Abstract
High-throughput phenotypic screening of chemical libraries has resulted in the identification of thousands of compounds with potent antimalarial activity, although in most cases, the mechanism(s) of action of these compounds remains unknown. Here we have investigated the mode of action of 90 antimalarial compounds derived from the Malaria Box collection using high-coverage, untargeted metabolomics analysis. Approximately half of the tested compounds induced significant metabolic perturbations in in vitro cultures of Plasmodium falciparum. In most cases, the metabolic profiles were highly correlated with known antimalarials, in particular artemisinin, the 4-aminoquinolines, or atovaquone. Select Malaria Box compounds also induced changes in intermediates in essential metabolic pathways, such as isoprenoid biosynthesis (i.e., 2-C-methyl-d-erythritol 2,4-cyclodiphosphate) and linolenic acid metabolism (i.e., traumatic acid). This study provides a comprehensive database of the metabolic perturbations induced by chemically diverse inhibitors and highlights the utility of metabolomics for triaging new lead compounds and defining specific modes of action, which will assist with the development and optimization of new antimalarial drugs.
INTRODUCTION
Malaria remains a major global health problem, and there is a pressing need to discover and develop new antimalarials. Traditional antimalarial drugs have been severely undermined by the emergence of drug-resistant strains and current treatments rely heavily on artemisinin-based combination therapies. Alarmingly, artemisinin resistance has arisen in Southeast Asia during the last decade, highlighting the urgent need for new antimalarials (1). Extensive efforts to produce a malaria vaccine have met limited success and a pipeline of new antimalarial drugs will be required to mitigate extensive morbidity and mortality from malaria for the foreseeable future (2).
High-throughput phenotypic screening of chemical libraries against the malaria parasite, Plasmodium falciparum, has provided a major step forward in the discovery of novel antimalarial compounds. Almost 30,000 compounds that selectively inhibit growth of cultured P. falciparum asexual red blood cell (RBC) stages have been identified in these screens, providing excellent starting points for the discovery of new antimalarial drugs (3–5). A prioritized collection of these “hit” compounds, known as the Malaria Box, has been assembled by the Medicines for Malaria Venture (MMV) and provided free to the research community to facilitate this drug development pipeline (6).
A key limitation to the further optimization of many of these compounds is the lack of information on their mode of action. While not essential for registration, information on the mode of action of inhibitors discovered in phenotypic screens will significantly accelerate drug development by allowing structure-based drug design, monitoring of activity, toxicity and resistance, and facilitation of rational clinical usage in combination with other medicines. Furthermore, in addition to providing active chemical scaffolds for medicinal chemistry optimization, these compounds are ideal chemical probes to determine which biochemical pathways are both essential to the parasite and amenable to chemical inhibition, thus facilitating discovery of druggable targets for P. falciparum.
Only limited studies have been performed in an attempt to identify the targets of Malaria Box compounds in P. falciparum. Screens against specific parasite proteins have identified inhibitors of kinesin-5 (7), thioredoxin reductase (8), parasite aminopeptidases (9), dihydrofolate reductase-thymidylate synthase (10), and deoxyhypusine hydroxylase (11). Analysis of the activity of these compounds in the presence of isoprenoid precursors has identified compounds that exert their action by inhibition of isoprenoid biosynthesis in the apicoplast (12–14). A beta-hematin inhibition assay identified 10 compounds that inhibit the heme detoxification pathway (15), and the activity of one compound appears to be related to autophagy (16). Twenty-eight compounds in the Malaria Box were recently found to inhibit the parasite plasma membrane cation transporter, PfATP4, previously shown to be the target of the antimalarial, spiroindolone, which is currently in clinical trials (17). While these targeted approaches have been successful in identifying new chemical scaffolds that inhibit validated drug targets, there is clearly a need for complementary global, untargeted approaches that allow the identification of new drug targets and/or systematic triage and prioritization of antimalarial compound libraries.
Metabolomics has seen increasing application in the pharmaceutical industry in recent years and has proved useful in the drug discovery phase for investigating the mechanism of action for existing drugs and new drug candidates (18, 19). The system-wide nature of metabolomics has enabled discovery of the mechanism of action of several antiprotozoal compounds (19) and has demonstrated mechanisms of action for a number of antimalarial compounds. Targeted metabolomics approaches demonstrated the metabolic perturbations induced by inhibitors of the polyamine pathway (20), isoprenoid precursor biosynthesis (21), and the mitochondrial electron transport chain (22) in P. falciparum. A widely targeted metabolomics approach recently revealed unique metabolic phenotypes for several currently used antimalarials, including dihydroartemisinin, chloroquine, atovaquone, pyrimethamine, cycloguanil, and proguanil (23). These studies demonstrate that the metabolic perturbations induced by antimalarial compounds are related to their specific mechanisms of action and provide a basis for further extrapolation of this technique to the investigation of novel compounds.
Recent developments in high-resolution mass spectrometry and the associated data analysis tools have enabled the application of untargeted metabolomics to cell culture systems, including P. falciparum (24, 25). These untargeted metabolomics techniques have already been used to identify mechanisms of action for novel drug candidates in other protozoan pathogens (26–28), providing an unbiased, hypothesis-free approach to reveal the actions of novel antiparasitic compounds with no known target, such as those found in the Malaria Box.
This study applies untargeted metabolomics to reveal the metabolic perturbations induced by 100 compounds in P. falciparum-infected red blood cells (iRBCs) in vitro. These data allow clustering of compounds based on similarities with existing antimalarial drugs that have characterized mechanisms of action and identify several novel metabolic pathways associated with specific compounds in the Malaria Box.
MATERIALS AND METHODS
Cell culture and drug incubations for LC-MS metabolomics analysis.
Asexual P. falciparum (3D7) parasites were cultured by the method of Trager and Jensen (29), with minor modifications, using human RBCs (Australian Red Cross Blood Service) at 3% hematocrit in modified RPMI medium containing hypoxanthine and 0.5% (wt/vol) Albumax (Gibco) at 37°C under a defined atmosphere (95% N2, 4% CO2, 1% O2). Parasites were synchronized with 5% (wt/vol) sorbitol twice at an interval of 14 h and cultured for a further 58 h to ensure that all experiments were performed on mid-trophozoite stage cultures (∼30 h postinfection) at 7 to 8% parasitemia. Cultures (200 μl) were incubated with test compounds (1 μM) for a further 5 h (i.e., from 30 to 35 h postinvasion) in 96-well plates. Four replicate incubations of each compound were conducted and analyzed. Untreated controls contained equivalent amounts of dimethyl sulfoxide (DMSO; as a vehicle), and additional “quench test” controls were prepared whereby the test compounds were added after the quenching step to allow detection of test compound-derived liquid chromatography-mass spectrometry (LC-MS) features that did not arise from biochemical metabolism within the cells. Each plate contained 10 or 20 test compounds in addition to positive controls (artemisinin and/or atovaquone) and negative controls (untreated DMSO and quench test controls described above). Incubations and extractions for the 100 test compounds (10 compounds with known antimalarial activity and 90 of the highest-priority compounds from the Malaria Box, including all of plate A and the first row of plate B) were performed in three separate batches, with the known antimalarial compounds and first 10 Malaria Box compounds in the first batch, the next 40 Malaria Box compounds in the second batch, and the following 40 Malaria Box compounds in the third batch.
Metabolite extraction for LC-MS metabolomics analysis.
After 5 h of incubation, culture medium was removed by aspiration, and the metabolism of the settled iRBCs was quenched by the addition of ice-cold phosphate-buffered saline (PBS). Subsequent steps were performed on ice. Cells were pelleted by centrifugation for 5 min at 1,000 × g, and the PBS supernatant was removed prior to the addition of 135 μl methanol (containing the internal standard compounds CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, CAPS [N-cyclohexyl-3-aminopropanesulfonic acid], and PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]) and rapid mixing by pipetting three times to extract iRBC metabolites. Samples were left on ice with gentle agitation for 60 min and then centrifuged at 3,000 × g to remove the insoluble material. Supernatants were transferred to glass high-performance liquid chromatography (HPLC) vials and stored (<4 months) at −80°C until analysis. An aliquot (10 μl) of each sample was combined to generate a pooled biological quality control (PBQC) sample, which was used to monitor downstream sample stability and analytical reproducibility and for metabolite identification purposes.
LC-MS metabolomics analysis.
Metabolite analysis was performed by LC-MS, using hydrophilic interaction liquid chromatography (HILIC) and high-resolution (orbitrap) MS. Samples (10 μl) were injected onto a Dionex RSLC U3000 LC system (Thermo) fitted with a ZIC-pHILIC column (5 μm, 4.6 by 150 mm; Merck) and 20 mM ammonium carbonate (A) and acetonitrile (B) as the mobile phases (30). A 30 min gradient starting from 80% B to 40% B over 20 min, followed by washing at 5% B for 3 min and reequilibration at 80% B, was used (23). Mass spectrometry utilized a Q-Exactive MS (Thermo) with a heated electrospray source operating in positive and negative modes (rapid switching) and a mass resolution of 35,000 from m/z 85 to 1,050. The instrument was cleaned (source only) and calibrated on a weekly basis, with a mass accuracy of <2 ppm. Conditioning was performed before each batch by using 2 or 3 blanks, 5 mixtures of authentic standards (234 metabolites), and 2 pooled extracts, which were analyzed in a data-dependent tandem mass spectrometry (MS/MS) mode to facilitate downstream metabolite identification where necessary. PBQC samples were analyzed periodically throughout each batch and all batches were analyzed sequentially, with the exception of the batch containing compounds A2 to A11, which was analyzed ∼1 month after the other batches. Samples within each batch were sorted according to blocks of replicates and randomized to avoid any impact of systematic instrument drift on metabolite signals. Retention times for all authentic standards were checked manually for each batch to aid metabolite identification.
Data analysis for LC-MS metabolomics analysis.
Metabolomics data were analyzed using IDEOM. Briefly, this involves conversion of raw files to mzXML with msconvert (31), extraction of LC-MS peak signals with the Centwave algorithm in XCMS (32), alignment of samples and filtering artifacts with mzMatch (33), and additional data filtering and metabolite identification (34). All parameters are included in the supplemental IDEOM file (see Data Set S1 in the supplemental material). Manual inspection of total ion chromatograms allowed exclusion of outlier samples, which accounted for only 4% of the 500 samples analyzed. Additional manual data filtering was performed to remove features with very low quality chromatographic peaks, features that were not reliably detected across replicates, and signals that arose directly from the test compounds. Metabolite identification (level 1 confidence according to the Metabolomics Standards Initiative [MSI]; confidence score of 10 in the IDEOM file in Data Set S1 in the supplemental material) was based on accurate mass and retention time for metabolites that were present in the standard mixture. Other features were putatively annotated (MSI level 2) based on accurate mass and predicted retention time using the IDEOM database (35). The final data set consisted of 460 putatively identified metabolites, from a total of ∼1,500 unique mass features. The annotated metabolites were consistently detected across batches, whereas many of the unidentified features appear to be contaminant signals that were specific to certain batches or samples. Metabolite abundance was determined by LC-MS peak height and is normalized to the average for untreated samples from the same plate. Statistical analyses utilized Welch's t test (α = 0.05) and Pearson's correlation (Microsoft Excel), as well as hierarchical clustering analysis (HCA) and principal-component analysis (PCA) using the Metabolomics package in R (36). All LC-MS data are included in the supplemental material (see Data Set S1; the full IDEOM file is available at https://dx.doi.org/10.4225/03/57A80CE1503CD) and were deposited in the NIH Metabolomics Workbench under accession no. 650.
GC-MS analyses for Malaria Box screening.
Synchronized mid-trophozoite stage (∼32 h postinvasion) cultures (2 ml containing 10% parasitemia at 4% hematocrit) were treated with test compounds (1 μM), chloroquine (1 μM, positive control), and DMSO (0.1% [vol/vol], negative control) in triplicate 24-well plates and incubated for 12 h. After incubation, cells were washed with cold HEPES-buffered saline (20 mM HEPES, 20 mM MES [morpholineethanesulfonic acid], 154 mM NaCl) and the metabolites were extracted as described previously (37). Polar metabolite extracts were transferred and dried in a gas chromatography (GC) vial insert, washed twice with methanol, and derivatized by methoximation and trimethylsilylation prior to GC-MS analysis (37). GC-MS data were analyzed using IDEOM with default settings for GC-MS analysis with low-resolution MS (34) (see Data Set S2 in the supplemental material). Seventy metabolites were identified based on the unique fragmentation profiles and retention time and validated using authentic standards. All metabolite peak intensities were subjected to natural logarithmic transformation (36), and the data were normalized to data for untreated samples in each plate. Pearson's correlation and Student's t test (Microsoft Excel; α = 0.05), as well as HCA (Metabolomics package in R), were performed for statistical analyses.
Bicarbonate labeling of pyrimidine pathway intermediates.
Parasite enrichment was performed using a custom-made magnetic apparatus (38) and LD columns (Miltenyi Biotec, Bergisch Gladbach, Germany) to obtain >95% purity of mid-trophozoite stage-infected RBCs, followed by 30 min of recovery in fresh medium prior to experimentation. Aliquots of purified infected RBCs (1 × 108 cells) were treated with either tested compounds (5× 50% inhibitory concentration [IC50]), atovaquone (10 nM, positive control), or DMSO (0.1% [vol/vol], negative control) for 2 h at 37°C and then incubated for an additional 30 min with [13C1]bicarbonate (2 g/liter). Cells were washed with cold PBS and extracted with acetonitrile (80% [vol/vol], 200 μl). Insoluble material was removed by centrifugation, and the supernatant was transferred to an LC glass insert prior to LC-MS analysis (23). LC-MS data were processed using MAVEN (39). Metabolite peaks were identified by accurate mass and retention time, and the area top was used to calculate the abundance and fractional labeling. Each compound was tested in duplicate within experiments and independently repeated at least twice.
Analysis of activity in yeast DHODH mutant strain.
The dihydroorotate dehydrogenase (DHODH) coding sequence from Saccharomyces cerevisiae (yDHODH) was amplified from the plasmid pUF1-Cas9 (40) with the primers DHODH_F (ATATCAGCTCGAGGTCCCATGGCAGCCAGTTTAACT) and DHODH_R (TTAAATCTGCAGTTAAATGCTGTTCAACTTCCCA), digested with XhoI and PstI, and ligated into the human dihydrofolate reductase (hDHFR)-selectable plasmid pEF-Luc-GFP-HA (41) digested with the same enzymes. The yDHODH plasmid was then electroporated into erythrocytes that were fed to P. falciparum 3D7 parasites and selected with 2.5 nM WR99210 as described previously (42). The transgenic yDHODH-expressing parasites were maintained under drug pressure (2.5 mM WR99210) and washed once with RPMI medium to remove WR99210 prior to drug activity assays. A fluorescence-based assay was performed to compare the levels of inhibition of parasite growth on wild-type 3D7 and transgenic yDHODH-expressing clones, using the SYBR green I assay described previously (43), with minor modifications as follows: synchronized ring stage cultures (0.5% parasitemia at 2% haematocrit) were seeded into 96-well plates containing 2-fold serial dilutions of tested compounds, DMSO (0.1% [wt/vol], untreated control), and chloroquine (250 nM, positive control) and incubated for 48 h. After incubation, cells were lysed and stained with SYBR green I (0.1 μl/ml). Fluorescence was assessed with a plate reader (FLUOstar Omega; BMG Labtech), with excitation and emission wavelengths of 485 and 590 nm, respectively. Each compound was tested in duplicate within experiments, and experiments were independently replicated at least three times.
RESULTS AND DISCUSSION
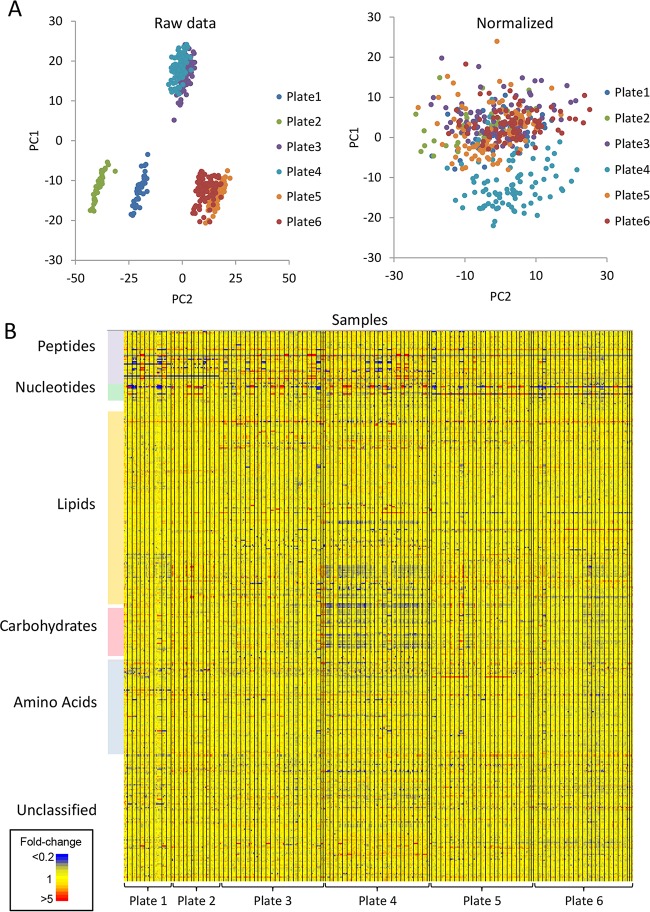
We have established a robust, medium-throughput, high-coverage metabolomics screen to investigate the mode of action of novel antimalarials and used this approach to analyze 90 compounds from the MMV Malaria Box collection (6). The key features of the screen are the use of the 96-well format, the analysis of unfractionated cultures, and the use of high-sensitivity LC-MS to reproducibly detect 460 putative metabolites from a range of metabolic pathways. Initial multivariate analysis of the raw data revealed significant batch effects, which are largely unavoidable in large metabolomics studies (36) and may be due to a combination of factors, including the batch of erythrocytes used for culture, subtleties of the cell preparation and incubation conditions (although this was tightly controlled), and the response of the LC-MS system. Normalization of data based on the untreated control samples in each plate minimized the impact of batch effects on the overall structure of the data (Fig. 1; see Fig. S1 in the supplemental material). After normalization, systematic variation was still observed in a small subset of drug treatments (Malaria Box compounds D2 to D10 in plate A) as a result of the decreased abundance of ∼10% of the metabolites. The reason for this systematic variation is not clear but may reflect slight differences in culture incubation conditions, as the analytical quality control procedures indicated no systematic change in LC-MS response for these samples. These data have been retained in the data set, as they did not obscure the detection of drug-induced perturbations. The majority of metabolites in the final normalized data set showed consistent responses across the 500 samples, which allowed treatment-specific metabolic perturbations to be observed (Fig. 1B).
FIG 1.
LC-MS metabolomics data from P. falciparum-infected erythrocytes treated with 100 antimalarial compounds (n = 4). Plate 1 contained 10 known drugs and metabolic inhibitors; plates 2 to 6 contained 90 Malaria Box compounds. (A) Principal-component analysis (PCA) of metabolite abundances from all samples before and after normalization. Samples from plates 3 and 4 were prepared and analyzed as a single batch on the same day. Samples from plates 5 and 6 were also prepared and analyzed as a single batch. (B) Heat map of metabolite (y axis) abundances in all samples (x axis; replicates are adjacent and different treatments are separated by vertical black lines). Yellow features indicate no significant perturbation of metabolite levels relative to untreated controls. Increased and decreased metabolite levels in treated cells are represented by red and blue, respectively.
Known drugs and inhibitors.
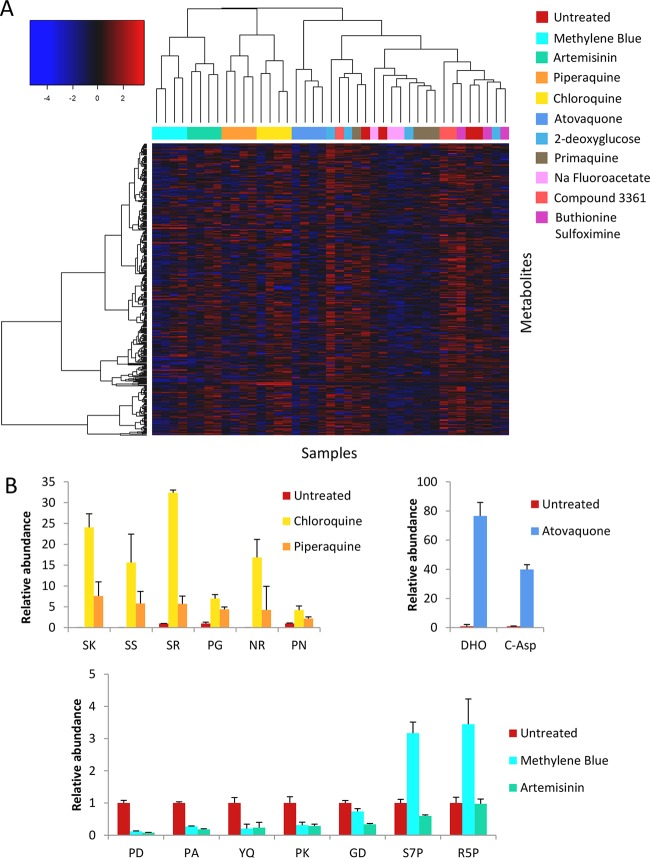
Trophozoite stage P. falciparum cultures were incubated with test compounds (1 μM) for 5 h, in order to identify early drug-induced changes in metabolism that preceded nonspecific cell death. Hierarchical clustering analysis (HCA) of LC-MS metabolite data from samples treated with the known drugs revealed clustering of replicates for each of the compounds with submicromolar activity (Fig. 2A). In particular, chloroquine and piperaquine clustered closely together, confirming a conserved mechanism of action for these related 4-aminoquinoline compounds. The major perturbation induced by the quinolines was the accumulation of a subset of dipeptides (Fig. 2B). Artemisinin also induced significant changes in a number of small peptides, although these changes were clearly different from the perturbations induced by the quinolines (Fig. 2B). Methylene blue revealed a metabolic fingerprint similar to that of artemisinin but also induced accumulation of the pentose phosphate pathway metabolites, ribose 5-phosphate and sedoheptulose 7-phosphate, consistent with the proposal that this compound may induce oxidative stress (44). As expected, atovaquone induced selective changes in metabolites involved in pyrimidine biosynthesis (22, 23). The increase in levels of N-carbamoyl-l-aspartate and dihydroorotate in atovaquone-treated cultures reflects the indirect inhibition of dihydroorotate dehydrogenase activity following primary inhibition of ubiquinol oxidation by the cytochrome bc1 complex (22). Interestingly, primaquine and the metabolic inhibitors, buthionine sulfoximine, 2-deoxyglucose, compound 3361 (45), and sodium fluoroacetate did not elicit reproducible metabolic fingerprints under these conditions (Fig. 2A). However, the growth inhibition IC50s for each of these compounds against P. falciparum are all greater than the 1 μM concentration used for this screen (see Table S1 in the supplemental material). These results confirm the capability of this methodology to detect relevant metabolic changes resulting from drug treatment rather than nonspecific metabolic variance at subtherapeutic drug concentrations.
FIG 2.
(A) Hierarchical clustering analysis (HCA) with a heat map of log-transformed relative metabolite abundances from LC-MS analysis of P. falciparum-infected erythrocytes treated with known antimalarial drugs and inhibitors (n = 3 or 4). Clustering of replicates was observed for the most potent antimalarials: chloroquine (yellow), piperaquine (orange), artemisinin (green), methylene blue (aqua), and atovaquone (dark blue). Other samples included primaquine (brown), 2-deoxyglucose (light blue), sodium fluoroacetate (pink), buthionine sulfoximine (purple), compound 3361 (red), and DMSO controls (maroon). (B) Relative abundance (mean peak height ± standard deviation, expressed relative to an untreated control; n = 3 or 4) of significantly perturbed metabolites (α = 0.05) following treatment with chloroquine, piperaquine, atovaquone, methylene blue, or artemisinin (note that SK, SS, and NR were not detected in the untreated control and are expressed relative to a minimum detectable intensity of 10,000). DHO, dihydroorotate; C-Asp, N-carbamoyl-l-aspartate; S7P, sedoheptulose 7-phosphate; R5P, ribose 5-phosphate; other metabolites are dipeptides as indicated.
Multivariate analysis reveals compounds that induce artemisinin-like metabolic disruption.
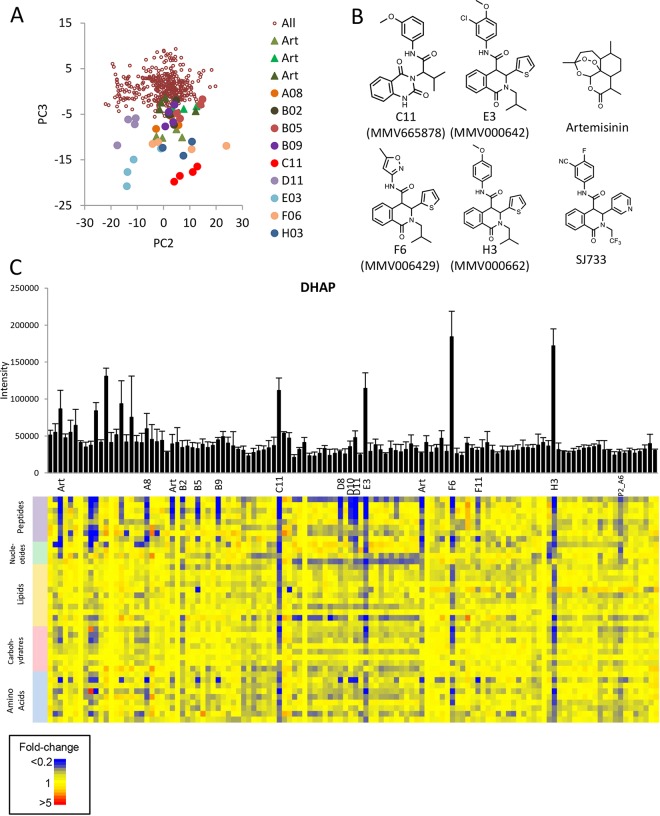
Ninety representative Malaria Box compounds were screened using untargeted metabolomics under the same conditions as described above (1 μM for 5 h). This compound collection includes 40 “probe-like” compounds and 40 “drug-like” compounds deemed as high priority during compilation of the Malaria Box (6). A multivariate analysis of the LC-MS metabolomics data set using principal-component analysis (PCA) did not reveal any treatment-specific perturbations in the first two principal components, indicating that stochastic biological and experimental variation accounted for the greatest component of variance in the full data set. Interestingly, the plot of scores revealed grouping of replicates from 13 Malaria Box compounds with artemisinin in the third principal component (PC3) (Fig. 3A). Four of the Malaria Box compounds (C11, MMV665878; E3, MMV000642; F6, MMV006429; H3, MMV000662) (Fig. 3B) that clustered along this third principal component induced greater metabolic perturbation than artemisinin (Fig. 3C). Nine other Malaria Box compounds induced the same metabolic phenotype, albeit with less-pronounced changes to metabolite levels, as seen by clustering in PC3 and a positive Pearson correlation coefficient for a metabolome-wide correlation with artemisinin (Table 1). The 40 metabolites with highest loadings in PC3 revealed significant depletion of diverse metabolites, including peptides, nucleotides, lipids, and sugars (Fig. 3C). The greatest decreases were observed for specific dipeptides (annotated as PD, PA, PK, YQ, and GD), which likely arise from disruption of hemoglobin breakdown, as well as the metabolites pipecolate and lactate. No metabolites were significantly increased in abundance for any of the compounds in this cluster, although choline phosphate accumulated significantly in D11 (MMV665831) and artemisinin-treated parasites (see Fig. S2 in the supplemental material) and dihydroxyacetone phosphate (DHAP) levels were higher for the four most potent compounds (C11, E3, H3, and F6). The perturbations observed for compounds in this cluster are consistent with, and expand on, the pathways observed in our previous metabolic analysis of dihydroartemisinin action (23), reflecting the increased feature detection achieved with untargeted high-mass-resolution MS.
FIG 3.
(A) Principal-component analysis (PCA) scatterplot of PC2 versus PC3 built using all LC-MS metabolite data from all samples (excluding pooled biological quality controls and blanks). Artemisinin samples, from three separate experiments, are represented by triangles, and other compounds with low scores on PC3 are represented by large filled circles. (B) Structures of artemisinin, SJ733, and the four Malaria Box compounds that showed the greatest impact on metabolism in this study. (C) Relative intensities (mean peak height + standard deviation) of DHAP in all samples and a heat map of the 40 most significantly perturbed metabolites in principal component 3 (loadings > 0.06).
TABLE 1.
Key LC-MS and GC-MS data demonstrating classification of Malaria Box compounds according to the three major mechanistic clusters based on correlation of all metabolite levels (LC-MS) and levels of selected metabolites (GC-MS)
| Mechanistic cluster and compound | HEOS identifier | LC-MS dataa correlation (r) with: |
GC-MS data (abundance relative to that of ctrl)b |
|||||
|---|---|---|---|---|---|---|---|---|
| Artemisinin | Quinoline | Atovaquone | Pipecolate | Putrescine | Fumarate | GABA | ||
| Artemisinin-like | ||||||||
| Artemisinin (plate 1) | 0.77 | −0.32 | −0.23 | 0.10 | 0.49 | 1.15 | 0.32 | |
| Artemisinin (plate 3) | 0.73 | −0.10 | −0.17 | |||||
| Artemisinin (plate 5) | 0.72 | −0.16 | −0.23 | |||||
| A8 | MMV019406 | 0.47 | −0.31 | 0.004 | 0.40 | 0.81 | 0.86 | 1.05 |
| B2 | MMV006427 | 0.57 | −0.17 | −0.20 | 0.21 | 0.50 | 1.01 | 1.63 |
| B5 | MMV000570 | 0.48 | −0.17 | −0.15 | 0.25 | 0.77 | 0.96 | 1.08 |
| B9 | MMV085583 | 0.42 | −0.17 | −0.06 | 0.16 | 0.67 | 0.74 | 0.82 |
| C11 | MMV665878 | 0.52 | −0.10 | −0.12 | 0.16 | 1.02 | 1.06 | 0.43 |
| D8 | MMV020788 | 0.34 | −0.15 | −0.08 | 0.16 | 0.42 | 0.92 | 0.40 |
| D10 | MMV665785 | 0.39 | −0.10 | −0.08 | 0.13 | 0.55 | 0.99 | 0.40 |
| D11 | MMV665831 | 0.47 | −0.13 | −0.03 | 0.11 | 0.65 | 1.18 | 0.38 |
| E3 | MMV000642 | 0.32 | −0.06 | −0.02 | 0.09 | 0.34 | 1.34 | 0.49 |
| F6 | MMV006429 | 0.50 | −0.12 | −0.17 | 0.09 | 0.37 | 1.40 | 0.63 |
| F11 | MMV665841 | 0.61 | −0.19 | −0.31 | 0.07 | 0.32 | 1.58 | 0.42 |
| H3 | MMV000662 | 0.46 | −0.09 | −0.19 | 0.10 | 0.41 | 1.34 | 0.71 |
| MB2_A6 | MMV007564 | 0.40 | −0.18 | −0.19 | ND3 | ND | ND | ND |
| Quinoline-like | ||||||||
| Chloroquine | −0.29 | 0.98 | −0.04 | 0.08 | 0.11 | 1.14 | 0.41 | |
| Piperaquine | −0.08 | 0.78 | −0.03 | 0.11 | 0.14 | 1.71 | 0.69 | |
| B8 | MMV019871 | −0.26 | 0.44 | 0.004 | 0.76 | 1.01 | 0.74 | 0.99 |
| C4 | MMV020549 | −0.14 | 0.50 | −0.04 | 0.19 | 0.43 | 1.15 | 0.38 |
| C9 | MMV000448 | −0.26 | 0.57 | −0.06 | 0.27 | 0.60 | 1.08 | 0.42 |
| C10 | MMV020500 | −0.32 | 0.58 | −0.04 | 0.13 | 0.77 | 1.04 | 0.39 |
| E5 | MMV006172 | −0.36 | 0.66 | −0.02 | 0.15 | 0.14 | 1.56 | 0.48 |
| E7 | MMV006087 | −0.35 | 0.65 | −0.03 | 0.09 | 0.09 | 1.26 | 0.33 |
| Pyrimidine synthesis inhibitors | ||||||||
| Atovaquone (plate 1) | −0.26 | −0.04 | 0.97 | 1.06 | 0.84 | 2.23 | 2.43 | |
| Atovaquone (plate 2) | −0.27 | −0.06 | 0.99 | |||||
| Atovaquone (plate 4) | −0.29 | −0.03 | 0.99 | |||||
| A4 | MMV396680 | −0.21 | −0.16 | 0.94 | 0.89 | 0.92 | 1.23 | 1.02 |
| A6 | MMV008294 | −0.27 | −0.06 | 0.99 | 1.15 | 0.97 | 2.59 | 2.26 |
| A7 | MMV011259 | −0.26 | −0.05 | 0.997 | 1.21 | 1.20 | 5.01 | 6.00 |
| B6 | MMV020439 | −0.28 | −0.05 | 0.90 | 0.67 | 0.93 | 1.67 | 2.15 |
| B11 | MMV665874 | −0.28 | −0.03 | 0.98 | 1.21 | 1.22 | 4.86 | 5.37 |
| C3 | MMV665977 | −0.29 | −0.05 | 0.96 | 0.74 | 0.78 | 3.06 | 1.26 |
| C8 | MMV007695 | −0.10 | −0.11 | 0.81 | 1.10 | 1.09 | 1.28 | 1.34 |
| D3 | MMV666596 | −0.29 | −0.03 | 0.99 | 1.17 | 1.09 | 1.58 | 1.47 |
| D4 | MMV396679 | −0.26 | −0.03 | 0.98 | 1.12 | 0.98 | 1.75 | 1.28 |
| D9 | MMV666691 | −0.29 | −0.04 | 0.98 | 0.96 | 0.98 | 2.96 | 1.49 |
| E2 | MMV011099 | −0.29 | −0.03 | 0.996 | 0.95 | 0.91 | 3.36 | 3.58 |
| E6 | MMV006309 | −0.26 | −0.04 | 0.97 | 0.89 | 0.94 | 2.97 | 3.89 |
| E11 | MMV665876 | −0.28 | −0.03 | 0.98 | 1.13 | 0.99 | 3.48 | 3.89 |
| F2 | MMV666023 | −0.18 | −0.05 | 0.74 | 0.79 | 0.69 | 2.24 | 2.34 |
| G2 | MMV007116 | −0.20 | −0.04 | 0.76 | 1.24 | 0.85 | 3.32 | 3.15 |
| G5 | MMV019258 | −0.21 | −0.03 | 0.72 | 1.16 | 1.06 | 3.64 | 3.18 |
| G7 | MMV011256 | −0.21 | −0.03 | 0.72 | 1.17 | 1.04 | 4.49 | 3.85 |
| G8 | MMV666693 | −0.21 | −0.03 | 0.72 | 1.03 | 0.94 | 3.79 | 3.52 |
| G10 | MMV665827 | −0.21 | −0.03 | 0.71 | 1.17 | 0.96 | 2.82 | 3.19 |
| H6 | MMV006457 | −0.22 | −0.08 | 0.97 | 1.05 | 1.01 | 1.40 | 1.38 |
| H7 | MMV396693 | −0.20 | −0.12 | 0.93 | 0.99 | 0.93 | 2.59 | 1.43 |
| H11 | MMV666021 | −0.25 | −0.10 | 0.96 | 1.06 | 1.01 | 1.21 | 1.03 |
| MB2_A4 | MMV666103 | −0.27 | −0.03 | 0.95 | ND | ND | ND | ND |
| MB2_A9 | MMV000563 | −0.28 | −0.03 | 0.98 | ND | ND | ND | ND |
Pearson correlation between relative metabolite abundances for compounds compared to artemisinin (using average values from the three independent artemisinin experiments), quinolines (using average values from chloroquine and piperaquine experiments), and atovaquone (using average values from the three independent atovaquone experiments). Positive correlations (r > 3) are shown in bold.
Abundance of metabolites based on GC-MS peak area relative to that of untreated control. Statistically significant changes are shown in bold (Student's t test; α = 0.05, n = 4). ctrl, control; ND, not determined.
The specific mode of action of the artemisinins is a matter of debate but is thought to involve iron-mediated activation of the peroxide bond to produce free radicals that induce nonspecific parasite damage (46). The significant impact of artemisinin on intracellular levels of hemoglobin peptides and other metabolites is consistent with specific inhibition of hemoglobin digestion and nonspecific inhibition of a number of pathways. However, it is notable that the 13 Malaria Box compounds that induced a metabolic phenotype similar to that of the artemisinins are structurally unrelated and do not possess the endoperoxide moiety that is critical for artemisinin activity. Interestingly, the four compounds in this PC3 cluster that had the greatest impact on metabolism are structurally related to each other and are analogues of the dihydroisoquinolone SJ733, which targets the cation ATPase PfATP4 and is currently undergoing preclinical development (Fig. 3B) (47). These four compounds, in addition to B2 (MMV006427; which also clusters in PC3), were previously shown to inhibit PfATP4, suggesting a conserved mode of action (17). Interestingly, two structurally distinct PfATP4 inhibitors, H8 (MMV011567) and H10 (MMV665805) (chemotype II according to Lehane et al. [17]), did not induce significant metabolic disruption under the conditions tested here.
Both the artemisinins and the PfATP4 inhibitors are potent, rapid-acting antimalarials. While it is possible that the Malaria Box compounds that cluster with artemisinin in PC3 share a similar mode of action, we propose that this metabolic profile may be a feature of compounds that rapidly mediate cell death rather than an indicator of a specific drug target or pathway. It is also notable that the PC3 score of these compounds did not correlate with the published EC50 for these compounds against asexual P. falciparum iRBCs in vitro. However, standard 50% effective concentration (EC50) assays measure parasite growth over 48 or 72 h, and our assay involving a 5-h treatment will likely exhibit a bias toward detecting changes induced by fast-acting parasiticidal compounds. We therefore propose that all of the compounds in this “artemisinin-like” cluster lead to rapid, significant, and widespread disruption of parasite metabolism and may therefore represent promising starting points for the discovery of novel, rapid-acting parasiticidal drug candidates.
A complementary GC-MS-based metabolomics analysis was performed after incubation with test compounds for 12 h, which allowed analysis of 70 metabolites from central metabolic pathways. Although this analysis did not reveal the extent of metabolic perturbation observed in the LC-MS study, distinct clustering was still observed in HCA analysis (see Fig. S3 in the supplemental material), with pipecolate and putrescine depletion contributing to the clustering of many of these artemisinin-like compounds in the GC-MS data set (Table 1). Unlike the LC-MS data set, this cluster also included quinoline-like compounds (see below) and chemotype II PfATP4 inhibitors (H8 and H10), which may reflect a slower onset of antiparasitic activity detected following the longer incubation in this study design.
Malaria Box compounds with a quinoline-like profile.
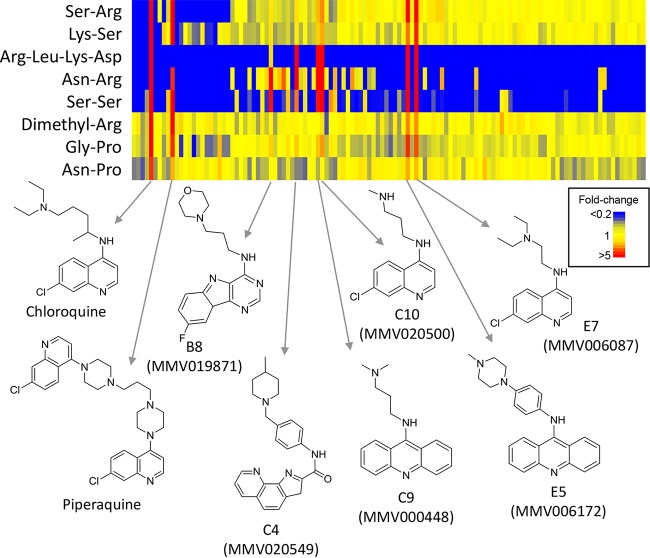
Univariate analysis of the LC-MS data set was systematically performed across all treatments to identify unique metabolic perturbations associated with specific compounds. Six of the Malaria Box compounds resulted in extensive accumulation of a subset of peptides, which were also significantly accumulated in chloroquine- and piperaquine-treated cultures (Fig. 4). These peptides were putatively identified as SK, SS, SR, PG, NR, PN, and DRLK. As data-dependent MS-MS confirmation was not obtained due to the low absolute abundance of these peptides, other isomeric sequences are possible. Interestingly, chloroquine resistance has previously been shown to be associated with perturbations in hemoglobin-derived peptides (48). However, none of the peptides reported in the chloroquine resistance study accumulated in the quinoline-treated cultures. Unexpectedly, only two of the unique peptides detected following quinoline treatment could have been derived from hemoglobin, SK (or KS) and NP (or PN). The precise source of each peptide cannot be determined from such short sequences, but neither SS, SR, GP, nor NR (or their isomers) is present in the sequence of hemoglobin. The accumulation of these peptides suggests that the quinolines and related Malaria Box compounds are selectively inhibiting specific aspects of protein degradation and that hemoglobin metabolism itself does not appear to be the primary target. The possibility that these compounds interfere with protein degradation was supported by increased levels of the modified amino acid dimethylarginine (DMA) following treatment. Methylation of arginine normally only occurs on intact proteins, and increased levels of DMA most likely reflects increased protein turnover. The accumulation of these seven peptides and DMA was specific to only 6 of the 90 tested Malaria Box compounds, and these metabolites were either not detected or not significantly perturbed in samples from the other 84 compounds. The high correlation of metabolite abundances for these six compounds (E7, E5, C9, C10, B8 and C4) (Table 1) with chloroquine and piperaquine suggests that they act by a conserved mechanism. The quinoline-like activity of these six compounds is not surprising, as two of these structures are quinolines, two contain tricyclic quinoline-like moieties, and the other two possess substituted planar heteroaromatic ring systems that may align with the quinoline pharmacophore (Fig. 4).
FIG 4.
Heat map of metabolites from the LC-MS data set that were significantly perturbed by chloroquine and the quinoline-like compounds (α = 0.05). Data shown are the average for 4 replicates for all compounds (horizontal axis) and are expressed relative to the average for the untreated controls in the same plate (or relative to a minimum detectable intensity of 10,000 if not detected in untreated controls).
Malaria Box compounds that inhibit pyrimidine biosynthesis.
A significant number of Malaria Box compounds induced changes in pyrimidine biosynthesis. Specifically, nearly one quarter of the compounds tested increased intracellular levels of pyrimidine intermediates N-carbamoyl-l-aspartate and dihydroorotate (Fig. 5A). These two metabolites are present in P. falciparum at concentrations near the lower limit of detection in untreated cultures, but the corresponding LC-MS peaks increased 2- to 100-fold following incubation with each of these specific compounds. This metabolic perturbation is strikingly similar to that observed for atovaquone and is consistent with inhibition of dihydroorotate dehydrogenase (DHODH) (Fig. 5B). A significant increase in 4-aminobutanoate (GABA) was also observed, but few other metabolites were affected by these compounds. Pearson correlation of all metabolite abundances confirmed the high correlations between these Malaria Box compounds and atovaquone (r > 0.7) (Table 1). This correlation was predominantly associated with the N-carbamoyl-l-aspartate and dihydroorotate levels.
FIG 5.
(A) Relative abundance (peak height) of dihydroorotate and N-carbamoyl-l-aspartate in P. falciparum-infected erythrocytes after treatment with all compounds (mean ± standard deviation; n = 4). Compounds that induced significant (α = 0.05) accumulation are labeled (see Table 1 for MMV identifiers). (B) Simplified schematic of the de novo pyrimidine biosynthesis pathway indicating the targets of atovaquone and DSM265. (C) 13C enrichment from [13C1]bicarbonate in pyrimidine intermediates following 2 h of treatment with atovaquone, G5, G7, and G8, expressed as the percent difference from drug-free controls. Metabolite abundance in these samples, relative to drug-free controls, is shown in the lower panels. (D) Concentration-response profiles for representative pyrimidine biosynthesis inhibitors and atovaquone in 48-h growth inhibition assays using wild-type 3D7 (blue) and yDHODH-expressing (red) P. falciparum iRBCs.
The identification of these compounds with metabolic phenotypes indicative of pyrimidine synthesis inhibition was further confirmed in the GC-MS data set. Although GC-MS is not suited to the detection of N-carbamoyl-l-aspartate or dihydroorotate directly, the accumulation of GABA was confirmed. In addition, atovaquone-induced fumarate accumulation was observed in the GC-MS data, consistent with our previous report (23) (note that the fumarate signal was discarded from this LC-MS data set due to coelution with an isobaric malate fragment). Significant accumulation of fumarate and/or GABA was observed by GC-MS for 17 of the pyrimidine synthesis inhibitors that were identified with the LC-MS platform (Table 1).
The mechanism of inhibition of pyrimidine biosynthesis was further confirmed by stable-isotope labeling of enriched trophozoite stage iRBCs with [13C1]bicarbonate for 2 h. In agreement with our previous work (23), incubation with atovaquone induced significantly lower 13C incorporation into UMP than was observed in untreated iRBCs, while labeling of the accumulated precursors N-carbamoyl-l-aspartate and dihydroorotate was not inhibited (Fig. 5C). Analysis of representative pyrimidine biosynthesis inhibitors from the Malaria Box revealed the same specific reduction in labeling of UMP and confirmed the accumulation of N-carbamoyl-l-aspartate and dihydroorotate, demonstrating functional inhibition of DHODH activity by structurally diverse compounds G8 (MMV666693, an atovaquone analogue), G7 (MMV011256, a DSM265 analogue), and G5 (MMV019258, a unique chemotype).
Additional growth inhibition assays in P. falciparum expressing yeast DHODH (yDHODH) were performed in order to determine whether inhibition of pyrimidine biosynthesis is responsible for the antimalarial activity of compounds in this cluster. Unlike DHODH in P. falciparum, the yDHODH is located in the cytosol and is not dependent on reduced ubiquinone derived from the mitochondrial electron transport chain (49). Therefore, parasites expressing yDHODH are resistant to the antimalarial compounds that act by inhibition of DHODH, either directly or via mitochondrial inhibition. Indeed, these transfectant parasites were resistant to most of the Malaria Box compounds that were predicted to act by inhibition of pyrimidine biosynthesis (Fig. 5D; see Table S2 in the supplemental material), whereas no change in the EC50 was observed for representative compounds from the “artemisininlike” and “quinoline-like” clusters (see Table S2). This finding provides independent validation that the metabolomics screening approach was able to successfully identify the mode of action for compounds that target pyrimidine biosynthesis. Interestingly, two compounds from the “atovaquone-like” cluster (F2 and H7) exhibited similar EC50s in the wild-type and yDHODH strains (see Table S2), suggesting that inhibition of pyrimidine synthesis is not the primary mode of action for these two compounds. F2 (MMV666023) was also identified in independent screens for inhibitors of deoxyhypusine hydroxylase (11) and metalloaminopeptidase activity (9). This compound is very hydrophobic (logP, 8.7) and contains an azo group, which has previously been identified as a problematic feature in high-throughput screens (50). Although pyrimidine biosynthesis inhibition is the predominant metabolic impact of F2, it is likely that the chelating ability of this compound is responsible for inhibition of multiple targets within the cell, indicating a pleiotropic mechanism of action for F2 that cannot be rescued by yDHODH alone.
The high prevalence of the atovaquone-like metabolic phenotype in the Malaria Box collection may be a reflection that pyrimidine biosynthesis is a critical aspect of metabolism in P. falciparum and is an attractive drug target, either through direct or indirect inhibition. As for atovaquone, it is likely that this metabolic phenotype will be observed for all compounds that inhibit early complexes in the electron transport chain or potentially other aspects of mitochondrial metabolism. Direct inhibition of DHODH is a validated drug target, and the triazolopyrimidine DHODH inhibitor DSM265 is currently in early stage clinical testing (51). Three of the compounds in the first plate of the Malaria Box are close structural analogues of DSM265 (A7, B11, and G7) (see Fig. S4). All three of these triazolopyrimidines induced this atovaquone-like metabolic phenotype, confirming that direct inhibition of DHODH induces the same metabolic perturbation as the indirect inhibition observed with atovaquone. In addition, four other compounds in this cluster appear structurally related to DSM265, including another triazolopyrimidine (E2), a triazolothiadiazole (E11), and two pyrazolopyrazines (A4 and D4), which may also target DHODH directly. No close analogues of atovaquone are present among these Malaria Box compounds, but eight compounds in this pyrimidine synthesis inhibitor cluster are structurally related benzoxazinones (G8 and E6), oxoquinolines (C3 and G10), hydroxyquinolines (G2 and H6), oxoquinazolines (MB2_A4), or benzodiazoles (B6), and further studies are warranted to determine whether these analogues also target cytochrome bc1. Importantly, several unrelated chemotypes are also present in this cluster, indicating that pyrimidine biosynthesis can be inhibited, either directly or indirectly, by unique molecules that would be unlikely to be affected by mutations in the target proteins that confer resistance to atovaquone or DSM265 (51, 52).
Identification of a specific inhibitor of isoprenoid precursor biosynthesis.
In addition to the alignment of metabolic profiles with those induced by treatment with existing antimalarial drugs, unique metabolic disturbances were observed that might reveal novel mechanisms of action. Treatment with D6 (MMV008138) led to the selective decrease in abundance of a metabolite annotated as 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (cMEPP). Decreased cMEPP levels were not induced by other Malaria Box compounds, with the exception of the four compounds that induced widespread metabolic disruption (C11, E3, H3, and F6) (Fig. 6A). cMEPP is an intermediate in isopentenyl diphosphate (IPP) biosynthesis and is synthesized by the apicoplast-targeted enzyme 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (IspF), the fifth enzyme in the IPP pathway. The depletion of cMEPP indicates that D6 may be an inhibitor of IspF itself or may affect other steps of the IPP pathway. Enzymes involved in IPP biosynthesis are validated drug targets and existing IPP inhibitors, such as fosmidomycin (inhibitor of 1-deoxyxylulose 5-phosphate reductoisomerase), also lead to depletion of cMEPP (21). Recent studies have provided independent confirmation that D6 inhibits IPP biosynthesis (12–14). Bowman et al. (12) utilized an isoprenoid-supplemented culture medium to test the activity of Malaria Box compounds and identified D6 as the only compound that displayed reduced activity in the supplemented medium compared to that in standard culture conditions. Wu et al. (13) replicated this and went on to show that D6 directly inhibits 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase (IspD), the third enzyme in the IPP pathway, and that IspD mutation confers resistance to this compound. Imlay et al. (14) confirmed these findings and showed that D6 competes with the CTP-binding site of IspD. These studies confirm that D6 targets the production of IPP intermediates in the apicoplast and provide independent evidence for the mechanism of cMEPP depletion observed in this metabolomics screen.
FIG 6.
Relative abundance (LC-MS peak height on log scale) of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (A), traumatic acid (B), and linoleic acid (C) in P. falciparum-infected erythrocytes after treatment with each compound (mean ± standard deviation, n = 4). The standard deviation is not shown if it was beyond the lower axis limit).
Inhibition of unique aspects of parasite fatty acid metabolism.
Treatment with C7 (MMV665915) also resulted in a unique metabolic signature. Specifically, treatment with C7 led to a 10-fold depletion of a metabolite (Fig. 6B) with the molecular formula C12H20O4. The accurate mass and LC retention time of this metabolite are consistent with it being traumatic acid, which has previously been identified in P. falciparum (53). Traumatic acid is an end product of the plant-like alpha-linolenic acid (ALA) pathway in P. falciparum, although the function of traumatic acid in Plasmodium is not known (53). The specific depletion of traumatic acid in parasites treated with C7 suggests that enzymes in this pathway are potential drug targets. Few other metabolites were perturbed in C7-treated samples. The significant (∼2-fold) decreases in polyunsaturated lipids [phosphatidylcholines PC(38:4) and PC(36:3), phosphatidylethanolamines PE(36:4) and PE(38:5), and phosphatidylserine PS(20:4)] may be related to perturbations in the ALA pathway but should not be overinterpreted, as these represent only a small fraction of the polyunsaturated phospholipids in the data set and the phospholipid responses exhibit some inherent variability as they elute in the solvent front of this HILIC analytical method. On the other hand, the C7-induced depletion of traumatic acid was consistent between the four replicates and unique to C7. E4 (MMV666600) was the only other compound to induce significant depletion of traumatic acid, albeit to a lesser extent. The abundances of the ALA precursor for this pathway [annotated as FA(18:3) in the IDEOM file in Data Set S1 in the supplemental material] and the alternative end product of ALA metabolism, jasmonic acid, were not altered, indicating that C7 specifically inhibits the traumatic acid branch of the ALA pathway. Further targeted lipidomic studies are required to elucidate the impact of C7 (and E4) on ALA metabolism in greater detail.
The Malaria Box compound P2_A3 (MMV000634) also induced specific depletion of polyunsaturated fatty acids, in this case FA(18:2) (linoleic acid) (Fig. 6C). This perturbation was unique to P2_A3 and was not associated with changes to levels of other metabolites. It is difficult to determine the mechanism responsible for this effect, but it is possible that P2_A3 inhibits fatty acid uptake, or phospholipase activity, as P. falciparum appears to lack the specific desaturase enzyme responsible for biosynthesis of linoleic acid (54).
Impact of other compounds on metabolism.
Of the 90 Malaria Box compounds tested, approximately half induced metabolic profiles that correlated with either artemisinins, quinolines, or atovaquone. Aside from the IPP and ALA pathways mentioned above, few perturbations unique to other pathways were observed. The remaining compounds did not induce detectable metabolic changes that might be associated with their antimalarial activity. There are several reasons that might explain the lack of observable effects for those compounds: (i) the mechanism of antimalarial activity may be unrelated to metabolism, as drugs that target the cell through DNA, RNA, or protein synthesis, protein-protein interactions, signaling, structure, ion channels, or trafficking may not directly impact levels of endogenous metabolites in the short term; (ii) the duration (5 h) and concentration (1 μM) may not be sufficient to induce metabolic changes; (iii) the compound may act predominantly at a stage of the cell cycle different from that of the trophozoite stage tested here; or (iv) metabolites perturbed by test compounds may have been below the limit of detection (or were not reproducibly detected) with these experimental methods. The selected conditions with a predefined drug concentration and duration of incubation, in cultures with 8% parasitemia and 3% hematocrit in 96-well plates, provided the optimal balance of throughput and sensitivity for this screening approach. Nevertheless, further work utilizing a more extensive experimental methodology involving time course, dose-response studies of enriched parasites with stable isotope labeling and by using methods such as those described recently (23) may reveal additional metabolic effects of the Malaria Box compounds. Furthermore, the addition of reversed-phase LC-MS to the HILIC platform described here would enhance the detection of lipid metabolites, providing additional information about the metabolic effects of these compounds.
Conclusions.
The medium-throughput, high-coverage metabolomics screen described here provides extensive information regarding the impact of Malaria Box compounds on parasite metabolism. Interestingly, half of the tested compounds elicited metabolic profiles analogous to three known classes of antimalarials, namely, the artemisinins, the quinolines, and atovaquone. These findings suggest that many compounds identified in these phenotypic screens may directly or indirectly inhibit a disproportionately small number of cellular processes. The finding of a potential metabolic signature for artemisinin-like fast-acting drugs is of particular interest in terms of triaging new compounds. A number of new chemotypes which were linked to highly specific metabolic perturbations were observed, implicating them as inhibitors of nucleotide and lipid metabolism pathways. These data are freely available (in the supplemental material and in the NIH Metabolomics Workbench under accession no. 650) and should assist further mechanistic studies and prioritization of these compounds for the development of new antimalarial drugs.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Medicines for Malaria Venture (MMV) for providing access to the MMV Malaria Box and the Australian Red Cross Blood Bank for the provision of human blood. We thank Jonathan Baell (Monash University) for constructive comments regarding the chemical classification of compounds.
M.J.M. is an NHMRC Principal Research Fellow (APP1059530). S.A.R. is supported by an NHMRC R. D. Wright Biomedical Fellowship (APP1062504). D.J.C. is supported by an NHMRC R. D. Wright Biomedical Fellowship (1088855). This work was supported by funding from the Victorian Operational Infrastructure Support Program received by the Burnet Institute.
Funding Statement
This work was funded by Australian National Health and Medical Research Council grants to Malcolm J. McConville and Stuart A. Ralph (APP106024), Darren J. Creek (APP1088855; APP), and Stuart A. Ralph (APP1062504).
M.J.M. is an NHMRC Principal Research Fellow, D.J.C. is an NHMRC Career Development Fellow, and S.A.R. is an NHMRC R. D. Wright Fellow.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01226-16.
For a companion article on this topic, see doi:10.1128/AAC.01224-16.
REFERENCES
- 1.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, et al. . 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells TNC, Hooft van Huijsduijnen R, Van Voorhis WC. 2015. Malaria medicines: a glass half full? Nat Rev Drug Discov 14:424–442. doi: 10.1038/nrd4573. [DOI] [PubMed] [Google Scholar]
- 3.Gamo F-J, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera J-L, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. 2010. Thousands of chemical starting points for antimalarial lead identification. Nature 465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 4.Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jimenez-Diaz MB, Martinez MS, Wilson EB, Tripathi AK, Gut J, Sharlow ER, Bathurst I, El Mazouni F, Fowble JW, Forquer I, McGinley PL, Castro S, Angulo-Barturen I, Ferrer S, Rosenthal PJ, Derisi JL, Sullivan DJ, Lazo JS, Roos DS, Riscoe MK, Phillips MA, Rathod PK, Van Voorhis WC, Avery VM, Guy RK. 2010. Chemical genetics of Plasmodium falciparum. Nature 465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plouffe D, Brinker A, McNamara C, Henson K, Kato N, Kuhen K, Nagle A, Adrian F, Matzen JT, Anderson P, Nam TG, Gray NS, Chatterjee A, Janes J, Yan SF, Trager R, Caldwell JS, Schultz PG, Zhou Y, Winzeler EA. 2008. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci U S A 105:9059–9064. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spangenberg T, Burrows JN, Kowalczyk P, McDonald S, Wells TNC, Willis P. 2013. The open access Malaria Box: a drug discovery catalyst for neglected diseases. PLoS One 8:e62906. doi: 10.1371/journal.pone.0062906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Richard J, Kim S, Wojcik EJ. 2014. Small molecule screen for candidate antimalarials targeting Plasmodium Kinesin-5. J Biol Chem 289:16601–16614. doi: 10.1074/jbc.M114.551408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiwari N, Reynolds P, Calderón A. 2016. Preliminary LC-MS based screening for inhibitors of Plasmodium falciparum thioredoxin reductase (PfTrxR) among a set of antimalarials from the Malaria Box. Molecules 21:424. doi: 10.3390/molecules21040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paiardini A, Bamert RS, Kannan-Sivaraman K, Drinkwater N, Mistry SN, Scammells PJ, McGowan S. 2015. Screening the Medicines for Malaria Venture “Malaria Box” against the Plasmodium falciparum aminopeptidases, M1, M17 and M18. PLoS One 10:e0115859. doi: 10.1371/journal.pone.0115859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aroonsri A, Akinola O, Posayapisit N, Songsungthong W, Uthaipibull C, Kamchonwongpaisan S, Gbotosho GO, Yuthavong Y, Shaw PJ. 2016. Identifying antimalarial compounds targeting dihydrofolate reductase-thymidylate synthase (DHFR-TS) by chemogenomic profiling. Int J Parasitol 46:527–535. doi: 10.1016/j.ijpara.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 11.von Koschitzky I, Gerhardt H, Lämmerhofer M, Kohout M, Gehringer M, Laufer S, Pink M, Schmitz-Spanke S, Strube C, Kaiser A. 2015. New insights into novel inhibitors against deoxyhypusine hydroxylase from Plasmodium falciparum: compounds with an iron chelating potential. Amino Acids 47:1155–1166. doi: 10.1007/s00726-015-1943-z. [DOI] [PubMed] [Google Scholar]
- 12.Bowman JD, Merino EF, Brooks CF, Striepen B, Carlier PR, Cassera MB. 2014. Antiapicoplast and gametocytocidal screening to identify the mechanisms of action of compounds within the Malaria Box. Antimicrob Agents Chemother 58:811–819. doi: 10.1128/AAC.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W, Herrera Z, Ebert D, Baska K, Cho SH, DeRisi JL, Yeh E. 2015. A chemical rescue screen identifies a Plasmodium falciparum apicoplast inhibitor targeting MEP isoprenoid precursor biosynthesis. Antimicrob Agents Chemother 59:356–364. doi: 10.1128/AAC.03342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imlay LS, Armstrong CM, Masters MC, Li T, Price KE, Edwards RL, Mann KM, Li LX, Stallings CL, Berry NG, O'Neill PM, Odom AR. 2015. Plasmodium IspD (2-C-methyl-d-erythritol 4-phosphate cytidyltransferase), an essential and druggable antimalarial target. ACS Infect Dis 1:157–167. doi: 10.1021/id500047s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong KY, Sandlin RD, Wright DW. 2015. Identification of β-hematin inhibitors in the MMV Malaria Box. Int J Parasitol Drugs Drug Resist 5:84–91. doi: 10.1016/j.ijpddr.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hain AUP, Bartee D, Sanders NG, Miller AS, Sullivan DJ, Levitskaya J, Meyers CF, Bosch J. 2014. Identification of an Atg8-Atg3 protein–protein interaction inhibitor from the Medicines for Malaria Venture Malaria Box active in blood and liver stage Plasmodium falciparum parasites. J Med Chem 57:4521–4531. doi: 10.1021/jm401675a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehane AM, Ridgway MC, Baker E, Kirk K. 2014. Diverse chemotypes disrupt ion homeostasis in the malaria parasite. Mol Microbiol 94:327–339. doi: 10.1111/mmi.12765. [DOI] [PubMed] [Google Scholar]
- 18.Kwon YK, Lu W, Melamud E, Khanam N, Bognar A, Rabinowitz JD. 2008. A domino effect in antifolate drug action in Escherichia coli. Nat Chem Biol 4:602–608. doi: 10.1038/nchembio.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creek DJ, Barrett MP. 2014. Determination of antiprotozoal drug mechanisms by metabolomics approaches. Parasitology 141:83–92. doi: 10.1017/S0031182013000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Brummelen AC, Olszewski KL, Wilinski D, Llinas M, Louw AI, Birkholtz LM. 2009. Co-inhibition of Plasmodium falciparum S-adenosylmethionine decarboxylase/ornithine decarboxylase reveals perturbation-specific compensatory mechanisms by transcriptome, proteome, and metabolome analyses. J Biol Chem 284:4635–4646. doi: 10.1074/jbc.M807085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Watts KM, Hodge D, Kemp LM, Hunstad DA, Hicks LM, Odom AR. 2011. A second target of the antimalarial and antibacterial agent fosmidomycin revealed by cellular metabolic profiling. Biochemistry 50:3570–3577. doi: 10.1021/bi200113y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biagini GA, Fisher N, Shone AE, Mubaraki MA, Srivastava A, Hill A, Antoine T, Warman AJ, Davies J, Pidathala C, Amewu RK, Leung SC, Sharma R, Gibbons P, Hong DW, Pacorel B, Lawrenson AS, Charoensutthivarakul S, Taylor L, Berger O, Mbekeani A, Stocks PA, Nixon GL, Chadwick J, Hemingway J, Delves MJ, Sinden RE, Zeeman A-M, Kocken CHM, Berry NG, O'Neill PM, Ward SA. 2012. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc Natl Acad Sci U S A 109:8298–8303. doi: 10.1073/pnas.1205651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobbold SA, Chua HH, Nijagal B, Creek DJ, Ralph SA, McConville MJ. 2016. Metabolic dysregulation induced in Plasmodium falciparum by dihydroartemisinin and other front-line antimalarial drugs. J Infect Dis 213:276–286. doi: 10.1093/infdis/jiv372. [DOI] [PubMed] [Google Scholar]
- 24.Park YH, Shi YP, Liang B, Medriano CAD, Jeon YH, Torres E, Uppal K, Slutsker L, Jones DP. 2015. High-resolution metabolomics to discover potential parasite-specific biomarkers in a Plasmodium falciparum erythrocytic stage culture system. Malar J 14:122. doi: 10.1186/s12936-015-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sana TR, Gordon DB, Fischer SM, Tichy SE, Kitagawa N, Lai C, Gosnell WL, Chang SP. 2013. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PLoS One 8:e60840. doi: 10.1371/journal.pone.0060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trochine A, Creek DJ, Faral-Tello P, Barrett MP, Robello C. 2014. Benznidazole biotransformation and multiple targets in Trypanosoma cruzi revealed by metabolomics. PLoS Negl Trop Dis 8:e2844. doi: 10.1371/journal.pntd.0002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali JAM, Creek DJ, Burgess K, Allison HC, Field MC, Mäser P, De Koning HP. 2013. Pyrimidine salvage in Trypanosoma brucei bloodstream forms and the trypanocidal action of halogenated pyrimidines. Mol Pharmacol 83:439–453. doi: 10.1124/mol.112.082321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alkhaldi AA, Creek DJ, Ibrahim H, Kim DH, Quashie NB, Burgess KE, Changtam C, Barrett MP, Suksamrarn A, de Koning HP. 2015. Potent trypanocidal curcumin analogs bearing a monoenone linker motif act on Trypanosoma brucei by forming an adduct with trypanothione. Mol Pharmacol 87:451–464. doi: 10.1124/mol.114.096016. [DOI] [PubMed] [Google Scholar]
- 29.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 30.Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. 2012. Evaluation of coupling reversed phase, aqueous normal phase, and hydrophilic interaction liquid chromatography with Orbitrap mass spectrometry for metabolomic studies of human urine. Anal Chem 84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- 31.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotech 30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tautenhahn R, Bottcher C, Neumann S. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. 2011. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal Chem 83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- 34.Creek DJ, Jankevics A, Burgess KEV, Breitling R, Barrett MP. 2012. IDEOM: an Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 28:1048–1049. doi: 10.1093/bioinformatics/bts069. [DOI] [PubMed] [Google Scholar]
- 35.Creek DJ, Jankevics A, Breitling R, Watson DG, Barrett MP, Burgess KEV. 2011. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal Chem 83:8703–8710. doi: 10.1021/ac2021823. [DOI] [PubMed] [Google Scholar]
- 36.De Livera AM, Dias DA, De Souza D, Rupasinghe T, Pyke J, Tull DL, Roessner U, McConville MJ, Speed TP. 2012. Normalising and integrating metabolomics data. Anal Chem 84:10768–10776. doi: 10.1021/ac302748b. [DOI] [PubMed] [Google Scholar]
- 37.Saunders EC, Ng WW, Chambers JM, Ng M, Naderer T, Kramer JO, Likic VA, McConville MJ. 2011. Isotopomer profiling of Leishmania mexicana promastigotes reveals important roles for succinate fermentation and aspartate uptake in tricarboxylic acid cycle (TCA) anaplerosis, glutamate synthesis, and growth. J Biol Chem 286:27706–27717. doi: 10.1074/jbc.M110.213553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, Fishbaugher M, Healer J, Cowman AF, Kappe SHI, Llinás M. 2013. Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J Biol Chem 288:36338–36350. doi: 10.1074/jbc.M113.503557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current Protoc Bioinformatics Chapter 14:Unit 14.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio J-J. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 41.de Azevedo MF, Gilson PR, Gabriel HB, Simoes RF, Angrisano F, Baum J, Crabb BS, Wunderlich G. 2012. Systematic analysis of FKBP inducible degradation domain tagging strategies for the human malaria parasite Plasmodium falciparum. PLoS One 7:e40981. doi: 10.1371/journal.pone.0040981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasenkamp S, Russell KT, Horrocks P. 2012. Comparison of the absolute and relative efficiencies of electroporation-based transfection protocols for Plasmodium falciparum. Malar J 11:210. doi: 10.1186/1475-2875-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchholz K, Schirmer RH, Eubel JK, Akoachere MB, Dandekar T, Becker K, Gromer S. 2008. Interactions of methylene blue with human disulfide reductases and their orthologues from Plasmodium falciparum. Antimicrob Agents Chemother 52:183–191. doi: 10.1128/AAC.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joët T, Eckstein-Ludwig U, Morin C, Krishna S. 2003. Validation of the hexose transporter of Plasmodium falciparum as a novel drug target. Proc Natl Acad Sci U S A 100:7476–7479. doi: 10.1073/pnas.1330865100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klonis N, Creek DJ, Tilley L. 2013. Iron and heme metabolism in Plasmodium falciparum and the mechanism of action of artemisinins. Curr Opin Microbiol 16:722–727. doi: 10.1016/j.mib.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Jimenez-Diaz MB, Ebert D, Salinas Y, Pradhan A, Lehane AM, Myrand-Lapierre ME, O'Loughlin KG, Shackleford DM, Justino de Almeida M, Carrillo AK, Clark JA, Dennis AS, Diep J, Deng X, Duffy S, Endsley AN, Fedewa G, Guiguemde WA, Gomez MG, Holbrook G, Horst J, Kim CC, Liu J, Lee MC, Matheny A, Martinez MS, Miller G, Rodriguez-Alejandre A, Sanz L, Sigal M, Spillman NJ, Stein PD, Wang Z, Zhu F, Waterson D, Knapp S, Shelat A, Avery VM, Fidock DA, Gamo FJ, Charman SA, Mirsalis JC, Ma H, Ferrer S, Kirk K, Angulo-Barturen I, Kyle DE, DeRisi JL, Floyd DM, Guy RK. 2014. (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proc Natl Acad Sci U S A 111:E5455–5462. doi: 10.1073/pnas.1414221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis IA, Wacker M, Olszewski KL, Cobbold SA, Baska KS, Tan A, Ferdig MT, Llinas M. 2014. Metabolic QTL analysis links chloroquine resistance in Plasmodium falciparum to impaired hemoglobin catabolism. PLoS Genet 10:e1004085. doi: 10.1371/journal.pgen.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. 2007. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature 446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 50.Baell JB, Holloway GA. 2010. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 51.Phillips MA, Lotharius J, Marsh K, White J, Dayan A, White KL, Njoroge JW, El Mazouni F, Lao Y, Kokkonda S, Tomchick DR, Deng X, Laird T, Bhatia SN, March S, Ng CL, Fidock DA, Wittlin S, Lafuente-Monasterio M, Benito FJG, Alonso LMS, Martinez MS, Jimenez-Diaz MB, Bazaga SF, Angulo-Barturen I, Haselden JN, Louttit J, Cui Y, Sridhar A, Zeeman A-M, Kocken C, Sauerwein R, Dechering K, Avery VM, Duffy S, Delves M, Sinden R, Ruecker A, Wickham KS, Rochford R, Gahagen J, Iyer L, Riccio E, Mirsalis J, Bathhurst I, Rueckle T, Ding X, Campo B, Leroy D, Rogers MJ, Rathod PK, Burrows JN, Charman SA. 2015. A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci Transl Med 7:296ra111. doi: 10.1126/scitranslmed.aaa6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siregar JE, Kurisu G, Kobayashi T, Matsuzaki M, Sakamoto K, Mi-ichi F, Watanabe Y, Hirai M, Matsuoka H, Syafruddin D, Marzuki S, Kita K. 2015. Direct evidence for the atovaquone action on the Plasmodium cytochrome bc1 complex. Parasitol Int 64:295–300. doi: 10.1016/j.parint.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Lakshmanan V, Rhee KY, Wang W, Yu Y, Khafizov K, Fiser A, Wu P, Ndir O, Mboup S, Ndiaye D. 2012. Metabolomic analysis of patient plasma yields evidence of plant-like α-linolenic acid metabolism in Plasmodium falciparum. J Infect Dis 206:238–248. doi: 10.1093/infdis/jis339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi-Ichi F, Kita K, Mitamura T. 2006. Intraerythrocytic Plasmodium falciparum utilize a broad range of serum-derived fatty acids with limited modification for their growth. Parasitology 133:399–410. doi: 10.1017/S0031182006000540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.