The virus entry machinery and the RdRp complex were identified as molecular targets of a large proportion of developmental drug candidates.
2.1 The RSV Entry Machinery
The RSV envelope is coated with two glycoproteins, the attachment glycoprotein (G) and the fusion (F) protein [18]. In contrast to attachment proteins of the paramyxovirinae, the RSV G protein is not essential for viral entry but contributes to the efficiency of entry and viral pathogenesis [19]. However, RSV F is critical for entry and facilitates fusion of the viral envelope with target cell membranes and of the plasma membranes of infected cells displaying F on the cell surface and uninfected bystander cells (Figure 2) [20].
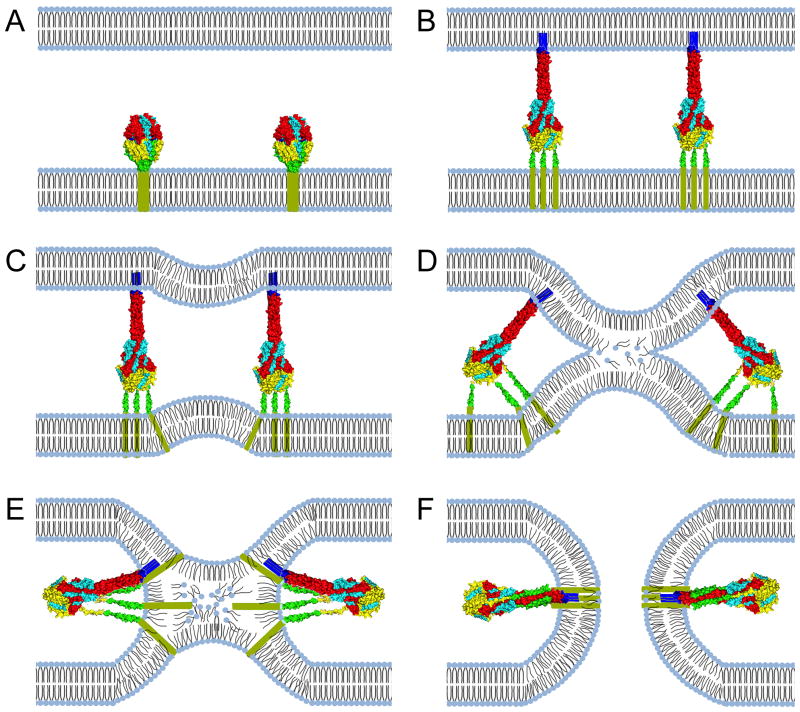
Figure 2.
Fusion pore formation through class I viral fusion proteins. (A) The F protein initially folds into a metastable prefusion conformation. (B) Once proteolytically matured and activated, the prefusion F head domain undergoes a series of major conformational changes, resulting in the assembly of an elongated trimeric coil (HR-A), which propels the fusion peptide towards the target membrane. (C) Heptad repeats proximal to the transmembrane domain (HR-B) start to fold onto the HR-A central helix during F hairpin formation, generating fusion dimples in the lipid bilayers. (D & E) Continued hairpin formation leads to the assembly of the 6HB through zippering of the HR-B domains into the grooves of the HR-A triple helix. Extreme negative curvature is introduced into the approaching bilayers, resulting in the disarray of lipids in the outer monolayers and ultimately merger of the outer layers in a hemifusion intermediate. (F) Transmembrane domains and fusion peptides are brought into close proximity, leading to the opening of a fusion pore. Creation of a productive fusion pore by paramyxovirus F proteins is not mandatorily dependent on full closure of the 6HB [100]. Images of protein structures were generated in Pymol. PDB codes: prefusion F:4MMS; postfusion F:3RRT
Paramyxovirus F proteins are class I fusion proteins that are synthesized as inactive F0 precursors (Figure 3). Proteolytic maturation, in most cases carried out by furin-type host proteases in late Golgi compartments, generates a larger transmembrane F1 fragment and exposes the membrane attack group, the fusion peptide, which is located at the newly liberated N-terminus of F1. The shorter F2 fragment remains associated with the F1 ectodomain and is an integral part of the F ectodomain structure [21].
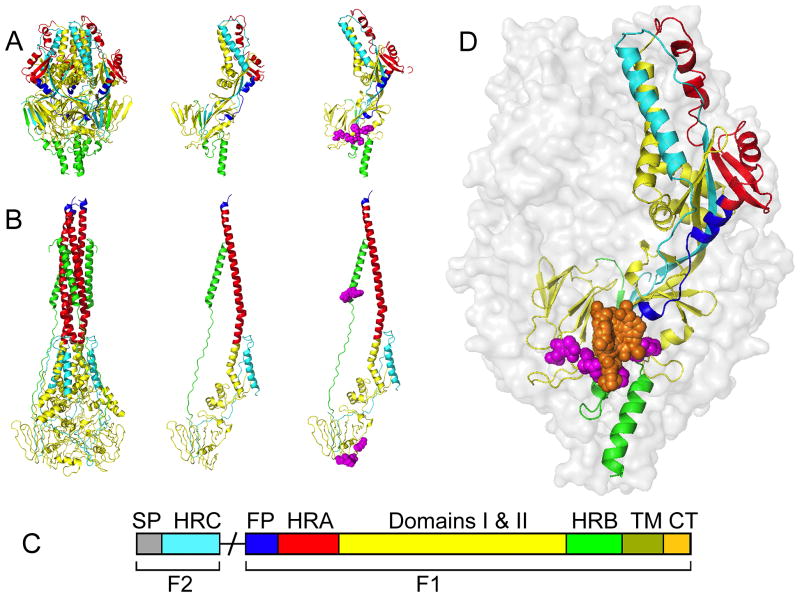
Figure 3.
Structural model of the RSV fusion protein. (A) The RSV F protein in the prefusion conformation. Shown are a ribbon model of the native prefusion F trimer (left), a single monomer (center), and a monomer with known resistance mutations (magenta) in the 392–401 and 486–489 microdomains (right). (B) The F protein in the thermodynamically stable postfusion conformation. Fields of view as described for (A). (C) Schematic of the domain architecture of the RSV fusion protein. Proteolytic maturation of newly synthesized F results in the liberation of the F1 and F2 subunits. F1 harbors the fusion peptide (FP; blue), heptad-repeat A (HR-A; red), connecting domains 1 and 2 (yellow), heptad-repeat B (HR-B; green), a transmembrane domain (TM; olive), and a short cytoplasmic or luminal tail (CP; tan). F2 remains an integral structural component of the trimer. This subunit contains the signal peptide (SP; grey) and a heptad-repeat C (HR-C; cyan) domain. (D) Schematic of the prefusion RSV F trimer overlaid with entry inhibitor binding locations (orange) and resistance hot spots (magenta spheres). For clarity, only one monomer was color-coded by domain structure as outlined in (C). Images of protein structures were generated in Pymol. PDB codes: prefusion F:4MMS; postfusion F:3RRT; Inhibitor bound F: 5EA3, 5EA4, 5EA5, 5EA6, 5EA7.
Triggering of metastable prefusion F is thought to be mediated by receptor binding, indicating the presence of a target membrane (Figure 2). Once activated, the prefusion F complex passes through a cascade of deep-seated conformational changes. The membrane distal part of the prefusion F head domain refolds into an extended trimeric heptad repeat (HR-A) coiled-coil helix, which propels the fusion peptide positioned at the tip of the coiled-coil towards the target membrane [18]. The resulting F pre-hairpin intermediate fold back onto itself, resulting in the assembly of a thermodynamically highly stable six helix bundle (6HB) structure consisting of the trimeric HR-A coiled coil and shorter HR-B domains that are positioned adjacent to the transmembrane domain. Zippering of the 6HB brings the transmembrane domain and fusion peptide, and hence donor and target membranes, into close proximity, ultimately fostering lipid merger at the fusion tip through the introduction of extreme local negative curvature into the bilayers (Figure 2).
2.2 Entry Inhibitors
Interference with RSV F proteolytic maturation, triggering of refolding, the refolding cascade, or 6HB zippering can all be anticipated to efficiently prevent viral entry. Indeed, numerous small molecule compounds have been discovered that inhibit RSV fusion in vitro [22–30]. These compounds typically show very high potency combined with low toxicity and have a tendency to emerge readily in anti-RSV drug screening campaigns [22–30]. One of the entry inhibitors is the compound GS-5806 that has advanced to clinical trials and has reduced viral loads in human volunteers [15].
Surprisingly, viral resistance mutations to all of these entry blockers emerged in two distinct microdomains of the RSV F protein: residues 392–401 and 486–489 (Figure 3) [28]. Considering the diverse chemical and structural nature of these compound classes, overlapping resistance profiles were rather unexpected. Originally, it was assumed that the compounds may interfere with 6HB zippering based on biochemical assays and photo-crosslinking studies that were carried out for a subset of developmentally advanced entry inhibitor classes in this panel [31]. While residues in the 486–489 microdomain are part of the 6HB structure in postfusion F, this mechanistic proposal could not account for resistance mediated by mutations in the 392–401 microdomain, however, which is located at the opposite end to the 6HB in postfusion F [31](Figure 3).
A solution to this conundrum was presented in a recent study, showing that residues in both resistance domains are located in immediate proximity of each other near the base of the prefusion F head domain [28]. This observation provided a basis for an alternative model of fusion inhibition, in which current RSV entry inhibitor classes may target and stabilize the prefusion F structure, preventing the initiation of F refolding rather than blocking 6HB zippering. Support for this view comes from the subsequently released high-resolution crystal structures of prefusion RSV F complexed with several of the entry inhibitor classes [32]. In all of these structures, the electron density for the small molecule inhibitors was located in the three-fold symmetry axis at the base of the prefusion F head (Figure 3). Additionally, all inhibitors are proposed to interact with the aromatic side chains of F140 and F488, which are are located within the fusion peptide and HR-B, respectively. This suggests that all of these diverse inhibitor classes may indeed block RSF F fusion activity by stabilizing F in its prefusion form. In concurrence with crystallographic data, binding of all compounds to the prefusion conformation of F was confirmed by isothermal titration calorimetry (ITC) [32].
2.3 Developmental Implications of RSV Pan-Resistance to Fusion Inhibitors
Mutations in the 392–401 and 486–489 F microdomains likely facilitate RSV escape from inhibition through different resistance mechanism, however, depending on the microdomain mutated and the nature of the amino acid substitution. F proteins harboring a signature mutation in the 486–489 domain, D489Y, were shown to maintain membrane fusion activities similar to those of standard F [32], whereas several resistance mutations in the 392–401 microdomain and a D489E substitution are associated with strong F hyperfusogenicity [28, 32]. Modeling of the D489Y mutation into the RSV F prefusion structure suggest a direct resistance mechanism, since a tyrosine side chain at position 489 is predicted to protrude into the binding pocket occupied by the fusion inhibitors (Figure 3), abolishing drug binding. By contrast, the D489E and D401E resistance mutations do not directly populate the drug target site but may lead to secondary resistance, resulting from a narrowed window of opportunity for productive drug binding due to accelerated refolding of the hyperfusogenic F mutants [28, 32]. Naturally, these direct and indirect (kinetic) escape mechanisms are not mutually exclusive, but several substitutions may mediate viral resistance through a combination of both effects.
However, the existence of different escape mechanisms from inhibition by all currently identified RSV fusion inhibitors with known resistance profiles raises concerns about the long-term value of therapeutic targeting of the RSV F protein. While it may be possible to address direct resistance through proactive, structure-guided drug design, overcoming kinetic resistance synthetically poses a substantially greater challenge. Unless kinetic resistance is directly associated with a penalty in RSV fitness and pathogenicity, widespread clinical implementation of RSV entry inhibitors must be considered at high risk to introduce preexisting resistance into circulating virus strains, comparable for instance to the experience with the use of the adamantes for the treatment of influenza A virus (IAV) infections. While originally efficacious, the use of amantadine against IAV is no longer recommended by the Centers for Disease Control and Prevention, since all seasonal strains tested in the 2008/2009 influenza season were fully resistant [33].
The effect of hyperfusogenic resistance mutations on RSV pathogenicity is unknown at present in the human host. In a mouse model of RSV infection that recapitulates several features of human disease [34], however, only recombinant RSV harboring the D489E mutation were found to be attenuated, while virus load and clinical symptoms associated with a D401E strain were identical to those of the non-resistant parent virus [28]. The mouse model certainly does not fully recapitulate the dynamics of human RSV infection, but the available data caution against the use of currently tested entry inhibitors as stand-alone therapeutics against RSV infection.
Considering the substantial up-front investments required for bringing a novel therapeutic to market, future anti-RSV drug discovery and development campaigns should be designed to avoid the entry inhibitor pan-resistance trap. Next-generation RSV reporter strains for drug screening campaigns were recently described that include a signature mutation mediating pan-resistance against entry inhibitors [28, 35]. Use of these strains at the primary or counterscreening stage should reliably eliminate compounds sensitive to this escape mechanism early from the hit pool, most likely shifting the focus of the campaign towards blockers of downstream stages of the viral life cycle, such as inhibitors of the viral polymerase complex, particle assembly, or virion release. Alternatively, mechanistically novel and highly interesting RSV entry inhibitors may be identified that are insensitive to the pan-resistance mechanism.
3.1 The RSV RdRp Complex
As a member of the order mononegavirales, RSV contains a non-segmented negative-sense RNA genome, which requires the virus-encoded RdRp complex for both genome replication and transcription of viral RNA. The RdRp complex is responsible for a variety of distinct enzymatic activities, highly specific protein-protein interfaces are required for activity, and there is no cellular equivalent to the RdRp complex in mammalian cells, making it overall an attractive target for antiviral therapies. In this section, we will discuss the role of different viral protein components required for RdRp activity, summarize the current structural insight into the organization of the complex, and evaluate the prospects of therapeutic targeting of specific RdRp substructures.
3.1.1 Nucleocapsid and Nucleocapsid Protein
Throughout the RSV replication cycle, the RNA genome exists as a distinct ribonucleoprotein complex, the nucleocapsid (NC), that is comprised of the genomic RNA fully encapsidated by the viral nucleocapsid (N) protein. Only the assembled NC can act as a template for RNA synthesis by the viral RdRp complex, which consists of the viral large (L) and phosphoprotein (P) in addition to host co-factors. The L protein performs all the enzymatic activities of RNA synthesis, while P acts as an indispensable cofactor. After RSV enters the host cell, the NC along with the attached RdRp complex is released into the cytoplasm. Once there, the NC serves as the sole template for both transcription and replication (Figure 4).
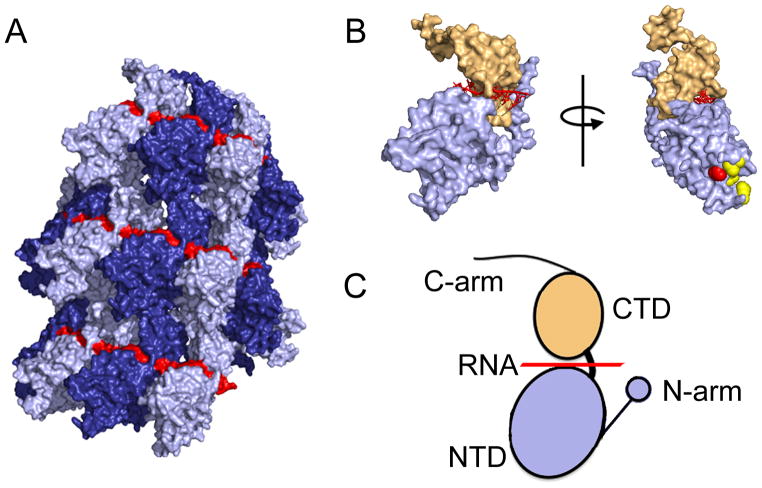
Figure 4.
Structural models of the RSV N protein and the nucleocapsid. (A) Structure of the helical RSV nucleocapsid showing the encapsidated RNA in red and individual N protein protomers in dark and light blue. (B) Surface representation of a RSV N monomer (left). Rotation of the N monomer reveals the RSV P protein binding site (red sphere) and resistance mutations against RSV604 (yellow). (C) The RSV N protein is composed of two core domains, NTD (blue) and CTD (tan), which enclose the viral RNA genome (red). Terminal extensions from the NTD and CTD domains, the N- and C-arm, respectively, interact with adjacent N subunits in the helical nucleocapsid structure. Images of protein structures were generated in Pymol. PDB codes:4BKK(RSV N), 4UCB(RSV NTD-P).
N subunits in the RSV NC are assembled side-by-side and parallel along the length of the RSV RNA genome, entirely sequestering the viral RNA (Figure 4) [36]. In electron micrographs, the RSV NC displayed a helical structure that is characteristic for members of the Paramyxoviridae family. A crystal structure of the RSV N composed of parallel layers of RSV N:RNA rings was recently solved [37] (Figure 4), and revealed an organization of each N protomer into two core domains, a N-terminal (NTD) and C-terminal (CTD) domain, which are linked via a hinge region (Figure 4). Both the NTD and CTD subunits of each N protomer contain N- and C-terminal extensions, designated N-arm (residues 1–28) and C-arm (residues 360–375), respectively, which interact with neighboring N protomers in a chain-link like arrangement [36]. In the crystal structure, RSV N protomers participate in weak top-to-bottom interactions between NTD originating from one helix layer and the CTD of the adjacent layer [37]. The RNA is positioned in a central groove at the NTD/CTD interface, forming a continuous RNA belt along the outside of the NC (Figure 4).
3.1.2 Phosphoprotein
While the RSV phospho- (P) protein lacks any catalytic activity, it serves as an indispensable cofactor for RdRp actvity. P performs a range of different tasks in the virus replication cycle and interacts with multiple viral proteins including L and N, in addition to cellular proteins [38–40]. P is thought to correctly position the L protein for RNA synthesis by interacting with the NC template [41, 42]. In addition, P also chaperones newly synthesized, RNA-free N protein (N0) to the nascent genomic and antigenomic viral RNA during replication [43]. Fitting to the multifunctional nature of paramyxovirus P bioactivity, the proteins displays a modular organization consisting of a central tetramerization domain flanked by N-terminal (P-NTD) and C-terminal (P-CTD) domains separated by flexible linker regions [44–46]. Structures for individual domains of several paramyxovirus P proteins have been previously solved [46, 47], but very little structural information is currently available for RSV P [48].
Due to the inability of the RSV L protein to bind to the NC, a critical function of P is to position the RdRp complex on the NC and maintain continued contact between the RdRp and the NC as the RdRp progresses along its template [49]. It has been shown that the RSV P C-terminal domain binds to a region in the NTD of RSV N (Figure 4) [50]. Furthermore, it was demonstrated that the extreme C-terminal residue of RSV P, F241, plays a critical role in the recognition of the RSV N by inserting into a hydrophobic binding pocket in the N-NTD (Figure 4) [48]. Importantly, an F241A substitution completely abolished RSV minigenome activity [50], revealing a critical role for P docking into this pocket for bioactivity. Although the precise mechanism of how the RdRp accesses the RNA encapsidated within the NC is unknown, it is thought that a hinge movement between the N NTD and CTD allows for the transient exposure of the encapsidated RNA as the RdRp complex progresses. In fact, transient interaction with the P protein or the L-P complex may directly induce this conformational change as suggested for the related P protein of Mumps virus [42]. If the RSV P binding domain in the NTD of the N subunits of the NC serve as anchor points for the attachment of the RdRp complex [41, 51], RNA synthesis will require continuous release and reattachment of these contacts as the RdRp complex advances along the NC [51].
3.1.3 Large Polymerase Protein
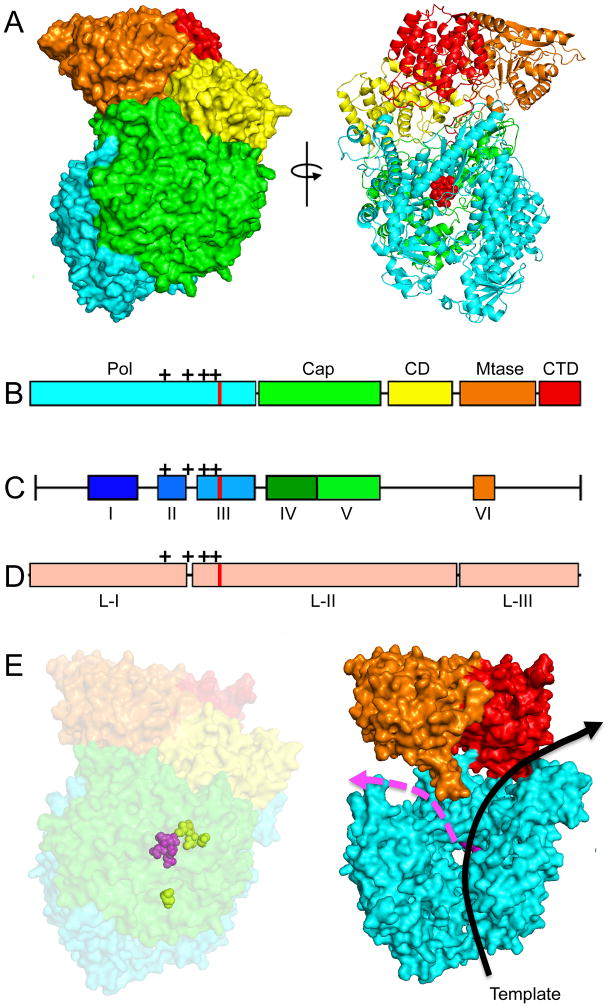
The L protein performs all catalytic activities necessary for RNA synthesis, mRNA capping, and mRNA polyadenylation [52–54]. Bioinformatics analyses have shown that the L protein can be divided into six conserved regions (CR I to CR VI) that are connected by flexible linker regions (Figure 5) [55, 56], suggesting a modular organization of the protein. Biochemical studies on RSV and MeV L protein confirmed that L can be divided into two distinct fragments that are capable of restoring RdRp bioactivity via trans-complementation when co-expressed [57]. This finding revealed that the L proteins are comprised of at least two independently folding-competent domains. In agreement with these data, bioinformatics analysis previously identified three large regions (LR I to LR III) separated by two linker domains [58]. LR I harbors CR I and II, LR II contains CR III-CR V, and CR VI is located in LR III. Consistent with at least a two-domain organization, insertion analysis on the L proteins of MeV and rinderpest virus showed that even large insertions were tolerated in the LR II/LR III junction, but not the LR I/LR II junction [58, 59].
Figure 5.
Structure of a mononegavirales L protein and homology models of the RSV L protein generated based on the coordinates reported for the related VSV L structure [65]. (A) Structure of the VSV L protein shown as surface (left) and ribbon (right) models. Red spheres in the ribbon model highlight the active site for phosphodiester bond formation. (B) The VSV L structure displayed five main structural units: polymerase (cyan), capping (green), connector domain (yellow), methyltransferase (orange), and C–terminal domain (red). (C) Sequence analysis revealed six conserved regions (CR I–VI) in the L proteins. (D) Insertion analysis has shown that paramyxovirus L proteins can be further divided into three large regions (LRI-III). The location of the polymerase active site is shown in (B–D) as red lines, mutations that confer resistance to the RSV nucleoside analog inhibitor ALS-8176 are shown as black crosses. (E) Homology models of the RSV L protein based off the VSV L structure. A surface representation (left) and a model of the internal organization (right) are shown. The active site for phosphodiester bond formation and the ALS-8176 resistance mutations are highlighted as magenta and yellow spheres, respectively. Arrows indicate channels for the template (black) and newly synthesized (magenta) RNA strands. Images of protein structures were generated in Pymol; PDB code:5A22 (VSV L). Homology models were generated using SWISS-MODEL.
Whereas the roles for each of the conserved L domains in RdRp activity remain not entirely understood, some functionalities have been assigned. CR I has been implicated in L–P interactions [60–63] and L-L oligomerization [60, 61, 64]. In fact, oligomer formation has been proposed for several related paramyxovirus L proteins [57, 61–64]. These studies show that L–L interaction domains are located in N-terminal regions of the L protein and supposedly function independent of P protein binding [57, 61, 62]. While no native structural data is available for RSV L or any other paramyxovirus L protein, a cryoEM structure of the related vesicular stomatitis virus was recently solved at near-atomic resolution (Figure 5) [65], showing a monomeric organization. If this structure is exemplary for a mononegavirales L protein, the actual physiological role, if any, that paramyxovirus L oligomerization would serve is unclear.
In agreement with L having a modular organization of functional domains, the VSV L structure displays a linear domain architecture (Figure 5) The N-terminal region of L encompassing CRs I-III assumes a polymerase fold, in which the catalytic center for phosphodiester bond formation is located in CR III [66]. In addition to two linker domains, the C-terminal L regions contain two enzymatic centers required for mRNA capping. In VSV L, a conserved GXXTnHR motif in CR V facilitates an unusual mechanism of capping the viral mRNAs through transfer of 5′-monophosphate-mRNA onto GDP [67]. However, RSV L may possess traditional guanylyltransferase activity, since a related paramyxovirus RdRp complexes from Rinderpest virus reportedly forms covalent guanosine monophosphate-L intermediates in vitro [68]. Finally, CR VI contains methyltransferase (MTase) activity [55, 69]. Recently, the structure of the closely related human metapneumovirus (HMPV) L methyltransferase domain was solved [70], which like RSV, belongs to the pneumovirinae subfamily within the paramyxoviruses. This high-resolution structure has strong potential to guide the future design of novel MTase inhibitor candidates.
Due to this multidomain architecture and the concentration of multiple essential enzymatic centers, the L protein offers an abundance of potentially druggable targets. The guanylyltransferase domain presents a case in point, since inhibiting the viral capping machinery using guanosine nucleotide analogs constitutes a proven antiviral approach against pathogens of other RNA virus families [71]. In addition, it may also be possible to target the proposed S-adenosyl-l-methionine transferase domain responsible for 5′-cap methylation [69, 72]. The therapeutic potential of S-adenosyl-l-homocysteine derivatives for use as potential antivirals has likewise been proven for other RNA virus families [73].
3.2 Targeting the RSV RdRp Complex: Nucleoside and Nucleotide Analogs
Nucleoside analog inhibitors contain modified nucleic acids and typically act, after intracellular conversion to the triphosphate state, as chain terminators in nucleic acid polymerization [74, 75]. Although numerous nucleoside analogs have shown great clinical success, ribavirin is the only substrate analog currently licensed to treat RSV disease. Ribavirin is a purine-analog with the ability to base pair with either cytosine or uracil [76]. Unlike typical nucleoside analogs, the drug is thought to cause hyper-mutations in the newly synthesized strands, stopping virus replication by error catastrophe [77]. However, Ribavirin causes severe adverse side effects and the clinical efficacy of ribavirin against RSV disease is uncertain [78]. Favipiravir (T-705), a nucleotide analog investigated for the treatment of multiple different virus infections, such as Ebola virus, influenza A, and foot-and-mouth-disease virus [79], showed activity against RSV in cell culture. However, effective concentrations were too high for clinical use [79, 80].
In contrast, ALS-8176, a newly developed nucleoside analog, is currently in phase II clinical trials for use against RSV infection [17]. ALS-8176 was well tolerated in initial trials and significantly reduced viral load when administered at the onset of infection [17], but the toxicity profile in particular for pediatric use is unclear at present. Nevertheless, these findings are promising and offer proof-of-concept for the clinical benefit of competitive polymerase inhibitors for RSV therapy. While ALS-8176 was demonstrated to block virus replication through chain termination and resistance sites are located in the L polymerase domain [81], an unrelated nucleoside showed a remarkably distinct resistance profile [82]. Viral escape from inhibition by this compound, identified in a high-throughput anti-RSV screening campaign, mapped to the RSV P protein instead of the L polymerase [82], suggesting a distinct and unexpected mechanism of antiviral activity other than chain termination and error catastrophe.
3.3 Non-Nucleoside RdRp Inhibitors
Allosteric RdRp inhibitors block polymerase activity by binding to regulatory sites, which may or may not be located in proximity to actual catalytic centers. Inhibitors of this class may structurally alter the active site through long-range effects or sterically prevent substrate access to the catalytic center. Compounds may also disrupt the assembly of protein complexes (protein-protein interaction (PPI) inhibitors) necessary for correct enzymatic function. Extensive clinical experience with allosteric polymerase inhibitors was gained with blockers of HIV reverse transcriptase (RT). Implementation of the first generation non-nucleoside RT inhibitors Nevirapine and Efavirenz [83, 84] has revealed that viral resistance emerges rapidly, making these compounds unsuitable for monotherapy [85]. In the precedence set by Nevirapine and Efavirenz, two main resistance mutations emerged, K103N for efavirenz and Y181C for nevirapine. These mutations prevented inhibitor binding to a hydrophobic binding pocket. However, development of second-generation non-nucleoside RT inhibitors such as Etravirine and Rilpivirine incorporated structure-based design to overcome these resistance mechanisms, allowing for the use of Etravirine in patients containing multidrug-resistant HIV [83]. These second-generations compounds utilized a diarylpyrimidine (DAPY) motif, which introduced much greater conformational flexibility, allowing for the inhibitors to bind in multiple conformations [86]. This flexibility allowed the molecules to breathe and overcome the previous resistance mutations towards the first-generation compounds [86]. The development of Etravirine and Rilpivirine offers a prime example of how structural knowledge can further the generation of new and highly potent small molecule inhibitors.
The RSV L protein doubtlessly offers a multitude of druggable targets for allosteric polymerase inhibitors. Indeed, drug discovery campaigns have identified distinct compounds that specifically block RdRp activity in cell culture and show high potential for lead development [82, 87–89]. It remains to be explored whether a rapid onset of viral resistance will undermine the clinical use of allosteric RdRp inhibitors as experienced with the first generation of HIV RT blockers. We can anticipate that vastly improved structural insight into the organization of the RSV polymerase complex will be available in the near future, which may open novel avenues for the proactive design of second generation allosteric blockers with improved escape profile. These may be suitable for solo-therapy, or could be used in a combination therapy approach with nucleoside analog blockers, forcing the virus to accumulate multiple mutations in the polymerase protein to mount resistance.
In addition to de novo drug screening campaigns for hit discovery, the growing body of L structural information makes feasible the structure-guided redesign against RSV polymerase of successful drug candidates developed against related RdRp complexes. For instance, an orally available blocker of related morbillivirus (i.e. MeV and canine distemper virus) polymerases with demonstrated efficacy in small-animal models [90] has high clinical potential, but lacks activity against RSV RdRp. Resistance profiling of this compound class showed escape hot-spots clustered in the polymerase domain of MeV L near the substrate binding site (Figure 5). Since the overall structural organization of this region is predicted to be highly conserved between MeV and RSV L, the MeV inhibitor class may serve as a viable point of entry for the synthetic adaptation of the MeV blocker scaffold to RSV L or highlight a fruitful molecular target for in silico drug discovery campaigns.
In addition to directly targeting the enzymatic activities of the L protein, the large number of dynamic and distinct PPIs available in the RdRp complex represents a promising but presently underappreciated strategy for the design of antiviral therapeutics. PPIs were once thought to be unviable targets for interference by small molecules due to typically large interface areas (1,000–2000 Å2) and the flat geometry of a traditional PPI interface [91]. However, natural small-molecule drugs such as cyclosporine and rapamycin underscored the fact that a PPI inhibitor need only block a small number of residues in critical regions that confer most of the binding energy in order to efficiently prevent protein-protein contact [92].
Currently, over 40 PPIs have been successfully blocked by small molecule compounds [93–95] and a variety of candidates have advanced to clinical testing. Previous campaigns have shown that PPIs most suitable for small molecule inhibition should focus on binding partners with short interacting sequences and contact hot-spots in well defined areas of less than 900 Å2. [94, 96].
Druggability of the RSV N-P interface has been explored in a recent study characterizing RSV inhibitor RSV604 [97] and a report examining 1-benzyl-1H-pyrazole-3,5-dicarboxylate (BPdC) based compounds [48]. The BPdC based compounds are thought to inhibit N-P interactions by mimicking the terminal F241 residue (Figure 3) and inhibit viral RdRp-NC contacts through the block of N-P interactions [48]. However, BPdC based compounds discussed in the study displayed unacceptable toxicity [48], and it is unclear whether lead development will be able to overcome this liability. Resistance mutations to RSV604 affected residues proximal to the P binding site on N-NTD [97]. RSV604 displays potent antiviral efficacy, uses a novel mechanism of action, and is also orally bioavailable. Moreover, RSV604 was safe and well tolerated by healthy volunteers in early clinical trials [97–99].
Importantly, the resistance profile of RSV604, as well as the co-crystal structures of RSV N-NTD with BPdC compounds, established proof-of-concept for the feasibility of small molecule interference with PPIs critically needed for RSV RdRp activity [48, 97]. As our structural knowledge into the organization of the RSV RdRp complex and the molecular nature of its PPIs advances, a combination of targeted high throughput screening campaigns directed against individual interfaces in the RdRp complex and structure-guided in silico evaluation and optimization of hit candidates should yield a new crop of developable RSV RdRp scaffolds.
4.1 Expert Opinion
The high contagiousness of RSV and the lack of vaccine protection against infection create an urgent need for the development of efficacious and cost-effective antiviral therapeutics that are capable of preventing infection if given prophylactically to high risk patients and/or improve disease management through suppression of progression to severe LRI. Based on the overlapping resistance profiles of all RSV entry inhibitors currently considered for human use, the likely existence of both direct and kinetic resistance pathways to overcome entry blockage, and the alarming observation that some pan-resistant RSV strains remained fully pathogenic in a small animal model of RSV infection, we believe that small-molecule RSV entry inhibitors will have little long-term impact if used for monotherapy. The same limitation conceptually applies, however, to blockers of RSV RdRp activity, whether they act through allosteric or competitive mechanisms.
Combination therapy, for instance consisting of a nucleoside analog and an allosteric RdRp blocker or an entry inhibitor, will therefore likely be required to counteract the rapid introduction of preexisting resistance into circulating RSV strains. Of these options, we consider combining different classes of RdRp inhibitors to be most promising, since the accumulation of multiple distinct resistance mutations in a single protein complex complex has inherently higher potential to be mandatorily linked to a viral fitness and pathogenesis penalty than mutations in distinct viral proteins, especially since it is already known that pan-resistance to entry inhibitors can be achieved without viral attenuation in the mouse model. Since paramyxoviruses including RSV cause predominantly acute disease and require continuous transmission to sustain presence in a population, even a slight reduction in viral fitness or transmission success caused by multiple mutations in the RdRp complex may prevent that strains resistant to multiple RdRp blocker classes become predominant in the field.
The use of next-generation RSV reporter strains engineered to eliminate compounds from the hit candidate pool that are sensitive to entry inhibitor pan-resistance will likely result in a shift of hits towards RdRp blockers. In addition, recent advances in our structural understanding of the spatial organization of the mononegavirales RdRp complexes and an increasing number of native crystal structures of RSV RdRp fragments are expected to provide a solid foundation for the structure-guided design of drug discovery campaigns aimed at both direct blockers of enzymatic activity and PPI inhibitors. High-resolution structures of RdRp components have also high prospect to open the field for a molecular understanding of polymerase blocker activity, the mechanistic evaluation of viral escape mechanisms, and structure-guided synthetic scaffold optimization campaigns of lead candidates. Considering the tremendous progress experienced in our structural understanding of RSV biology in recent years and encouraging early results of a nucleoside analog inhibitor in clinical trials, there is high prospect that new generations of much needed effective anti-RSV therapeutics will become available for clinical use in the foreseeable future.
Acknowledgments
This work was supported, in part, by Public Health Service grants AI071002 and HD079327 from the NIH/NIAID and NIH/NICHD (to R.K.P.).
References
- 1.Collins PLCJ., Jr . Respiratory Syncytial Virus and Metapneumoviruses. In: Knipe DMHP, editor. Fields Virology. Lippincott, Williams, & Wilkins; Philadelphia: 2007. pp. 1601–1645. [Google Scholar]
- 2.Shay DK, Holman RC, Roosevelt GE, Clarke MJ, Anderson LJ. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979–1997. J Infect Dis. 2001;183:16–22. doi: 10.1086/317655. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falsey AR. Respiratory syncytial virus infection in elderly and high-risk adults. Exp Lung Res. 2005;31(Suppl 1):77. [PubMed] [Google Scholar]
- 6.Lee N, Lui GC, Wong KT, Li TC, Tse EC, Chan JY, Yu J, Wong SS, Choi KW, Wong RY, Ngai KL, Hui DS, Chan PK. High morbidity and mortality in adults hospitalized for respiratory syncytial virus infections. Clin Infect Dis. 2013;57:1069–1077. doi: 10.1093/cid/cit471. [DOI] [PubMed] [Google Scholar]
- 7.DeVincenzo JP, Wilkinson T, Vaishnaw A, Cehelsky J, Meyers R, Nochur S, Harrison L, Meeking P, Mann A, Moane E, Oxford J, Pareek R, Moore R, Walsh E, Studholme R, Dorsett P, Alvarez R, Lambkin-Williams R. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med. 2010;182:1305–1314. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeVincenzo JP, El Saleeby CM, Bush AJ. Respiratory syncytial virus load predicts disease severity in previously healthy infants. J Infect Dis. 2005;191:1861–1868. doi: 10.1086/430008. [DOI] [PubMed] [Google Scholar]
- 9.El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis. 2011;204:996–1002. doi: 10.1093/infdis/jir494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahadevia PJ, Malinoski FJ. Cost-effectiveness of respiratory syncytial virus prophylaxis with palivizumab. Arch Pediatr Adolesc Med. 2007;161:519–520. doi: 10.1001/archpedi.161.5.519. author reply 520. [DOI] [PubMed] [Google Scholar]
- 11.Weiner LB, Masaquel AS, Polak MJ, Mahadevia PJ. Cost-effectiveness analysis of palivizumab among pre-term infant populations covered by Medicaid in the United States. J Med Econ. 2012;15:997–1018. doi: 10.3111/13696998.2012.672942. [DOI] [PubMed] [Google Scholar]
- 12.Anderson LJ, Parker RA, Strikas RL. Association between respiratory syncytial virus outbreaks and lower respiratory tract deaths of infants and young children. J Infect Dis. 1990;161:640–646. doi: 10.1093/infdis/161.4.640. [DOI] [PubMed] [Google Scholar]
- 13.Groothuis JR, Woodin KA, Katz R, Robertson AD, McBride JT, Hall CB, McWilliams BC, Lauer BA. Early ribavirin treatment of respiratory syncytial viral infection in high-risk children. J Pediatr. 1990;117:792–798. doi: 10.1016/s0022-3476(05)83347-5. [DOI] [PubMed] [Google Scholar]
- 14.Johnson D. Croup BMJ Clin Evid 2009. 2009 [PMC free article] [PubMed] [Google Scholar]
- 15.DeVincenzo JP, Whitley RJ, Mackman RL, Scaglioni-Weinlich C, Harrison L, Farrell E, McBride S, Lambkin-Williams R, Jordan R, Xin Y, Ramanathan S, O’Riordan T, Lewis SA, Li X, Toback SL, Lin SL, Chien JW. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med. 2014;371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Deval J, Hong J, Dyatkina N, Prhavc M, Taylor J, Fung A, Jin Z, Stevens SK, Serebryany V, Liu J, Zhang Q, Tam Y, Chanda SM, Smith DB, Symons JA, Blatt LM, Beigelman L. Discovery of 4′-chloromethyl-2′-deoxy-3′,5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J Med Chem. 2015;58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- 17.DeVincenzo J, Fathi H, Mcclure M, Westland C, Chanda S, Lambkin-Williams R, Smith P, Harrison L, Symons J, Scaglioni-Weinlich C, Zhang Q, Nieforth K, Beigelman L, Blatt L, Fry Ja. Treatment with Oral ALS-008176, a Nucleoside Analog, Rapidly Reduces RSV Viral Load and Clinical Disease Severity in a Healthy Volunteer Challenge Study. Open Forum Infectious Diseases. 2014;1:S66–S69. [Google Scholar]
- 18.McLellan JS, Ray WC, Peeples ME. Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol. 2013;372:83–104. doi: 10.1007/978-3-642-38919-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh EE, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983;47:171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Razinkov V, Gazumyan A, Nikitenko A, Ellestad G, Krishnamurthy G. RFI-641 inhibits entry of respiratory syncytial virus via interactions with fusion protein. Chem Biol. 2001;8:645–659. doi: 10.1016/s1074-5521(01)00042-4. [DOI] [PubMed] [Google Scholar]
- 23.Andries K, Moeremans M, Gevers T, Willebrords R, Sommen C, Lacrampe J, Janssens F, Wyde PR. Substituted benzimidazoles with nanomolar activity against respiratory syncytial virus. Antiviral Res. 2003;60:209–219. doi: 10.1016/j.antiviral.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Wyde PR, Chetty SN, Timmerman P, Gilbert BE, Andries K. Short duration aerosols of JNJ 2408068 (R170591) administered prophylactically or therapeutically protect cotton rats from experimental respiratory syncytial virus infection. Antiviral Res. 2003;60:221–231. doi: 10.1016/j.antiviral.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 25.McKimm-Breschkin J. VP-14637 ViroPharma. Curr Opin Investig Drugs. 2000;1:425–427. [PubMed] [Google Scholar]
- 26.Cianci C, Genovesi EV, Lamb L, Medina I, Yang Z, Zadjura L, Yang H, D’Arienzo C, Sin N, Yu KL, Combrink K, Li Z, Colonno R, Meanwell N, Clark J, Krystal M. Oral efficacy of a respiratory syncytial virus inhibitor in rodent models of infection. Antimicrob Agents Chemother. 2004;48:2448–2454. doi: 10.1128/AAC.48.7.2448-2454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglas JL, Panis ML, Ho E, Lin KY, Krawczyk SH, Grant DM, Cai R, Swaminathan S, Cihlar T. Inhibition of respiratory syncytial virus fusion by the small molecule VP-14637 via specific interactions with F protein. J Virol. 2003;77:5054–5064. doi: 10.1128/JVI.77.9.5054-5064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan D, Lee S, Thakkar VD, Luo M, Moore ML, Plemper RK. Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc Natl Acad Sci U S A. 2014;111:E3441–3449. doi: 10.1073/pnas.1405198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cianci C, Yu KL, Combrink K, Sin N, Pearce B, Wang A, Civiello R, Voss S, Luo G, Kadow K, Genovesi EV, Venables B, Gulgeze H, Trehan A, James J, Lamb L, Medina I, Roach J, Yang Z, Zadjura L, Colonno R, Clark J, Meanwell N, Krystal M. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob Agents Chemother. 2004;48:413–422. doi: 10.1128/AAC.48.2.413-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonfanti JF, Meyer C, Doublet F, Fortin J, Muller P, Queguiner L, Gevers T, Janssens P, Szel H, Willebrords R, Timmerman P, Wuyts K, van Remoortere P, Janssens F, Wigerinck P, Andries K. Selection of a respiratory syncytial virus fusion inhibitor clinical candidate. 2. Discovery of a morpholinopropylaminobenzimidazole derivative (TMC353121) J Med Chem. 2008;51:875–896. doi: 10.1021/jm701284j. [DOI] [PubMed] [Google Scholar]
- 31.Roymans D, De Bondt HL, Arnoult E, Geluykens P, Gevers T, Van Ginderen M, Verheyen N, Kim H, Willebrords R, Bonfanti JF, Bruinzeel W, Cummings MD, van Vlijmen H, Andries K. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci U S A. 2010;107:308–313. doi: 10.1073/pnas.0910108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battles MB, Langedijk JP, Furmanova-Hollenstein P, Chaiwatpongsakorn S, Costello HM, Kwanten L, Vranckx L, Vink P, Jaensch S, Jonckers TH, Koul A, Arnoult E, Peeples ME, Roymans D, McLellan JS. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat Chem Biol. 2015 doi: 10.1038/nchembio.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centers for Disease C and Prevention. Update: drug susceptibility of swine-origin influenza A (H1N1) viruses, April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:433–435. [PubMed] [Google Scholar]
- 34.Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS, Jr, Moore ML. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol. 2011;85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan D, Weisshaar M, Lamb K, Chung HK, Lin MZ, Plemper RK. Replication-Competent Influenza Virus and Respiratory Syncytial Virus Luciferase Reporter Strains Engineered for Co-Infections Identify Antiviral Compounds in Combination Screens. Biochemistry. 2015;54:5589–5604. doi: 10.1021/acs.biochem.5b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 37.El Omari K, Dhaliwal B, Ren J, Abrescia NG, Lockyer M, Powell KL, Hawkins AR, Stammers DK. Structures of respiratory syncytial virus nucleocapsid protein from two crystal forms: details of potential packing interactions in the native helical form. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1179–1183. doi: 10.1107/S1744309111029228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khattar SK, Yunus AS, Samal SK. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J Gen Virol. 2001;82:775–779. doi: 10.1099/0022-1317-82-4-775. [DOI] [PubMed] [Google Scholar]
- 39.Garcia J, Garcia-Barreno B, Vivo A, Melero J. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22 K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira AP, Simabuco FM, Tamura RE, Guerrero MC, Ribeiro PG, Libermann TA, Zerbini LF, Ventura AM. Human respiratory syncytial virus N, P and M protein interactions in HEK-293T cells. Virus Res. 2013;177:108–112. doi: 10.1016/j.virusres.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Kingston RL, Hamel DJ, Gay LS, Dahlquist FW, Matthews BW. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc Natl Acad Sci U S A. 2004;101:8301–8306. doi: 10.1073/pnas.0402690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox R, Pickar A, Qiu S, Tsao J, Rodenburg C, Dokland T, Elson A, He B, Luo M. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci U S A. 2014;111:15208–15213. doi: 10.1073/pnas.1413268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yabukarski F, Lawrence P, Tarbouriech N, Bourhis JM, Delaforge E, Jensen MR, Ruigrok RW, Blackledge M, Volchkov V, Jamin M. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 44.Llorente MT, Garcia-Barreno B, Calero M, Camafeita E, Lopez JA, Longhi S, Ferron F, Varela PF, Melero JA. Structural analysis of the human respiratory syncytial virus phosphoprotein: characterization of an alpha-helical domain involved in oligomerization. J Gen Virol. 2006;87:159–169. doi: 10.1099/vir.0.81430-0. [DOI] [PubMed] [Google Scholar]
- 45.Karlin D, Ferron F, Canard B, Longhi S. Structural disorder and modular organization in Paramyxovirinae N and P. J Gen Virol. 2003;84:3239–3252. doi: 10.1099/vir.0.19451-0. [DOI] [PubMed] [Google Scholar]
- 46.Cox R, Green TJ, Purushotham S, Deivanayagam C, Bedwell GJ, Prevelige PE, Luo M. Structural and functional characterization of the mumps virus phosphoprotein. J Virol. 2013;87:7558–7568. doi: 10.1128/JVI.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruhn JF, Barnett KC, Bibby J, Thomas JM, Keegan RM, Rigden DJ, Bornholdt ZA, Saphire EO. Crystal structure of the nipah virus phosphoprotein tetramerization domain. J Virol. 2014;88:758–762. doi: 10.1128/JVI.02294-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouizougun-Oubari M, Pereira N, Tarus B, Galloux M, Lassoued S, Fix J, Tortorici MA, Hoos S, Baron B, England P, Desmaele D, Couvreur P, Bontems F, Rey FA, Eleouet JF, Sizun C, Slama-Schwok A, Duquerroy S. A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of Human Respiratory Syncytial Virus. J Virol. 2015;89:11129–11143. doi: 10.1128/JVI.01612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology. 2004;318:463–473. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 50.Galloux M, Tarus B, Blazevic I, Fix J, Duquerroy S, Eleouet JF. Characterization of a viral phosphoprotein binding site on the surface of the respiratory syncytial nucleoprotein. J Virol. 2012;86:8375–8387. doi: 10.1128/JVI.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kingston RL, Baase WA, Gay LS. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J Virol. 2004;78:8630–8640. doi: 10.1128/JVI.78.16.8630-8640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 53.Gupta AK, Mathur M, Banerjee AK. Unique capping activity of the recombinant RNA polymerase (L) of vesicular stomatitis virus: association of cellular capping enzyme with the L protein. Biochem Biophys Res Commun. 2002;293:264–268. doi: 10.1016/S0006-291X(02)00217-6. [DOI] [PubMed] [Google Scholar]
- 54.Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J Biol Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- 55.Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71(Pt 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 56.Svenda M, Berg M, Moreno-Lopez J, Linne T. Analysis of the large (L) protein gene of the porcine rubulavirus LPMV: identification of possible functional domains. Virus Res. 1997;48:57–70. doi: 10.1016/s0168-1702(96)01426-8. [DOI] [PubMed] [Google Scholar]
- 57.Dochow M, Krumm SA, Crowe JE, Jr, Moore ML, Plemper RK. Independent structural domains in the paramyxovirus polymerase protein. J Biol Chem. 2012 doi: 10.1074/jbc.M111.325258. [DOI] [PMC free article] [PubMed]
- 58.Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J Virol. 2002;76:7322–7328. doi: 10.1128/JVI.76.14.7322-7328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown DD, Rima BK, Allen IV, Baron MD, Banyard AC, Barrett T, Duprex WP. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase. J Virol. 2005;79:14330–14338. doi: 10.1128/JVI.79.22.14330-14338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cevik B, Holmes DE, Vrotsos E, Feller JA, Smallwood S, Moyer SA. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology. 2004;327:297–306. doi: 10.1016/j.virol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 61.Cevik B, Smallwood S, Moyer SA. The L-L oligomerization domain resides at the very N-terminus of the sendai virus L RNA polymerase protein. Virology. 2003;313:525–536. doi: 10.1016/s0042-6822(03)00342-8. [DOI] [PubMed] [Google Scholar]
- 62.Holmes DE, Moyer SA. The phosphoprotein (P) binding site resides in the N terminus of the L polymerase subunit of sendai virus. J Virol. 2002;76:3078–3083. doi: 10.1128/JVI.76.6.3078-3083.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horikami SM, Smallwood S, Bankamp B, Moyer SA. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology. 1994;205:540–545. doi: 10.1006/viro.1994.1676. [DOI] [PubMed] [Google Scholar]
- 64.Smallwood S, Moyer SA. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology. 2004;318:439–450. doi: 10.1016/j.virol.2003.09.045. [DOI] [PubMed] [Google Scholar]
- 65.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malur AG, Gupta NK, De Bishnu P, Banerjee AK. Analysis of the mutations in the active site of the RNA-dependent RNA polymerase of human parainfluenza virus type 3 (HPIV3) Gene Expr. 2002;10:93–100. [PMC free article] [PubMed] [Google Scholar]
- 67.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 68.Gopinath M, Shaila MS. RNA triphosphatase and guanylyl transferase activities are associated with the RNA polymerase protein L of rinderpest virus. J Gen Virol. 2009;90:1748–1756. doi: 10.1099/vir.0.010975-0. [DOI] [PubMed] [Google Scholar]
- 69.Ferron F, Longhi S, Henrissat B, Canard B. Viral RNA-polymerases -- a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci. 2002;27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 70.Paesen GC, Collet A, Sallamand C, Debart F, Vasseur JJ, Canard B, Decroly E, Grimes JM. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat Commun. 2015;6:8749. doi: 10.1038/ncomms9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Issur M, Picard-Jean F, Bisaillon M. The RNA capping machinery as an anti-infective target. Wiley Interdiscip Rev RNA. 2011;2:184–192. doi: 10.1002/wrna.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy AM, Grdzelishvili VZ. Identification of sendai virus L protein amino acid residues affecting viral mRNA cap methylation. J Virol. 2009;83:1669–1681. doi: 10.1128/JVI.01438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim SV, Rahman MB, Tejo BA. Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinformatics. 2011;12(Suppl 13):S24. doi: 10.1186/1471-2105-12-S13-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Clercq E, Neyts J. Antiviral agents acting as DNA or RNA chain terminators. Handb Exp Pharmacol. 2009:53–84. doi: 10.1007/978-3-540-79086-0_3. [DOI] [PubMed] [Google Scholar]
- 75.Soriano V, Vispo E, de Mendoza C, Labarga P, Fernandez-Montero JV, Poveda E, Trevino A, Barreiro P. Hepatitis C therapy with HCV NS5B polymerase inhibitors. Expert Opin Pharmacother. 2013;14:1161–1170. doi: 10.1517/14656566.2013.795543. [DOI] [PubMed] [Google Scholar]
- 76.Wright PJ, Crameri G, Eaton BT. RNA synthesis during infection by Hendra virus: an examination by quantitative real-time PCR of RNA accumulation, the effect of ribavirin and the attenuation of transcription. Arch Virol. 2005;150:521–532. doi: 10.1007/s00705-004-0417-5. [DOI] [PubMed] [Google Scholar]
- 77.Crotty S, Andino R. Implications of high RNA virus mutation rates: lethal mutagenesis and the antiviral drug ribavirin. Microbes Infect. 2002;4:1301–1307. doi: 10.1016/s1286-4579(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 78.Gilbert BE, Knight V. Biochemistry and clinical applications of ribavirin. Antimicrob Agents Chemother. 1986;30:201–205. doi: 10.1128/aac.30.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deval J, Hong J, Wang G, Taylor J, Smith LK, Fung A, Stevens SK, Liu H, Jin Z, Dyatkina N, Prhavc M, Stoycheva AD, Serebryany V, Liu J, Smith DB, Tam Y, Zhang Q, Moore ML, Fearns R, Chanda SM, Blatt LM, Symons JA, Beigelman L. Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2′-Fluoro-4′-Chloromethyl-Cytidine Triphosphate. PLoS Pathog. 2015;11:e1004995. doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laganas VA, Dunn EF, McLaughlin RE, Tiong-Yip CL, Yuzhakov O, Isabella VM, Hill P, Yu Q. Characterization of novel respiratory syncytial virus inhibitors identified by high throughput screen. Antiviral Res. 2014;115C:71–74. doi: 10.1016/j.antiviral.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 83.de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009) Antiviral Res. 2010;85:75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 84.Basavapathruni A, Anderson KS. Reverse transcription of the HIV-1 pandemic. FASEB J. 2007;21:3795–3808. doi: 10.1096/fj.07-8697rev. [DOI] [PubMed] [Google Scholar]
- 85.Usach I, Melis V, Peris JE. Non-nucleoside reverse transcriptase inhibitors: a review on pharmacokinetics, pharmacodynamics, safety and tolerability. Journal of the International AIDS Society. 2013;16:1–14. doi: 10.7448/IAS.16.1.18567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Das K, Clark AD, Jr, Lewi PJ, Heeres J, De Jonge MR, Koymans LM, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, De Bethune MP, Boyer PL, Clark P, Hughes SH, Janssen PA, Arnold E. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 87.Matharu DS, Flaherty DP, Simpson DS, Schroeder CE, Chung D, Yan D, Noah JW, Jonsson CB, White EL, Aube J, Plemper RK, Severson WE, Golden JE. Optimization of potent and selective quinazolinediones: inhibitors of respiratory syncytial virus that block RNA-dependent RNA-polymerase complex activity. J Med Chem. 2014;57:10314–10328. doi: 10.1021/jm500902x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, Massariol MJ, Do F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tiong-Yip CL, Aschenbrenner L, Johnson KD, McLaughlin RE, Fan J, Challa S, Xiong H, Yu Q. Characterization of a respiratory syncytial virus L protein inhibitor. Antimicrob Agents Chemother. 2014;58:3867–3873. doi: 10.1128/AAC.02540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoon JJ, Krumm SA, Ndungu JM, Hoffman V, Bankamp B, Rota PA, Sun A, Snyder JP, Plemper RK. Target analysis of the experimental measles therapeutic AS-136A. Antimicrob Agents Chemother. 2009;53:3860–3870. doi: 10.1128/AAC.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hwang H, Vreven T, Janin J, Weng Z. Protein-protein docking benchmark version 4.0. Proteins. 2010;78:3111–3114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Labbe CM, Laconde G, Kuenemann MA, Villoutreix BO, Sperandio O. iPPI-DB: a manually curated and interactive database of small non-peptide inhibitors of protein-protein interactions. Drug Discov Today. 2013;18:958–968. doi: 10.1016/j.drudis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 94.Basse MJ, Betzi S, Bourgeas R, Bouzidi S, Chetrit B, Hamon V, Morelli X, Roche P. 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic acids research. 2013;41:D824–827. doi: 10.1093/nar/gks1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Higueruelo AP, Schreyer A, Bickerton GR, Pitt WR, Groom CR, Blundell TL. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem Biol Drug Des. 2009;74:457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 96.Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev Mol Med. 2012;14:e16. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chapman J, Abbott E, Alber DG, Baxter RC, Bithell SK, Henderson EA, Carter MC, Chambers P, Chubb A, Cockerill GS, Collins PL, Dowdell VC, Keegan SJ, Kelsey RD, Lockyer MJ, Luongo C, Najarro P, Pickles RJ, Simmonds M, Taylor D, Tyms S, Wilson LJ, Powell KL. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob Agents Chemother. 2007;51:3346–3353. doi: 10.1128/AAC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Challa S, Scott AD, Yuzhakov O, Zhou Y, Tiong-Yip CL, Gao N, Thresher J, Yu Q. Mechanism of Action for Respiratory Syncytial Virus Inhibitor RSV604. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.04119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marty F. A double-blind, randomized, placebo-controlled study to evaluate the safety and efficacy of RSV604 in adults with respiratory syncytial virus infection following stem cell transplantation. IXth International Symposium on Respiratory Viral Infections.2007. [Google Scholar]
- 100.Brindley MA, Plattet P, Plemper RK. Efficient replication of a paramyxovirus independent of full zippering of the fusion protein six-helix bundle domain. Proc Natl Acad Sci U S A. 2014;111:E3795–3804. doi: 10.1073/pnas.1403609111. [DOI] [PMC free article] [PubMed] [Google Scholar]