Abstract
Benzothiazinones (BTZs) are a class of compounds found to be extremely potent against both drug-susceptible and drug-resistant Mycobacterium tuberculosis strains. The potency of BTZs is explained by their specificity for their target decaprenylphosphoryl-d-ribose oxidase (DprE1), in particular by covalent binding of the activated form of the compound to the critical cysteine 387 residue of the enzyme. To probe the role of C387, we used promiscuous site-directed mutagenesis to introduce other codons at this position into dprE1 of M. tuberculosis. The resultant viable BTZ-resistant mutants were characterized in vitro, ex vivo, and biochemically to gain insight into the effects of these mutations on DprE1 function and on M. tuberculosis. Five different mutations (C387G, C387A, C387S, C387N, and C387T) conferred various levels of resistance to BTZ and exhibited different phenotypes. The C387G and C387N mutations resulted in a lower growth rate of the mycobacterium on solid medium, which could be attributed to the significant decrease in the catalytic efficiency of the DprE1 enzyme. All five mutations rendered the mycobacterium less cytotoxic to macrophages. Finally, differences in the potencies of covalent and noncovalent DprE1 inhibitors in the presence of C387 mutations were revealed by enzymatic assays. As expected from the mechanism of action, the covalent inhibitor PBTZ169 only partially inhibited the mutant DprE1 enzymes compared to the near-complete inhibition with a noncovalent DprE1 inhibitor, Ty38c. This study emphasizes the importance of the C387 residue for DprE1 activity and for the killing action of covalent inhibitors such as BTZs and other recently identified nitroaromatic inhibitors.
INTRODUCTION
Mycobacterium tuberculosis is the etiological agent of tuberculosis (TB), an infectious disease which is a leading cause of death worldwide and poses a major threat to global health. The World Health Organization estimates that in 2014, 9.6 million people contracted TB, and 1.5 million people died (1). In addition, the emergence and worldwide spread of multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB) are alarming. With MDR-TB strains being resistant to the frontline drugs isoniazid and rifampin and XDR-TB strains being resistant to frontline and additionally second-line drugs, there is an urgent need for new drugs for TB.
1,3-Benzothiazin-4-ones (BTZs) were discovered in 2009, with the lead compound BTZ043 having high potency (MIC of 1 ng/μl) against M. tuberculosis strain H37Rv (2) and demonstrating efficacy against MDR and XDR clinical isolates (3). Piperazine-containing BTZ (PBTZ) derivatives were then designed with improved pharmacological properties (4), and the optimized lead compound PBTZ169 is currently in clinical trials (5).
Genetic analysis of resistant mutants and enzymology have identified the target of BTZs as decaprenylphosphoryl-β-d-ribose oxidase (DprE1), an essential flavoenzyme in M. tuberculosis involved in cell wall synthesis (2). DprE1 acts in concert with DprE2 to catalyze the epimerization of decaprenyl-phosphoribose (DPR) to decaprenyl-phospho-d-arabinofuranose (DPA), which is the sole precursor for the synthesis of arabinogalactan and lipoarabinomannan (LAM) in the mycobacterium cell wall (6).
BTZ behaves as a suicide substrate for the reduced form of DprE1 by undergoing nitroreduction to form a nitroso derivative, which specifically forms a covalent adduct with C387 in the DprE1 active site (7–10). The C387 residue of DprE1 is highly conserved in orthologous enzymes in actinobacteria, except in Mycobacterium avium and M. aurum, where cysteine is replaced by alanine and serine, respectively. These mutations confer natural resistance to BTZ (2). Spontaneous mutants resistant to BTZ that were raised in Mycobacterium smegmatis and M. tuberculosis revealed that glycine or serine substitutions at C387 increased the MIC by at least 1,000-fold (2). The clinical importance of the C387 residue of DprE1 was confirmed as well when 240 M. tuberculosis clinical isolates were tested, since all these isolates were found to be BTZ sensitive and had the conserved cysteine codon.
The vulnerability of DprE1 lies in its essentiality in mycobacteria and its localization in the cell wall (11), accounting for the fact that DprE1 has been identified as the target of several structurally distinct compounds in recent drug screens. These compounds can be classified as covalent or noncovalent DprE1 inhibitors. Covalent inhibitors such as BTZ, the nitroquinoxaline VI-9376 (12), and the nitroimidazole 377790 (13) are nitroaromatic compounds possessing the necessary nitro group required for covalent adduct formation at C387 on DprE1. Noncovalent inhibitors such as TCA1 (14), 1,4-azaindoles (15), pyrazolopyridones (16), 4-aminoquinolone piperidine amides (17), and Ty38c (18) block enzyme activity by forming hydrophobic, electrostatic, and van der Waals interactions with particular residues in the DprE1 active site.
Given the pivotal role played by the C387 residue of DprE1 in the efficacy of nitroaromatic compounds, the aim of this study was to identify mutations at C387 that are tolerated and confer resistance to (P)BTZ in order to understand the underlying mechanisms of resistance involved as well as the overall influence of these mutations on the DprE1 enzyme and on the pathogen M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and chemicals.
M. tuberculosis H37Rv, M. smegmatis mc2155, and merodiploid strains were grown at 37°C in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% albumin-dextrose-catalase (ADC) or on Middlebrook 7H10 agar (Difco) supplemented with 0.2% glycerol and 10% oleic acid-albumin-dextrose-catalase (OADC). For cloning procedures, One Shot TOP10 chemically competent Escherichia coli cells (Invitrogen) were grown in Luria-Bertani (LB) broth or on LB agar containing kanamycin (50 μg/ml) or hygromycin (200 μg/ml). All chemicals were purchased from Sigma-Aldrich unless otherwise stated.
Generation of randomly mutated dprE1 in merodiploid M. tuberculosis strains.
The dprE1 gene under the control of its natural promoter, located upstream of Rv3789, was amplified together with Rv3789 by using primers rv3790-fwd and rv3790-rev and cloned in the pCR-Blunt II-TOPO vector (Invitrogen). The resulting plasmid was used to generate random mutations in the TGC codon encoding Cys387. Site-directed mutagenesis was carried out by using the Stratagene QuikChange II site-directed mutagenesis kit with primers containing random bases at the site of interest. The mutated fragments were ligated into the pND255 vector, kindly provided by N. Dhar, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, which harbors a hygromycin resistance cassette. Six pools of randomly mutated plasmids were obtained (pBLX 1 to pBLX 6), and each pool was screened independently. The resulting integrative vectors were then transformed and integrated at the L5-attB site of M. smegmatis mc2155 and M. tuberculosis H37Rv. Transformants were selected on 7H10 agar plates with or without BTZ043 at 400 ng/ml. A specific primer was used to repeat the site-directed mutagenesis to obtain the DprE1C387G mutant, which was selected on BTZ043-containing medium. All primers are listed in Table S1 in the supplemental material.
Determination of MICs.
MICs were determined by using the resazurin reduction microplate assay (REMA) as previously described (19). M. tuberculosis strains were grown in 7H9 medium to log phase (optical density at 600 nm [OD600] of 0.4 to 0.8) and diluted to an OD600 of 0.0001. One hundred microliters of the bacterial suspension (3 × 103 cells) was pipetted into wells of a 96-well plate. Compounds (BTZ043, PBTZ169, Ty38c, moxifloxacin, and rifampin) were added to the first column, and subsequently, 2-fold serial dilutions were made. After 6 days of incubation at 37°C, 10 μl of 0.025% (wt/vol) resazurin was added to each well. The fluorescence intensity was read after 24 h of incubation by using an Infinite F200 Tecan plate reader, and MIC values were determined by nonlinear fitting of the data to the Gompertz equation (35) using GraphPad Prism.
Site-directed mutagenesis of dprE1 at C387 in M. tuberculosis H37Rv.
Generation of point mutations in M. tuberculosis H37Rv was done by a recombineering method (20, 21). H37Rv/pJV53 was grown to log phase (OD600 of 0.5) in 7H9 medium containing 25 μg/ml of kanamycin before being induced with 0.2% acetamide overnight. Competent cells were transformed with 100 ng of 70-bp single-stranded oligonucleotides (leading and lagging strands) (see Table S1 in the supplemental material) containing the desired mutations, and transformants were selected on 7H10 agar plates either with or without 400 ng/ml BTZ043. Single-nucleotide polymorphisms (SNPs) in resistant colonies were confirmed by colony PCR.
Fitness assessment of dprE1 mutant strains in liquid culture.
All strains were diluted to an initial OD600 of 0.05, and OD600 measurements were taken every 24 h to monitor growth over a period of 2 weeks. The generation time for each strain was calculated by using the equation G = t/[3.3 × log10(b/B)], where G is the generation time, t is the time interval of two measurements in the exponential phase, b is the final OD600, and B is the initial OD600. Two independent cultures for each strain were used for growth rate measurements.
Expression and purification of wild-type and mutant M. tuberculosis DprE1.
M. tuberculosis dprE1 was cloned into plasmid pET28a, and the recombinant protein was coexpressed in E. coli BL21(DE3) along with the M. tuberculosis GroEL2 (Rv0440) and E. coli GroES chaperones in a modified version of the pGro7 plasmid (TaKaRa Bio Inc.). DprE1 was purified as described previously (4) to obtain pure protein with bound flavin adenine dinucleotide (FAD). BTZ-resistant C387G, C387S, C387A, C387T, and C387N mutants were obtained by site-directed mutagenesis with the pET28a-dprE1 plasmid by using the Stratagene QuikChange II site-directed mutagenesis kit. Expression and purification of the mutant proteins were carried out as described above for the wild-type (WT) protein. Protein concentrations were determined by using the bicinchoninic acid (BCA) assay (Pierce BCA protein assay kit; Thermo Scientific).
DprE1 enzymatic activity assays.
Enzyme activities of wild-type and mutant DprE1 proteins were determined in a two-step coupled assay (9). Reactions were carried out in black 96-well half-area plates (catalog number 3686; Corning) in a final volume of 25 μl per well. The reaction mixture consisted of the DprE1 protein (protein concentrations were adapted to obtain similar fluorescence signals [1.5 μM for the WT, 1.5 μM for the C387S mutant, 7.5 μM for C387G, 3 μM for C387A, 10.5 μM for C387T, and 10.5 μM for C387N]), FAD (1 μM), horseradish peroxidase (HRP) (0.2 μM), Amplex Red (50 μM) (Life Technologies), and farnesyl-phosphoryl-β-d-ribofuranose (FPR) (0 μM, 0.2 μM, 0.4 μM, 0.6 μM, or 0.8 μM) in assay buffer (50 mM glycyl glycine [pH 8.0], 200 mM potassium glutamate, 0.002% Brij 35). A standard curve was obtained with a serial dilution of resorufin sodium salt. FPR was used to start the reaction, and the conversion of Amplex Red to resorufin was immediately monitored by fluorescence measurement (excitation/emission wavelength of 560/590 nm) in the kinetic mode on a Tecan M200 instrument at 30°C. To determine 50% inhibitory concentrations (IC50s), the reaction mixture consisting of DprE1, FAD, HRP, and Amplex Red was first incubated with the test compound (with 2-fold serial dilution starting with dimethyl sulfoxide [DMSO] [1% final DMSO concentration]) for 10 min at 30°C before the addition of FPR (0.3 μM). The background fluorescence intensity from the reaction mixture without FPR was subtracted from the values for all reactions. Fluorescence units were converted to resorufin concentrations by using a standard curve. Reaction rates at each FPR concentration or compound concentration were determined and fitted to either the Hill equation for non-Michaelis-Menten kinetics to obtain steady-state kinetic constants or a log[inhibitor]-versus-normalized response (with 100% activity of each enzyme being defined under steady-state conditions in the absence of the inhibitor and 0% activity being defined as the full inhibition of WT DprE1 in the presence of 40 μM the inhibitor) to obtain IC50s by using GraphPad Prism.
Infection of THP-1 macrophages.
Human monocytic THP-1 cells were grown in RPMI medium supplemented with 10% FBS. A total of 2 × 104 cells/well of a 96-well plate in 50 μl RPMI medium were differentiated by using phorbol-12-myristate-13-acetate (PMA) (final concentration of 4 nM). Plates were sealed with gas-permeable sealing films and incubated at 37°C with 5% CO2. Cells were infected the following day. RPMI medium containing PMA was removed from the wells, and cells were washed and incubated with RPMI medium alone. H37Rv, dprE1 mutant strains, and H37RvΔRD1 were grown to log phase (OD600 of between 0.4 and 0.8), washed in 7H9 medium, and resuspended to an OD600 of 1 (3 × 108 bacteria/ml). THP-1 cells were infected at a multiplicity of infection (MOI) of 5 in 50 μl RPMI medium and incubated for 3 days before the addition of 5 μl PrestoBlue cell viability reagent (Life Technologies). After 1 h of incubation at room temperature (RT), fluorescence was measured by using a Tecan M200 instrument (excitation/emission wavelength of 560/590 nm).
Structural studies of WT and mutant DprE1 proteins.
DprE1 mutants were modeled based on the M. tuberculosis DprE1 structure (PDB accession number 4NCR) (9), using the “Position Scan” function in the FolDX plug-in (22) implemented in YASARA View molecular graphics software (23). Illustrations were made by using PyMOL (24), and the prediction of the functional effect of amino acid substitutions was computed by using the Provean Web server (25, 26).
RESULTS
Five DprE1 mutations confer resistance to BTZ043 in M. tuberculosis.
In an initial screen to determine which mutations at the C387 residue of DprE1 confer resistance to BTZ043, the gene with a randomly mutated codon at position 387 was expressed under the control of its natural promoter in M. tuberculosis H37Rv. Since previous reports described BTZ-resistant substitutions at C387 as being dominant (2), a partially diploid strain was used. The promoter of the Rv3789-dprE1-dprE2 operon (27) was exploited for the construction of partially diploid strains carrying random mutations at the C387 residue of DprE1. In doing so, potential artifacts derived from any unbalanced expression of the wild-type and mutant alleles were avoided.
After selection on 400 ng/ml BTZ043, transformants were screened by PCR to identify mutations associated with resistance, and their MICs for BTZ043 were determined. Various mutations were identified in colonies from BTZ043-containing plates, of which 28 were serine (C387S), 18 were threonine (C387T), 5 were alanine (C387A), 3 were glycine (C387G), and 7 were asparagine (C387N) substitutions. Random mutagenesis proved successful, as several different codons for each mutation were found, and BTZ043 MIC values of the colonies established their resistance levels (Table 1). Out of 9 possible codons that can be derived from single mutations at C387, which are most likely to occur clinically, only codon TCT coding for C387S appeared in the screen. The C387G mutation resulted in slower-growing colonies on solid medium, although this effect was not observed in liquid culture (see Fig. S1 and S2 in the supplemental material). The finding that the observed phenotype was not due to additional spontaneous mutations was confirmed by growing M. tuberculosis cells transformed with a plasmid (pBLG) (see Table S2 in the supplemental material) harboring the C387G substitution, suggesting that the growth defect of M. tuberculosis on solid medium was attributed to the glycine mutation. Although the same strategy was adopted to select for BTZ-resistant mutants in M. smegmatis expressing the M. tuberculosis dprE1 locus, this was unsuccessful (data not shown).
TABLE 1.
DprE1 C387 mutations conferring resistance to BTZ043 in M. tuberculosis
| Residue or mutation | Source or organism in which mutation was previously identified | No. of isolatesb | BTZ043 MIC (μg/ml) | MXFc MIC (μg/ml) | Codon(s) identified by sequencing |
|---|---|---|---|---|---|
| C387 (WT) | Wild type | 0.002 | 0.031 | TGC | |
| C387G | M. tuberculosis, M. smegmatisa | 3 | 12.5 | 0.063 | GGA, GGC |
| C387S | M. tuberculosis, M. smegmatis, M. auruma | 28 | >100 | 0.031 | TCA, TCC, TCG, TCT, AGC |
| C387A | M. aviuma | 5 | >100 | 0.031 | GCC, GCG |
| C387T | This study | 18 | >100 | 0.031 | ACA, ACC, ACT |
| C387N | This study | 7 | 6.25 | 0.031 | AAT |
See reference 2.
Colonies were isolated on plates containing 400 ng/ml of BTZ043.
MXF, moxifloxacin.
To further exclude any effects of having two copies of dprE1 due to the partially diploid strain used in the initial screen, site-directed mutagenesis was performed directly on chromosomal dprE1 of H37Rv at the C387 residue. C387S, C387A, and C387T mutations were successfully introduced into dprE1 without additional undesired mutations, and normal-sized colonies were selected on BTZ043-containing plates (Table 2). The introduction of C387G and C387N substitutions gave rise to two different colony sizes, both of which occurred in the absence of BTZ043. Small colonies were found to harbor solely the desired mutation (either C387G or C387N), whereas normal-sized colonies were found to have either a C387G V388I double mutation or C387T instead of the desired C387N mutation (Table 2). Resistance of these strains with dprE1 C387 mutations to BTZ043 was confirmed by REMA (Table 2).
TABLE 2.
Characterization of different H37Rv dprE1 mutantsa
| Mutation | Codon | Colony size | Presence of BTZ in plate from which colonies were picked | Mutation(s) identified by sequencing | BTZ043 MIC (μg/ml) | RIF MIC (μg/ml) |
|---|---|---|---|---|---|---|
| C387 | TGC (WT) | 0.0008 | 0.0016 | |||
| C387S | TCC | Normal | BTZ043 | C387S | ≥10 | 0.0016 |
| C387A | GCC | Normal | BTZ043 | C387A | >10 | 0.0016 |
| C387T | AAT | Normal | BTZ043 | C387T | >10 | 0.0016 |
| C387G | GGC | Small | C387G | Undet. | Undet. | |
| Normal | C387G V388I | 5 | 0.0008 | |||
| C387N | ACC | Small | C387N | 1.25 | 0.0008 | |
| Normal | C387T* | >10 | 0.0016 |
RIF, rifampin; Undet., undetermined. * indicates the C387T mutation with an ACT codon.
Fitness assessment of dprE1 mutant strains.
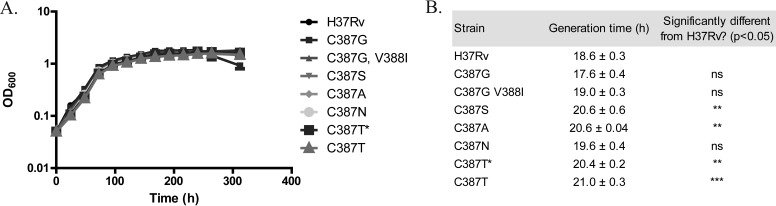
The growth of H37Rv and dprE1 mutant strains was monitored in liquid 7H9 medium in the absence of BTZ043 over 2 weeks. Growth curves did not reflect any striking differences in fitness between H37Rv and mutant strains during the exponential or the stationary phase (Fig. 1A). The generation time for each strain was calculated in the exponential phase of growth. For certain mutants (C387S, C387A, and C387T), their generation times significantly differ from that of H37Rv (Fig. 1B), although these differences appear to be too small to be reflected in the growth curves of the mutant strains. Notably, the growth defect of the C387G and C387N mutants on solid medium was not observed in liquid medium.
FIG 1.
Fitness of H37Rv and dprE1 mutant strains in 7H9 medium. (A) Growth curves of H37Rv and dprE1 mutant strains obtained by measuring the OD600 of bacterial cultures at 24-h intervals over a period of 14 days, with an initial OD600 of 0.05. (B) The generation time for each strain was calculated by using the equation G = t/[3.3 × log10(b/B)], where G is the generation time; t is the time interval between two measurements within the exponential phase, here 0 to 48 h; b is the final OD600; and B is the initial OD600. Data from two independent experiments are presented as means ± standard deviations. Statistical analysis was performed by using one-way analysis of variance with Dunnett's posttest (**, P < 0.01; ***, P < 0.001). C387T* indicates a mutation containing the ACT codon, compared to C387T, which has the AAT codon instead. “ns” indicates no significance.
Effect of DprE1 C387 mutations on M. tuberculosis cytotoxicity.
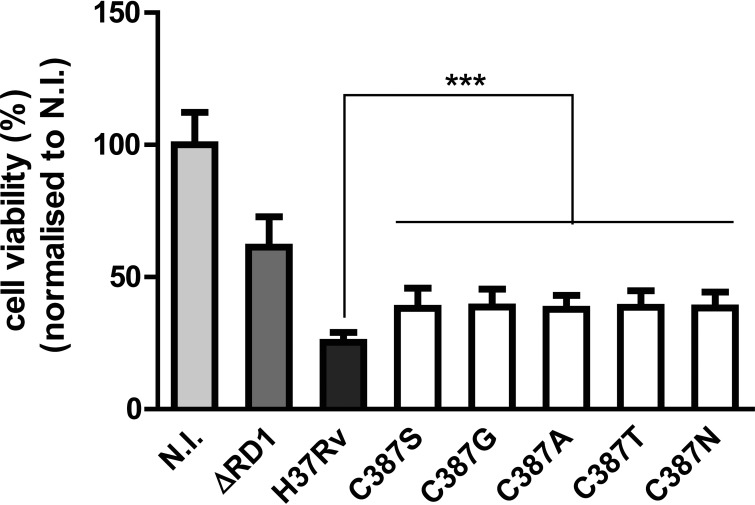
The influence of C387 mutations on the cytotoxicity of the mycobacterium was examined ex vivo by infecting THP-1 macrophages with the H37Rv and H37Rv dprE1 C387 mutant strains. As a control, an attenuated strain with reduced virulence, H37RvΔRD1 (28), was used, and this showed decreased cytotoxicity, with macrophage viability being around 60% of that of noninfected macrophages. About 40% of macrophages remained viable after infection with each of the five dprE1 mutants, compared to about 25% of macrophages remaining viable after being infected with H37Rv (Fig. 2), indicating that the C387 mutations decrease the cytotoxicity of M. tuberculosis.
FIG 2.
DprE1 C387 mutations affect the cytotoxicity of M. tuberculosis. THP-1 cells were infected with H37Rv, H37RvΔRD1, and H37Rv dprE1 C387 mutant strains at an MOI of 5 or left untreated (not infected [N.I.]). Macrophage viability was measured at 3 days postinfection. Data from two independent experiments were normalized to data under noninfected conditions and are presented as means ± standard deviations. Statistical analysis was performed by using one-way analysis of variance with Dunnett's posttest (***, P < 0.001).
Steady-state enzymatic activity of DprE1 mutants.
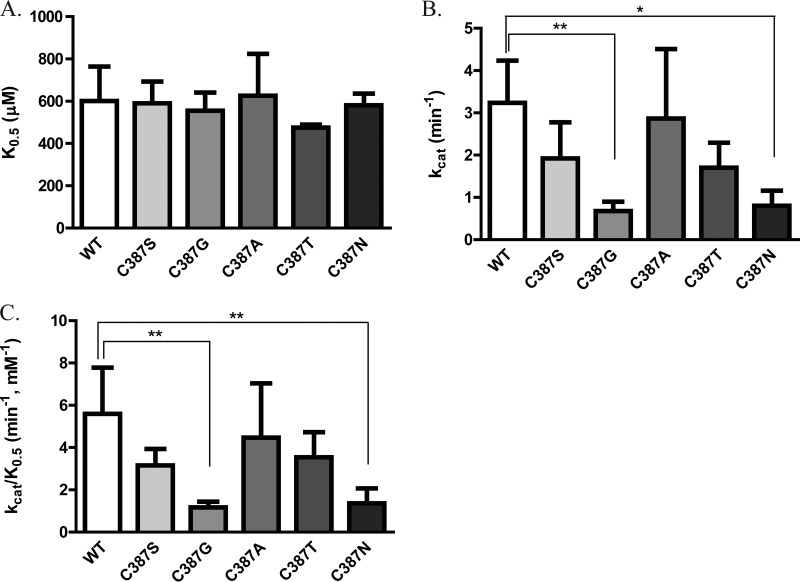
To investigate whether DprE1 C387 mutations could affect the proper functioning of the enzyme, the steady-state enzymatic activity of purified WT and mutant DprE1 proteins was measured by using a two-step coupled enzymatic assay. For each protein, reaction rates at four substrate concentrations were fitted to the Hill equation for non-Michaelis-Menten kinetics to obtain parameters of substrate binding affinity (K0.5), turnover number (kcat), and catalytic efficiency (kcat/K0.5). None of the mutations at C387 of DprE1 affected the binding affinity of the substrate for the enzyme, as seen from their K0.5 values, which were similar to that of the WT enzyme (Fig. 3A). C387G and C387N mutations significantly decreased the turnover rate of the enzyme (Fig. 3B), and consequently, these two mutations reduced the overall catalytic efficiency of DprE1 by about 4-fold (Fig. 3C).
FIG 3.
Steady-state enzymatic activity of WT and DprE1 mutants. Shown are substrate binding affinity (K0.5) (A), turnover number (kcat) (B), and catalytic efficiency (kcat/K0.5) (C) steady-state parameters obtained for the WT and DprE1 mutants by fitting enzyme activity at various FPR concentrations (0.2 to 0.8 mM) to the equation Y = Vmax × Xh/(Kprime + Xh), where Y is enzyme activity, X is the substrate concentration, Kprime equals (K0.5)h, and h is the Hill coefficient, by using GraphPad Prism. Data from at least three independent experiments are presented as means ± standard deviations. Statistical analysis was performed by using one-way analysis of variance with Dunnett's posttest (*, P < 0.05; **, P < 0.01).
Effect of DprE1 C387 mutations on the potency of DprE1 inhibitors.
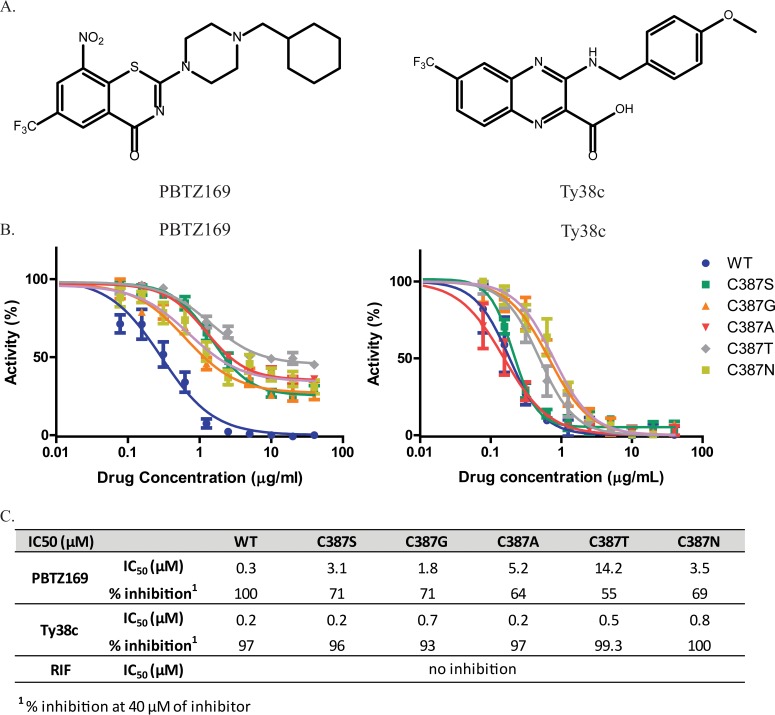
IC50 values of covalent (PBTZ169) and noncovalent (Ty38c) DprE1 inhibitors (Fig. 4A) were determined for WT and DprE1 mutant proteins. As BTZ043 showed behavior similar to that of PBTZ169 in this assay (data not shown), and the MIC of PBTZ169 was 3-fold lower than that of BTZ043 in M. tuberculosis, PBTZ169 was used in this study. Unlike the wild-type enzyme, whose activity is completely inhibited at high PBTZ169 concentrations, mutant enzymes are only partially inhibited at the highest PBTZ169 concentration (40 μM), apparently reaching a plateau at 30 to 40% activity (Fig. 4B). In contrast, this concentration of the noncovalent inhibitor Ty38c was sufficient to inhibit almost all activity of both WT and mutant enzymes (Fig. 4B and C).
FIG 4.
Effects of DprE1 C387 mutations on potency of DprE1 inhibitor activity. (A) Structures of the DprE1 inhibitors PBTZ169 and Ty38c. PBTZ169 inhibits DprE1 via a covalent bond between the reduced form of its nitro group and the C387 residue of DprE1, whereas Ty38c, which lacks the nitro group, is a noncovalent inhibitor of DprE1. (B) Curves of enzyme activity with increasing inhibitor (PBTZ169 or Ty38c) concentrations for WT and mutant enzymes. A total of 40 μM the inhibitor was used for the highest concentration, with subsequent 2-fold serial dilutions. Enzymatic activities at each inhibitor concentration were normalized to steady-state enzymatic activity in the absence of any inhibitor. (C) PBTZ169 and Ty38c IC50 values and maximum inhibition for WT and mutant enzymes. IC50 values were obtained by fitting the curves in panel B to the log[inhibitor]-versus-normalized response by using GraphPad Prism. Maximum percent inhibition of the WT or mutant enzyme was determined with 40 μM the inhibitor. Data from at least two independent experiments are presented as means ± standard deviations.
This finding confirms the importance of the C387 residue of DprE1 in the mode of inhibition of the covalent inhibitor PBTZ169 and indicates that the replacement of the Cys residue hardly affects the binding of noncovalent inhibitors to DprE1. This can be further seen from the IC50 values of PBTZ169 and Ty38c obtained for WT and mutant enzymes (Fig. 4C). All mutant enzymes showed increased PBTZ169 IC50 values compared to that for the WT, thus accounting for the BTZ resistance of the mutants. Ty38c IC50 values for the C387S and C387A mutants were identical to that for the WT, while the C387G, C387T, and C387N mutants had slightly higher values than that for the WT (Fig. 4C), indicating that substitutions at C387 have a more modest effect on Ty38c activity. PBTZ169 and Ty38c MIC values were also determined for H37Rv and dprE1 mutant strains (Table 3). C387 mutations cause an increase in the PBTZ169 MIC compared to H37Rv, with strains harboring C387S, C387A, and C387T mutations having higher levels of resistance (MIC of >1 μg/ml) and those with C387G and C387N mutations having lower levels of resistance (MIC of 0.5 to 0.6 μg/ml) to PBTZ169. In the case of Ty38c, mutant strains had MIC values ranging from 0.1 to 0.8 μg/ml, and the trend was similar to that seen with PBTZ169.
TABLE 3.
PBTZ169 and Ty38c MICs for H37Rv and H37Rv dprE1 C387 mutant strains
| Strain | PBTZ169 MIC (μg/ml) | Ty38c MIC (μg/ml) | RIFa MIC (μg/ml) |
|---|---|---|---|
| H37Rv | 0.0001 | 0.1 | 0.001 |
| C387S | >1 | 0.5 | 0.001 |
| C387G | 0.5 | 0.04 | 0.0005 |
| C387A | >1 | 0.1 | 0.001 |
| C387T | >1 | 0.8 | 0.001 |
| C387N | 0.6 | 0.1 | 0.001 |
| NTB1b | >1 | 0.7 | 0.001 |
RIF, rifampin.
BTZ-resistant strain with a C387S mutation (control).
Taken together, these data demonstrate that C387 mutations decrease the potency of covalently binding DprE1 inhibitors such as PBTZ169 to a much greater extent than noncovalent DprE1 inhibitors, represented here by Ty38c. As the MICs of lower-resistance mutants could be measured, one of the mutants (C387N) was used to investigate the effect of a C387 mutation on the interaction between BTZ and bedaquiline (BDQ), as synergism between these two classes of compounds in vitro (29) and in vivo (4) was previously described. Using a checkerboard assay, the partial synergism of PBTZ169 and BDQ observed in H37Rv at 0.5× MIC was not significantly altered by the DprE1 C387N mutation (see Fig. S3 in the supplemental material).
Modeling the effect of C387 mutations on the active site of DprE1.
To gain further insight into the mechanism of resistance of DprE1 mutants to BTZ and to predict the effect of each mutation on the functionality of DprE1, substitution at C387 with every amino acid was modeled in silico based on the structure of M. tuberculosis DprE1.
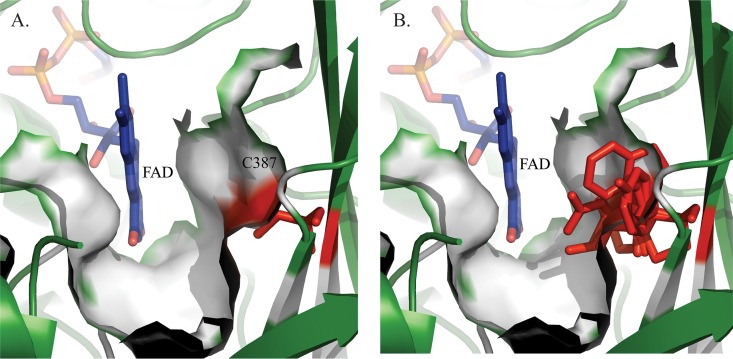
Modeling showed that all the substitutions are tolerated in terms of steric hindrance. Indeed, amino acid 387 is localized on a beta sheet where the side chain could easily be accommodated since it is exposed to the solvent toward the substrate binding pocket (Fig. 5A). Substitutions at this position will lead to a modification of the shape and volume of the substrate binding pocket (Fig. 5B). It is evident from these models that some substitutions will lead to nonfunctional proteins. More precisely, only substitutions by small and/or polar residues such as asparagine, aspartate, serine, threonine, alanine, and glycine would lead to functional enzymes, whereas amino acids with larger side chains (arginine, tryptophan, phenylalanine, glutamate, glutamine, tyrosine, methionine, lysine, and histidine) would block access to the substrate, resulting in nonfunctional enzymes. Furthermore, exposure of the side chain to the solvent excludes the possibility of a hydrophobic residue occupying this position (leucine, isoleucine, and valine), and the proline substitution is not favored in beta sheet structures, as it cannot complete the hydrogen-bonding network.
FIG 5.
Closeup views of the substrate binding pocket of M. tuberculosis WT DprE1 compared to mutant models. (A) Substrate binding pocket in the crystal structure of M. tuberculosis DprE1 (PDB accession number 4NCR). FAD is represented in blue, and cysteine 387 is presented in red. The solvent-accessible surface is represented by the white surface. (B) Superposition of crystal structures of M. tuberculosis WT DprE1 and 19 substitution models at residue 387 (represented by sticks in red). The side chains of some amino acids likely block the substrate binding site, leading to a nonfunctional enzyme.
To confirm these observations, a prediction of the functional effects of amino acid substitutions was computed by using the Provean Web server (25, 26). Interestingly, only alanine, serine, and threonine were found to be nondeleterious. Altogether, these results are in agreement with the data obtained from the screen.
The superposition of crystal structures of WT DprE1 and the C387A, C387S, C387G, C387T, and C387N mutants in complex with PBTZ169 or Ty38c revealed that these five substitutions do not block the access of both inhibitors to the binding pocket (see Fig. S4 in the supplemental material). As expected, the five mutants are not able to form the critical covalent adduct with PBTZ169. In these cases, PBTZ169 is stabilized only unspecifically by van der Waals interactions in the binding pocket and behaves as an inefficient noncovalent inhibitor, resulting in a dramatic reduction of PBTZ169 potency. On the other hand, C387 is not directly implicated in the binding of the noncovalent inhibitor Ty38c. Although C387 mutations do not influence the main interaction with Ty38c, they could still affect the environment in the substrate binding pocket, leading to a modest impairment of the inhibitor activity.
DISCUSSION
DprE1 plays an essential role in the DPA pathway for cell wall synthesis in M. tuberculosis. Having been identified as the target of at least five structurally distinct inhibitors in recent years, DprE1 is an extremely vulnerable target of the pathogen (30, 31). Among the inhibitors, PBTZ169 is a highly potent covalent inhibitor of DprE1 and is currently in phase I trials. We have studied the critical C387 residue of DprE1 required for the covalent interaction with PBTZ169 and with other nitroaromatic inhibitors by identifying and characterizing substitutions at this residue that are tolerated and confer resistance to BTZs.
The data obtained confirmed the dominant nature of the mutations introduced (2) and identified C387S, C387A, C387T, C387G, and C387N as DprE1 mutations viable for the bacteria while conferring various levels of resistance to BTZ043 and PBTZ169. The C387S, C387A, and C387T mutations confer high levels of resistance of H37Rv to BTZs, while C387G and C387N confer intermediate levels of resistance. As previously reported, C387S and C387A substitutions are naturally occurring mutations found in M. aurum and M. avium, and C387G and C387S were found in colonies spontaneously resistant to BTZ043 (2). C387N and C387T are novel mutations identified here from this screen. To our knowledge, these mutations have hitherto remained unidentified in DprE1 in mutants spontaneously resistant to BTZ043, likely because two or three bases of the cysteine codon (TGC) need to be mutated to generate a threonine (ACA, ACC, and ACT) or an asparagine (AAT) codon.
The fact that C387S and C387A mutations do not result in M. tuberculosis growth defects or in reduced enzymatic capabilities and are predicted to be nondeleterious mutations that do not block the access of the substrate to the binding pocket are consistent with their occurrence in nature. Although C387G and C387N mutations appear to allow substrate access in structural models, it appears that the nonoptimal size of their side chains (i.e., either too small or too large) strongly impacts the resulting shape of the substrate binding pocket and would affect the stability and binding of the substrate, resulting in reduced enzymatic activity. Furthermore, these mutations are also predicted to be deleterious and would have a detrimental impact on the biological function of DprE1. As seen experimentally, growth defects were observed on solid medium in the presence of these mutations. This could be a consequence of the impaired catalytic capability of DprE1, which would imply that the epimerization of DPR to DPA and, by extension, the formation of the arabinogalactan layer in the mycobacterium cell wall occur at lower rates.
DprE1 C387 mutations are less cytotoxic than the WT in macrophages. Regardless of the substitution at C387, there is an impaired ability of M. tuberculosis to induce host cell death. The exact mechanism remains unclear; however, one might speculate that these mutations result in slower replication of the mycobacterium in an ex vivo context. Another possibility is that the M. tuberculosis cell wall component LAM is formed at a reduced rate as a result of decreased DPA synthesis. Since LAM has been found to inhibit phagosome maturation within macrophages (32), this capability could be reduced in the presence of C387 mutations, with a consequent decrease in the survival of the pathogen in the host cell.
The importance of the C387 residue in the binding of covalent DprE1 inhibitors is illustrated biochemically by the increase in PBTZ169 IC50 values in the presence of C387 substitutions and the fact that even at high concentrations of PBTZ169, there is no full inhibition in all the mutant enzymes. This residual enzymatic activity explains the ability of the mutant to survive in vitro even in the presence of the compound at concentrations much higher than the MIC. In the case of the noncovalent DprE1 inhibitor, C387G and C387N mutations modestly affect its activity although to a considerably lesser extent than that of PBTZ169. These findings are further supported by WT and mutant DprE1 structures docked with either PTBZ169 or Ty38c, where the effects of C387 mutations on the covalent and noncovalent mechanisms of inhibition are modeled at the atomic level.
With the initial BTZ-resistant mutant selection procedure, mutants conferring lower-level resistance may have been missed due to growth defects. An explanation for the unsuccessful selection of BTZ-resistant mutants in M. smegmatis may be due to the differences in the regulation of arabinogalactan or lipoarabinomannan biosynthesis in the two species or to the different genomic organization of M. smegmatis. Furthermore, it was reported previously that the level of activity of the M. tuberculosis embA promoter was markedly lower in M. smegmatis (33), suggesting that control of gene expression may be organized differently in this nonpathogenic mycobacterium. Therefore, it appears that M. smegmatis may not be the best model for studying genes implicated in arabinan-derived cell wall components expressed under the control of M. tuberculosis promoters.
In conclusion, we have identified five mutations in DprE1 that do not affect the viability of M. tuberculosis, which give rise to a functional enzyme and generate BTZ resistance. As these five BTZ-resistant mutants appear less cytotoxic in macrophages, they may also be less fit should these mutations arise in humans. From a clinical standpoint, an understanding of DprE1-mediated BTZ resistance will facilitate clinical trials of the drug candidate PBTZ169 by screening M. tuberculosis for C387 mutations over the course of treatment. In the case of the C387N mutant with an intermediate level of resistance to PBTZ169, it appears that the synergy between PBTZ169 and BDQ does not depend on the covalent bond of C387 with PBTZ169. This is consistent with the finding that noncovalent DprE1 inhibitors, the 1,4-azaindoles, also display synergism with BDQ (34). The lack of cross-resistance of BTZ-resistant mutants to Ty38c as reported in this work would also be an important consideration when deciding which and how many DprE1 inhibitors to develop clinically, as a noncovalent DprE1 inhibitor could be used after resistance to the covalent one develops, or vice versa. Altogether, this work provides timely insight into the mechanisms of resistance of M. tuberculosis to DprE1 inhibitors as PBTZ169 progresses into clinical trials.
Supplementary Material
ACKNOWLEDGMENTS
We thank Neeraj Dhar for providing plasmid pND255, Vadim Makarov for BTZ043 and PBTZ169, Maria-Paola Costi for Ty38c, and Anthony Vocat for his technical assistance in REMAs.
Funding Statement
This work was supported by the European Community's Seventh Framework Program FP7/2007-2013 under grant agreement 260872. Benoit Lechartier was a recipient of a grant from the Fondation Jacqueline Beytout. João Neres was awarded a Marie Curie fellowship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01523-16.
REFERENCES
- 1.WHO. 2015. Global tuberculosis report 2015. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianiskova P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST. 2009. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324:801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasca MR, Degiacomi G, de Jesus Lopes Ribeiro AL, Zara F, De Mori P, Heym B, Mirrione M, Brerra R, Pagani L, Pucillo L, Troupioti P, Makarov V, Cole ST, Riccardi G. 2010. Clinical isolates of Mycobacterium tuberculosis in four European hospitals are uniformly susceptible to benzothiazinones. Antimicrob Agents Chemother 54:1616–1618. doi: 10.1128/AAC.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarov V, Lechartier B, Zhang M, Neres J, van der Sar AM, Raadsen SA, Hartkoorn RC, Ryabova OB, Vocat A, Decosterd LA, Widmer N, Buclin T, Bitter W, Andries K, Pojer F, Dyson PJ, Cole ST. 2014. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol Med 6:372–383. doi: 10.1002/emmm.201303575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Working Group on New TB Drugs. 2015. Drug pipeline. Stop TB Partnership, Geneva, Switzerland: http://www.newtbdrugs.org/pipeline.php Accessed 22 November 2015. [Google Scholar]
- 6.Mikusova K, Huang H, Yagi T, Holsters M, Vereecke D, D'Haeze W, Scherman MS, Brennan PJ, McNeil MR, Crick DC. 2005. Decaprenylphosphoryl arabinofuranose, the donor of the d-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J Bacteriol 187:8020–8025. doi: 10.1128/JB.187.23.8020-8025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trefzer C, Rengifo-Gonzalez M, Hinner MJ, Schneider P, Makarov V, Cole ST, Johnsson K. 2010. Benzothiazinones: prodrugs that covalently modify the decaprenylphosphoryl-β-d-ribose 2′-epimerase DprE1 of Mycobacterium tuberculosis. J Am Chem Soc 132:13663–13665. doi: 10.1021/ja106357w. [DOI] [PubMed] [Google Scholar]
- 8.Trefzer C, Škovierová H, Buroni S, Bobovská A, Nenci S, Molteni E, Pojer F, Pasca MR, Makarov V, Cole ST, Riccardi G, Mikušová K, Johnsson K. 2012. Benzothiazinones are suicide inhibitors of mycobacterial decaprenylphosphoryl-β-d-ribofuranose 2′-oxidase DprE1. J Am Chem Soc 134:912–915. doi: 10.1021/ja211042r. [DOI] [PubMed] [Google Scholar]
- 9.Neres J, Pojer F, Molteni E, Chiarelli LR, Dhar N, Boy-Rottger S, Buroni S, Fullam E, Degiacomi G, Lucarelli AP, Read RJ, Zanoni G, Edmondson DE, De Rossi E, Pasca MR, McKinney JD, Dyson PJ, Riccardi G, Mattevi A, Cole ST, Binda C. 2012. Structural basis for benzothiazinone-mediated killing of Mycobacterium tuberculosis. Sci Transl Med 4:150ra121. doi: 10.1126/scitranslmed.3004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batt SM, Jabeen T, Bhowruth V, Quill L, Lund PA, Eggeling L, Alderwick LJ, Futterer K, Besra GS. 2012. Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proc Natl Acad Sci U S A 109:11354–11359. doi: 10.1073/pnas.1205735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brecik M, Centárová I, Mukherjee R, Kolly GS, Huszár S, Bobovská A, Kilacsková E, Mokošová V, Svetlikova Z, Šarkan M, Neres J, Korduláková J, Cole ST, Mikušová K. 29 April 2015. DprE1 is a vulnerable tuberculosis drug target due to its cell wall localization. ACS Chem Biol doi: 10.1021/acschembio.5b00237. [DOI] [PubMed] [Google Scholar]
- 12.Magnet S, Hartkoorn RC, Székely R, Pató J, Triccas JA, Schneider P, Szántai-Kis C, Őrfi L, Chambon M, Banfi D, Bueno M, Turcatti G, Kéri G, Cole ST. 2010. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis 90:354–360. doi: 10.1016/j.tube.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai H, Zhang L, Hung DT. 2012. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol 7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, Ahmad I, Yang P, Zhang Y, Kim J, Hassani M, Huszar S, Trefzer C, Ma Z, Kaneko T, Mdluli KE, Franzblau S, Chatterjee AK, Johnsson K, Mikusova K, Besra GS, Fütterer K, Robbins SH, Barnes SW, Walker JR, Jacobs WR, Schultz PG. 2013. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci U S A 110:E2510–E2517. doi: 10.1073/pnas.1309171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirude PS, Shandil R, Sadler C, Naik M, Hosagrahara V, Hameed S, Shinde V, Bathula C, Humnabadkar V, Kumar N, Reddy J, Panduga V, Sharma S, Ambady A, Hegde N, Whiteaker J, McLaughlin RE, Gardner H, Madhavapeddi P, Ramachandran V, Kaur P, Narayan A, Guptha S, Awasthy D, Narayan C, Mahadevaswamy J, Vishwas KG, Ahuja V, Srivastava A, Prabhakar K, Bharath S, Kale R, Ramaiah M, Choudhury NR, Sambandamurthy VK, Solapure S, Iyer PS, Narayanan S, Chatterji M. 2013. Azaindoles: noncovalent DprE1 inhibitors from scaffold morphing efforts, kill Mycobacterium tuberculosis and are efficacious in vivo. J Med Chem 56:9701–9708. doi: 10.1021/jm401382v. [DOI] [PubMed] [Google Scholar]
- 16.Panda M, Ramachandran S, Ramachandran V, Shirude PS, Humnabadkar V, Nagalapur K, Sharma S, Kaur P, Guptha S, Narayan A, Mahadevaswamy J, Ambady A, Hegde N, Rudrapatna SS, Hosagrahara VP, Sambandamurthy VK, Raichurkar A. 2014. Discovery of pyrazolopyridones as a novel class of non-covalent dpre1 inhibitor with potent anti-mycobacterial activity. J Med Chem 57:4761–4771. doi: 10.1021/jm5002937. [DOI] [PubMed] [Google Scholar]
- 17.Naik M, Humnabadkar V, Tantry SJ, Panda M, Narayan A, Guptha S, Panduga V, Manjrekar P, Jena LK, Koushik K, Shanbhag G, Jatheendranath S, Manjunatha MR, Gorai G, Bathula C, Rudrapatna S, Achar V, Sharma S, Ambady A, Hegde N, Mahadevaswamy J, Kaur P, Sambandamurthy VK, Awasthy D, Narayan C, Ravishankar S, Madhavapeddi P, Reddy J, Prabhakar K, Saralaya R, Chatterji M, Whiteaker J, McLaughlin B, Chiarelli LR, Riccardi G, Pasca MR, Binda C, Neres J, Dhar N, Signorino-Gelo F, McKinney JD, Ramachandran V, Shandil R, Tommasi R, Iyer PS, Narayanan S, Hosagrahara V, Kavanagh S, Dinesh N, Ghorpade SR. 2014. 4-Aminoquinolone piperidine amides: noncovalent inhibitors of DprE1 with long residence time and potent antimycobacterial activity. J Med Chem 57:5419–5434. doi: 10.1021/jm5005978. [DOI] [PubMed] [Google Scholar]
- 18.Neres J, Hartkoorn RC, Chiarelli LR, Gadupudi R, Pasca MR, Mori G, Venturelli A, Savina S, Makarov V, Kolly GS, Molteni E, Binda C, Dhar N, Ferrari S, Brodin P, Delorme V, Landry V, de Jesus Lopes Ribeiro AL, Farina D, Saxena P, Pojer F, Carta A, Luciani R, Porta A, Zanoni G, De Rossi E, Costi MP, Riccardi G, Cole ST. 2015. 2-Carboxyquinoxalines kill Mycobacterium tuberculosis through noncovalent inhibition of DprE1. ACS Chem Biol 10:705–714. doi: 10.1021/cb5007163. [DOI] [PubMed] [Google Scholar]
- 19.Palomino J-C, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat Rev Microbiol 6:851–857. doi: 10.1038/nrmicro2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybniker J, Vocat A, Sala C, Busso P, Pojer F, Benjak A, Cole ST. 2015. Lansoprazole is an antituberculous prodrug targeting cytochrome bc1. Nat Commun 6:7659. doi: 10.1038/ncomms8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Durme J, Delgado J, Stricher F, Serrano L, Schymkowitz J, Rousseau F. 2011. A graphical interface for the FoldX forcefield. Bioinformatics 27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- 23.Krieger E, Vriend G. 2014. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrödinger LLC. 2015. The PyMOL molecular graphics system, version 1.8. Schrödinger, LLC, Cambridge, MA. [Google Scholar]
- 25.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y, Chan AP. 2015. PROVEAN Web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolly GS, Mukherjee R, Kilacsková E, Abriata LA, Raccaud M, Blaško J, Sala C, Dal Peraro M, Mikušová K, Cole ST. 2015. GtrA protein Rv3789 is required for arabinosylation of arabinogalactan in Mycobacterium tuberculosis. J Bacteriol 197:3686–3697. doi: 10.1128/JB.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechartier B, Hartkoorn RC, Cole ST. 2012. In vitro combination studies of benzothiazinone lead compound BTZ043 against Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:5790–5793. doi: 10.1128/AAC.01476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riccardi G, Pasca MR, Chiarelli LR, Manina G, Mattevi A, Binda C. 2013. The DprE1 enzyme, one of the most vulnerable targets of Mycobacterium tuberculosis. Appl Microbiol Biotechnol 97:8841–8848. doi: 10.1007/s00253-013-5218-x. [DOI] [PubMed] [Google Scholar]
- 31.Mikusova K, Makarov V, Neres J. 2013. DprE1—from the discovery to the promising tuberculosis drug target. Curr Pharm Des 20:4379–4403. doi: 10.2174/138161282027140630122724. [DOI] [PubMed] [Google Scholar]
- 32.Vergne I, Fratti RA, Hill PJ, Chua J, Belisle J, Deretic V. 2004. Mycobacterium tuberculosis phagosome maturation arrest: mycobacterial phosphatidylinositol analog phosphatidylinositol mannoside stimulates early endosomal fusion. Mol Biol Cell 15:751–760. doi: 10.1091/mbc.E03-05-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amin AG, Goude R, Shi L, Zhang J, Chatterjee D, Parish T. 2008. EmbA is an essential arabinosyltransferase in Mycobacterium tuberculosis. Microbiology 154:240–248. doi: 10.1099/mic.0.2007/012153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterji M, Shandil R, Manjunatha MR, Solapure S, Ramachandran V, Kumar N, Saralaya R, Panduga V, Reddy J, Prabhakar KR, Sharma S, Sadler C, Cooper CB, Mdluli K, Iyer PS, Narayanan S, Shirude PS. 2014. 1,4-Azaindole, a potential drug candidate for treatment of tuberculosis. Antimicrob Agents Chemother 58:5325–5331. doi: 10.1128/AAC.03233-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert RJW, Pearson J. 2000. Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and noninhibitory concentration (NIC) values. Journal of Applied Microbiology 88:784–790. doi: 10.1046/j.1365-2672.2000.01017.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.