Abstract
Carbapenemase-producing Enterobacteriaceae (CPE) are increasing globally; here we report on the investigation of CPE in Canada over a 5-year period. Participating acute care facilities across Canada submitted carbapenem-nonsusceptible Enterobacteriaceae from 1 January 2010 to 31 December 2014 to the National Microbiology Laboratory. All CPE were characterized by antimicrobial susceptibilities, pulsed-field gel electrophoresis, multilocus sequence typing, and plasmid restriction fragment length polymorphism analysis and had patient data collected using a standard questionnaire. The 5-year incidence rate of CPE was 0.09 per 10,000 patient days and 0.07 per 1,000 admissions. There were a total of 261 CPE isolated from 238 patients in 58 hospitals during the study period. blaKPC-3 (64.8%) and blaNDM-1 (17.6%) represented the highest proportion of carbapenemase genes detected in Canadian isolates. Patients who had a history of medical attention during international travel accounted for 21% of CPE cases. The hospital 30-day all-cause mortality rate for the 5-year surveillance period was 17.1 per 100 CPE cases. No significant increase in the occurrence of CPE was observed from 2010 to 2014. Nosocomial transmission of CPE, as well as international health care, is driving its persistence within Canada.
INTRODUCTION
Gram-negative organisms account for 5 of the top 10 most common bacterial organisms isolated from patients in Canadian hospitals, with Enterobacteriaceae representing 33.8% of isolates (1). Enterobacteriaceae are implicated in both community- and hospital-acquired infections. Their plasticity and persistence can be attributed to the ease in which they are transferred (contaminated food, water, environmental surfaces, and hand carriage). The increase in carbapenem-resistant Enterobacteriaceae (CRE) is predominantly due to the acquisition of carbapenemase genes residing on mobile genetic elements (2).
Most carbapenemases hydrolyze penicillins, cephalosporins, and carbapenems. Additionally, carbapenemases are often associated with multidrug resistance, as they are commonly found on plasmids containing multiple determinants of resistance to other classes of antimicrobials, making treatment options limited and the use of antimicrobials such as tigecycline and colistin more common (3).
The occurrence of carbapenemase-producing Enterobacteriaceae (CPE) in Canada is rare. The first Canadian Nosocomial Infection Surveillance Program report on the occurrence of carbapenem-resistant Gram-negative bacilli from 2009 to 2010 indicated that only 0.1% of Enterobacteriaceae were resistant to carbapenems, and of those, 16.9% harbored a carbapenemase gene (4). In addition, in a national point prevalence survey, it was determined that CPE were identified in only 7% of Canadian hospitals in 2012 (5). Currently, Canadian reports of CPE from single hospital sites are becoming more common, including the emergence of blaKPC-2/-3 organisms among Klebsiella pneumoniae (6, 7) and more broadly among Enterobacteriaceae (8). Additionally, the emergence of blaNDM-1 (9, 10) and blaOXA-48/-181 (11, 12) organisms as well as outbreaks of blaNDM-1 (13) and blaKPC-3 (14) organisms have been described.
This report describes the incidence, risk factors for acquisition, epidemiology, and molecular mechanism of carbapenem-nonsusceptible Enterobacteriaceae over 5 years (2010 to 2014) from selected centers in a national hospital surveillance network.
MATERIALS AND METHODS
Surveillance period and surveillance population.
Surveillance was carried out between 1 January 2010 and 31 December 2014 and included in- and outpatients from Canadian acute care hospitals participating in the Canadian Nosocomial Infection Surveillance Program (CNISP). Participating hospitals increased from 33 in 2010 to 58 in 2014. The 58 CNISP hospitals were distributed among 10 Canadian provinces: 10 (17%) in the east (Prince Edward Island, New Brunswick, Nova Scotia, and Newfoundland and Labrador), 28 (48%) in central Canada (Quebec and Ontario), and 20 (35%) in the west (Manitoba, Saskatchewan, Alberta, and British Columbia).
Study eligibility criteria.
Any Enterobacteriaceae collected from a patient (colonized or infected) admitted to a CNISP participating hospital, emergency department, or outpatient clinic that exhibited nonsusceptibility to imipenem, meropenem (≥2 mg/liter from 2010 to 2014), or ertapenem (≥0.5 mg/liter in 2010 to 2011 and ≥1 mg/liter in 2012 to 2014) in accordance with the Clinical and Laboratory Standards Institute (15) was considered eligible for inclusion. Primary laboratory detection and identification were conducted by either the hospital or the provincial laboratories using standard diagnostic laboratory procedures. All eligible isolates were sent to the National Microbiology Laboratory (NML; Winnipeg, Canada) for MIC confirmation using ertapenem, meropenem, and imipenem Etest strips (bioMérieux, St. Laurent, QC, Canada).
Patient questionnaires.
The following were submitted with all isolates: age, sex, date of admission, date of positive culture, organism, ward, and anatomical site of isolation. From 2011 to 2014 a questionnaire was required for all patients with CPE which provided detailed patient history regarding travel, antimicrobial use, underlying medical conditions, and patient outcome. Patients were monitored for a maximum of 30 days after the date of the first positive CPE culture to determine all-cause deaths and deaths attributable to CPE infection (as judged by local case review).
Molecular characterization of isolates.
Detection of the genes for the following carbapenemases by PCR was conducted as previously described (4): NDM, KPC, IMP, VIM, GES, and SME. Detection further included genes for OXA-48-type (16) and NMC/IMI-type (NMC-1 5′-TGGTGTCTACGCTTTAGAC-3′ NMC-2 5′-ACCATGTCTGATAGGTTTCC-3′) enzymes. PCR mapping of the Tn4401 element was conducted using previously described primers (17). Multilocus sequence typing (MLST) (http://bigsdb.pasteur.fr/klebsiella/ and http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) and macrorestriction analysis using pulsed-field gel electrophoresis (PFGE) as previously described (4) were conducted on all CPE. BioNumerics software (version 3.5; Applied Maths, Saint Lartens-Latem, Belgium) was used to analyze fingerprints. Plasmid restriction fragment length polymorphism (pRFLP) analysis was conducted as previously described using electroporation to transfer a carbapenemase gene-harboring plasmid to Escherichia coli DH10B (4) in which NDM-type plasmids were digested with BglII and all other carbapenemase gene-containing plasmids with EcoRI. Plasmid-based replicon typing (PBRT) was conducted as previously described (18, 19). Antimicrobial susceptibility testing was performed using Vitek2 (AST-GN25 or AST-N219; bioMérieux, St. Laurent, Canada) using 2014 CLSI breakpoints (20). Tigecycline breakpoints were based on FDA breakpoints for Enterobacteriaceae (susceptible [S], ≤2 mg/liter; intermediate, 4 mg/liter; resistant [R], ≥8 mg/liter). Etest (bioMérieux) was used to determine colistin MICs with EUCAST breakpoints for Enterobacteriaceae (S ≤ 2 mg/liter; R > 2 mg/liter). Antimicrobial testing on plasmids was done on representatives from large pRFLP clusters along with all plasmids with unique pRFLP profiles.
Statistical analyses.
Annual incidence rates for CPE and CRE were calculated from the number of microorganisms tested divided by the number of patient days multiplied by 10,000 and number of microorganisms divided by patient admissions multiplied by 1,000. The numbers of microorganisms are provided by the submitting hospitals yearly. The numerators of these rates consists of the numbers of cases reported and therefore may include individuals more than once. Rates exclude cases identified in emergency departments and outpatient clinics.
Descriptive statistics and bivariate analysis were conducted on all molecular data. These data exceed the number of individual cases, as patients may have more than one microorganism identified. In addition, antimicrobial resistance rates were subjected to the Cochran-Armitage test in order to assess significance, at the 5% level, of year-related trends. Individual patient cases (inpatients, emergency patients, and outpatients) were used to derive patient demographics and risk factors (underlying condition, age, sex, type of carbapenemase, etc.). Comparisons were conducted using Fisher's exact two-sided test at the 5% significance level. Statistical analyses were performed using SAS EG version 5.1.
RESULTS
Patient demographics for CPE.
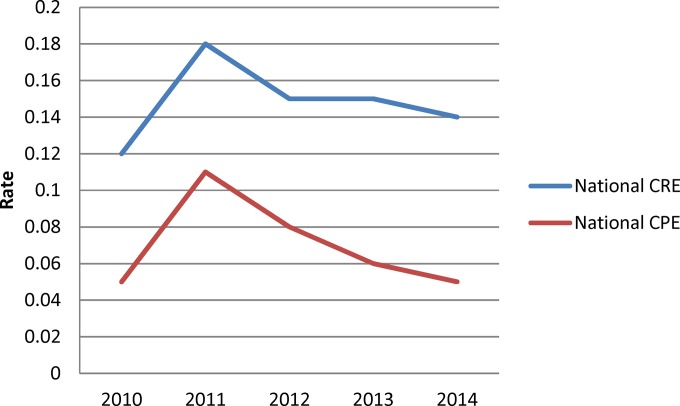
A total of 823 carbapenem-intermediate or resistant Enterobacteriaceae were eligible for study inclusion. Of those, 613 (74.5%) were CRE and 261 (31.7%) were CPE as determined by PCR. Only 10 CPE (3.8%) were carbapenem intermediate by Etest, of which 7 produced KPC, 2 VIM, and 1 OXA-48. There were a total of 261 CPE isolated from 238 patients during the study period. From 2010 to 2014, the overall CNISP incidence rates of CRE and CPE per 10,000 patient days were 0.19 and 0.09 and per 1,000 admissions were 0.15 and 0.07, respectively (Fig. 1). Overall, the rates did not significantly increase over the 5 years. The increase in CRE and CPE in 2011 is most likely attributable to a KPC outbreak at a single hospital site.
FIG 1.
Overall national carbapenem-resistant Enterobacteriaceae (CRE) and carbapenemase-producing Enterobacteriaceae (CPE) rates per 1,000 patient admissions from 2010 to 2014.
Table 1 summarizes demographic data available for patients with CPEs. The majority of CPE cases (84%) were isolated from inpatients, while the remainder were identified from the outpatient setting (16%). Male patients contributed 61.9% of isolates. The most common sites of isolation were the rectum (48.7%), urine (26.1%), and respiratory tract (12.6%). Colonizations represented 59.4% of CPE cases. Although nosocomial transmission was observed in 55% of cases, approximately 80% of these cases were part of outbreaks at three hospital sites. Data on underlying medical conditions were available for 211 (88.7%) of the CPE cases, of which 89.1% (n = 188) included at least one underlying medical condition. The most commonly reported conditions were heart disease (36.7%), diabetes (29.8%), active cancer (25.5%), and lung disease (21.8%).
TABLE 1.
Summary of available demographics from patients harboring carbapenemase-producing Enterobacteriaceae
| Characteristica | No. (%) of cases |
|---|---|
| Gender (n = 236) | |
| Male | 146 (61.9) |
| Female | 90 (38.1) |
| Age, yrs (n = 238) | |
| 0–18 | 9 (3.8) |
| 19–64 | 88 (37.0) |
| ≥65 | 141 (59.2) |
| Hospital ward (n = 238) | |
| Medical | 93 (39.1) |
| Emergency room | 32 (13.4) |
| Intensive care unit | 50 (21.0) |
| Surgical | 34 (14.3) |
| Other | 25 (10.5) |
| Isolate site (n = 238) | |
| Stool/rectal swab | 116 (48.7) |
| Urine | 62 (26.1) |
| Sputum | 30 (12.6) |
| Blood | 20 (8.4) |
| Skin/soft tissue | 25 (10.5) |
| Surgical site | 11 (4.6) |
| Other | 9 (3.8) |
| Infections/colonizations (n = 212) | |
| Infection | 86 (40.6) |
| Colonization | 126 (59.4) |
| Nosocomial transmission (n = 164) | |
| Yes | 91 (55.5) |
| No | 73 (44.5) |
Demographic denominators vary depending on data available from hospital sites. Patients can report multiple hospital wards and isolation sites, which explains why the number of isolation site exceeds the number of patients. Patients can report organisms which can represent either infections or colonizations. However, cases that reported both a colonization and an infection would be classified only as infection, since infection is considered more important than colonization.
Data on international travel were collected from 201 (84.5%) patients with CPE cases. Table 2 describes the 49 cases (24.3%) involving persons who reported international travel within the 12 months prior to diagnosis; India was the most common travel destination, associated with 15 of the 49 cases (30.6%). Among these cases, 42 (85.7%) individuals reported that they had sought medical care while on international travel. Of the blaNDM-type-associated cases involving known travel, 87.5% were linked to patients who had sought medical attention within the Indian subcontinent. Similarity, 75% of blaOXA-48-type-associated cases involved patients who had sought medical attention in India.
TABLE 2.
Distribution of CPE isolated from patients with international travel history within 12 months of date of positive culture
| Country | Carbapenemase(s) | Organism(s) | No. of cases | Medical attention receiveda |
|---|---|---|---|---|
| Bahamas | KPC-2 | K. pneumoniae | 1 | Y |
| Bangladesh | NDM-7 | E. coli | 1 | Y |
| China, Shanghai | KPC- | K. pneumoniae | 1 | Y |
| Croatia | VIM-1 | Enterobacter spp. | 1 | Y |
| United States | KPC-3 | K. pneumoniae | 1 | Unknown |
| Enterobacter spp. | 2 | Y | ||
| Ecuador | KPC-2 | Enterobacter cloacae | 1 | Unknown |
| Egypt | OXA-48 | E. coli | 1 | Y |
| Greece | KPC-2, VIM-1 | K. pneumoniae | 1 | Y |
| KPC-2 | K. pneumoniae | 1 | Y | |
| India | NDM-1 | Providencia rettgeri | 1 | Y |
| K. pneumoniae | 4 | Y (3), N (1) | ||
| E. coli | 1 | Y | ||
| Enterobacter spp. | 1 | Y | ||
| NDM-5 | E. coli | 1 | Y | |
| NDM-7 | E. coli | 2 | Y | |
| OXA-181 | K. pneumoniae | 2 | Y (1), N (1) | |
| E. coli | 1 | Y | ||
| OXA-232 | K. pneumoniae | 1 | Y | |
| NDM-1, OXA-232 | K. pneumoniae | 1 | Y | |
| Israel | KPC-3 | K. pneumoniae | 1 | Y |
| KPC-2 | K. pneumoniae | 1 | Y | |
| Italy | KPC-3 | K. pneumoniae | 1 | Y |
| Oman | NDM-1 | K. pneumoniae | 1 | Y |
| Pakistan | NDM-2 | K. pneumoniae | 1 | Y |
| Puerto Rico | KPC-3 | K. pneumoniae | 1 | Y |
| Saudi Arabia | OXA-48 | K. pneumoniae | 1 | Y |
| E. coli | 1 | Y | ||
| Serbia | NDM-1, OXA-48 | K. pneumoniae | 1 | Y |
| Sri Lanka | NDM-1 | K. pneumoniae | 1 | Y |
| Not specified | KPC-3 | Enterobacter spp. | 1 | Unknown |
| K. pneumoniae | 2 | Unknown (1), Y (1) | ||
| Kluyvera spp. | 1 | Unknown | ||
| NDM-type | K. pneumoniae | 3 | Y | |
| Enterobacter spp. | 1 | Y | ||
| Providencia rettgeri | 1 | Y | ||
| E. coli | 1 | Y | ||
| S. marcesens | 1 | Unknown | ||
| OXA-48 | K. pneumoniae | 1 | Y | |
| OXA-181 | K. pneumoniae | 1 | Unknown |
Y, yes; N, no.
Treatment data were available for 147 CPE cases (61.8%) and were collected only from 2010 to 2013. Antimicrobial treatment was administered to 73.5% (n = 108) of CPE cases within 2 weeks of their diagnosis. The most commonly prescribed antimicrobials were glycopeptides (29.5%), β-lactams (32%), fluoroquinolones (22.1%), and carbapenems (19.7%). Forty-nine percent of patients received >1 antimicrobial.
Thirty-day outcomes were available for 82 cases, with 14 (17.1%) deaths reported. Of total bloodstream infections (n = 19), nine (47.4%) deaths were reported. Death was reported as attributable to a CPE infection in four (28.6%) cases; three were bacteremic events (S. marcescens harboring blaGES-5 and K. pneumoniae and Enterobacter spp. harboring blaKPC-3) and one case involved a surgical site infection (K. pneumoniae harboring blaOXA-232).
CPE.
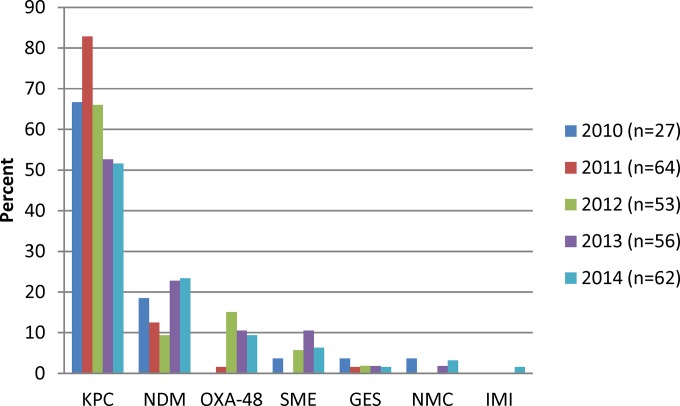
A total of 261 CPE were collected over the study period as follows: 2010, 26 (10.3%); 2011, 64 (24.5%); 2012, 53 (20.3%); 2013, 56 (21.5%); and 2014, 62 (23.8%). The proportion of carbapenemases observed per year is shown in Fig. 2. Over the 5 years of study, blaKPC-type carbapenemases had the highest incidence, with an average of 66.9% of total CPE per year, followed by blaNDM-1 carbapenemases at 17.3% per year. Overall, there was a significant decrease in blaKPC carbapenemases (P = 0.0458), whereas blaSME (P = 0.0494) and blaOXA-48-type (P < 0.0001) carbapenemases increased significantly.
FIG 2.
Proportion of carbapenemases by year (n = 265). Note that there were 261 CPE with 265 carbapenemases identified.
All carbapenemases were sequenced to confirm variant type. There were 169 blaKPC (156 blaKPC-3, 9 blaKPC-2, 2 blaKPC-4, and 2 unknown), 46 blaNDM (36 blaNDM-1, 1 blaNDM-3, 2 blaNDM-5 6 blaNDM-7, and 1 unknown), 21 blaOXA-48-type (10 blaOXA-48, 9 blaOXA-181, and 2 blaOXA-232), 4 blaVIM (3 blaVIM-1 and 1 blaVIM-2), 5 blaGES-5, 14 blaSME (10 blaSME-1 and 4 blaSME-2), 1 blaIMP-13, and 4 blaNMC/IMI (2 blaNMC-A, 1 blaIMI-1, and 1 blaIMI-5) carbapenemases. Four K. pneumoniae isolates contained multiple carbapenemases: 1 blaKPC-2 and blaVIM-1 carbapenemases, and 3 blaNDM and blaOXA-48-type carbapenemases. In addition, CPE coproduced the β-lactamases SHV (56.3%), TEM (49.0%), CTX-M (24.9%), OXA-1 (15.7%), and/or CMY-2 (8.0%). Table 3 describes the distribution of CPE among Enterobacteriaceae. K. pneumoniae, Enterobacter spp., and E. coli were the top three carbapenemase producers.
TABLE 3.
Distribution of 261 CPE harboring 265 carbapenemasesa
| Pathogen | No. of isolates with indicated type of carbapenemase |
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| NDM | KPC | OXA-48-type | VIM | GES | IMP | SME | NMC/IMI | ||
| K. pneumoniae | 28 | 88 | 15 | 1 | 0 | 0 | NA | NA | 132 |
| E. coli | 10 | 12 | 6 | 0 | 2 | 0 | NA | NA | 30 |
| Enterobacter sp. | 2 | 36 | 0 | 3 | 0 | 1 | NA | 4 | 46 |
| Serratia sp. | 1 | 14 | 0 | 0 | 3 | 0 | 14 | NA | 32 |
| Citrobacter sp. | 1 | 10 | 0 | 1 | 0 | 0 | NA | NA | 12 |
| Klebsiella oxytoca | 0 | 6 | 0 | 0 | 0 | 0 | NA | NA | 6 |
| Morganella morganii | 2 | 0 | 0 | 0 | 0 | 0 | NA | NA | 2 |
| P. rettgeri | 2 | 0 | 0 | 0 | 0 | 0 | NA | NA | 2 |
| Pantoea sp. | 0 | 1 | 0 | 0 | 0 | 0 | NA | NA | 1 |
| Kluyvera sp. | 0 | 2 | 0 | 0 | 0 | 0 | NA | NA | 2 |
| Total | 46 | 169 | 21 | 5 | 5 | 1 | 14 | 4 | 265 |
Four isolates contained multiple carbapenemases; a K. pneumoniae isolate (blaKPC-2 and blaVIM-1) and three K. pneumoniae isolates (blaNDM-1 and blaOXA-48). NA, not applicable.
Antimicrobial susceptibility testing of CPE.
Antimicrobial susceptibilities for all CPE over the 5 years of study can be found in Table S1 in the supplemental material. CPEs were commonly resistant to ciprofloxacin (59.8%), tobramycin (59.4%), and trimethoprim-sulfamethoxazole (70.1%). Resistance to tigecyline (18.4%) and colistin (5%) was observed. Over 5 years of study, CPEs demonstrated significantly increased resistance to piperacillin-tazobactam (P < 0.0001), cefazolin (P = 0.0094), meropenem (P = 0.0011), and gentamicin (P = 0.0336). There was a significant decrease in resistance to ciprofloxacin (P = 0.022), amikacin (P = 0.0031), and nitrofurantoin (P = 0.0029). These resistance patterns reflected the emergence of blaOXA-48-type and blaSME-type carbapenemase producers, which tended to have resistance to fewer non-β-lactam antimicrobials (Table 4).
TABLE 4.
Antimicrobial resistance observed in CPE from 2010 to 2014
| Antimicrobial | % of isolates producing indicated carbapenemase and resistant to antimicrobial |
|||||||
|---|---|---|---|---|---|---|---|---|
| NDM (n = 43) | OXA-48 (n = 18) | KPC (n = 169) | GES (n = 5) | VIM (n = 4) | OXA-48 + NDM (n = 3) | SME (n = 14) | NMC (n = 4) | |
| Ampicillin | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Piperacillin-tazobactam | 93 | 100 | 98.2 | 100 | 100 | 100 | 14.3 | 50 |
| Cefazolin | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 75 |
| Ceftriaxone | 100 | 83.3 | 99.4 | 100 | 100 | 100 | 0 | 50 |
| Ciprofloxacin | 86 | 72.2 | 58.6 | 40 | 75 | 66.7 | 0 | 0 |
| Ertapenem | 100 | 100 | 96.4 | 100 | 75 | 100 | 100 | 100 |
| Meropenem | 90.7 | 61.1 | 88.8 | 100 | 75 | 100 | 92.9 | 75 |
| Amikacin | 60.5 | 11.1 | 29 | 20 | 25 | 66.7 | 0 | 0 |
| Tobramycin | 83.7 | 61.1 | 57.4 | 100 | 75 | 100 | 0 | 0 |
| Gentamicin | 76.7 | 72.2 | 33.1 | 80 | 50 | 100 | 0 | 0 |
| Nitrofurantoin | 79.1 | 72.2 | 55 | 80 | 50 | 66.7 | 100 | 0 |
| Trimethoprim-sulfamethoxazole | 76.7 | 77.8 | 72.8 | 80 | 100 | 100 | 0 | 0 |
| Tigecylcine | 18.6 | 22.2 | 20.1 | 0 | 0 | 66.7 | 0 | 0 |
| Colistin | 7 | 5.6 | 5.3 | 0 | 0 | 0 | NA | 0 |
Table 4 describes the antimicrobial resistance associated with each carbapenemase. Significantly higher rates of resistance were observed for ciprofloxacin (P < 0.001) and all aminoglycosides (P ≤ 0.001) in NDM producers than in KPC producers and for amikacin (P < 0.001) than in OXA-48-type producers. Nonsusceptibilities to tigecycline were observed in 47.3%, 43.5%, and 35.5% of blaNDM, blaOXA-48-type, and blaKPC carbapenemase producers, respectively. Colistin resistance was observed in 7%, 5.6%, and 5.3% of blaNDM, blaOXA-48-type, and blaKPC carbapenemase producers, respectively. One isolate, a K. pneumoniae ST512 blaKPC carbapenemase producer, was resistant to all classes of antimicrobials tested, including tigecycline and colistin. A recent publication describes the interim recommendation of reporting extensively drug-resistant (XDRO) and pan-drug-resistant (PDRO) Gram-negatives (21). According to these recommendations, we found 122 (46.7%) and 53 (20.3%) of CPE to be XDRO and PDRO, respectively. The proportions of CPE with a PDRO phenotype were 41.0% of NDM, 16% of OXA-48-type, and 13% of KPC carbapenemase producers.
DNA macrorestriction of CPE.
Pulsed-field gel electrophoresis (PFGE) was conducted on all carbapenemase-producing (CP) E. coli, Serratia marcescens, Enterobacter sp., and K. pneumoniae isolates.
Of the 46 CP Enterobacter spp., 36 (78.3%) produced blaKPC-type enzymes. A single hospital site reported an ongoing outbreak that began in 2010 (14) and accounted for 4 KPC-3 clusters representing 41.3% of Enterobacter spp. (data not shown). The remaining Enterobacter spp. (15 producing blaKPC-3 carbapenemase, 1 blaKPC-2, 2 blaKPC-4, 2 blaNDM-1, 3 blaVIM-1, 1 blaIMP-13, and 3 blaNMC/IMI) all had unique pattern types and represent 11 different hospital sites.
K. pneumoniae isolates represented the majority of CPE (126/261 [48.3%]) and were shown to produce blaKPC-type (n = 85), blaNDM-1 (n = 25), and blaOXA-48-type (n = 12) carbapenemases as well as coproduce blaKPC-2 plus blaVIM-1 (n = 1) and blaNDM-1 plus blaOXA-48-type (n = 3) carbapenemases (see Fig. S1 in the supplemental material). Of 126 CP K. pneumoniae isolates, 53.2% fall into 1/3 clonal clusters (I, n = 7; II, n = 48; and III, n = 12). Cluster I isolates are all sequence type 14 (ST14) isolates harboring blaOXA-181 (n = 5), blaNDM-1 (n = 1), or blaNDM-1 plus blaOXA-252 (n = 1) from either a western (n = 6) or central (n = 1) hospital site. Cluster II isolates are blaKPC-3 Tn4401a-like-carbapenemase-producing K. pneumoniae isolates mainly from a single hospital site in central Canada (38/48 [79.2%]). These isolates belonged to either ST258 or ST512 (a single base pair change in one allele from ST258). Cluster III isolates are blaNDM-1 carbapenemase-producing K. pneumoniae ST147 isolates from four sites in central Canada, one of which was associated with an outbreak due to nosocomial transmission in 2011 (13).
Among 30 CP E. coli (2 blaGES-5, 6 blaOXA-48/-181, 10 blaNDM-1/-5/-7, and 12 blaKPC-3) 28 unique PFGE fingerprints and 17 sequence types were observed (data not shown). The global endemic strain ST131 was observed in 5 blaKPC-3 carbapenemase-producing E. coli isolates.
Of 32 CP S. marcescens isolates identified, 12 (37.5%) clustered with >95% similarity (data not shown). These isolates were all blaKPC-3 carbapenemase producers that contained a Tn4401b-like element upstream of the blaKPC-3 gene. With one exception, all were from a single site in central Canada and were isolated in 2010 (n = 4), 2011 (n = 6), and 2012 (n = 2), suggesting a persistent blaKPC-3 carbapenemase-harboring clone at this site. A cluster of blaSME-1 carbapenemase producers (n = 5) were isolated from 3 hospital sites in western and central Canada between 2013 and 2014. This pulsotype has been previously observed across Canada (east, central, and west Canada), in 2010 to 2012 (22).
Plasmid analysis.
Plasmids harboring carbapenemases were successfully isolated from 226 CP Enterobacteriaceae, and their pRFLP patterns were analyzed. Forty-one blaNDM-1/3/5/7-carrying plasmids were isolated (see Fig. S2 in the supplemental material). NDM plasmid patterns were mainly diverse, and only three small clusters were observed: a cluster with an unknown Inc group isolated from K. pneumoniae ST11, an IncR cluster from K. pneumoniae ST147 from an outbreak in 2011 (13), and a cluster with an unknown Inc group isolated from multiple species harboring blaNDM-7 observed in five different hospital sites across Canada. NDM plasmids also carried markers of resistance to aminoglycosides (82.5%) and trimethoprim-sulfamethoxazole (25%).
Of pRFLP patterns from 165 blaKPC-type, 4 blaGES-5, 3 blaVIM-1, and 11 blaOXA-48-type carbapenemases, 101 (54.6%) fell into one of seven clusters (see Fig. S3 in the supplemental material). The largest cluster (II [n = 42]) belonged to IncFllA and consisted predominantly of K. pneumoniae (40/42 [95%]) blaKPC-3 producers. IncFllA plasmids contained additional markers of resistance to aminoglycosides (6.7%) and trimethoprim-sulfamethoxazole (20%). Though predominantly from a single hospital site (n = 32), these plasmids were also isolated from six other sites (n = 12). Conversely, the second largest cluster (IV [n = 24]) belonged to IncP and IncL/M, which all harbored blaKPC-3 and were isolated from 5 different bacterial species from a single hospital site. Plasmid type IncN (n = 50) was observed in several clusters (I, VI, and VII) as well as among numerous other pattern types. The IncN group has much more diverse pRFLP pattern types than the IncFllA or IncP/IncL/M groups. IncN plasmids were isolated from 6 different bacterial species, and all harbored blaKPC-type, with the exception of one blaOXA-48. IncN plasmids carried additional markers of resistance to the aminoglycosides (38.9%) and trimethoprim-sulfamethoxazole (86.1%).
Six of seven blaOXA-48 harboring plasmids clustered into an unknown incompatibility group (cluster V). These isolates came from 5 different hospital sites in 2012, 2013, and 2014, suggesting that a successful OXA-48 plasmid is circulating in Canada.
Isolation of IMI/NMC plasmids was attempted and not successful, suggesting chromosomal location of the genes.
DISCUSSION
The global emergence of CPE is a growing concern. Although the first carbapenemase was described >20 years ago, extensive reporting of CPE has only occurred in the last 10 years (23). Reports of endemic KPC have been described from Latin America, the eastern United States, Europe, and Southeast Asia. In addition, the global dissemination of NDM since its discovery in 2009 (24) and the current emergence of OXA-48-type carbapenemase outside regions where it is endemic since its discovery in 2003 (16) have highlighted the global emergence and dissemination of CPE and the need for surveillance. In spite of increased global incidence of CRE, the current study from a national surveillance network in Canada reports a rate of 0.07 CPE/1,000 admissions or 0.09 CPE per 10,000 admissions, with no significant increase over the 5 years.
Limitations to the study include the lack of information regarding laboratory protocols for CPE screening and the criteria for screening patients for CPE. Some sites maybe underreporting CPE occurrence by identifying only clinical cases. A national survey on CPE laboratory practices and infection prevention surveillance is under way.
Common reservoirs have been described for specific carbapenemases: the United States, Israel, Greece, and Italy for blaKPC carbapenemases; Turkey and North Africa for blaOXA-48 carbapenemases; and the Indian subcontinent is commonly associated with blaNDM, blaKPC, and blaOXA-181 carbapenemases (23). In the current study, we found that nearly a quarter (24.3%) of carbapenemases were associated with international travel, including health care. A large proportion of cases (48%) were first identified from rectal cultures performed for high-risk patient groups, reflecting the value of this type of screening in identifying CPE carriers. We found the blaOXA-48 and blaNDM carbapenemase producers associated with patients that had traveled to India and sought medical attention, which is consistent with previous reports (25–27). Although blaKPC carbapenemase producers were identified from a number of patients with history of travel to regions where blaKPC carbapenemase producers are endemic (Greece, Italy, and the eastern United States), infection or colonization with blaKPC carbapenemase producers was significantly less likely to be associated with patients who had traveled internationally than was infection or colonization with CPE producing either blaNDM or blaOXA-48-type carbapenemases (P < 0.0001). This is most likely attributed to a large outbreak of K. pneumoniae ST512 producing blaKPC at a single hospital site over the course of this study (14).
Although blaKPC represented the most common carbapenemase gene, there was a significant decrease in the number of blaKPC carbapenemase producers over the study (P = 0.0458). This finding is most likely again due to the control of a large outbreak at a single site. Conversely, there was a significant increase in the number of cases involving blaSME (P = 0.0494) and blaOXA-48-like (P < 0.0001) carbapenemase producers. blaSME was the first carbapenemase gene identified in a member of the Enterobacteriaceae originally isolated from a patient in London in 1982 (28). Current cases of organisms carrying blaSME are only sporadically reported. The emergence of organisms carrying blaSME in Canada is interesting, as there was no travel history associated with the cases and little similarity in macrorestriction patterns, making the potential reservoir for these cases unclear.
Higher mortality rates have been described for CPE infections, with inappropriate antimicrobial therapy suggested as a contributing factor (29). The current study revealed an all-cause mortality rate of 17.1/100 CPE infections, with 47.4% of bacteremias resulting in death. Most of the patients had multiple comorbidities, and attributable mortality could not be determined.
Molecular characterization of CPE revealed a great diversity in Canada not only in the types of carbapenemases but also in the species diversity and molecular subtypes. However, in some cases, large clonal clusters were observed, specifically in K. pneumoniae; indeed, over half of the K. pneumoniae isolates fell into one of three major clonal clusters: ST14, ST258/512, and ST147. Our blaOXA-181 findings with K. pneumoniae ST14 are similar to findings in a report from Finland (30); however, ST14 has been well described for other CPEs, like blaNDM-1 producers from India, Sweden, and the United Kingdom (27, 31). All patients in this cluster that had indicated recent international travel had sought medical attention within the Indian subcontinent, where blaNDM-1 and blaOXA-181 have been well documented (26). The predominance of the ST258/512 clone in Canada is not surprising, as the association of blaKPC with K. pneumoniae ST258 has been well documented and this organism has even been suggested as one of the most highly successful multiresistant nosocomial pathogens known to date (32). Although ST147 has been previously reported for blaNDM-type carbapenemase producers (31), which is what we report in this study, historically this sequence type was commonly reported as harboring blaCTX-M-15 and then later blaKPC-type (33–35) and may represent the transfer of blaNDM-types plasmids to a current successful clone, similar to the success of blaKPC in K. pneumoniae ST258.
From the data we have presented in this report, the dissemination of blaNDM in Canada is complex and may represent multiple independent introductions as opposed to a successful plasmid or clonal strain dissemination. Conversely, the association of IncFllA plasmids harboring blaKPC with the successful K. pneumoniae ST258/512 has been well documented (4, 36–39) and represented ∼30% of KPC plasmids in this study. The more recently observed plasmids IncP/IncL/M and IncN have a much more diverse host range than the IncFllA plasmids and may contribute to the spread of the blaKPC-3 and blaOXA-48-type genes between species and represent the current status of KPC and OXA-48 dissemination in Canada.
Apart from travel (with or without health care), the main force driving the spread of organisms producing carbapenemases in Canada has shifted from clonal outbreaks which were observed at two hospital sites (13, 14) to an increase in cases that are not linked by clonal lineage and include multiple Enterobacteriaceae species. These nonclonal cases in Canada represent the spread of many carbapenemase genes due to successful plasmids. Notably, many CPE in Canada are detected through surveillance cultures rather than clinically driven testing. It is thus critical for infection control that hospitals have programs for CPE surveillance to rapidly identify these organisms to minimize the risk of transmission. In addition, CPE surveillance programs encourage antibiotic stewardship by providing the data required to optimize appropriate therapy, improve patient outcome, ensure cost-effective therapy, and reduce adverse effects associated with inappropriate antimicrobial use.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Ken Fakharuddin and Sean Ahmed for technical assistance and the DNA Core Facility National Microbiology Laboratory for oligonucleotides and Sanger sequencing. The members of CNISP are Alice Wong, Royal University Hospital, Saskatoon, SK; Allison McGeer, Mount Sinai Hospital, Toronto, ON; Andrew Simor, Sunnybrook Health Sciences Centre, Toronto, ON; Bonita Lee, Stollery Children's Hospital, Edmonton, AB; Camille Lemieux, University Health Network, Toronto, ON; Caroline Quach, Montreal Children's Hospital, Montreal, QC; Charles Frenette, McGill University Health Centre, Montreal, QC; Chelsea Ellis, The Moncton Hospital, Moncton, NB; Deanna Hembroff, University Hospital of Northern BC, Prince George, BC; Dominik Mertz, Hamilton Health Sciences Corporation, Hamilton, ON; Dorothy Moore, Montreal Children's Hospital, Montreal, QC; Elizabeth Bryce, Vancouver Coastal Health Authority, Vancouver, BC; Elizabeth Henderson, Alberta Health Services, Calgary, AB; Geoffrey Taylor, University of Alberta Hospital, Edmonton, AB; Gerald Evans, Kingston General Hospital, Kingston, ON; Gregory German, Queen Elizabeth Hospital, Charlottetown, PEI; Ian Davis, Queen Elizabeth II Health Sciences Centre, Halifax, NS; Janice de Heer, Interior Health Authority, Kelowna, BC; Jessica Minion, Regina Qu'Appelle Health Region, Regina, SK; Joanne Embree, Health Sciences Centre, Winnipeg, MB; Joanne Langley, IWK Health Centre, Halifax, NS; Jocelyn Srigley, Children and Women's Hospital of British Columbia, Vancouver, BC; John Conly, Foothills Medical Centre, Calgary, AB; John Embil, Health Sciences Centre, Winnipeg, MB; Joseph Vayalumkal, Alberta Children's Hospital, Calgary, AB; Karl Weiss, Maisonneuve-Rosemont Hospital, Montreal, QC; Kathryn Suh, The Ottawa Hospital, Ottawa, ON; Kevin Katz, North York General Hospital, Toronto, ON; Lynn Johnston, Queen Elizabeth II Health Sciences Centre, Halifax, NS; Marie-Astrid Lefebvre, Montreal Children's Hospital, Montreal, QC; Mark Loeb, Hamilton Health Sciences Corporation, Hamilton, ON; Mary Vearncombe, Sunnybrook Health Sciences Centre, Toronto, ON; Michael John, London Health Sciences Centre, London, ON; Natalie Bridger, Eastern Health-HSC, St. John's, NFLD; Nathalie Turgeon, CHUQ-Hôtel-Dieu, Quebec, QC; Nisha Thampi, Children's Hospital of Eastern Ontario, Ottawa, ON; Pamela Kibsey, Royal Jubilee Hospital, Victoria, BC; Paula Stagg, Western Memorial Hospital, Corner Brook, NL; Stephanie Smith, University of Alberta Hospital, Edmonton, AB; Susan Richardson, Hospital for Sick Children, Toronto, ON; Suzanne Pelletier, Health Sciences North, Sudbury, ON; Virginia Roth, The Ottawa Hospital, Ottawa, ON; Yves Longtin, SMBD-Jewish General Hospital, Montreal, QC.
We have no conflicts of interest to declare.
All funding was through the Public Health Agency of Canada.
Funding Statement
All funding was through the Public Health Agency of Canada.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01359-16.
REFERENCES
- 1.Zhanel GG, Adam HJ, Baxter MR, Fuller J, Nichol KA, Denisuik AJ, Lagace-Wiens PR, Walkty A, Karlowsky JA, Schweizer F, Hoban DJ, Canadian Antimicrobial Resistance Alliance. 2013. Antimicrobial susceptibility of 22746 pathogens from Canadian hospitals: results of the CANWARD 2007-11 study. J Antimicrob Chemother 68(Suppl 1):i7–i22. [DOI] [PubMed] [Google Scholar]
- 2.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falagas ME, Karageorgopoulos DE, Nordmann P. 2011. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol 6:653–666. doi: 10.2217/fmb.11.49. [DOI] [PubMed] [Google Scholar]
- 4.Mataseje LF, Bryce E, Roscoe D, Boyd DA, Embree J, Gravel D, Katz K, Kibsey P, Kuhn M, Mounchili A, Simor A, Taylor G, Thomas E, Turgeon N, Mulvey MR, Canadian Nosocomial Infection Surveillance Program. 2012. Carbapenem-resistant Gram-negative bacilli in Canada 2009-10: results from the Canadian Nosocomial Infection Surveillance Program (CNISP). J Antimicrob Chemother 67:1359–1367. doi: 10.1093/jac/dks046. [DOI] [PubMed] [Google Scholar]
- 5.Williams V, Simor AE, Kiss A, McGeer A, Hirji Z, Larios OE, Moore C, Weiss K, Infection Prevention and Control—Canada. 2015. Is the prevalence of antibiotic-resistant organisms changing in Canadian hospitals? Comparison of point-prevalence survey results in 2010 and 2012. Clin Microbiol Infect 21:553–559. [DOI] [PubMed] [Google Scholar]
- 6.Goldfarb D, Harvey SB, Jessamine K, Jessamine P, Toye B, Desjardins M. 2009. Detection of plasmid-mediated KPC-producing Klebsiella pneumoniae in Ottawa, Canada: evidence of intrahospital transmission. J Clin Microbiol 47:1920–1922. doi: 10.1128/JCM.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mataseje LF, Boyd DA, Willey BM, Prayitno N, Kreiswirth N, Gelosia A, Poutanen SM, Low DE, Jenkins SG, Katz K, Mulvey MR. 2011. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring bla(KPC) from New York City and Toronto. J Antimicrob Chemother 66:1273–1277. doi: 10.1093/jac/dkr092. [DOI] [PubMed] [Google Scholar]
- 8.Leung V, Loo VG, Frenette C, Domingo MC, Bourgault AM, Mulvey MR, Robson HG. 2012. First Canadian outbreak of Enterobacteriaceae-expressing Klebsiella pneumoniae carbapenemase type 3. Can J Infect Dis Med Microbiol 23:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed-Bentley J, Chandran AU, Joffe AM, French D, Peirano G, Pitout JD. 2013. Gram-negative bacteria that produce carbapenemases causing death attributed to recent foreign hospitalization. Antimicrob Agents Chemother 57:3085–3091. doi: 10.1128/AAC.00297-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. 2011. New Delhi metallo-beta-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg Infect Dis 17:103–106. doi: 10.3201/eid1701.101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis C, Chung C, Tijet N, Patel SN, Desjardins M, Melano RG, Toye B. 2013. OXA-48-like carbapenemase-producing Enterobacteriaceae in Ottawa, Canada. Diagn Microbiol Infect Dis 76:399–400. doi: 10.1016/j.diagmicrobio.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Mataseje LF, Boyd DA, Hoang L, Imperial M, Lefebvre B, Miller M, Poutanen SM, Roscoe D, Willey BM, Mulvey MR. 2013. Carbapenem-hydrolyzing oxacillinase-48 and oxacillinase-181 in Canada, 2011. Emerg Infect Dis 19:157–160. doi: 10.3201/eid1901.120706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe CF, Kus JV, Salt N, Callery S, Louie L, Khan MA, Vearncombe M, Simor AE. 2013. Nosocomial transmission of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Toronto, Canada. Infect Control Hosp Epidemiol 34:49–55. doi: 10.1086/668778. [DOI] [PubMed] [Google Scholar]
- 14.Haraoui LP, Levesque S, Lefebvre B, Blanchette R, Tomkinson M, Mataseje L, Mulvey MR, Miller MA. 2013. Polyclonal outbreak of KPC-3-producing Enterobacter cloacae at a single hospital in Montreal, Quebec, Canada. J Clin Microbiol 51:2406–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI supplement M100S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 19.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.German GJ, Jamieson FB, Gilmour M, Almohri H, Bullard J, Domingo MC, Fuller J, Girouard G, Haldane D, Hoang L, Levett PN, Longtin J, Melano R, Needle R, Patel SN, Rebbapragada A, Reyes RC, Mulvey MR. 2016. Interim recommendations of the reporting of extensively drug resistant and pan-drug resistant isolates of Enterobacteriaceae, Pseudomonas aeruginosa, Acinetobacter spp. and Stenotrophomonas maltophilia. Can Commun Dis Rep 42:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mataseje LF, Boyd DA, Delport J, Hoang L, Imperial M, Lefebvre B, Kuhn M, Van Caeseele P, Willey BM, Mulvey MR. 2014. Serratia marcescens harbouring SME-type class A carbapenemases in Canada and the presence of blaSME on a novel genomic island, SmarGI1-1. J Antimicrob Chemother 69:1825–1829. [DOI] [PubMed] [Google Scholar]
- 23.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 24.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother 55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 28.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. [DOI] [PubMed] [Google Scholar]
- 29.Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. 2014. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 20:862–872. doi: 10.1111/1469-0691.12697. [DOI] [PubMed] [Google Scholar]
- 30.Österblad M, Kirveskari J, Hakanen AJ, Tissari P, Vaara M, Jalava J. 2012. Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008–11). J Antimicrob Chemother 67:2860–2864. doi: 10.1093/jac/dks299. [DOI] [PubMed] [Google Scholar]
- 31.Giske CG, Froding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR. 2012. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob Agents Chemother 56:2735–2738. doi: 10.1128/AAC.06142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damjanova I, Toth A, Paszti J, Hajbel-Vekony G, Jakab M, Berta J, Milch H, Fuzi M. 2008. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new ‘MRSAs’? J Antimicrob Chemother 62:978–985. doi: 10.1093/jac/dkn287. [DOI] [PubMed] [Google Scholar]
- 34.Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. 2011. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J Antimicrob Chemother 66:1510–1513. doi: 10.1093/jac/dkr166. [DOI] [PubMed] [Google Scholar]
- 35.Richter SN, Frasson I, Franchin E, Bergo C, Lavezzo E, Barzon L, Cavallaro A, Palu G. 2012. KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009–December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog 4:7–15. doi: 10.1186/1757-4749-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Zabicka D, Kania-Pudlo M, Mlynarczyk G, Zak-Pulawska Z, Hryniewicz W, Gniadkowski M, KPC-PL Study Group. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob Agents Chemother 55:5493–5499. doi: 10.1128/AAC.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leavitt A, Carmeli Y, Chmelnitsky I, Goren MG, Ofek I, Navon-Venezia S. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob Agents Chemother 54:3002–3006. doi: 10.1128/AAC.01818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. 2010. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother 65:243–248. doi: 10.1093/jac/dkp417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.