ABSTRACT
The marine bacterium Vibrio alginolyticus has a single polar flagellum, the number of which is regulated positively by FlhF and negatively by FlhG. FlhF is intrinsically localized at the cell pole, whereas FlhG is localized there through putative interactions with the polar landmark protein HubP. Here we focused on the role of HubP in the regulation of flagellar number in V. alginolyticus. Deletion of hubP increased the flagellar number and completely disrupted the polar localization of FlhG. It was thought that the flagellar number is determined primarily by the absolute amount of FlhF localized at the cell pole. Here we found that deletion of hubP increased the flagellar number although it did not increase the polar amount of FlhF. We also found that FlhG overproduction did not reduce the polar localization of FlhF. These results show that the absolute amount of FlhF is not always the determinant of flagellar number. We speculate that cytoplasmic FlhG works as a quantitative regulator, controlling the amount of FlhF localized at the pole, and HubP-anchored polar FlhG works as a qualitative regulator, directly inhibiting the activity of polar FlhF. This regulation by FlhF, FlhG, and HubP might contribute to achieving optimal flagellar biogenesis at the cell pole in V. alginolyticus.
IMPORTANCE For regulation of the flagellar number in marine Vibrio, two proteins, FlhF and FlhG, work as positive and negative regulators, respectively. In this study, we found that the polar landmark protein HubP is involved in the regulation of flagellar biogenesis. Deletion of hubP increased the number of flagella without increasing the amount of pole-localizing FlhF, indicating that the number of flagella is not determined solely by the absolute amount of pole-localizing FlhF, which is inconsistent with the previous model. We propose that cytoplasmic FlhG and HubP-anchored polar FlhG negatively regulate flagellar formation through two independent schemes.
INTRODUCTION
The bacterial flagellum is a filamentous locomotory organ protruding from the cell surface of bacteria. The flagellum is composed of tens of thousands of protein molecules, and its biogenesis is regulated by various genes and proteins (1, 2). Although the mechanism of flagellar biogenesis is well conserved in all flagellated bacteria, the number and position of flagella vary among bacterial species (3, 4). For example, Escherichia coli and Bacillus subtilis have multiple peritrichous flagella, Campylobacter jejuni has bipolar flagella, and Vibrio cholerae and Pseudomonas aeruginosa have a single polar flagellum. Vibrio alginolyticus and Vibrio parahaemolyticus also have a single polar flagellum, whereas multiple peritrichous flagella are formed under high-viscosity conditions (5). Because defects in the regulation of the number and position of flagella inhibit bacterial cell motility, the formation of adequate flagella is an important factor in the survival strategies of bacteria in nature.
In the marine bacterium V. alginolyticus, which has a single polar flagellum, the number of flagella is tightly regulated by two cytoplasmic proteins, FlhF and FlhG (6, 7). FlhF is a GTPase protein and is similar to the signal recognition particle receptor FtsY (8, 9). The GTPase motif of FlhF influences the intracellular location of this protein (10, 11), and GTPase activity is stimulated through interactions with FlhG (12–14). FlhG is an ATPase protein and is similar to MinD, a cell division regulator in prokaryotes (15–17). It was reported that FlhG interacts with the flagellar basal body protein FliM-FliY-FliG complex, in an ATP- and lipid-dependent manner, in B. subtilis and Shewanella putrefaciens (16). In bacteria with polar flagella, FlhF overproduction increases the number of flagella and depletion of flhF causes a decrease in flagellation (6, 18). In contrast, overproduction of FlhG causes a decrease in flagellation, and depletion of flhG increases the number of flagella (6, 19, 20). Therefore, FlhF and FlhG work antagonistically, acting as positive and negative regulators, respectively, of flagellar biogenesis. FlhF intrinsically localizes at the cell pole, without other flagellar proteins (7), and may promote the assembly of the scaffold proteins necessary for flagellar formation (FliF and/or FlhA). Deletion of flhF occasionally causes peritrichous flagellation (7, 10, 21, 22), which may suggest that FlhF determines not only the number but also the position of flagella. We have shown that, in V. alginolyticus, FlhG also localizes at the cell pole but at lower levels, compared with FlhF. Because FlhF intensely localizes at the cell pole when FlhG is absent (7), it is thought that FlhG captures FlhF in the cytoplasm and inhibits the polar localization of FlhF. ATPase activity and/or ATP binding of FlhG is involved in the polar localization of FlhG (17). In Vibrio species, the DnaJ family protein SflA works as a putative negative regulator of flagellar biogenesis (23, 24). Deletion of sflA does not confer any phenotypic change affecting motility, but it causes flagellation at random positions in the absence of both FlhF and FlhG. It was suggested that SflA inhibits flagellation in an FlhF/FlhG-independent manner and regulates the number and position of flagella through a different pathway (23, 24).
In V. cholerae, HubP was identified as a polar landmark protein that anchors three ParA-like proteins (ParA1, ParC, and FlhG) to the cell pole (25). HubP is conserved in all Vibrio and Photobacterium species and several other gammaproteobacteria. It is a single-pass transmembrane protein whose N and C termini show similarity to FimV, a positive regulator of type IV pilus formation (26, 27). HubP has an N-terminal periplasmic peptidoglycan-binding motif (LysM) and a large cytoplasmic domain. The LysM domain is important for the polar localization of HubP, and the large cytoplasmic domain interacts with ParA-like proteins (25). It was suggested that HubP is involved in the regulation of chromosome partitioning and chemoreceptor positioning but is not strongly involved in the regulation of flagellar number and positioning in V. cholerae, because deletion of hubP had few effects on the flagellar number (25). In S. putrefaciens, HubP plays roles similar to those in V. cholerae (14).
In this study, we investigated the involvement of HubP in the formation of the flagellum in the marine bacterium V. alginolyticus. We showed that HubP affects the number of flagella, and we proposed a novel model for the regulation of flagellar formation in bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. V. alginolyticus was cultured at 30°C in VC medium (0.5% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.2% [wt/vol] glucose) or VPG medium (1% [wt/vol] polypeptone, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.5% [wt/vol] glycerol). If needed, chloramphenicol and kanamycin were added at final concentrations of 2.5 μg ml−1 and 100 μg ml−1, respectively. E. coli was cultured at 37°C in LB medium (1% [wt/vol] bactotryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl). If needed, chloramphenicol and ampicillin were added at final concentrations of 25 μg ml−1 and 100 μg ml−1, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− λ− recA1 hsdR17 endA1 supE44 thi-1 relA1 gyrA96 Δ(argF-lacZYA)U169 ϕ80dlacZΔM15 (recipient in cloning experiments) | 32 |
| S17-1 | recA hsdR thi pro ara RP-4 2-Tc::Mu-Km::Tn7 (Tpr Smr) (recipient in conjugational transfer of pMMB206) | 33 |
| β3914 | β2163 gyrA462 zei-298::Tn10 (Kmr Emr Tcr) (recipient in conjugational transfer of pSW7848) | 34 |
| BL21(DE3) | Host for overexpression | Stratagene |
| V. alginolyticus strains | ||
| VIO5 | Rifr Pof+ Laf− | 35 |
| LPN1 | VIO5 ΔflhF | 7 |
| KK148 | VIO5 flhG | 6 |
| LPN2 | VIO5 ΔflhFG | 7 |
| NMB303 | VIO5 ΔhubP | This study |
| NMB304 | LPN1 ΔhubP | This study |
| NMB305 | KK148 ΔhubP | This study |
| NMB308 | VIO5 hubP-egfp | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning vector, Ampr | Promega |
| pTSK117-1 | pGEM-T Easy-hubP* | This study |
| pTSK117 | pGEM-T Easy-hubP*-egfp | This study |
| pSW7848 | Suicide vector (oriVR6Kγ oriTRP4 araC-PBAD-ccdB), Cmr | 36 |
| pTSK118 | pSW7848-hubP*-egfp | This study |
| pSJ2 | pSW7848-ΔhubP | This study |
| pBAD33 | Cmr PBAD | 31 |
| pTY200 | pBAD33-egfp | 37 |
| pTSJ3 | pBAD33-hubP | This study |
| pTSJ4 | pBAD33-hubP-His6 | This study |
| pAK520 | pBAD33-flhG | 6 |
| pAK325 | pBAD33-flhF-egfp | 7 |
| pAK541 | pBAD33-flhG-egfp | 7 |
| pTY60 | Kmr PBAD | 38 |
| pTY60-flhF-gfp | pAK325, Cms, Kmr | Akiko Kusumoto |
| pMMB206 | Cmr Ptac Plac UV5 | 39 |
| pNT16 | pMMB206-flhG | This study |
| pTrcHisB | Ampr Ptrc | Invitrogen |
| pTrc-flhG | pTrcHisB-His6-tev-flhG | 17 |
Tpr, trimethoprim resistant; Smr, streptomycin resistant; Kmr, kanamycin resistant; Emr, erythromycin resistant; Tcr, tetracycline resistant; Rifr, rifampin resistant; Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Cms, chloramphenicol sensitive; Pof+, normal polar flagellar formation; Laf−, defective in lateral flagellar formation; Ptac, tac promoter; Plac, lac promoter; PBAD, arabinose promoter; Ptrc, trc promoter; hubP*-egfp, ∼1.7-kb DNA fragment containing the hubP 3′ coding region and its downstream sequence, with an in-frame insertion of the egfp sequence at the 3′ end of hubP.
Construction of HubP-GFP-expressing strain and hubP deletion mutants.
The HubP-green fluorescent protein (HubP-GFP)-expressing strain NMB308 was generated from the VIO5 (wild-type) strain by homologous recombination with the hubP*-egfp sequence (∼1.7 kbp), which was composed of 500 bp of the 3′-terminal sequence of hubP fused to the egfp coding sequence and followed by 500 bp of the noncoding downstream sequence of hubP, using the suicide vector pSW7848, as described previously (28) with slight modifications. A 1,000-bp DNA fragment around the 3′ end of hubP on the chromosome was PCR amplified using an upstream sense primer (5′-CGAGCTCGTATGGATTTTGAAACCATG-3′) and a downstream antisense primer containing a SacI site (5′-CGAGCTCCCAGGTCGTAAAGCATGATTG-3′), cloned into pGEM-T Easy Vector (Promega, Tokyo, Japan), and named pTSK117-1. The egfp gene in pTY200 and the whole sequence of pTSK117-1 were PCR amplified using the following primers: for egfp, 5′-GGTGGTGGTGGTTCTATGGTGAGCAAGGGC-3′ and 5′-GCTACCCTTTTCGATTTACTTGTACAGCTC-3′, and for pTSK117-1, 5′-GAGCTGTACAAGTAAATCGAAAAGGGTAGC-3′ and 5′-AGAACCACCACCACCTCGGCCATTAATGGC-3′. Amplified PCR fragments were ligated to generate pTSK117, which contained hubP*-egfp flanked by SacI sites at both ends. This hubP*-egfp was subcloned into the SacI site of the suicide vector pSW7848 to generate pTSK118, which was introduced into V. alginolyticus by conjugational transfer from E. coli β3914, as described previously (29). Spontaneous homologous recombination was performed on a VC medium-1.25% agar plate containing 300 μM 2,6-diaminopimelic acid; positive selection for loss of the plasmid, which was facilitated by ccdB-encoded toxin under the arabinose-inducible promoter in pTSK118, was performed on a VC medium-1.25% agar plate containing 0.2% (wt/vol) arabinose. The insertion of egfp at the 3′ end of hubP on the chromosome was confirmed by DNA sequencing. The resulting HubP-enhanced GFP (HubP-eGFP) fusion protein contained a 5-residue linker (Gly-Gly-Gly-Gly-Ser) between the C-terminal end of HubP and the N-terminal initiation Met of eGFP.
The hubP deletion mutants were generated from the VIO5 (wild-type), LPN1 (ΔflhF), and KK148 (flhG) strains by homologous recombination with the ΔhubP sequence (1,000 bp), which was composed of 500 bp upstream and 500 bp downstream of hubP, by using methods similar to those described above. The ΔhubP DNA fragment was PCR amplified using an upstream sense primer (5′-CGAATTCCGTGTGCCTGTATTCTATGG-3′) and a downstream antisense primer containing a SacI site (5′-GGAATTCCCAGGTCGTAAAGCATGATTG-3′). pSW7848 and the ΔhubP fragment were ligated using the SacI site to generate pSJ2. By using pSJ2, ΔhubP strains were obtained. The deletion was confirmed by colony PCR, using a 500-bp upstream sense primer and a 500-bp downstream antisense primer.
Construction of HubP, HubP-His, and FlhG expression plasmids.
The chromosomal hubP gene of V. alginolyticus was PCR amplified using an upstream sense primer (5′-GGGGTACCAAATTGTATTTAGCACAAAAC-3′) and a downstream antisense primer containing a KpnI site (5′-GGGGTACCTTATCGGCCATTAATGGCATC-3′). PCR-amplified DNA fragments and pBAD33, a plasmid vector, were digested with KpnI, and the purified fragments, prepared as described above, were ligated in order to generate pTSJ3. For construction of pTSJ4, we introduced a His6 sequence into the 3′ end of the hubP gene on pTSJ3 through site-directed mutagenesis, using the QuikChange method as described by Stratagene. The flhG gene on pNT16 was subcloned from pAK520. pAK520 and pMMB206, which are plasmid vectors, were digested with BamHI and HindIII, and the purified fragments, prepared as described above, were ligated to generate pNT16. All plasmids were checked by DNA sequencing. Transformation of V. alginolyticus with plasmids for which the vectors were pBAD33 or pTY60 was carried out using the electroporation method, as described previously (30). Transformation of V. alginolyticus with the pNT16 plasmid was carried out using the conjugation method, as described previously (31).
Electron microscopy.
Freshly formed single colonies of V. alginolyticus on VC medium-1.25% agar plates were picked and negatively stained with 2% (wt/vol) potassium phosphotungstate (pH 7). Subsequently, they were placed on a carbon-coated copper grid and observed using a transmission electron microscope (JEM1011; JEOL, Tokyo, Japan).
Dark-field microscopy.
Overnight cultures of V. alginolyticus in VC medium were diluted 1:100 in VPG medium containing 0.02% (wt/vol) arabinose and were cultured for 4 h at 30°C. The cells with flagella were observed using a high-intensity dark-field microscope (BX50; Olympus, Tokyo, Japan) equipped with a 100-W high-pressure mercury lamp (BH2-RFL-T3; Olympus). The videos were recorded using a charge-coupled device (CCD) camera (SSC-M420; Sony, Tokyo, Japan) and imaging software (IC Capture 2.3; The Imaging Source Europe GmbH, Bremen, Germany). The number of flagella per cell was determined, and the phenotypes were categorized into three types, i.e., no flagellum, single flagellum, or multiple flagella.
Motility assay in soft agar plates.
Two microliters of an overnight culture were spotted on VPG soft agar plates (VPG medium containing 0.25% [wt/vol] Bacto agar [Difco]) with 0.02 or 0.2% (wt/vol) arabinose and were incubated at 30°C for the appropriate time, as indicated in the figure legends.
Fluorescence microscopy.
Overnight cultures of V. alginolyticus cells in VC medium were diluted 1:100 in VPG medium (for NMB308) or VPG medium containing 0.02% (wt/vol) arabinose (for all other plasmid-borne GFP fusions) and were cultured for 4 h at 30°C. The cultures were harvested by centrifugation, suspended in TMN500 (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose, 500 mM NaCl), loaded onto poly-l-lysine-coated tunnel slides, washed with TMN500, and observed using a fluorescence microscope (BX50; Olympus) equipped with a 100-W high-pressure mercury lamp (BH2-RFL-T3; Olympus). Images exposed for 1 s were recorded using a CCD camera (ORCA-Flash4.0; Hamamatsu Photonics) and imaging software (High-speed Recording Software, version 1.7.1.0; Hamamatsu Photonics). The fluorescence intensities at the pole and in the cytoplasm were measured with analytical software (ImageJ 1.48u4; National Institutes of Health). The pole/cytoplasm (P/C) ratio was calculated as described previously (7).
For the staining of polar flagella, the cells in the tunnel slide were treated as described previously (31). In brief, cells were fixed on the poly-l-lysine-coated tunnel slide as described above, and then TMN500 containing antibody against the polar flagellum was applied to the tunnel slide. After 5 min of incubation, the tunnel slide was washed with TMN500, and then TMN500 containing rhodamine-conjugated anti-rabbit IgG antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was applied. After 5 min of incubation, the tunnel slide was washed with TMN500 and then observed under a microscope, as described above. Because the polar flagellum is covered with a lipid sheath connected to the outer membrane, the cell body is also stained.
Detection of proteins by immunoblotting.
Overnight cultures grown in VC medium were inoculated, at a 100-fold dilution, into VPG medium containing 0.02% or 0.2% (wt/vol) arabinose and were cultured at 30°C for 4 h. Cells were harvested by centrifugation, suspended to an optical density at 660 nm of 10 in sodium dodecyl sulfate (SDS) loading buffer containing 5% (vol/vol) β-mercaptoethanol, and boiled at 95°C for 5 min. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes, and immunoblotting was performed using anti-FlhF (FlhF B0239) (7), anti-FlhG (FlhG B0728) (17), and anti-His antibodies (Medical & Biological Laboratories Co., Ltd., Nagoya, Japan).
Immunoprecipitation.
FlhG protein was purified from E. coli cells expressing the protein from plasmid pTrc-flhG as described previously (17), with slight modifications. E. coli BL21(DE3) cells harboring pTrc-flhG were inoculated into LB medium containing 1.75% (wt/vol) d-lactose monohydrate and were cultured at 30°C for 16 h. Cells were harvested by centrifugation, suspended in TN buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl) containing 5 to 10 mg of lysozyme and 0.5 mM protease inhibitor phenylmethylsulfonyl fluoride (PMSF), and disrupted by sonication. The supernatant from ultracentrifugation at 160,000 × g for 30 min was loaded on a HiTrap Talon column connected to an AKTAprime system (GE Healthcare, Chicago, IL, USA). After washing with TN buffer, His-tagged FlhG was eluted using an imidazole gradient (from 5 mM to 300 mM), and the purity was checked by SDS-PAGE Coomassie brilliant blue staining.
HubP-His protein was purified from V. alginolyticus cells. NMB303 cells harboring plasmid pTSJ4 were inoculated into VPG medium containing 0.2% (wt/vol) arabinose and were cultured for 4 h at 30°C. Cells were harvested by centrifugation, suspended in TMN500 buffer, and disrupted by sonication. The precipitate from ultracentrifugation at 100,000 × g for 30 min was suspended in TMN500 buffer containing 1% (wt/vol) n-dodecyl-β-d-maltoside (DDM) and was incubated at 4°C for 1 h. The supernatant from ultracentrifugation at 100,000 × g for 30 min was mixed with Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen, Hilden, Germany). After washing with TMN500 buffer containing 0.1% DDM and 5 mM imidazole, HubP-His was eluted with buffer containing 500 mM imidazole. Purified His-FlhG (wild type), His-FlhG(D171A), HubP-His, and/or ATP (final concentration of 0.5 mM, with 5 mM MgCl2) was added to protein A–Sepharose CL-4B beads (GE Healthcare) that had been equilibrated with TMN500 containing 0.1% (wt/vol) DDM and anti-FlhG antibody, and the mixture was incubated for 60 min at room temperature. The beads were washed with TMN500 containing 0.1% (wt/vol) DDM, and bound proteins were eluted from the beads by boiling at 95°C in SDS loading buffer for 10 min. Samples were separated using SDS-PAGE and transferred to PVDF membranes, and immunoblotting was performed using an anti-His antibody.
RESULTS
Increased flagellar number in V. alginolyticus with deletion of hubP.
In V. alginolyticus, HubP is composed of 1,444 amino acids, its molecular size is estimated to be 159 kDa, and it contains an N-terminal signal peptide (26 amino acids) (see Fig. S1 in the supplemental material). As in its orthologs, a repeat sequence was found seven times in the cytoplasmic region (10 times in V. cholerae and nine times in S. putrefaciens) (see Fig. S1). At first, we observed the subcellular localization of HubP. We fused the egfp gene to the 3′ end of hubP on the chromosome of the V. alginolyticus VIO5 strain, which has a wild-type polar flagellum, and observed the cells by fluorescence microscopy. HubP-GFP mainly localized at the flagellated cell pole; however, some of the cells had a fluorescence dot at the pole without flagella, and the fluorescence intensities seemed to be weaker than those of flagellated poles (Fig. 1A). This result is consistent with the previous results for V. cholerae and S. putrefaciens (14, 25). We confirmed that GFP fusion to the C terminus of HubP did not affect flagellation and motility (Fig. 1). In order to test the contribution of HubP to the regulation of the flagellar number in V. alginolyticus, hubP was deleted in the V. alginolyticus VIO5 strain. The ΔhubP strain was observed using transmission electron microscopy (TEM), and it was shown that multiple flagella existed at the cell pole (Fig. 2A). These cells were also observed using a high-intensity dark-field microscope. The number of flagella per cell was determined, and the phenotypes were categorized into three classes (i.e., no flagellum, single flagellum, or multiple flagella) (Fig. 2B). More than 80% of the wild-type cells had a single flagellum, and more than 80% of the flhG mutant cells had multiple flagella. In ΔhubP mutants, about 50% of the cells formed multiple flagella, and about 20% of the cells formed a single flagellum. When the HubP expression plasmid was introduced into the ΔhubP strain, the cells formed a single flagellum at the cell pole, similar to VIO5 (wild-type) cells. More than 80% of flhG ΔhubP mutant cells had multiple flagella, similar to flhG mutant cells. We also observed the flagellar number by electron microscopy and confirmed a tendency similar to the result shown in Fig. 2B, that is, the flhG mutant had more flagella per cell than the ΔhubP mutant and the hubP deletion from the flhG mutant did not increase the flagellar number (see Fig. S2 in the supplemental material).
FIG 1.
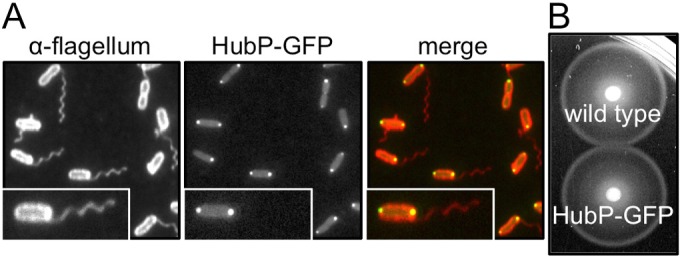
Polar localization of HubP in V. alginolyticus. (A) Fluorescence images of cells expressing HubP-GFP (NMB308). Left, cells treated with anti-polar flagellum antibody followed by anti-rabbit IgG conjugated to rhodamine; middle, GFP fluorescence image; right, merged GFP and rhodamine images. Insets are enlarged images of representative cells. (B) Motility of VIO5 (wild-type) and NMB308 cells in soft agar plates. The incubation time was 3.5 h.
FIG 2.
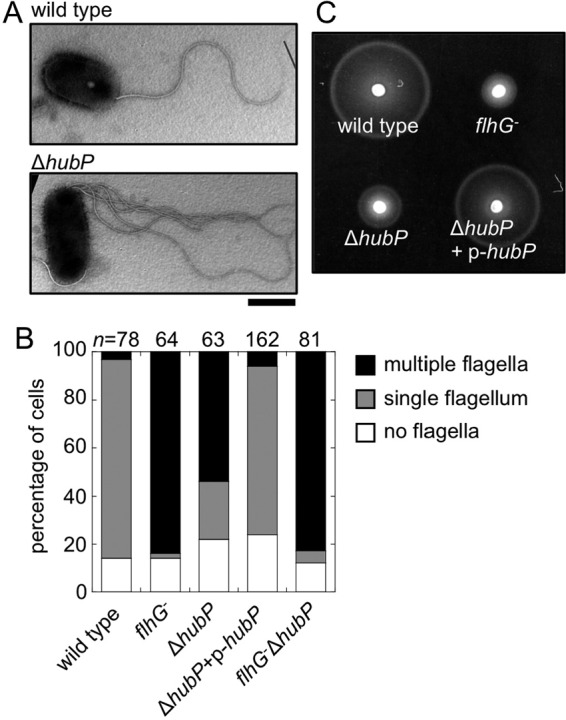
Increased flagellar number in V. alginolyticus with deletion of hubP. (A) Electron micrographs of a wild-type cell and a ΔhubP cell. Representative examples are shown. Scale bar, 1 μm. (B) Flagellar number per cell observed for wild-type cells, flhG mutant cells, ΔhubP mutant cells, ΔhubP mutant cells complemented with the plasmid-expressed hubP gene, and flhG ΔhubP mutant cells. The cells were observed using dark-field microscopy, and the flagellar number per cell was determined. (C) Soft agar plate cell motility. Overnight cultures (2 μl) were spotted onto VPG medium–0.25% agar plates containing 0.02% (wt/vol) arabinose and incubated at 30°C for 4 h.
The motility of mutant strains was monitored in soft agar plates (Fig. 2C). It was demonstrated previously that flhG-depleted mutant cells showed reduced motility in soft agar plates (6). Here, the deletion of hubP led to decreased motility in soft agar plates, similar to the flhG mutant, and the hubP-expressing plasmid complemented this motility defect (Fig. 2C). The deletion of hubP did not strongly affect chromosomal expression of FlhF and FlhG (see Fig. S3A in the supplemental material).
Subcellular localization of FlhF and FlhG in ΔhubP mutant cells.
Because deletion of hubP led to an increase in the flagellar number, it was speculated that the localization of FlhF or FlhG would be affected by hubP deletion in V. alginolyticus. We fused GFP to FlhF and FlhG and observed the intracellular localization of these proteins in cells with or without hubP. We confirmed that the expression of FlhG-GFP and FlhF-GFP was not affected by the deletion of hubP (see Fig. S3B and C in the supplemental material). In cells with hubP expression, FlhG-GFP localized in a dot pattern at the cell poles in about 60% of the cells (Fig. 3), as described previously (7, 17). In cells without hubP expression, however, polar dots of FlhG-GFP were almost not visible, indicating that HubP is essential for polar localization of FlhG (Fig. 3), which is consistent with the results obtained previously in V. cholerae and S. putrefaciens (14, 25). FlhG(D171A), an ATPase-active FlhG mutant that localizes strongly at the cell pole (17), also lost its polar localization in the absence of HubP (Fig. 3). FlhF-GFP is known to localize at the cell poles in wild-type cells, and here we found that the FlhF-GFP position was not affected by the deletion of hubP (Fig. 4B), which is also consistent with the results obtained in V. cholerae and S. putrefaciens (14, 25). We analyzed the intensity of FlhF-GFP polar dots in the investigated cells (Fig. 4). Fluorescent signal intensities were measured at the cell poles and in the cytoplasm (Fig. 4A), and the P/C ratios were calculated and plotted in a histogram (Fig. 4B). In wild-type cells, the polar dot intensity (P/C ratio) for FlhF-GFP was 3.6 ± 1.6 (mean ± SD [n = 326]). In the ΔflhG mutant, the P/C ratio for FlhF-GFP was 18.2 ± 19.4 (mean ± SD [n = 264]), which was higher than that for the wild-type cells, consistent with previously reported results (7). In the ΔhubP mutant, the P/C ratio for FlhF-GFP was 3.4 ± 1.7 (mean ± SD [n = 232]), which was similar to that for the wild-type cells. These results indicate that the deletion of hubP did not affect the efficiency of the polar localization of FlhF, unlike in the flhG-depleted mutant cells.
FIG 3.
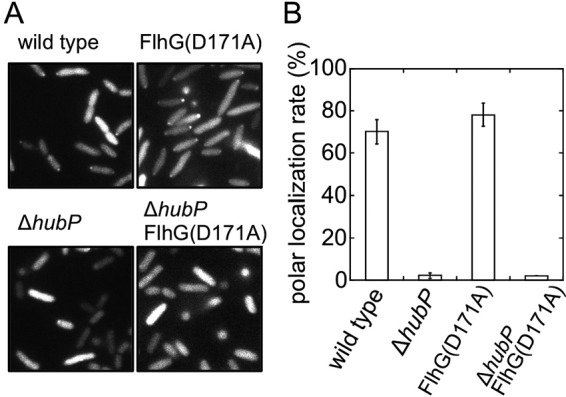
Effects of hubP deletion on polar localization of FlhG. The intracellular localization of FlhG-GFP was observed using a fluorescence microscope (A), and the rate of polar dot formation was determined by calculating the ratio of the number of cells with a fluorescent dot at the cell pole to the number of cells seen in the image (B). FlhG-GFP with or without a D171A mutation was expressed from a plasmid in a flhG-depleted mutant (wild type) and a flhG ΔhubP mutant (ΔhubP).
FIG 4.
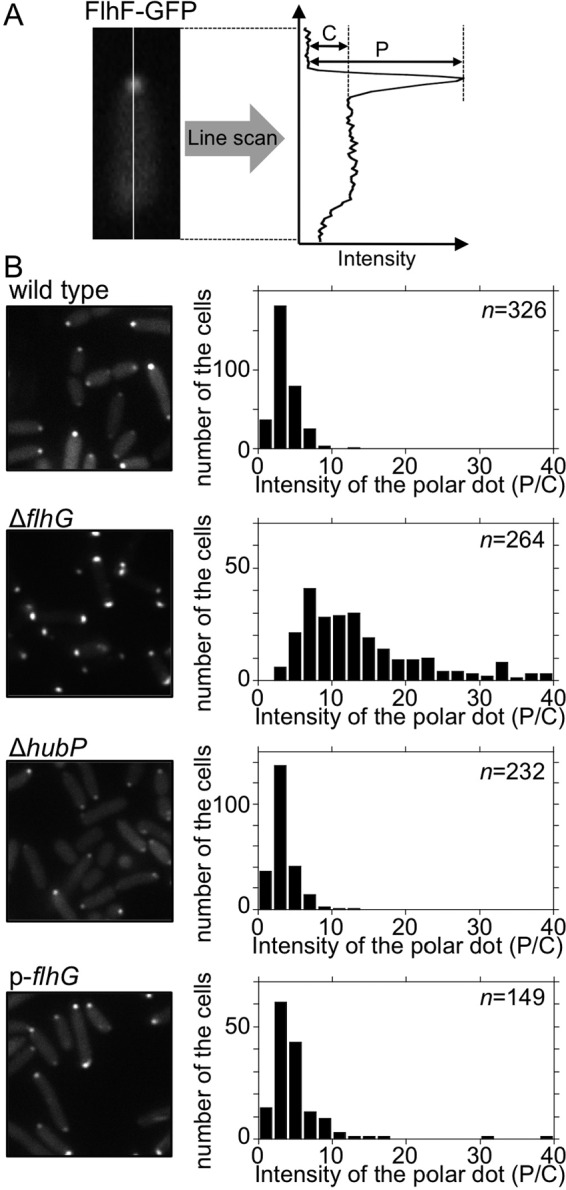
Effects of hubP deletion or FlhG overproduction on the polar localization of FlhF. (A) Schematic of the quantitative analysis of the polar localization of FlhF-GFP. The fluorescence intensities at the pole (P) and in the cytoplasm (C) were determined by subtracting the background intensity. (B) Intracellular localization of FlhF-GFP and intensity of fluorescent dots. FlhF-GFP was expressed in ΔflhF cells (wild type), ΔflhF ΔflhG cells (ΔflhG), ΔflhF ΔhubP cells (ΔhubP), and ΔflhF ΔflhG cells overproducing FlhG from the plasmid pAK520 (p-flhG). Left, fluorescence images; right, P/C ratio histograms.
Effects of FlhG overproduction on localization of FlhF.
In P. aeruginosa, overproduction of FleN, a FlhG homologue, produces a nonflagellated phenotype, with a few cells having a single flagellum (19). Similar results were reported for V. alginolyticus (17). In addition, the introduction of a plasmid (pNT16) expressing FlhG from the uncontrolled leaky promoter into the flhG-depleted strain completely inhibits flagellation and the motility of cells in soft agar plates (see Fig. S4 in the supplemental material). We observed the localization of FlhF-GFP under these FlhG-overproducing conditions. We introduced two different plasmids, one expressing FlhF-GFP from the arabinose-inducible promoter (pAK325) and pNT16. Unexpectedly, FlhF-GFP localized at the cell pole at similar levels, compared with the wild-type cells (Fig. 4), indicating that FlhF-GFP can localize at the cell pole independent of FlhG overproduction.
DISCUSSION
Flagella are the largest organelles in bacteria, and their numbers are tightly controlled in some bacterial species. The marine bacterium V. alginolyticus has a single flagellum at the cell pole, and two proteins, FlhF and FlhG, regulate the flagellar number. FlhF is localized at the cell pole and is thought to promote flagellation. FlhG was thought to inhibit the polar localization of FlhF by capturing the protein in the cytoplasm. In this study, we analyzed the contribution of HubP, a large, pole-localizing, single-pass, transmembrane protein involved in the polar localization of FlhG, to the regulation of flagellar number in V. alginolyticus. In the absence of HubP, cells increase the number of their flagella, indicating that HubP is involved in the inhibition of flagellar formation. These results are inconsistent with the results obtained in V. cholerae, which showed that deletion of hubP did not strongly affect flagellar number (25). This might suggest that V. cholerae has more robust mechanisms to control flagellar formation, independent of HubP and distinct from those in V. alginolyticus. Plasmid-based complementation of hubP restored the flagellar number of the ΔhubP mutant to the wild-type level. Because the level of expression of HubP from a plasmid under the control of the araBAD promoter, induced with 0.02% (wt/vol) arabinose, should be higher than chromosomal expression, the overproduction of HubP did not produce a nonflagellated phenotype, in contrast to the overproduction of FlhG. There may be a threshold level of HubP that can localize at the cell pole, because of the largeness of HubP as a membrane protein. The deletion of hubP decreased the motility of cells in soft agar plates, which may be attributed to the inhibition of motility through an undue increase in flagellar number, similar to that in the flhG-depleted mutant, and to a defect in the chemotactic response that may be caused by mislocalization of the chemoreceptor cluster as a consequence of hubP deletion, as previously described for other species (14, 25).
The deletion of hubP disrupted the localization of FlhG at the cell pole almost completely and FlhG was diffused in the cytoplasm, which agrees with the results obtained previously in other species (14, 25). This indicates that HubP, which is a pole-localizing protein, anchors FlhG to the cell pole. A direct interaction between HubP and FlhG in V. cholerae was suggested by a bacterial two-hybrid assay (25). We performed a pulldown assay to detect the direct interaction; however, HubP was not copurified with FlhG (see Fig. S5 in the supplemental material). We also tried to detect the interaction with a bacterial two-hybrid assay using a similar fragment of V. cholerae; however, we could not achieve any positive results. Those findings may suggest that the interaction between HubP and FlhG is not as strong as that in V. cholerae or that other proteins are required to support the interaction in V. alginolyticus.
In contrast, deletion of hubP did not affect the localization of FlhF. In other words, both hubP deletion and flhG depletion increased flagellar number, whereas the polar localization of FlhF was increased only with flhG depletion. In the past, it was thought that the absolute amount of FlhF molecules at the cell pole determined the number of flagella and cytoplasmic FlhG molecules influenced the downregulation of the polar assembly of FlhF (7). However, here we found that deletion of hubP increased the number of flagella without increasing the polar assembly of FlhF. This suggests that the flagellar number is determined not only by the amount of polar FlhF but also by another factor combined with HubP. Furthermore, we analyzed the localization of FlhF in FlhG-overproducing cells and found that the polar localization of FlhF was not strongly affected by the excess FlhG, although the cells were not able to form flagella. This may indicate that FlhF strongly localizes at the cell pole, and overproduced FlhG cannot completely take FlhF away from the cell pole. Moreover, because the deletion of hubP in the flhG-depleted mutant cells did not cause an additional increase in flagellar number, compared with the flhG-depleted mutant (Fig. 2B), HubP should inhibit flagellation through a FlhG-mediated mechanism, probably by giving FlhG a binding platform at the cell pole. This is consistent with our previous findings showing that the ATP-dependent polar localization of FlhG is crucial for its ability to downregulate the number of polar flagella (17).
Based on these results, we suggest a new model for the regulation of flagellar number in V. alginolyticus by FlhF and FlhG (Fig. 5). In wild-type cells, FlhG may inhibit the flagellation in two ways, i.e., (i) through cytoplasmic FlhG, which captures FlhF in the cytoplasm and prevents the polar localization of FlhF, and (ii) through inhibition by ATP-dependent, pole-localizing FlhG anchored by HubP, which directly prevents flagellation without causing mislocalization of FlhF. In the flhG-depleted mutant cells, both inhibition pathways regulated by FlhG are lost and multiflagellar formation is strongly induced by a large amount of pole-localizing FlhF. In the hubP deletion mutant, FlhF is captured by cytoplasmic FlhG and the amount of FlhF localized at the pole is controlled. However, FlhG cannot localize at the pole through interactions with HubP, and the inhibition of flagellation by polar FlhG is impaired. It is still unclear how polar FlhG downregulates FlhF at the pole. Since the ATPase-active mutant of FlhG, which localizes strongly at the pole, severely inhibits flagellation (17), it is possible that the activities of FlhG (ATP hydrolysis and FlhF inhibition) are correlated and are activated at the pole. HubP may be involved in the regulation of the amount of activated FlhG at the pole. We conclude that not only the ratio of protein expression levels but also the ratio of pole-localizing FlhF and FlhG levels regulate the flagellar number in V. alginolyticus. HubP may have a role in the formation of a specific “field” necessary for polar assembly of the appropriate amount of FlhG, in order for it to be able to regulate flagellar biogenesis.
FIG 5.
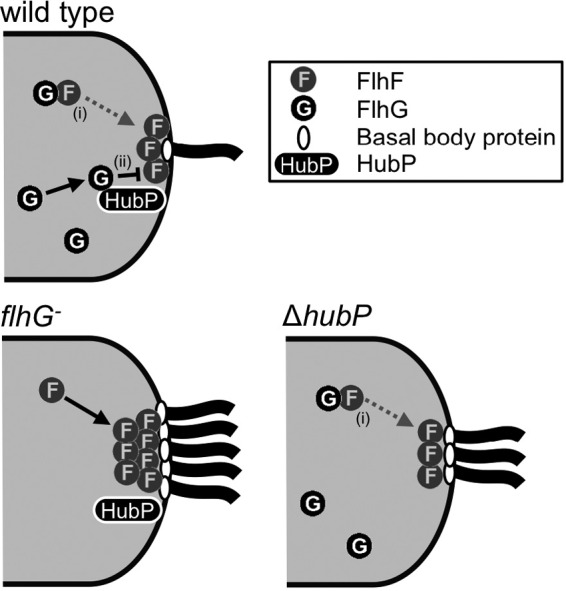
Model of the regulation of flagellar number by FlhF, FlhG, and HubP in V. alginolyticus cells. In wild-type cells, FlhF intrinsically localizes at the cell pole and promotes the assembly of the flagellar basal body protein (FliF). FlhG inhibits flagellation in two ways, i.e., (i) cytoplasmic FlhG captures FlhF in the cytoplasm and prevents the polar localization of FlhF and (ii) polar FlhG, which is anchored by HubP, directly inhibits the activity of the polar FlhF. Polar localization of FlhG depends on ATP as well. In the flhG-depleted mutant, both FlhG inhibition pathways were impaired and strong activation of multiflagellar formation by polar FlhF was induced. In the hubP deletion mutant, the amount of polar FlhF was controlled by cytoplasmic FlhG but direct inhibition by polar FlhG was impaired and multiflagellar formation was induced, even though the flagellar number was reduced, compared with the flhG mutant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Toshiaki Gotoh for assistance with electron microscopy, Shiwei Zhu and Akiko Abe for technical support in making deletion mutants, and Hikaru Hirata for purification of the FlhG protein.
This work was supported in part by JSPS KAKENHI grants (grants JP24117004 and JP23247024 to M. Homma and grants JP26115705 and JP16H04774 to S. Kojima) and by a JSPS fellowship (grant JP13J02161 to N. Takekawa). N. Takekawa was supported in part by the Integrative Graduate Education and Research Program of Nagoya University.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00462-16.
REFERENCES
- 1.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 2.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazmierczak BI, Hendrixson DR. 2013. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol Microbiol 88:655–663. doi: 10.1111/mmi.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuhmacher JS, Thormann KM, Bange G. 2015. How bacteria maintain location and number of flagella? FEMS Microbiol Rev 39:812–822. doi: 10.1093/femsre/fuv034. [DOI] [PubMed] [Google Scholar]
- 5.McCarter LL. 2004. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 6.Kusumoto A, Kamisaka K, Yakushi T, Terashima H, Shinohara A, Homma M. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem 139:113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- 7.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390–1399. doi: 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter PB, Hanlon DW, Ordal GW. 1992. flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol Microbiol 6:2705–2713. doi: 10.1111/j.1365-2958.1992.tb01447.x. [DOI] [PubMed] [Google Scholar]
- 9.Bange G, Petzold G, Wild K, Parlitz RO, Sinning I. 2007. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc Natl Acad Sci U S A 104:13621–13625. doi: 10.1073/pnas.0702570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green JC, Kahramanoglou C, Rahman A, Pender AM, Charbonnel N, Fraser GM. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J Mol Biol 391:679–690. doi: 10.1016/j.jmb.2009.05.075. [DOI] [PubMed] [Google Scholar]
- 11.Kusumoto A, Nishioka N, Kojima S, Homma M. 2009. Mutational analysis of the GTP-binding motif of FlhF which regulates the number and placement of the polar flagellum in Vibrio alginolyticus. J Biochem 146:643–650. doi: 10.1093/jb/mvp109. [DOI] [PubMed] [Google Scholar]
- 12.Bange G, Kummerer N, Grudnik P, Lindner R, Petzold G, Kressler D, Hurt E, Wild K, Sinning I. 2011. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat Struct Mol Biol 18:1376–1380. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 13.Gulbronson CJ, Ribardo DA, Balaban M, Knauer C, Bange G, Hendrixson DR. 2016. FlhG employs diverse intrinsic domains and influences FlhF GTPase activity to numerically regulate polar flagellar biogenesis in Campylobacter jejuni. Mol Microbiol 99:291–306. doi: 10.1111/mmi.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossmann F, Brenzinger S, Knauer C, Dorrich AK, Bubendorfer S, Ruppert U, Bange G, Thormann KM. 2015. The role of FlhF and HubP as polar landmark proteins in Shewanella putrefaciens CN-32. Mol Microbiol 98:727–742. doi: 10.1111/mmi.13152. [DOI] [PubMed] [Google Scholar]
- 15.Lutkenhaus J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 16.Schuhmacher JS, Rossmann F, Dempwolff F, Knauer C, Altegoer F, Steinchen W, Dorrich AK, Klingl A, Stephan M, Linne U, Thormann KM, Bange G. 2015. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci U S A 112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono H, Takashima A, Hirata H, Homma M, Kojima S. 2015. The MinD homolog FlhG regulates the synthesis of the single polar flagellum of Vibrio alginolyticus. Mol Microbiol 98:130–141. doi: 10.1111/mmi.13109. [DOI] [PubMed] [Google Scholar]
- 18.Pandza S, Baetens M, Park CH, Au T, Keyhan M, Matin A. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol Microbiol 36:414–423. doi: 10.1046/j.1365-2958.2000.01859.x. [DOI] [PubMed] [Google Scholar]
- 19.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 182:357–364. doi: 10.1128/JB.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray TS, Kazmierczak BI. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol 188:6995–7004. doi: 10.1128/JB.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao T, Shi M, Ju L, Gao H. 2015. Investigation into FlhFG reveals distinct features of FlhF in regulating flagellum polarity in Shewanella oneidensis. Mol Microbiol 98:571–585. doi: 10.1111/mmi.13141. [DOI] [PubMed] [Google Scholar]
- 23.Kojima M, Nishioka N, Kusumoto A, Yagasaki J, Fukuda T, Homma M. 2011. Conversion of mono-polar to peritrichous flagellation in Vibrio alginolyticus. Microbiol Immunol 55:76–83. doi: 10.1111/j.1348-0421.2010.00290.x. [DOI] [PubMed] [Google Scholar]
- 24.Kitaoka M, Nishigaki T, Ihara K, Nishioka N, Kojima S, Homma M. 2013. A novel dnaJ family gene, sflA, encodes an inhibitor of flagellation in marine Vibrio species. J Bacteriol 195:816–822. doi: 10.1128/JB.01850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. 2012. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semmler AB, Whitchurch CB, Leech AJ, Mattick JS. 2000. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146:1321–1332. doi: 10.1099/00221287-146-6-1321. [DOI] [PubMed] [Google Scholar]
- 27.Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL. 2011. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol 193:540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu S, Kumar A, Kojima S, Homma M. 2015. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol 98:101–110. doi: 10.1111/mmi.13103. [DOI] [PubMed] [Google Scholar]
- 29.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol Microbiol 62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 30.Kawagishi I, Okunishi I, Homma M, Imae Y. 1994. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology 140:2355–2361. doi: 10.1099/13500872-140-9-2355. [DOI] [Google Scholar]
- 31.Yorimitsu T, Mimaki A, Yakushi T, Homma M. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J Mol Biol 334:567–583. doi: 10.1016/j.jmb.2003.09.052. [DOI] [PubMed] [Google Scholar]
- 32.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. 1983. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [PubMed] [Google Scholar]
- 34.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okunishi I, Kawagishi I, Homma M. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol 178:2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Val ME, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. 2012. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet 8:e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima S, Nonoyama N, Takekawa N, Fukuoka H, Homma M. 2011. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na+-driven flagella of Vibrio alginolyticus. J Mol Biol 414:62–74. doi: 10.1016/j.jmb.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


