Abstract
T-705 (favipiravir) is a new antiviral agent in advanced clinical development for influenza therapy. It is supposed to act as an alternative substrate for the viral polymerase, causing inhibition of viral RNA synthesis or virus mutagenesis. These mechanisms were also proposed for ribavirin, an established and broad antiviral drug that shares structural similarity with T-705. We here performed a comparative analysis of the effects of T-705 and ribavirin on influenza virus and host cell functions. Influenza virus-infected cell cultures were exposed to T-705 or ribavirin during single or serial virus passaging. The effects on viral RNA synthesis and infectious virus yield were determined and mutations appearing in the viral genome were detected by whole-genome virus sequencing. In addition, the cellular nucleotide pools as well as direct inhibition of the viral polymerase enzyme were quantified. We demonstrate that the anti-influenza virus effect of ribavirin is based on IMP dehydrogenase inhibition, which results in fast and profound GTP depletion and an imbalance in the nucleotide pools. In contrast, T-705 acts as a potent and GTP-competitive inhibitor of the viral polymerase. In infected cells, viral RNA synthesis is completely inhibited by T-705 or ribavirin at ≥50 μM, whereas exposure to lower drug concentrations induces formation of noninfectious particles and accumulation of random point mutations in the viral genome. This mutagenic effect is 2-fold higher for T-705 than for ribavirin. Hence, T-705 and ribavirin both act as purine pseudobases but profoundly differ with regard to the mechanism behind their antiviral and mutagenic effects on influenza virus.
INTRODUCTION
The global burden of influenza is estimated at 250,000 to 500,000 deaths per year (1). On a global average, influenza A virus accounts for ∼75% of seasonal infections; the other ∼25% is caused by influenza B virus (2). In addition, zoonotic influenza A viruses of swine or avian species create a risk for pandemics, such as occurred in 2009 (3). Influenza vaccination is a highly effective preventive measure, yet it suffers from some limitations: unsatisfactory immune response in elderly or immunodeficient persons, poor efficacy in the case of vaccine mismatch, and a lag time of >6 months to prepare a novel vaccine in the case of a pandemic (4). Hence, antiviral drugs are indispensable as a complementary strategy to combat influenza virus infections. The recommended therapy with the neuraminidase inhibitors (NAI) oseltamivir and zanamivir is subject to concerns about potential drug resistance (particularly for oseltamivir) (5) and rather modest clinical effectiveness (6, 7). Drug resistance is even more problematic for the M2 channel blockers amantadine and rimantadine (8).
Given its pivotal role in virus genome replication, the influenza virus polymerase represents an excellent antiviral drug target (9). Nucleoside analogue-type viral polymerase inhibitors are a successful drug class for diverse virus infections, yet their development for influenza has been very slow. One prototypic inhibitor is T-705 (favipiravir), a nucleobase analogue first reported in 2002 (10) that possesses broad in vitro and in vivo activities against influenza A, B, and C viruses (10, 11); human and avian (12) viruses, and NAI-resistant virus strains (11). In addition, this molecule displays activity against a wide variety of unrelated RNA viruses (reviewed by Furuta and coworkers [13, 14]). T-705 was approved in Japan for restricted use against NAI-resistant pandemic influenza viruses (15) and is currently in phase 3 trials in Europe and the United States (https://clinicaltrials.gov/ct2/results?term=T-705&Search=Search). According to available clinical information, its antiviral effects are similar to those of oseltamivir (16), giving a statistically significant decrease in symptom duration (17). An important structural element in T-705 is the carboxamide moiety, which is also present in the broad RNA virus inhibitor ribavirin. The usefulness of ribavirin for influenza therapy seems limited since clinical trials, performed in the 1980s, indicated that it has rather modest efficacy against this virus (18, 19). In anecdotal studies, ribavirin served as a last resort to treat (multi)drug-resistant influenza infections in immunocompromised patients (20, 21). The known hematological toxicity of ribavirin seems not to occur during short-term influenza treatment (22).
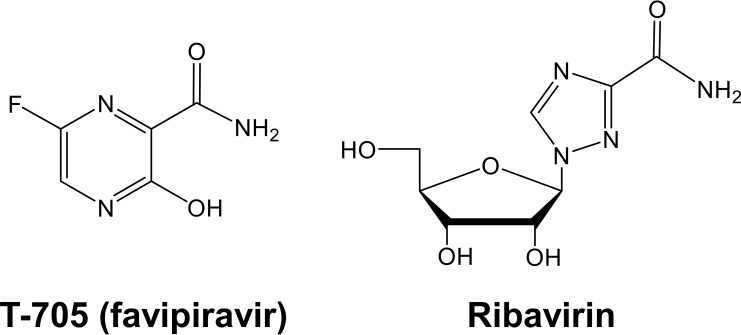
There are still unknowns regarding the precise anti-influenza mechanism of T-705 and the potential parallels with ribavirin, which has been studied more extensively. Inside the host cells, T-705 and ribavirin undergo conversion to their ribosyl-5′-mono-, di-, and triphosphate metabolites (23). The high doses required for T-705 (on the order of 800 to 2,400 mg per day) could, at least partially, be related to low efficiency of its metabolic activation (24). Two structural elements are shared by the ribosyl-5′-triphosphate forms of T-705 (T-705-RTP) and ribavirin (RBV-TP). First, the rotating carboxamide moiety (Fig. 1) makes their base a mimic of guanine as well as adenine and provides them with ambiguous base-pairing properties. Second, the presence of a natural ribose implies that in theory, chain elongation is possible in the case that T-705-RTP or RBV-TP is used as an alternative substrate by the influenza virus polymerase. These two factors could explain recent observations that cell culture passaging of influenza virus in the presence of T-705 (25) or ribavirin (26, 27) leads to virus mutagenesis; this mutagenic effect was also seen in influenza virus-infected mice receiving T-705 therapy (28). Enzymatic studies showed that influenza virus polymerase recognizes T-705-RTP as an alternative substrate versus GTP and ATP (29, 30). It was found that incorporation of a single T-705-RMP into viral RNA slows down but does not halt viral RNA extension, whereas two consecutive T-705-RMP incorporation events result in nonimmediate chain termination of viral RNA synthesis (29). The first mechanism implies that at lower concentrations, T-705 might be incorporated into nascent or full-length viral RNA, thereby inducing mutations in the viral genome. In contrast, a direct inhibitory effect of RBV-TP on the viral polymerase was seen only at relatively high compound concentrations (31, 32). Alternatively, the virus mutagenic effect of ribavirin may be related to a reduced cellular GTP pool (33) due to the inhibitory action of ribavirin 5′-monophosphate on the cellular IMP dehydrogenase (IMPDH) enzyme (34).
FIG 1.
Chemical structures of T-705 and ribavirin.
The diverse mechanisms proposed for T-705 and ribavirin were largely investigated in separate studies, and the link between data from enzyme- and cell-based studies still needs to be firmly proven. To address this, we used all relevant methodologies to verify the proposed mechanisms, make a side-by-side comparison between T-705 and ribavirin, and correlate the biochemical and virological findings. Cell culture studies were performed to investigate their impact on influenza virus RNA synthesis and infectivity during one-cycle or multicycle replication, or after serial virus passaging. The latter allowed us to follow virus mutagenesis during prolonged compound exposure. In addition, we assessed the role of indirect mechanisms (i.e., alterations in the nucleotide pools) or direct effects on the influenza virus polymerase.
MATERIALS AND METHODS
Virus, cells, and chemical compounds.
The influenza virus A/X-31 strain (a reassortant virus that carries the hemagglutinin [HA] and neuraminidase [NA] genes from the A/Aichi/2/68 [A/H3N2 subtype] and all other genes from A/Puerto Rico/8/1934 [A/H1N1]) was expanded in embryonated hen eggs. Madin-Darby canine kidney (MDCK) cells were kindly donated by M. Matrosovich (Marburg, Germany), whereas human lung carcinoma A549 cells were obtained from the American Type Culture Collection (ATCC). Virus experiments were carried out with infection medium that consisted of Ultra MDCK medium (Lonza), supplemented with 0.02% sodium bicarbonate, 2 mM l-glutamine, and 2 μg/ml of tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma).
T-705 (favipiravir; 6-fluoro-3-hydroxy-2-pyrazinecarboxamide) was from YouChemicals (Shanghai, China), whereas ribavirin (Virazole) was from ICN Pharmaceuticals. T-705-RTP (35) was synthesized at Alios BioPharma, Inc., South San Francisco, CA. Ribavirin 5′-triphosphate (RBV-TP) was purchased from Carbosynth (Berkshire, UK). [8-3H]guanosine-5′-triphosphate (specific activity: 13.5 Ci/mmol) was acquired from PerkinElmer (Boston, MA), and [2,8-3H]hypoxanthine (specific radioactivity: 15 Ci/mmol) was from Moravek (Brea, CA). To remove free tritium label, the [2,8-3H]hypoxanthine was purified by reverse-phase high-performance liquid chromatography (HPLC) analysis with an ammonium formate buffer, followed by evaporation under vacuum.
Multicycle assays.
To perform the cytopathic effect (CPE) reduction assay (36), MDCK cells were seeded in 96-well plates at 7,500 cells per well. Sixteen hours later, the compounds were added, followed by the virus (multiplicity of infection [MOI]: 0.0004 PFU per cell). After 3 days of incubation at 35°C, virus-induced CPE and compound cytotoxicity were monitored by microscopy and then confirmed by the formazan-based MTS cell viability assay (Cell Titer 96 Aqueous One Solution cell proliferation assay from Promega). The antiviral 50% effective concentration (EC50) was defined as the concentration producing 50% inhibition of virus-induced CPE.
To determine the effect on virus yield, cells were infected as described above (MOI: 0.0004 PFU per cell) but at 1 h postinfection (p.i.), the supernatant was replaced by medium containing compound to remove the unbound virus. After 72 h of incubation, supernatants were collected. The infectious virus titer was determined by adding serial dilutions of these supernatants to fresh MDCK cells (four wells per dilution) and assessing the viral CPE at day 3 p.i. The titer, expressed as 50% cell culture infective dose (CCID50), was calculated by the method of Reed and Muench (37). In parallel, the supernatants were submitted to one-step reverse transcription-quantitative PCR (RT-qPCR) to determine the viral RNA (vRNA) copy number, using the CellsDirect One-Step qRT-PCR kit from Invitrogen. All details on sample preparation, PCR program, M-gene-directed primers, probe, and plasmid standard can be found elsewhere (38).
To perform the plaque assay, MDCK cells were seeded into 12-well plates at 125,000 cells per well in Opti-MEM I medium (Life Technologies). On the next day, the virus (MOI: 20 to 50 PFU per well) was added to the cells and allowed to adsorb for 1 h at 35°C. After removal of unbound virus, the compounds were added together with overlay medium to obtain a final concentration of 0.4% ultrapure agarose in Opti-MEM I. Three days later, the cell monolayers were fixated in 1.2% formaldehyde and then stained with 0.1% crystal violet.
One-cycle time-of-addition experiments.
MDCK cells were seeded in 24-well plates at 125,000 cells per well. After 16 h, X-31 influenza virus was added at an MOI of 0.0004 PFU per cell, and the compounds were added at −12, 0, 1, 2, 3, 4, 5, 6, 7, or 8 h p.i. At 10 h p.i., total cellular RNA extracts were prepared and the vRNA copy number was determined by two-step RT-qPCR (39). To quantify the vRNA in the inoculum, an RNA extract was prepared at time zero from virus-infected cells receiving no compound. To determine the kinetics of vRNA synthesis, virus-infected cells were lysed at 0 to 10 h p.i., and two-step RT-qPCR analysis was performed as described above.
Virus passage experiments and full virus genome sequencing.
Three passage experiments (at an MOI of 0.0004 to 0.004 PFU per cell) were performed which were separated by time. MDCK cells, seeded in 96-well plates, were infected with influenza virus and exposed to T-705, ribavirin, or medium. After 3 days of incubation, conditions showing obvious CPE were selected, and from these wells, the cells plus supernatants were harvested and frozen at −80°C. These yields were serially diluted and inoculated onto fresh MDCK cells, with new addition of T-705, ribavirin, or medium. The procedure was repeated for a total of 14 passages in experiment 1, 12 passages in experiment 2, and 14 passages in experiment 3.
For full virus genome sequencing on the 12th/14th or intermediate virus passages, the samples (cells plus supernatant) were processed, PCR amplified, and analyzed on an Illumina HiSeq2000 platform, followed by analysis of the deep-sequencing data (for details, see the supplemental material). Briefly, to assess the mutational load in the deep-sequenced virus samples, we first reconstructed the evolution of divergence in the presence or absence of T-705 or ribavirin, with a focus on the coding genome regions, by comparing the majority-rule consensus sequences obtained with CLC Bio software v. 7.5.1 (Qiagen). Next, we explored the overall genetic diversity by contrasting a site-specific measure of nucleotide diversity (the standard Shannon entropy) (40) for the populations obtained under T-705 or ribavirin or the controls that were passaged without compound. Finally, we compiled the nonsynonymous changes with respect to the parent virus stock in the consensus sequences of the viruses obtained from the last passages and, in case of experiment 3, the intermediate passages.
HPLC analysis of cellular NTP pools.
To quantify the NTP (i.e., ATP, CTP, GTP, and UTP) pools, MDCK cells in 25-cm2 flasks were exposed to T-705 or ribavirin for 1 to 72 h (specified in Results). To prepare extracts, the cells were washed three times with blank medium and then subjected to trypsinization, cell counting, and centrifugation. The cell pellet was resuspended in 500 μl of ice-cold methanol-water (2:1), and after standing for 10 min on ice, the extracts were clarified by centrifugation at 16,000 × g. HPLC analysis was performed on a Partisphere SAX strong-anion exchange column using the equipment and phosphate buffer gradient described previously (41). The retention times of ATP, CTP, GTP, and UTP were 20, 22, 23, and 27 min, respectively, and UV detection was done at 259 nm. To normalize the data, the absolute peak areas were converted into the peak area per cell.
Enzymatic assay with influenza virus polymerase.
Viral ribonucleoproteins (vRNPs) were isolated from virions and purified by gradient ultracentrifugation (42–45). These vRNPs contain the native PA-PB1-PB2 polymerase enzyme and, as the template for transcription, viral RNA complexed to nucleoprotein. To determine their inhibitory effects on viral RNA elongation in competition with GTP, the compounds were added to a reaction mixture (final volume: 25 μl) containing 50 mM Tris (pH 8.0), 100 mM KCl, 1 mM dithiothreitol, 5 mM MgCl2, 0.4 U/μl of recombinant RNasin (Promega), 200 μM ApG primer [adenyl (3′-5′)guanosine, obtained from Sigma], 100 μM UTP, 100 μM CTP, 500 μM ATP, 2.2 μM GTP, and 1.5 μM [8-3H]GTP. After addition of the isolated vRNPs, the samples were incubated for 1 h at 30°C. The amount of radioactivity incorporated into viral RNA was quantified by a filter-based method as described elsewhere (42, 43).
Cell-based method to measure inhibition of IMPDH.
To estimate the compounds' effects on the conversion of hypoxanthine to GMP (which requires catalysis by hypoxanthine guanine phosphoribosyltransferase (HGPRT) and IMPDH, consecutively), a tritium release assay was performed, which was a variation of an established method for suspension cells (46). A549 cells seeded into 24-well plates were preincubated with the compounds for 4 h at 37°C. Then [2,8-3H]hypoxanthine was added at 0.7 μCi per well and a total concentration of 4.4 μM. After 45 min of incubation, the supernatants were mixed 1:1 with an ice-cold suspension containing 150 mg per ml of activated charcoal and 7.5% trichloroacetic acid. After standing for 10 min on ice, followed by high-speed centrifugation, the clarified supernatants were collected for scintillation counting.
Statistical analysis.
To analyze differences between two conditions, the unpaired two-sided Student t test was used, setting a P value of ≤0.05 as the cutoff for statistical significance.
RESULTS
At high concentrations, T-705 and ribavirin shut off virus replication; at lower concentrations, T-705 gives abundant production of noninfectious virus particles.
There is some divergence in the literature regarding the antiviral activity of T-705 in influenza virus-infected MDCK cells. Furuta et al. (10) reported EC50s, based on a plaque reduction assay, in the range of 0.08 to 3 μM, whereas in the CPE reduction assay conducted by Baranovich et al. (25), T-705 had EC50s in the range of 11 to 17 μM. This concurs with the EC50 of 11 μM that we found in this study (Table 1) and a previous study (24). Importantly, in each of these investigations, T-705 was devoid of cytotoxicity at the highest concentration tested, i.e., 600 μM (Table 1), 1,000 μM (25), or 6,400 μM (10). The selectivity index of T-705 by far exceeds that of ribavirin which, in our CPE assay, displayed an EC50 for influenza virus of 8.1 μM yet visible cytostatic activity at 100 μM.
TABLE 1.
Antiviral activities of T-705 and ribavirin in influenza virus-infected MDCK cellsa
| Compound | Antiviral activity |
MCCd (μM) | ||
|---|---|---|---|---|
| CPE assay result, EC50b (μM) | RT-qPCR assay resultc |
|||
| EC90 (μM) | EC99 (μM) | |||
| T-705 | 11 ± 0 | 9.2 ± 1.1 | 13 ± 1 | >600 |
| Ribavirin | 8.1 ± 1.3 | 9.1 ± 1.9 | 14 ± 2 | 100 ± 0 |
Data are means ± SEMs from 3 independent experiments.
EC50, compound concentration giving 50% inhibition of virus-induced CPE, as determined by microscopy at 72 h postinfection.
EC90 and EC99, compound concentrations producing 1-log10 and 2-log10 reductions in virus yield, respectively, as determined by RT-qPCR analysis of vRNA copy number in the supernatant at 72 h postinfection.
MCC, minimum cytotoxic concentration, i.e., concentration giving minimal alterations in cell morphology, as determined by microscopy.
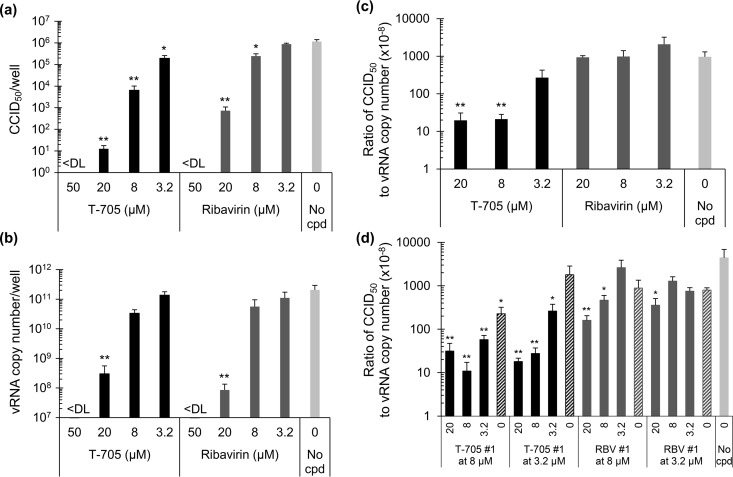
In parallel, we determined the virus yield in the supernatant after multiple replication rounds (i.e., at 72 h p.i.), using an infectious virus titration (CCID50) assay as well as RT-qPCR quantification of vRNA to estimate the number of virus particles. Both T-705 and ribavirin produced a dose-dependent reduction in infectious virus yield (Fig. 2a). This dose-dependent effect was also seen by RT-qPCR (Fig. 2b) with EC99s (corresponding to 100-fold reduction in vRNA copy number) of 13 μM for T-705 and 14 μM for ribavirin (Table 1). Strikingly, when the data were expressed as the ratio of CCID50 to vRNA copy number (Fig. 2c), the T-705-exposed cells were found to produce high quantities of vRNA that was not associated with infectious virus. Compared to the condition receiving no compound, the CCID50-to-vRNA ratio was 49-fold lower for virus exposed to 20 μM T-705. This effect was seen only at low T-705 concentrations (i.e., 20, 8, or 3.2 μM), since at 50 μM T-705, no virus was detected by either CCID50 or RT-qPCR assay (Fig. 2a and b). Complete inhibition was also seen with ribavirin at concentrations of ≥50 μM, but lower concentrations of ribavirin did not give the reduction in virus infectivity that was seen with T-705.
FIG 2.
Production of noninfectious virus particles at low concentrations of T-705. MDCK cells were exposed to influenza virus and T-705 or ribavirin, and the virus present in the supernatants at 72 h p.i. was quantified. (a) Infectious virus titer, expressed as CCID50; (b) vRNA copy number, determined by RT-qPCR; (c) ratio of CCID50 to vRNA copy number. (d) The passage 1 supernatants were used to infect fresh MDCK cells with, again, addition of T-705, ribavirin, or compound-free medium. After 72 h, the passage 2 supernatants were submitted to virus titration and RT-qPCR analysis. The graph shows the ratio of CCID50 value to vRNA copy number in function of compound concentration during passage 2; the hatched bars mark the conditions in which no compound was added. <DL, below the detection limit. Data are the means ± SEM (n = 4). **, P < 0.005; *, P ≤ 0.05 (versus the no-compound [cpd] condition; Student t test on log-transformed values).
Next, the virus-containing supernatants collected as described above were used to infect MDCK cells in the presence or absence of T-705 or ribavirin, and again, the virus yield at 72 h p.i. was determined by CCID50 and RT-qPCR analysis. In this passage, passage 2 (Fig. 2d), T-705 again produced a marked (up to 400-fold) decrease in the CCID50-to-vRNA ratio. When the T-705-exposed passage 1 virus was incubated without compound (see hatched bars in Fig. 2d), the CCID50-to-vRNA ratio at least partially returned to the level seen in the no-compound control.
The pronounced effect of T-705 was not seen with ribavirin. In passage 1, ribavirin at a concentration of 20, 8, or 3.2 μM gave CCID50-to-vRNA ratios similar to those of the no-compound control (Fig. 2c), and in passage 2, the reduction in particle infectivity was at maximum 28-fold (Fig. 2d).
In combination, these data indicate that T-705 interferes with influenza virus replication in a dual manner. The compound dose dependently inhibits viral replication and RNA synthesis, with complete shutoff at concentrations of ≥50 μM. At lower T-705 concentrations, the inhibition is only partial and nonviable viral genomes are abundantly produced. In the case of ribavirin, the formation of noninfectious particles is much less pronounced.
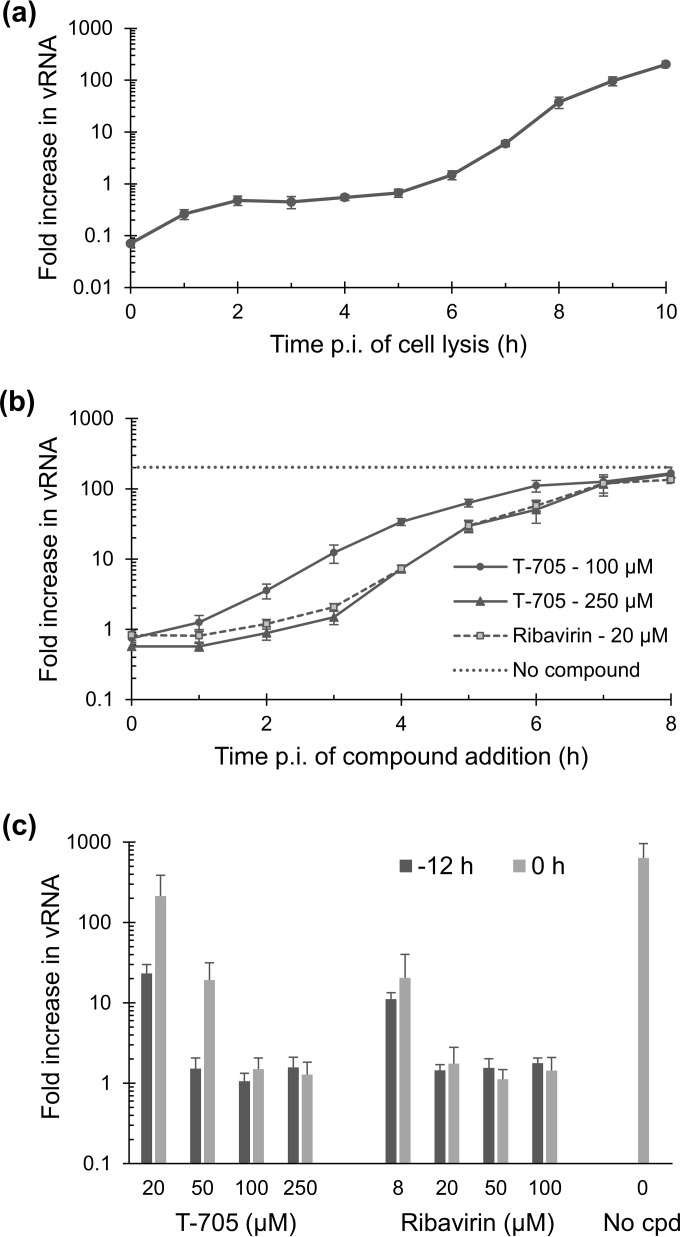
Both T-705 and ribavirin inhibit first-cycle viral RNA synthesis; for T-705, activation is slow at low compound concentrations.
A time-of-addition experiment was performed, in which the compounds were added at different time points, and first-cycle vRNA synthesis (of the M gene) was monitored at 10 h p.i. First, we analyzed the kinetics of vRNA synthesis during this one cycle by performing RNA extractions at 1-h intervals and quantifying the vRNA levels in the cells. As shown in Fig. 3a, vRNA synthesis started at 5 h and then showed an exponential phase between 6 and 8 h, after which it slowed down.
FIG 3.
Inhibitory effect on influenza virus vRNA synthesis within one replication cycle. (a) Time course for intracellular vRNA synthesis. MDCK cells were infected at time zero, and at several time points the vRNA level (for the M-gene) was determined by RT-qPCR. (b) Intracellular vRNA levels at 10 h p.i. after exposure to T-705 (at 100 or 250 μM) or ribavirin (at 20 μM), as a function of compound addition time. (c) Effects of the compounds when added at different concentrations, at either 12 h or 0 h before virus infection. The data in panels a to c are expressed as the fold increase in vRNA copy number relative to the virus input added at time zero. Data are means ± SEMs (n = 3).
When ribavirin (at 20 μM) or T-705 (at 250 μM) was added at the time of virus infection (i.e., time zero) or at 1, 2, or 3 h p.i., both compounds produced 100% inhibition of vRNA synthesis (i.e., increase in vRNA compared to virus input = 1 [Fig. 3b]). The inhibition was clearly lower when the compounds were added at 4 h p.i. or later. The curve of ribavirin fully overlapped that of T-705 at 250 μM; in both cases, the compound's activity was mainly situated between 3 and 5 h p.i. When T-705 was applied at the lower concentration of 100 μM, the time-of-addition curve shifted to the left (Fig. 3b). A plausible explanation is that the formation of T-705-RTP is slower at lower concentrations of T-705, as demonstrated by Smee et al. (47). Hence, an experiment was performed in which different concentrations of T-705 or ribavirin were added at either −12 or 0 h p.i. Compound preincubation gave a clear advantage for T-705 at 20 or 50 μM but had no impact for the 100 and 250 μM conditions (compare dark and light gray bars in Fig. 3c). This means that once activated, T-705 provided at least a 400-fold (2.6-log10) reduction in first-cycle vRNA synthesis at concentrations of 50 μM or more; at 20 μM, the reduction was 27-fold (1.4 log10). This dose-dependent inhibition of vRNA synthesis was also seen with ribavirin, but in this case, the effect was the same whether or not compound preincubation was applied.
T-705 and, to a lesser degree, ribavirin induce mutations in the influenza virus genome upon sustained drug exposure.
To investigate how influenza virus populations evolve when exposed to T-705 or ribavirin for longer periods, we performed three independent virus passage experiments. In each passage, virus was grown for 72 h in the absence or presence of compound, after which the virus was harvested and used to infect fresh cells. This virus subcultivation was limited to 12 or 14 passages. In the first two experiments, we applied T-705 at different concentrations (3.2, 8, or 20 μM) and selected the highest concentration condition showing viral CPE to perform each next passage (see Table S1 in the supplemental material). This approach ensured that each passage was done at the maximum mutagenic pressure tolerated by the virus and also increased the chance of selecting a mutant virus with altered fidelity. However, at 20 μM T-705, the infectivity tended to be so low that the subsequent passage required a reduction in compound concentration in order to recover the virus. Therefore, passage experiment 3 was conducted with a constant concentration of 8 μM T-705 or ribavirin, throughout passages 1 to 14.
After performing deep-sequencing on the 12th or 14th virus passage, we first analyzed the point mutations in their majority-rule consensus sequences. Clearly, the virus accumulated mutations when exposed to T-705 and this phenomenon was reproducible among the three passage experiments. In total, 37, 48, and 45 mutations were detected under T-705 (Table 2), compared to 2, 4, and 11 mutations under the no-compound condition, meaning that in the presence of T-705 the dominant nucleotide was replaced at 18, 12, and 4 times more positions in experiments 1, 2, and 3, respectively. In the case of ribavirin, the effect was more variable, with 11-fold and 1.6-fold increases in consensus nucleotide changes in experiments 2 and 3, respectively. For both compounds, G→A or C→U transitions represented the majority (i.e., 62 to 89%) of the total number of mutations. This is in line with the view that T-705 and ribavirin are ambiguous purine pseudobases that mimic both guanine and adenine.
TABLE 2.
Number of point mutations in the influenza virus genome after 12 or 14 virus passages under T-705 or ribavirin
| Compound | No. of specific point mutations (% of total no.)a |
Total no. of mutations | Fold increase in mutant frequency relative to no compound condition | % nonsynonymous mutationsa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G→A | C→U | A→G | U→C | A→C | C→A | G→U | A→U | U→A | ||||
| Expt 1 (passage 14)b | ||||||||||||
| T-705 | 18 (49) | 7 (19) | 8 (22) | 4 (11) | 0 | 0 | 0 | 0 | 0 | 37 | 18 | 51 |
| No compound | 0 | 0 | 1 (50) | 0 | 0 | 0 | 0 | 0 | 1 (50) | 2 | 50 | |
| Expt 2 (passage 12)b | ||||||||||||
| T-705 | 21 (44) | 12 (25) | 9 (19) | 4 (8) | 0 | 1 (2) | 1 (2) | 0 | 0 | 48 | 12 | 44 |
| Ribavirin | 24 (53) | 16 (36) | 3 (7) | 0 | 0 | 1 (2) | 0 | 1 (2) | 0 | 45 | 11 | 53 |
| No compound | 3 (75) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (25) | 0 | 4 | 75 | |
| Expt 3 (passage l14)b | ||||||||||||
| T-705 | 18 (40) | 11 (24) | 7 (16) | 8 (18) | 1 (2) | 0 | 0 | 0 | 0 | 45 | 4 | 36 |
| Ribavirin | 11 (61) | 4 (22) | 0 | 1 (6) | 0 | 1 (6) | 1 (6) | 0 | 0 | 18 | 1.6 | 56 |
| No compound | 5 (45) | 1 (9) | 2 (18) | 2 (18) | 0 | 0 | 0 | 1 (9) | 0 | 11 | 18 | |
Values are the numbers of mutations present in the entire virus genome. In parentheses, the proportion of the specific mutations versus the total number of mutations is given. This analysis of the deep-sequencing data took into account all mutations that were present in the majority-rule consensus sequences.
In experiments 1 and 2, the compounds were applied at different concentrations (3.2, 8, or 20 μM), and the highest concentration condition showing viral CPE was used to perform the next passage (see Table S1 in the supplemental material for details). This means that each passage was done at the maximum mutagenic pressure tolerated by the virus. However, at 20 μM T-705, the infectivity tended to be so low that subsequent passaging required a lower compound concentration to allow virus recovery. Hence, experiment 3 was performed with T-705 or ribavirin at 8 μM, kept constant throughout passages1 to 14.
Percent nonsynonymous mutations versus the total number of mutations.
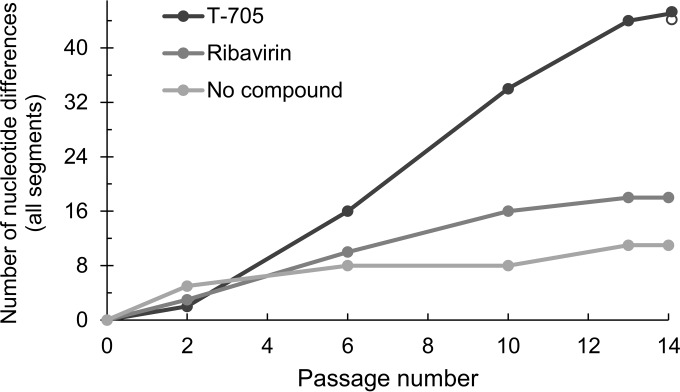
By sequencing the intermediate passages in experiment 3, the mutation rate was shown to be comparatively higher under the T-705 condition (Fig. 4). This trend was stable throughout the 14 passages, indicating that the virus population incorporated mutations at a rather constant rate.
FIG 4.
Mutation rate during serial virus passaging in the presence of T-705 or ribavirin. In passage experiment 3, deep-sequencing analysis was performed at passages 2, 6, 10, 13, and 14 (concentration of T-705 or ribavirin: 8 μM from passage 1 until passage 14). The y axis shows the total number of point mutations in the full viral genome. For the T-705 curve, the solid black circles denote the condition receiving T-705, whereas the open circle marks the condition in which T-705 was omitted during passage 14.
The mutations were distributed over all eight viral genome segments, with a trend toward somewhat higher mutation frequency in the NS gene (see Table S2 in the supplemental material). Nonsynonymous changes accounted for 43%, 54%, and 35% of the mutations detected in the conditions under T-705, ribavirin, and no compound, respectively. The amino acid substitutions were present in all viral proteins (including the PA, PB1, and PB2 subunits of the polymerase complex), but none was present in all three experiments. Hence, no particular substitution seemed to be associated with compound (T-705 or ribavirin) selection pressure or cell culture adaptation. The majority of the mutations (whether silent or not) remained during subsequent passaging; also, the two T-705 conditions that underwent passage 14 with or without T-705 had identical consensus nucleotide sequences (see last data point in Fig. 4). Since our data may help to assess the viability of certain amino acid changes, a complete list of all substitutions present at frequencies of >50% is given in Table S3 in the supplemental material.
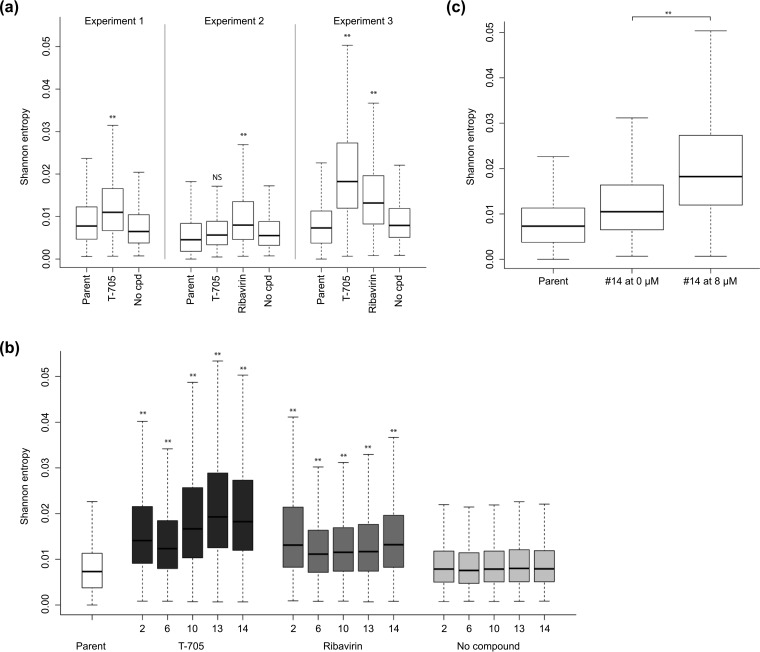
We also examined the overall diversity of the virus populations by contrasting the per-site standard Shannon entropy distributions for the compound-exposed conditions with those of the parent stock (used at the start of the experiments) and the condition of passage without compound (Fig. 5). Positions in the genome for which all variants in the population share the same nucleotide have a Shannon entropy of zero. This measure of uncertainty incorporates both the frequency information and the number of possible characters and has a maximum value when all possible characters are present at equal frequencies. Figure 5a shows that the per-site diversity distributions were generally shifted upwards in passages 12 and 14 under T-705 or ribavirin compared to those of the unexposed populations. This increase in entropy was particularly pronounced (P < 0.005) in experiment 3, in which the concentration of T-705 or ribavirin was kept constant at 8 μM. By analyzing the intermediate passages, we found that the increase in population diversity was already significant within two passages under T-705 or ribavirin. With T-705, the entropy seemed to reach a maximum plateau after 10 passages (Fig. 5b); the high level of population diversity induced by T-705 was not seen within 14 passages under ribavirin. The effect of T-705 was readily reversible since the entropy value practically normalized when T-705 was omitted for one single passage (Fig. 5c).
FIG 5.
Population sequence diversity of the viruses passaged under T-705 or ribavirin. (a) Shannon entropy analysis was performed on the deep-sequencing data from passage experiments 1 (12 virus passages), 2 (14 passages), and 3 (14 passages) (see Table S1 in the supplemental material for all details). (b) Entropy analysis for the intermediate virus passages collected in experiment 3. (c) In experiment 3, the 14th passage was done either in the presence of 8 μM T-705 (on the right) or in the absence of compound (in the middle). In panels a to c, the data on the left represent the parent virus that was used at the start of the passage experiment. **, P < 0.005; *, P ≤ 0.05; NS, not significant, i.e., P > 0.05 (two-sided Student t test) for comparison between the T-705 or ribavirin versus no-compound condition (a and b) and between passage 14 in the presence versus absence of T-705 (c).
Notably, there was no increase in the number of variable sites in the presence of T-705 or ribavirin, compared to the controls without compound. For T-705, between 88% and 94% of the variable sites (i.e., showing nonzero entropy) were shared, and for ribavirin this fraction was between 86% and 94%. These results suggest that the increased entropy in the virus populations under T-705 or ribavirin results from a higher proportion of variants harboring alternative nucleotides at the variable positions due to a mutagenic effect.
Viruses passaged under T-705 or ribavirin had reduced replication fitness and particle infectivity but unaltered antiviral susceptibility.
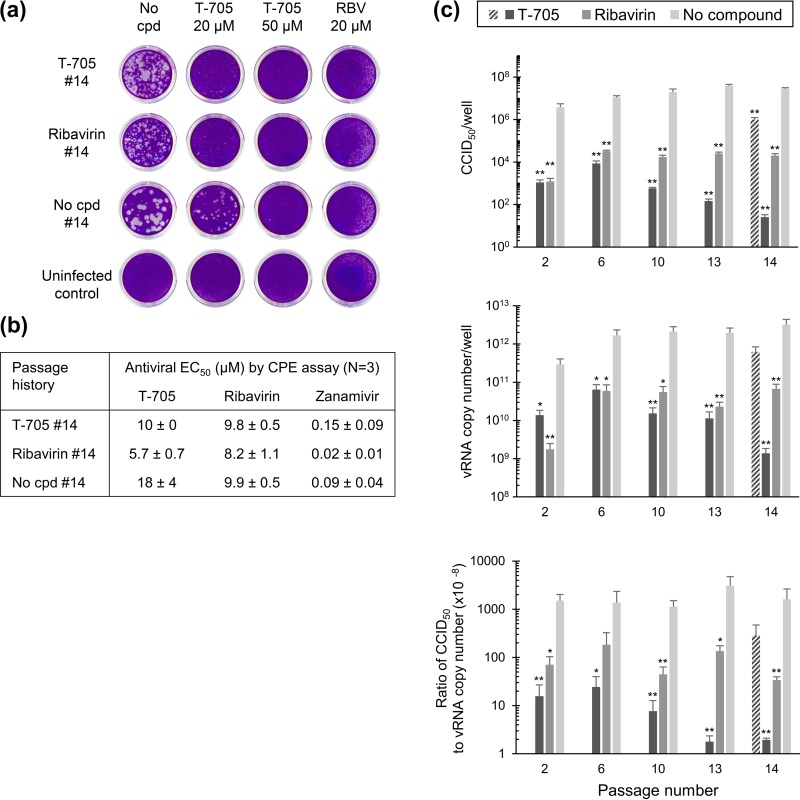
To characterize the phenotype of the viruses obtained after 14 passages, a plaque assay was performed (Fig. 6a). Each virus was fully inhibited by 20 μM ribavirin or 50 μM T-705, whereas 20 μM T-705 gave a clear reduction in plaque size and number. Compared to the virus grown in the absence of compound, the viruses passaged under T-705 or ribavirin displayed a smaller plaque size (Fig. 6a, left images), indicating reduced replication fitness. The sensitivity to T-705 or ribavirin was not reduced after 14 passages, as evident from the antiviral EC50s determined by CPE assay (Fig. 6b) and confirmed by a colorimetric formazan-based cell viability assay (data not shown). Finally, a virus yield study was done using the CCID50 and RT-qPCR assays (see above). The passage 14 viruses harvested under T-705 or ribavirin had impaired particle infectivity compared to the virus grown without compound, the reductions in CCID50-to-vRNA ratio being ∼800-fold for T-705 and ∼50-fold for ribavirin (Fig. 6c). For T-705, the CCID50-to-vRNA ratios were similar in the viruses harvested after 1 (Fig. 2c), 2, or 6 passages and then dropped at passages 13 and 14 (Fig. 6c). This means that the property of the virus to produce noninfectious particles when exposed to T-705 gradually increased during these 14 passages. The particle infectivity practically returned to the normal level when T-705 was omitted during one (i.e., the 14th) passage (see hatched bar in Fig. 6c, 14). In contrast, the condition grown in the absence of compound showed unaltered CCID50-to-vRNA ratios from passages 2 to 14. For ribavirin, the particle infectivity was 10- to 50-fold lower, with no downward trend during serial virus passaging.
FIG 6.
Fitness and antiviral sensitivity after 14 serial passages under T-705 or ribavirin. (a) Plaque assay with viruses obtained after 14 passages in the presence of 8 μM T-705 (first row), 8 μM ribavirin (second row), or no compound (third row) along with an uninfected control (fourth row). The plaque assay was performed in, from left to right, the absence of compound or the presence of 20 or 50 μM T-705 or 20 μM ribavirin (RBV). Note that the slight disturbance of the cell monolayer in the ribavirin-treated wells is due to its cytostatic effect and not to virus plaque formation. (b) Antiviral sensitivity of these passage 14 viruses to T-705, ribavirin, and zanamivir, expressed as the EC50 by CPE assay (means ± SEMs; n = 3). (c) Noninfectious particle production in the viruses harvested after 2, 6, 10, 13, or 14 passages under 8 μM T-705 or ribavirin. The viruses were subjected to infectious virus titration by CCID50 assay and vRNA quantification by RT-qPCR. The y axis shows the CCID50 (top graph), vRNA copy number (middle graph), or the ratio (bottom graph). The hatched bar represents the T-705 condition in which the last (14th) passage was done in the absence of compound. Data are means ± SEMs (n = 3) from three analyses.
Ribavirin but not T-705 causes fast and profound GTP depletion and NTP imbalance.
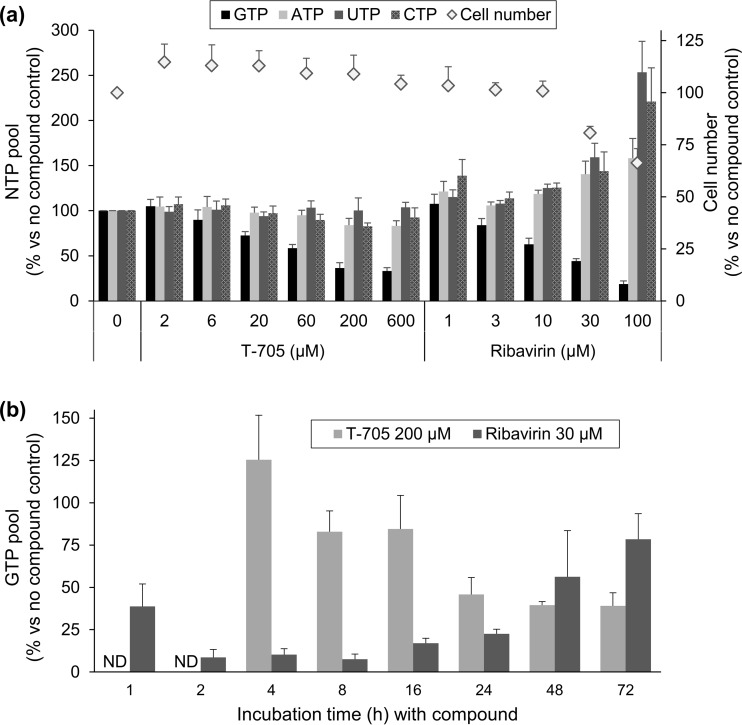
To define the biochemical basis for the mutagenic effect of T-705 and ribavirin upon influenza virus, we first measured their effects on the cellular NTP pools in MDCK cells. As shown in Fig. 7a, ribavirin caused manifest GTP depletion after 24 h, the reductions in the GTP pool being 81%, 56%, and 37% at 100, 30, and 10 μM ribavirin, respectively (P ≤ 0.06 for comparison of the GTP peak area in untreated cells versus the 30 or 100 μM ribavirin condition). This effect coincided with a slight increase in the ATP pool and pronounced increase in the pools of CTP and, in particular, UTP (i.e., P < 0.05 for comparison of the UTP peak area in the untreated versus 30 or 100 μM ribavirin condition). It was also accompanied by a cytostatic effect (i.e., 34% reduction in the cell number under the condition of 100 μM ribavirin). In the case of T-705, the GTP pool was reduced by 67% at 600 μM and 42% at 60 μM (Fig. 7a) (P > 0.2 for comparison of the GTP peak area in untreated cells versus any of the two T-705 conditions). No alterations were seen in the pools of ATP, CTP, or UTP, nor was there any cytostatic effect at 600 μM T-705.
FIG 7.
NTP pools in MDCK cells exposed to T-705 or ribavirin. (a) The compounds were added to uninfected MDCK cells, and 24 h later, cell extracts were prepared and analyzed by HPLC. The bars represent the GTP, ATP, UTP, and CTP pools (expressed as peak area per cell; left y axis) and the diamonds indicate the cell number (right y axis), both expressed as the percentage versus the no-compound condition. (b) The experiment was performed as described for panel a yet at one single concentration of T-705 (200 μM) or ribavirin (30 μM) and applying different compound incubation times (x axis). The y axis shows the GTP pool relative to that under the no-compound condition. Data are means ± SEMs (n = 3 to 10).
We next determined the kinetics of this GTP depletion (Fig. 7b). To our surprise, the effect of 30 μM ribavirin was already visible at 1 h and achieved its maximum at 2 h (i.e., 91% reduction in the GTP pool; P < 0.005 for comparison with the untreated condition). After 8 h, the GTP pool was gradually restored, and at 72 h, it was 78% that of untreated cells. These data indicate that (i) the conversion of ribavirin to its 5′-monophosphate is very fast in dividing MDCK cells, (ii) the GTP pool in these cells is very dynamic, and (iii) upon GTP depletion by ribavirin, a feedback mechanism is initiated to restore the GTP pool; a possible explanation is upregulation of the HGPRT purine salvage enzyme. With T-705 (concentration: 200 μM), the picture was entirely different. At 4 h, the GTP level was still normal; then it gradually declined to reach a plateau at 24 to 72 h, when the GTP pool was ∼40% of the control level. The GTP depletion by T-705 was not restored within the 72-h observation period.
Hence, the GTP depleting effects of ribavirin and T-705 clearly differ with regard to magnitude and kinetics, and only ribavirin causes an NTP imbalance. Although the GTP depletion by ribavirin has been documented in other studies (34, 48), we are the first to recognize the fast kinetics of this effect.
Ribavirin acts via the cellular IMPDH enzyme, whereas T-705 acts upon the influenza virus polymerase.
We next performed a cell-based IMPDH inhibition assay using [2,8-3H]hypoxanthine. Upon addition to cells, this purine base is converted by HGPRT to IMP, which is subsequently converted by IMPDH to GMP. The second reaction results in release of the tritium label at position 2 of the purine base, which can be detected in the supernatant. The clinically used IMPDH inhibitor mycophenolic acid was included as a control. A dose-dependent inhibition of the IMPDH reaction was seen with ribavirin and mycophenolic acid, their EC50s being 7.6 and 0.55 μM, respectively (see Fig. S1a in the supplemental material). No effect was seen with T-705 at up to 200 μM, the highest concentration tested. When all mechanistic data for ribavirin are combined, a striking correlation can be seen between the concentrations producing GTP depletion and IMPDH inhibition on the one hand and virus inhibition on the other hand.
Finally, to investigate whether the ribosyl-5′-triphosphate forms of T-705 and ribavirin have a direct effect on the viral polymerase, we conducted enzymatic studies with isolated influenza virus vRNPs. The 50% inhibitory concentrations (IC50s; for inhibition of [8-3H]GTP incorporation) were 27 μM for the obligate chain terminator 3′-dGTP and 4.6 μM for the nonobligate chain terminator 2′-fluoro-2′-dGTP (49) (see Fig. S1b in the supplemental material). An even stronger inhibition was seen with T-705-RTP, with an IC50 of 2.7 μM; this value is similar to that reported elsewhere (29). In contrast, RBV-TP manifested only weak inhibition of influenza virus polymerase-dependent RNA synthesis, since its IC50 was as high as 216 μM, which is in line with the values reported by others (31, 32).
DISCUSSION
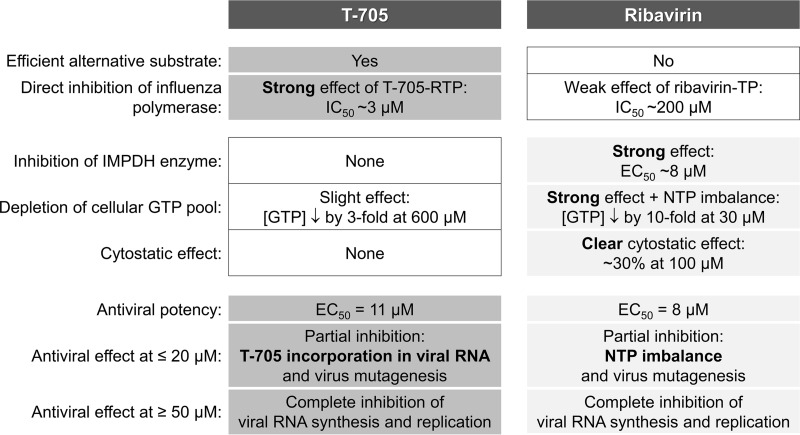
T-705 is the first influenza virus polymerase inhibitor achieving market approval (in Japan) or advanced clinical testing (in Europe and the United States). It has an excellent therapeutic index that is far superior to that of ribavirin, another purine pseudobase derivative. The enzymatic studies by Furuta and colleagues (23, 30) identified T-705-ribosyl-5′-triphosphate as a strong inhibitor of the influenza virus polymerase. For ribavirin, the cellular IMPDH enzyme was presented as the indirect and predominant target for antiviral activity (34, 50), although inhibition of the influenza virus polymerase was also reported (23, 31–33). Since both T-705 (25, 28) and ribavirin (26, 27) were recently reported as influenza virus mutagens, the aim of our study was to make a side-by-side comparison between these two molecules, using cell-based virus infection methods combined with biochemical assays to verify and, particularly, link all possible mechanisms of action. A schematic summary of our findings is presented in Fig. 8.
FIG 8.
Summary scheme. In the left column, the different mechanisms explored in our experiments are listed. T-705 and ribavirin have similar EC50s, and both shut off viral RNA synthesis at concentrations of ≥50 μM. T-705-RTP is an efficient alternative substrate inhibitor for the influenza virus polymerase; combined with its ambiguous base properties, this explains the strong virus mutagenic effect of T-705 at concentrations of ≤20 μM. T-705 does not affect the cellular IMPDH enzyme and has no cytostatic effect. Ribavirin acts via IMPDH inhibition; the resulting GTP depletion and NTP imbalance explain its virus mutagenic and clear cytostatic effects.
First-cycle viral vRNA synthesis is fully inhibited at T-705 concentrations of ≥50 μM and ribavirin concentrations of ≥20 μM, provided that they are added no later than 2 h prior to the onset of vRNA synthesis. For ribavirin, this is explained by the fact that its GTP-depleting effect reached its maximum after 2 h. For T-705, the requirement of early addition is related to the slow and concentration-dependent rate of its metabolic activation as shown by Smee et al. (47). Inhibition of viral RNA synthesis was also seen in the multicycle (72-h) assay, since both agents gave complete shutoff at 50 μM. At lower (8 or 20 μM) concentrations, a remarkable divergence was seen, since T-705 but not ribavirin gave rise to abundant production of noninfectious virus particles. When virus was passaged under T-705, the infectivity gradually declined to reach a minimum after 13 passages, when particle infectivity was 800-fold beyond that of control virus. With ribavirin, the reduction in particle infectivity was much less pronounced and did not evolve over 14 passages.
Deep-sequencing analysis of the passaged viruses demonstrated that both agents induced gradual accumulation of point mutations in the viral genome, of which 62 to 89% were G-to-A plus C-to-U transitions. The number of point mutations after 12 to 14 passages was increased 12-fold by T-705 and 6-fold by ribavirin, meaning that the mutagenic effect of T-705 was 2-fold more pronounced. The fact that we performed three independent passage experiments with whole-virus genome sequencing adds to the relevance of these estimated mutation frequencies. We observed interexperimental variations in the mutational burden for individual genome segments, meaning that a mutational analysis that is restricted to some specific segment(s) may give a biased result. Our deep-sequencing approach also enabled entropy analysis for sequence diversity, which confirmed that T-705 has a higher mutagenic effect upon influenza virus than ribavirin. In our study, the population sequence diversity increased within two passages under T-705 to reach a maximum after 10 to 14 passages and quickly declined when T-705 was no longer added.
Our investigation is the first in which the mutagenic effects of T-705 and ribavirin on influenza virus have been directly compared. In line with our data (49-fold-lower virus infectivity at 20 μM T-705), Baranovich et al. (25) found that T-705 reduces virus infectivity 18-fold at 10 μM and 740-fold at 100 μM. In their passage experiment with a low viral MOI similar to the one we used, the steepest decrease in viral infectivity was seen at ≥10 μM T-705, which is in the same concentration range as ours. T-705 (applied at gradually rising concentrations) increased the number of mutations by a factor of ∼3 after 24 passages (25). Cheung et al. (26) noted, after four passages under 35 μM ribavirin, an increase in the number of mutations by a factor of 4. Pauly and Lauring (27) observed a 1.7-fold increase in mutation number after one virus passage with 5 or 40 μM ribavirin. Experimental variations make it difficult to compare these separate reports, indicating the importance of side-by-side analyses like the one that we did. Besides variations in compound concentrations, the viral MOI seems particularly relevant, since Baranovich et al. (25) found that virus extinction by T-705 (due to reducing infectivity) occurs within 24 passages when a low viral MOI is used and 3 passages in the case of a high MOI, a condition that promotes formation of noninfectious particles (51). Also, laboratory strains (such as the X-31 strain that we used) have high intrinsic replication rates, and their stocks can contain high quantities of noninfectious particles after passaging in cell culture or eggs. These factors could render them more sensitive to lethal mutagens than low-passage-number clinical isolates.
The viruses that we obtained under T-705 or ribavirin contained many nonsynonymous mutations distributed over all eight genome segments. In contrast, Baranovich et al. (25) detected, in their viruses grown under T-705, only a few substitutions in the NA or HA protein. These authors concluded that the reduced fitness and subsequent virus extinction are explained by lethal mutagenesis. According to this concept, a mutagenic antiviral agent increases the mutation rate of the virus population above a threshold level, rendering the virus nonviable (52). Our results confirm that at low concentrations, T-705 and, to a lower degree, ribavirin induce a mutated and genetically diverse influenza virus population. However, in our experiments, the reduced replication fitness of these virus populations seems to be explained by accumulation of fitness-reducing amino acid substitutions. This variety of amino acid changes spread over different viral proteins was also seen in the ribavirin passaging experiments by Pauly and Lauring (27) and Cheung et al. (26). In the latter investigation, substitution V43I in PB1 was identified, after 17 passages, as conferring weak (2-fold) resistance to ribavirin, and this effect was attributed to slightly higher fidelity of the mutant polymerase. In our study, no resistance to T-705 or ribavirin emerged within 14 passages. We did not detect the PB1-V43I change despite deep-sequencing analysis. Likewise, the fact that the mutation rate remained stable during 14 passages under T-705 indicates that a mutant virus with increased fidelity was not selected under our experimental conditions. In another study, T-705 resistance did not develop within as many as 30 virus passages (14), and no signs of T-705 resistance were seen in a recently described patient study (17).
Regarding the biochemical mechanism, a clear distinction can be seen between T-705 and ribavirin. For ribavirin, the inhibitory effect upon viral RNA synthesis nicely correlates with its GTP-depleting effect, in both time and concentration dependency. Somewhat to our surprise, we observed that this GTP depletion, which is explained by inhibition of the IMPDH enzyme, is very fast and profound (90% reduction within 2 h) and is accompanied by a marked increase in the pools of UTP and CTP. Another study has shown that GTP depletion following IMPDH inhibition (in this case, by mycophenolic acid) coincides with an increase in the UTP pool (53). Thus, the virus mutagenic effect during sustained exposure to ribavirin seems to be explained by a skewed UTP/GTP balance. Ribavirin-5′-triphosphate is only marginally active in influenza virus polymerase enzyme assays, with IC50s of 70 μM (32), ∼100 μM (31), and 216 μM (our study). Hence, we consider it improbable that ribavirin could function as an alternative substrate for the polymerase and cause mutations in the influenza virus genome after its incorporation.
For T-705, the picture is entirely different, since it did not inhibit the IMPDH reaction in our cell-based assay or in the enzymatic study by Furuta et al. (23). We saw some drop in the GTP pool at high T-705 concentrations (i.e., 67% reduction at 600 μM, a concentration that is 60-fold higher than its antiviral EC50), but the levels of UTP or CTP were unchanged. Minor GTP depletion was also reported by Smee et al. (47), who saw a 36% reduction in the GTP pool in cells receiving 1,000 μM T-705. Possibly, T-705-ribosyl-5′-monophosphate requires conversion by GMP kinase, which at higher T-705 concentrations could interfere with the biosynthesis of GTP. It seems implausible that GTP depletion would be a factor in the anti-influenza virus effect of T-705. Rather, the strong inhibitory effect of T-705-RTP at the viral polymerase level implies that it functions as an alternative substrate for this enzyme. Consistent with the enzymatic experiments of Jin et al. (29), our cell culture data demonstrate that T-705 shuts off viral RNA synthesis at high concentrations. At lower concentrations, it is incorporated into the viral genome, giving either chain termination or chain elongation and inducing primarily G-to-A and C-to-U transitions due to its ambiguous base-pairing behavior. Incorporation of T-705 into the viral genome might be a relatively frequent event: the viral polymerase was shown to incorporate T-705-ribosyl-5′-triphosphate with an efficiency that is only 19- and 30-fold lower than those of GTP and ATP (29), respectively. This explains why the virus mutagenic effect is higher for T-705 than for ribavirin, since the impact of GTP depletion coupled to UTP/GTP imbalance is reasonably less pronounced.
One intriguing question is the nature of the noninfectious virus particles that are abundantly produced in T-705-exposed virus cultures. We exclude the possibility that these are defective viruses containing large internal deletions (54), since these would have been detected in our deep-sequencing analysis. A plausible hypothesis is that these are incomplete particles lacking one (or more) of the eight genome segments (55), due to chain termination by T-705 and, hence, the impossibility to generate the packaging signals at the 5′ and 3′ termini of the vRNA segment(s). The hypothesis that T-705 may enhance the production of incomplete influenza virus virions is supported by our observation that the virus readily returns to normal particle infectivity when T-705 is omitted from the culture. We do not exclude an alternative (and nonmutually exclusive) explanation that these nonviable particles contain T-705 incorporated into their genomes. To assess whether and to what degree T-705 can be incorporated into viral particles, the use of radiolabeled compound would be required. Our RT-qPCR and sequencing analyses probably ignored many (if not all) vRNA segments with incorporated T-705, since the RT reaction is likely slowed or terminated when faced with a T-705 molecule incorporated into the vRNA template.
To summarize (Fig. 8), T-705 and ribavirin have dual yet distinct effects on viral RNA synthesis. Both agents give complete inhibition at high concentrations. At lower concentrations, T-705 induces abundant production of noninfectious particles. Prolonged exposure to low concentrations of T-705 or ribavirin causes viral mutagenesis; this effect is 2-fold higher for T-705 than ribavirin and leads to reduced viral fitness associated with a wide range of amino acid substitutions. Whereas T-705 functions at the level of the viral polymerase, ribavirin acts by GTP depletion via IMPDH inhibition; this explains why the therapeutic index of T-705 by far exceeds that of ribavirin. Whether the mutagenic effect of these agents on influenza virus also plays in the clinical context is at this stage speculative; recent in vivo data obtained in a mouse influenza model (28) suggest that this may be possible for T-705. Quasispecies formation of hepatitis C virus following exposure to ribavirin has been observed in cell culture-based studies but not during long-term treatment of patients (56). In the case of influenza virus, T-705- or ribavirin-induced noninfectious or mutant viruses may be even less likely to appear, considering that influenza treatment is much shorter and that nonviable virus particles are probably cleared by the patient's immune response.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge excellent technical assistance from Stijn Stevens, Ria Van Berwaer, and Wim van Dam, and we acknowledge Wim Meert and Evelyne Luyten from the Genomics Core (KU Leuven) for performing the NGS experiments.
E.V. performed experiments and data analysis and wrote the manuscript; B.V. performed data analysis; J.V.H. performed experiments; S.G. and G.A. provided help with data analysis; P.L. provided help with data interpretation; V.K.R. performed chemical synthesis of test molecules; and L.N. performed study design and data interpretation and wrote the manuscript.
We have no conflicts of interest to declare.
This work was supported by the Flemish Fonds voor Wetenschappelijk Onderzoek (FWO grant no. 1509715N), the Bijzonder Onderzoeksfonds KU Leuven (BOF grant no. OT/14/115), and the European Union Seventh Framework Programme (FP7/2007-2013) under grant no. 278433-PREDEMICS and ERC grant no. 260864. The VIROGENESIS project receives funding from the European Union's Horizon 2020 research and innovation program under grant no. 634650.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01156-16.
REFERENCES
- 1.World Health Organization. March 2014. Influenza (seasonal). Fact sheet no. 211. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs211/en/. [Google Scholar]
- 2.Glezen WP, Schmier JK, Kuehn CM, Ryan KJ, Oxford J. 2013. The burden of influenza B: a structured literature review. Am J Public Health 103:e43–e51. doi: 10.2105/AJPH.2012.301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggerstaff M, Reed C, Swerdlow DL, Gambhir M, Graitcer S, Finelli L, Borse RH, Rasmussen SA, Meltzer MI, Bridges CB. 2015. Estimating the potential effects of a vaccine program against an emerging influenza pandemic—United States. Clin Infect Dis 60(Suppl 1):S20–S29. doi: 10.1093/cid/ciu1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samson M, Pizzorno A, Abed Y, Boivin G. 2013. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Jefferson T, Jones MA, Doshi P, Del Mar CB, Hama R, Thompson MJ, Spencer EA, Onakpoya I, Mahtani KR, Nunan D, Howick J, Heneghan CJ. 2014. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database Syst Rev 4:CD008965. doi: 10.1002/14651858.CD008965.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muthuri SG, Venkatesan S, Myles PR, Leonardi-Bee J, Al Khuwaitir TS, Al MA, Anovadiya AP, Azziz-Baumgartner E, Baez C, Bassetti M, Beovic B, Bertisch B, Bonmarin I, Booy R, Borja-Aburto VH, Burgmann H, Cao B, Carratala J, Denholm JT, Dominguez SR, Duarte PA, Dubnov-Raz G, Echavarria M, Fanella S, Gao Z, Gerardin P, Giannella M, Gubbels S, Herberg J, Iglesias AL, Hoger PH, Hu X, Islam QT, Jimenez MF, Kandeel A, Keijzers G, Khalili H, Knight M, Kudo K, Kusznierz G, Kuzman I, Kwan AM, Amine IL, Langenegger E, Lankarani KB, Leo YS, Linko R, Liu P, Madanat F, Mayo-Montero E, McGeer A, Memish Z, Metan G, Mickiene A, Mikic D, Mohn KG, Moradi A, Nymadawa P, Oliva ME, Ozkan M, Parekh D, Paul M, Polack FP, Rath BA, Rodriguez AH, Sarrouf EB, Seale AC, Sertogullarindan B, Siqueira MM, Skret-Magierlo J, Stephan F, Talarek E, Tang JW, To KK, Torres A, Torun SH, Tran D, Uyeki TM, Van ZA, Vaudry W, Vidmar T, Yokota RT, Zarogoulidis P, Nguyen-Van-Tam JS. 2014. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir Med 2:395–404. doi: 10.1016/S2213-2600(14)70041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moscona A. 2008. Medical management of influenza infection. Annu Rev Med 59:397–413. [DOI] [PubMed] [Google Scholar]
- 9.Stevaert A, Naesens L. 29 August 2016. The influenza virus polymerase complex: an update on its structure, functions and significance for antiviral drug design. Med Res Rev doi: 10.1002/med.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. 2002. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother 46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sleeman K, Mishin VP, Deyde VM, Furuta Y, Klimov AI, Gubareva LV. 2010. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob Agents Chemother 54:2517–2524. doi: 10.1128/AAC.01739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidwell RW, Barnard DL, Day CW, Smee DF, Bailey KW, Wong MH, Morrey JD, Furuta Y. 2007. Efficacy of orally administered T-705 on lethal avian influenza A (H5N1) virus infections in mice. Antimicrob Agents Chemother 51:845–851. doi: 10.1128/AAC.01051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. 2013. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res 100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. 2009. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res 82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyama Chemical Co, Ltd. 24 March 2014. The new drug application approval of “AVIGAN® tablet 200mg” in Japan for the anti-influenza virus drug. Toyama Chemical Co, Ltd, Tokyo, Japan: https://www.toyama-chemical.co.jp/eng/news/news140324e.html. [Google Scholar]
- 16.Zumla A, Memish ZA, Maeurer M, Bates M, Mwaba P, Al-Tawfiq JA, Denning DW, Hayden FG, Hui DS. 2014. Emerging novel and antimicrobial-resistant respiratory tract infections: new drug development and therapeutic options. Lancet Infect Dis 14:1136–1149. doi: 10.1016/S1473-3099(14)70828-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKimm-Breschkin JL, Fry AM. 2016. Meeting report: 4th ISIRV antiviral group conference: novel antiviral therapies for influenza and other respiratory viruses. Antiviral Res 129:21–38. doi: 10.1016/j.antiviral.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnussen CR, Douglas RG Jr, Betts RF, Roth FK, Meagher MP. 1977. Double-blind evaluation of oral ribavirin (Virazole) in experimental influenza A virus infection in volunteers. Antimicrob Agents Chemother 12:498–502. doi: 10.1128/AAC.12.4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein DS, Creticos CM, Jackson GG, Bernstein JM, Hayden FG, Schiff GM, Bernstein DI. 1987. Oral ribavirin treatment of influenza A and B. Antimicrob Agents Chemother 31:1285–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. 2009. Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients—Seattle, Washington, 2009. MMWR Morb Mortal Wkly Rep 58:893–896. [PubMed] [Google Scholar]
- 21.van der Vries E, Stelma FF, Boucher CA. 2010. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N Engl J Med 363:1381–1382. doi: 10.1056/NEJMc1003749. [DOI] [PubMed] [Google Scholar]
- 22.Kim WY, Young SG, Huh JW, Kim SH, Kim MJ, Kim YS, Kim HR, Ryu YJ, Han MS, Ko YG, Chon GR, Lee KH, Choi SH, Hong SB. 2011. Triple-combination antiviral drug for pandemic H1N1 influenza virus infection in critically ill patients on mechanical ventilation. Antimicrob Agents Chemother 55:5703–5709. doi: 10.1128/AAC.05529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K. 2005. Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother 49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naesens L, Guddat LW, Keough DT, van Kuilenburg AB, Meijer J, Vande Voorde J, Balzarini J. 2013. Role of human hypoxanthine guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir). Mol Pharmacol 84:615–629. doi: 10.1124/mol.113.087247. [DOI] [PubMed] [Google Scholar]
- 25.Baranovich T, Wong SS, Armstrong J, Marjuki H, Webby RJ, Webster RG, Govorkova EA. 2013. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol 87:3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung PP, Watson SJ, Choy KT, Fun SS, Wong DD, Poon LL, Kellam P, Guan Y, Malik Peiris JS, Yen HL. 2014. Generation and characterization of influenza A viruses with altered polymerase fidelity. Nat Commun 5:4794. doi: 10.1038/ncomms5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauly MD, Lauring AS. 2015. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J Virol 89:3584–3597. doi: 10.1128/JVI.03483-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marathe BM, Wong SS, Vogel P, Garcia-Alcalde F, Webster RG, Webby RJ, Najera I, Govorkova EA. 2016. Combinations of oseltamivir and T-705 extend the treatment window for highly pathogenic influenza A(H5N1) virus infection in mice. Sci Rep 6:26742. doi: 10.1038/srep26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. 2013. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS One 8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangawa H, Komeno T, Nishikawa H, Yoshida A, Takahashi K, Nomura N, Furuta Y. 2013. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob Agents Chemother 57:5202–5208. doi: 10.1128/AAC.00649-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eriksson B, Helgstrand E, Johansson NG, Larsson A, Misiorny A, Noren JO, Philipson L, Stenberg K, Stening G, Stridh S, Oberg B. 1977. Inhibition of influenza virus ribonucleic acid polymerase by ribavirin triphosphate. Antimicrob Agents Chemother 11:946–951. doi: 10.1128/AAC.11.6.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wray SK, Gilbert BE, Knight V. 1985. Effect of ribavirin triphosphate on primer generation and elongation during influenza virus transcription in vitro. Antiviral Res 5:39–48. doi: 10.1016/0166-3542(85)90013-0. [DOI] [PubMed] [Google Scholar]
- 33.Wray SK, Gilbert BE, Noall MW, Knight V. 1985. Mode of action of ribavirin: effect of nucleotide pool alterations on influenza virus ribonucleoprotein synthesis. Antiviral Res 5:29–37. [DOI] [PubMed] [Google Scholar]
- 34.Streeter DG, Witkowski JT, Khare GP, Sidwell RW, Bauer RJ, Robins RK, Simon LN. 1973. Mechanism of action of 1-b-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc Natl Acad Sci U S A 70:1174–1178. doi: 10.1073/pnas.70.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egawa H, Furuta Y, Murakami M, Takahashi K, Tsutsui Y, Uehara S. May 2004. Novel virus proliferation inhibition/virucidal method and novel pyradine nucleotide/pyradine nucleoside analogue. European patent EP 1417967 A1.
- 36.Vanderlinden E, Vanstreels E, Boons E, ter Veer W, Huckriede A, Daelemans D, Van LA, Roth E, Sztaricskai F, Herczegh P, Naesens L. 2012. Intracytoplasmic trapping of influenza virus by a lipophilic derivative of aglycoristocetin. J Virol 86:9416–9431. doi: 10.1128/JVI.07032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 38.Stevaert A, Dallocchio R, Dessi A, Pala N, Rogolino D, Sechi M, Naesens L. 2013. Mutational analysis of the binding pockets of the diketo acid inhibitor L-742,001 in the influenza virus PA endonuclease. J Virol 87:10524–10538. doi: 10.1128/JVI.00832-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanderlinden E, Goktas F, Cesur Z, Froeyen M, Reed ML, Russell CJ, Cesur N, Naesens L. 2010. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J Virol 84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Archer J, Baillie G, Watson SJ, Kellam P, Rambaut A, Robertson DL. 2012. Analysis of high-depth sequence data for studying viral diversity: a comparison of next generation sequencing platforms using Segminator II. BMC Bioinformatics 13:47. doi: 10.1186/1471-2105-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balzarini J, Gago F, Kulik W, van Kuilenburg AB, Karlsson A, Peterson MA, Robins MJ. 2012. Introduction of a fluorine atom at C3 of 3-deazauridine shifts its antimetabolic activity from inhibition of CTP synthetase to inhibition of orotidylate decarboxylase, an early event in the de novo pyrimidine nucleotide biosynthesis pathway. J Biol Chem 287:30444–30454. doi: 10.1074/jbc.M112.378091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meneghesso S, Vanderlinden E, Brancale A, Balzarini J, Naesens L, McGuigan C. 2013. Synthesis and biological evaluation of purine 2′-fluoro-2′-deoxyriboside ProTides as anti-influenza virus agents. ChemMedChem 8:415–425. doi: 10.1002/cmdc.201200562. [DOI] [PubMed] [Google Scholar]
- 43.Meneghesso S, Vanderlinden E, Stevaert A, McGuigan C, Balzarini J, Naesens L. 2012. Synthesis and biological evaluation of pyrimidine nucleoside monophosphate prodrugs targeted against influenza virus. Antiviral Res 94:35–43. doi: 10.1016/j.antiviral.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 45.Rochovansky OM. 1976. RNA synthesis by ribonucleoprotein-polymerase complexes isolated from influenza virus. Virology 73:327–338. doi: 10.1016/0042-6822(76)90394-9. [DOI] [PubMed] [Google Scholar]
- 46.Balzarini J, De Clercq E. 1992. Assay method for monitoring the inhibitory effects of antimetabolites on the activity of inosinate dehydrogenase in intact human CEM lymphocytes. Biochem J 287:785–790. doi: 10.1042/bj2870785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smee DF, Hurst BL, Egawa H, Takahashi K, Kadota T, Furuta Y. 2009. Intracellular metabolism of favipiravir (T-705) in uninfected and influenza A (H5N1) virus-infected cells. J Antimicrob Chemother 64:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paeshuyse J, Dallmeier K, Neyts J. 2011. Ribavirin for the treatment of chronic hepatitis C virus infection: a review of the proposed mechanisms of action. Curr Opin Virol 1:590–598. doi: 10.1016/j.coviro.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 49.Tisdale M, Ellis M, Klumpp K, Court S, Ford M. 1995. Inhibition of influenza virus transcription by 2′-deoxy-2′-fluoroguanosine. Antimicrob Agents Chemother 39:2454–2458. doi: 10.1128/AAC.39.11.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leyssen P, Balzarini J, De CE, Neyts J. 2005. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol 79:1943–1947. doi: 10.1128/JVI.79.3.1943-1947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak DP. 1980. Defective interfering influenza viruses. Annu Rev Microbiol 34:619–644. doi: 10.1146/annurev.mi.34.100180.003155. [DOI] [PubMed] [Google Scholar]
- 52.Graci JD, Cameron CE. 2008. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol 3:553–566. doi: 10.2217/17460794.3.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raab M, Daxecker H, Karimi A, Markovic S, Cichna M, Markl P, Muller MM. 2001. In vitro effects of mycophenolic acid on the nucleotide pool and on the expression of adhesion molecules of human umbilical vein endothelial cells. Clin Chim Acta 310:89–98. doi: 10.1016/S0009-8981(01)00527-7. [DOI] [PubMed] [Google Scholar]
- 54.Dimmock NJ, Easton AJ. 2014. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J Virol 88:5217–5227. doi: 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brooke CB. 2014. Biological activities of ‘noninfectious’ influenza A virus particles. Future Virol 9:41–51. doi: 10.2217/fvl.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quiles-Pérez R, Munoz-de-Rueda P, Maldonado AM, Martin-Alvarez A, Quer J, Salmeron J. 2014. Effects of ribavirin monotherapy on the viral population in patients with chronic hepatitis C genotype 1: direct sequencing and pyrosequencing of the HCV regions. J Med Virol 86:1886–1897. doi: 10.1002/jmv.24035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.