Abstract
The global dissemination and increasing incidence of carbapenem-resistant, Gram-negative organisms have resulted in acute public health concerns. Here, we present a retrospective multicenter study on molecular characterization of metallo-β-lactamase (MBL)-producing clinical Escherichia coli isolates recovered from extraintestinal infections in two hospitals in Pune, India. We screened a large sample size of 510 E. coli isolates for MBL production wherein we profiled their molecular determinants, antimicrobial resistance phenotypes, functional virulence properties, genomic features, and transmission dynamics. Approximately 8% of these isolates were MBL producers, the majority of which were of the NDM-1 (69%) type, followed by NDM-5 (19%), NDM-4 (5.5%), and NDM-7 (5.5%). MBL producers were resistant to all antibiotics tested except for colistin, fosfomycin, and chloramphenicol, which were effective to various extents. Plasmids were found to be an effective means of dissemination of NDM genes and other resistance traits. All MBL producers adhered to and invaded bladder epithelial (T24) cells and demonstrated significant serum resistance. Genomic analysis of MBL-producing E. coli isolates revealed higher resistance but a moderate virulence gene repertoire. A subset of NDM-1-positive E. coli isolates was identified as dominant sequence type 101 (ST101) while two strains belonging to ST167 and ST405 harbored NDM-5. A majority of MBL-producing E. coli strains revealed unique genotypes, suggesting that they were clonally unrelated. Overall, the coexistence of virulence and carbapenem resistance in clinical E. coli isolates is of serious concern. Moreover, the emergence of NDM-1 among the globally dominant E. coli ST101 isolates warrants stringent surveillance and control measures.
INTRODUCTION
Infections with Gram-negative multidrug-resistant (MDR) pathogens, particularly those caused by β-lactamase-producing Escherichia coli, constitute a major reason for the “global SOS” in public health (1). Carbapenems are often considered to be the last-resort antibiotics available for treating infections caused by extended-spectrum-β-lactamase (ESBL)- or AmpC-producing bacteria (2) and are categorized in three major types: the KPC types, the metallo-β-lactamases (MBLs), and the oxacillinases (3). The major geographically widespread MBLs consist of imipenemase (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and New Delhi metallo-β-lactamase (NDM); NDM has attracted significant worldwide attention as the NDM gene has been detected on different transferable plasmid types (both on typeable and untypeable) and has also been shown to be chromosomally integrated in other bacterial species (4, 5).
NDM belongs to the amber class B β-lactamase. Unlike other classes of β-lactamases (A, C, and D), NDM requires zinc ions to catalyze the hydrolysis of β-lactam antibiotics, and due to this, MBL activity can be inhibited in the presence of metal chelating agents such as EDTA (6). NDM-producing bacteria are mostly multidrug resistant and show resistance toward sulfonamides, fluoroquinolones, aminoglycosides, and macrolides but are susceptible to only a few antimicrobials, such as aztreonam, tigecycline, and colistin (7, 8). Recent reports suggest that this susceptibility is expected to be short-lived (1). Moreover, NDM enzymes are generally not inhibited by any of the known and available β-lactamase inhibitors such as sulbactam, clavulanate, and tazobactam (9).
NDM-producing bacteria have been implicated in both hospital- and community-acquired infections and have been recovered from several infection sites, including those associated with urinary tract infections, pneumonia, septicemia, and wound infections as well as from device-associated infections and companion animals (10–13). Although the exact geographical origin and the precise time for the emergence of the blaNDM gene are unknown, the first NDM-1-positive isolate was recovered in 2009 from a Swedish patient who previously (2007) had an episode of hospitalization in New Delhi, India (14). Later, in the year 2010, Kumarasamy et al. provided evidence that NDM-1-positive Enterobacteriaceae are widespread in India, Pakistan, and the United Kingdom (15). Recent reports further indicate that several variants of NDM (NDM-1 to NDM-14)-producing Enterobacteriaceae are currently spreading worldwide, including in the United States, Netherlands, Australia, Canada, France, and Oman (16, 17) (http://www.lahey.org/studies). Most of these case reports/epidemiological studies provide evidence of connections of patients to the Indian subcontinent in the form of migration/travel visits or medical treatment received there. However, certain reports indicate infection sources outside this region with patients without any obvious connection to this area (18). Nevertheless, the Indian subcontinent particularly offers several risk factors for the emergence and spread of NDM-producing superbugs, which include poor sanitation, easy access to broad-spectrum antibiotics over the counter without proper prescriptions, increased medical tourism for health care, and lack of stringent antibiotic policies.
Members of Enterobacteriaceae, particularly E. coli, are often disseminated globally through distinct and successful lineages, such as sequence types (ST) 131, 95, 10, etc. (19–22). Our previous studies have put forward strong evidence for the widespread occurrence of ST131 strains among extraintestinal infections in India and have also reported the existence of NDM-1-positive ST131 E. coli, thus representing a serious therapeutic challenge (23–27). This, together with the implication of NDM-producing strains globally, warrants serious attention of health care specialists and epidemiologists with respect to surveillance and resistance tracking in India and elsewhere so as to monitor threats locally and globally. Also, the data on the virulence traits associated with NDM-producing E. coli strains are currently scarce. With the advent of high-resolution, whole-genome sequencing methods, it has now become easy to decipher the genomes of the NDM-producing isolates and understand the underlying resistance and virulence mechanisms. The use of comparative genomics in infection tracking has enabled a comprehensive understanding of epidemiology, resistomes, virulomes, and the genetic background of NDM-producing MDR E. coli strains.
In the present study, we sought to determine the prevalence and phenotypes of MBL-producing strains from among a large number of E. coli isolates collected chronologically for 6 years (2009 to 2014) from a major tertiary hospital and to determine the antibiotic susceptibility patterns by taking into account the allelic diversity of blaNDM genes and the plasmid replicon types. Further, we determined the virulence traits of NDM-producing E. coli by various phenotypic assays in conjunction with whole-genome sequences of five MDR NDM-producing E. coli strains and consequent analyses of resistomes, virulomes, and genetic affinities as important parameters required to portray a correct epidemiological scenario. We believe that such exhaustive analyses potentially provide for a clear and solid epidemiological baseline to guide an effective antibiotic policy in a country of endemicity such as India.
MATERIALS AND METHODS
Bacterial isolates.
A total of 510 clinical isolates of E. coli were included in this study. The isolates were collected consecutively between January 2009 and December 2014 from in-patients and outpatients treated at two major tertiary care hospitals in Pune, India. The collection included clinical isolates from urine (n = 393), pus (n = 88), and other sources (n = 29). All the isolates were obtained as pure cultures and were cryopreserved at −80°C in 20% glycerol. The isolates were revived on LB agar plates and in LB broth prior to experiments. Collection of isolates was in accordance with the guidelines/approvals of Institutional Biosafety Committees at participating institutes/centers, where appropriate.
Detection of metallo-β-lactamase production and its molecular characterization.
A total of 510 E. coli isolates were screened for MBL production using meropenem (with and without EDTA) with the help of Ezy MIC Strips (HiMedia; India), as per the manufacturer's instructions. All positive E. coli strains were also screened for the presence of blaNDM-1 by a gene-specific PCR (28). E. coli strains confirmed as MBL producers were further analyzed by high-fidelity PCR (with the use of Q5 DNA polymerase from NEB) using flanking primers of the NDM gene, as described earlier (29). Custom sequencing of the resulting amplicons was performed, and the nucleotide and translated protein sequences were analyzed using MEGA6 (30). NDM types were determined in accordance with the sequences available in the NCBI database(s).
Testing for antimicrobial susceptibility and its molecular determinants.
Antimicrobial sensitivity of 39 strains against 23 different antibiotics was performed on Mueller-Hinton agar (MHA) plates by the Kirby-Bauer disc diffusion method using antibiotic discs (HiMedia) and Icosa urinary tract infection (UTI) rings (HiMedia), according to the manufacturer's protocols. Two in-house strains that were consistently resistant and susceptible to most of the antimicrobial agents were employed as positive and negative controls, respectively. Results were interpreted according to the criteria of the Clinical and Laboratory Standards Institute. For colistin, a zone of <11 mm was considered resistant. The MIC values of four frontline antibiotics, namely, tetracycline, co-trimoxazole, gentamicin, and ciprofloxacin, were determined by using HiComb MIC strips (HiMedia, India) as per the manufacturer's protocol. MIC values were determined and compared among a subset of 10 MBL-producing strains, 10 non-ESBL-producing strains, and 7 meropenem-resistant transconjugants. All MBL-producing E. coli strains were analyzed for molecular determinants of antibiotic resistance by PCR. DNA template was prepared using a standard heat lysis method. Thereafter, gene-specific primer sets were used to screen for the following ESBL genes: blaTEM, blaSHV, blaCTX-M-15, blaOXA-48, blaKPC, and blaNDM-1 (28, 31). Other antibiotic resistance genes, such as those conferring resistance to tetracycline [tet(A) and tet(C)], sulfonamides (sul1 and sul2), streptomycin (strA), and trimethoprim (dfr) and including the quinolone resistance determinants (carried on plasmids) such as aac(6′)-lb, qnrB, and qnrS, were amplified as per standard PCR protocols (32, 33). The amplicons were run on 1% agarose gels, and the images were documented using a gel documentation system (Major Sciences, USA). The correlation between the phenotypic and different genotypic determinants of resistance was calculated with the help of observed phenotypic resistance and the frequency of the genotypic determinant probed.
Conjugation, mating experiments, and plasmid analysis.
Conjugal transfer of resistance genes was performed by broth mating or filter mating experiments for a subset of nine carbapenem-resistant E. coli strains using the sodium azide-resistant recipient strain E. coli K-12 J53 (34). Transconjugants were selected on selection plates supplemented with a combination of meropenem (4 μg/ml) and sodium azide (50 μg/ml). In addition, other antibiotics and sodium azide combinations were used to select transconjugants. However, as meropenem-containing transconjugants were the focus of this study, the same were subjected to further analyses. Pure cultures of the transconjugants were preserved in 20% glycerol at −80°C. Antibiotic susceptibility and MICs were compared between the parental strains and their respective transconjugants. Plasmid classes were determined using PCR-based replicon typing (35).
Phenotypic assays.
A total of nine random isolates were analyzed for functional virulence traits as described below. Nine MBL-producing E. coli strains were tested for their adhesion and invasion efficiency. Human bladder epithelial cell lines (T24) were cultured and maintained in RPMI 1640 medium (Invitrogen, USA) with 10% fetal bovine serum (FBS). Cells were seeded in 24-well plates and grown until a monolayer was obtained. Infection was done at a multiplicity of infection (MOI) of 10. After 3 h of incubation at 37°C with 5% CO2, cells were washed three times with 1× phosphate-buffered saline (PBS), followed by lysis with 1 ml of 0.1% Triton X-100. Cell lysates were serially diluted, plated on LB agar plates, and incubated overnight at 37°C. For invasion assays, after 3 h of incubation, the cells were washed and further incubated for 1.5 h with 1 ml of RPMI medium containing 100 μg/ml of fosfomycin. After incubation, cells were washed three times with 1× PBS and processed in a way similar to that described above. Assays were performed three times with three technical replicates.
Serum resistance was determined for nine MBL-producing E. coli isolates by in vitro time-kill experiments as described earlier (22). Briefly, 5 μl of overnight-grown culture was added to 495 μl of LB broth and incubated for 1 h at 37°C in a shaker incubator. The culture was pelleted and resuspended in 1 ml of 1× PBS, and then 30 μl of this bacterial suspension was added to 96-well microtiter plate wells, each containing 270 μl of 50% sterile human serum (Pan Biotech, Germany) in 1× PBS. Thirty microliters of serum was then collected (0-h count), serially diluted, and plated on LB agar plates for CFU enumeration. The plate was then incubated for 3 h at 37°C. After the incubation time (3-h count), 30 μl of sample was pipetted for serial dilution and plated as described above. Growth was measured in terms of CFU counts. Samples were considered serum resistant if the 3-h CFU count equaled or exceeded the 0-h CFU count of the same sample. The serum assay was performed twice in triplicates.
Total siderophore production was tested by streaking all 39 MBL-positive E. coli samples on chrome azurol S (CAS) agar plates (blue medium). Color changes (haloes) from yellow to orange on the CAS agar medium plates, after overnight incubation at 37°C, were recorded. No color change in the medium corresponded to no siderophore production by the culture.
ERIC-PCR-based strain typing.
Enterobacterial intergenic repetitive element sequence (ERIC)-PCR-based fingerprint analysis was performed as described previously (36). Strain-specific DNA banding profiles obtained on 1.5% agarose gel images were compared using BioNumerics software (version 7.1; Applied Maths, Belgium). After the bands were scored on dice similarity indices based on the unweighted-pair group method using average linkages (UPGMA; arithmetic mean), a dendrogram was generated in order to analyze similarity/diversity within carbapenem-resistant E. coli strains. The definition of ERIC clusters was based on a Dice similarity index.
WGS, assembly, annotation, and genomic analysis.
Whole-genome sequencing (WGS) of five selected strains was carried out on an Illumina MiSeq system with an insert size of 400 to 500 bp. The paired-end sequence reads were filtered and trimmed using the NGS QC Toolkit (version 2.3.3) (37). These high-quality reads were assembled into contigs using SPAdes Genome Assembler (version 3.6.1) (38). The contigs were then ordered and scaffolded using an in-house workbench, Contig-Layout-Authenticator (39), and the resulting ordered scaffolds were joined by placing Ns between them to construct draft genomes. These were then submitted to the RAST (40) server for annotation, and the genome statistics were extracted using ARTEMIS (41). For all of the genomes, genes were predicted using GeneMarkS (42). Amino acid sequences of the products of the predicted genes from all strains were compared using blastp (43) against the database of E. coli virulence genes from the Virulence Factors Database (VFDB) (44). Only genes with 60% identity and 85% query coverage were considered present, and in this way the presence-absence status of each virulence gene in each of these strains was represented in the form of a heat map using R. A similar heat map was generated for putative resistance-related genes after comparison with the Comprehensive Antibiotic Resistance Database (CARD) (45), using blastp. Insertion sequence (IS) elements and putative phage sequences in the genome were identified using IS Finder (46) and PHAST (47), respectively. The sequence type (ST) of each of these strains was determined by submitting the contigs to the server of the Centre for Genomic Epidemiology (CGE) (https://cge.cbs.dtu.dk/services/MLST/).
Statistical analysis.
All statistical analyses were performed using GraphPad Prism (version 5.01). For adhesion and serum resistance, a nonparametric Mann-Whitney U test was performed while for the invasion assay Wilcoxon matched-pair tests were performed, and P values were determined.
Accession number(s).
The results of this whole-genome shotgun project have been deposited in the DDBJ/ENA/GenBank databases under the accession numbers LXFJ00000000 (NA703), LXFK00000000 (NA724), LXEZ00000000 (NA086), LXFA00000000 (NA099), and LWTZ00000000 (NA084). The versions described in this paper are LXFJ01000000, LXFK01000000, LXEZ01000000, LXFA01000000, and LWTZ01000000, respectively.
RESULTS
Prevalence of MBL producers was low with a high abundance of the NDM-1 genotype.
Of the 510 clinical E. coli isolates, 39 (7.6%) were found to be MBL producers. These 39 MBL-producing E. coli isolates were deemed to be of potential interest and were used further for in depth analyses, as described above. All of the 39 MBL-producing E. coli isolates were also found to be ESBL producers phenotypically. Most of the MBL producers (25/39) were from urine isolates, followed by those from pus (11/39) and other sources (3/39). The 39 MBL producers as identified at the phenotypic level were screened for the presence of major carbapenemase genes, including blaNDM, blaKPC, and blaOXA-48. With the exception of the blaKPC gene, which was not detected in any of the MBL producers, 36 (92%) MBL producers were blaNDM positive, and 6 (15%) of them were also blaOXA-48 positive. Sequence analysis of the full-length NDM genes from 36 MBL producers revealed that the majority of them carried the blaNDM-1 gene (25 isolates), followed by blaNDM-5 (7 isolates). Further, the gene blaNDM-4 was present in two isolates, and the gene blaNDM-7 was harbored by two other MBL producers.
All MBL-producing E. coli isolates were identified to be MDR and pan-drug resistant.
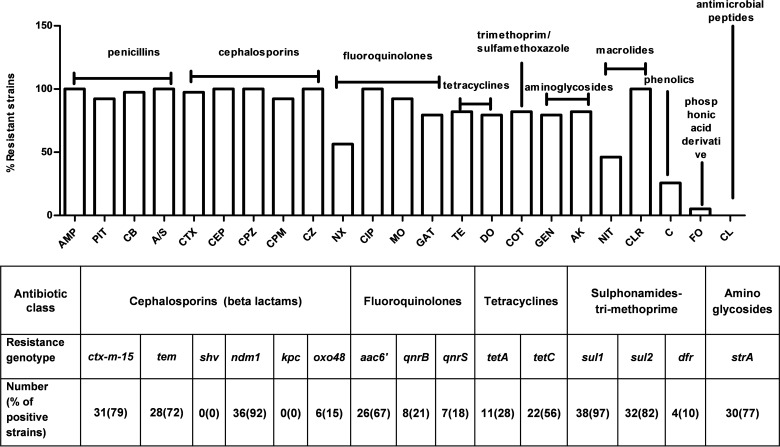
Of the 23 antimicrobial agents representing 10 different classes, our MBL-producing E. coli isolates showed an average resistance to nearly 18 antibiotics, with maximum resistance to 21 and minimum resistance to 10 antimicrobial agents. The frequencies of susceptibility/resistance are indicated in Fig. 1. Among all MBL-positive isolates, highest resistance was observed against the β-lactam antibiotic class while highest sensitivity (100%) was observed to the peptide antibiotic class, i.e., colistin. Each of the MBL-producing E. coli isolates was found to be at least 90% resistant to all the β-lactam and β-lactam/β-lactamase inhibitor combinations used, including ampicillin, carbenicillin, cefotaxime, cephalothin, cefoperazone, cefepime, cefazolin, piperacillin-tazobactam, and ampicillin-sulbactam. In contrast, each of the MBL-producing E. coli isolates was found to be sensitive to colistin. All MBL-positive isolates were highly susceptible to fosfomycin and chloramphenicol (Fig. 1). Other resistance patterns were as follows: 80% of isolates were resistant to each of the tetracycline and aminoglycoside classes and 82% were resistant to the trimethoprim/sulfonamide class. Notably, all MBL-positive isolates (100%) were identified to be MDR as they were resistant to three or more classes of antimicrobials. Moreover, all the MBL-producing E. coli isolates were found to be pan-drug resistant, meaning that they were resistant to as many as seven antimicrobial agents. The MIC values of MBL-ESBL-producing E. coli and non-ESBL E. coli isolates varied widely for different antibiotics used in the study (Table 1). Overall, the MBL-ESBL-producing E. coli isolates revealed higher MICs of all four frontline antimicrobial substances than non-ESBL strains. That the MIC values of the four antimicrobials for the meropenem transconjugants were higher than those of the recipient strain J53 E. coli indicated horizontal transmission of respective resistance genes. All 39 ESBL-MBL producers were negative for blaSHV and blaKPC. However, 31 isolates harbored the blaCTX-M-15 gene, and 28 isolates harbored blaTEM. The correlation among different resistance genotypes (presence or absence of resistance genes) and phenotypes (resistance or susceptibility) was found to be high for aminoglycosides (94% agreement) and sulfonamides (84% agreement), whereas the agreement between resistance to tetracycline and the presence of tet(A) and tet(C) was 69% and that for fluoroquinolone was (67%). The overall prevalences of all the major resistance genes in 39 MBL-producing E. coli isolates are presented in Fig. 1.
FIG 1.
(Top) Antibiogram of MBL-producing carbapenem-resistant E. coli strains as tested by the disc diffusion method for the following 23 different antibiotics: ampicillin (Amp), piperacillin-tazobactam (PIT), carbenicillin (CB), ampicillin/sulbactam (A/S), cefotaxime (CTX), cephalothin (CEP), cefoperazone (CPZ), cefepime (CPM), cefazolin (CZ), nalidixic acid (NX), ciprofloxacin (CIP), moxifloxacin (MO), gatifloxacin (GAT), tetracycline (TE), doxycycline HCl (DO), co-trimoxazole (COT), gentamicin (GEN), amikacin (AK), nitrofurantoin (NIT), clarithromycin (CLR), chloramphenicol (C), fosfomycin (FO), and colistin (CL). All MBL-producing isolates were detected to be pan-drug resistant. (Bottom) Identification of resistance determinants among 39 MBL-producing E. coli isolates by gene-specific PCRs.
TABLE 1.
MIC values of carbapenem-resistant strains, meropenem transconjugants, a recipient strain, and non-ESBL non-carbapenem-resistant E. coli isolates from the present study
| Strain group and no. | MIC (μg/ml)b |
|||
|---|---|---|---|---|
| Gentamicin | Ciprofloxacin | Co-trimoxazole | Tetracycline | |
| Carbapenem-resistant strains (n = 17)a | ||||
| NA084 | >240 | >240 | >240 | 3 |
| NA644 | >240 | >240 | >240 | 5 |
| NA708 | >240 | 60 | >240 | 3 |
| NA086 | >240 | >240 | >240 | 2 |
| NA086 T | >240 | 0.01 | 0.05 | 0.1 |
| NA099 | >240 | >240 | >240 | 0.1 |
| NA099T | 0.01 | 0.1 | 0.01 | 0.01 |
| NA316 | >240 | >240 | >240 | 3 |
| NA316T | 0.1 | ND | 0.05 | 0.1 |
| NA635 | >240 | 120 | >240 | 5 |
| NA635T | 24 | ND | >240 | 3 |
| NA703 | >240 | >240 | >240 | 0.001 |
| NA703T | 16 | ND | 0.05 | 0.001 |
| NA724 | >240 | 120 | >240 | 3 |
| NA724T | 30 | 0.01 | 0.05 | 0.1 |
| NA805 | 60 | 10 | >240 | 0.01 |
| NA805T | ND | 1 | 0.05 | 0.01 |
| Recipient strain (n = 1) | ||||
| K-12 J53 | 0.001 | 0.008 | 0.05 | 0.01 |
| Non-ESBL non-carbapenem-resistant strains (n = 10) | ||||
| NA433 | 0.1 | 60 | >240 | 10 |
| NA010 | 0.1 | 0.008 | 30 | 0.1 |
| NA295 | 0.01 | 0.01 | 120 | 0.1 |
| NA053 | 0.25 | 1 | >240 | 2 |
| NA057 | 0.1 | 30 | >240 | 0.1 |
| NA120 | 0.01 | 0.1 | >240 | 0.5 |
| NA140 | 0.1 | 0.01 | >240 | 30 |
| NA150 | 0.1 | 0.25 | 2 | 0.01 |
| NA203 | 0.01 | 0.01 | 0.1 | 0.01 |
| NA274 | 0.1 | 10 | >240 | 30 |
The total includes seven meropenem transconjugants (T suffix).
ND, not determined.
Metallo-β-lactamase resistance was naturally transferable via plasmids.
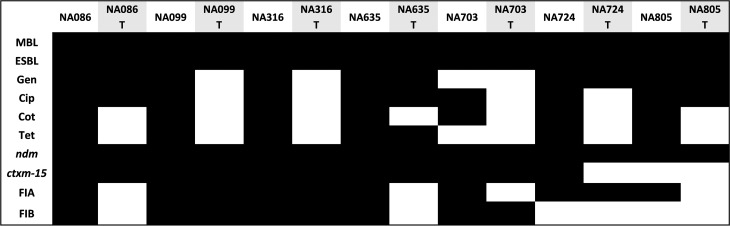
NDM genes are basically located on plasmids. In order to determine the capability of these MBL-positive E. coli strains to disseminate NDM genes horizontally, we carried out conjugation mating experiments with the sodium azide-resistant recipient strain E. coli K-12 J53. Out of the nine representative MBL-positive strains employed, we were able to obtain seven meropenem-resistant transconjugants alongside several other transconjugants. However, only meropenem transconjugants were further analyzed for this study. The transconjugants were analyzed for their phenotypic drug resistance to four different classes of antibiotics, along with MBL and ESBL detection. All transconjugants were identified to be MBL and ESBL positive, with coresistance varying widely for different antibiotics used in the study. The detailed results are shown in Fig. 2. Transconjugants were confirmed to be carrying the NDM gene by gene-specific PCR. PCR-based replicon typing revealed the presence of primarily two plasmid types, the FIA and FIB plasmids, among transconjugants. These observations reflect the role of transmissible plasmids in dissemination of NDM-based carbapenem resistance.
FIG 2.
In vitro conjugation experiment and comparative analysis between clinical strains and their respective transconjugants revealed transfer of resistances via plasmids. The T suffix designates transconjugants. Gen, gentamicin; Cip, ciprofloxacin; Cot, co-trimoxazole; Tet, tetracycline. ndm and ctxm-15 represent blaNDM and blaCTX-M-15 genes, respectively. FIA and FIB are the plasmid replicon types. Black and white boxes indicate presence and absence of corresponding traits, respectively.
MBL-producing E. coli isolates were moderately virulent.
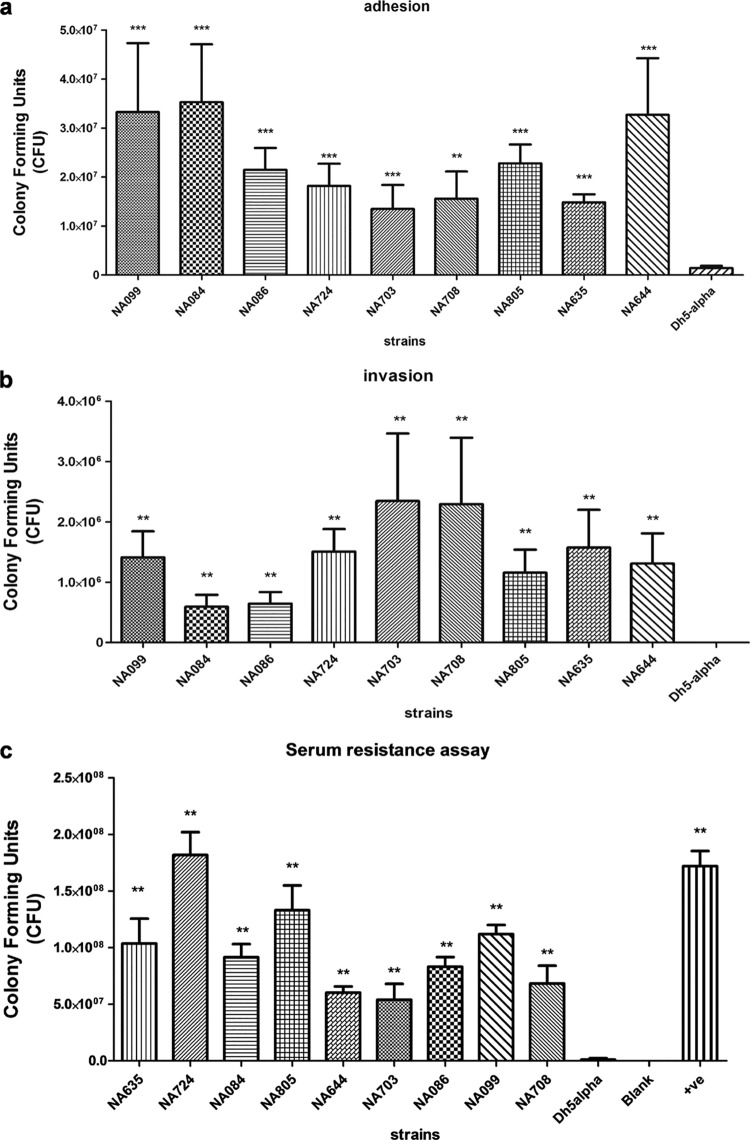
Phenotypic virulence properties of the MBL-positive E. coli isolates were assayed using adhesion, invasion, serum bactericidal, and siderophore production assays. The two former assays were carried out on nine randomly selected strains, and siderophore production was determined for all 39 strains. Adhesive and invasive virulence properties are important features during intestinal as well as extraintestinal infections. In order to evaluate this, we quantitatively assayed the pathogen's ability to adhere and invade the urinary bladder epithelial cells. Compared to the negative-control strain E. coli DH5α, all nine MBL-positive E. coli isolates showed significant adhesion to T24 cells with various efficiencies, as illustrated in Fig. 3a. Among the nine MBL-positive isolates tested, the NA084 isolate demonstrated highest adherence capability toward T24 cells. T24 cells were invaded by all investigated isolates, in contrast to the absence of invasion by the negative-control strain E. coli DH5α. Numbers of intracellular bacteria are depicted in Fig. 3b.
FIG 3.
A subset of nine MBL-producing E. coli isolates was tested for three virulence-associated phenotypes: adhesion to human bladder epithelial cells (T24) at an MOI of 10 (a), invasion of human bladder epithelial cells (T24) (b), and resistance to 50% human serum for a 3-h time period (c). Adhesion and invasion assays were repeated three times in triplicates while the serum resistance assay was repeated twice in triplicates. All nine MBL-producing E. coli isolates demonstrated ExPEC-associated phenotypes. **, P < 0.01; ***, P < 0.001, for results compared to those with the control (DH5α).
Resistance to serum is quite essential for the pathogens to survive and cause invasive infections; we found all nine tested isolates to be resistant (100%) to serum bactericidal activity compared to results with the control DH5α E. coli. Bacterial survival in serum over a period of 3 h is depicted in Fig. 3c. Though the virulence genes tested were not prevalent in these isolates, bacterial resistance toward the bactericidal effect of serum could result from individual or combined effects of capsular polysaccharide, surface proteins, and toxin secretions.
Siderophore production, as visualized by an iron mobilization zone around the bacterial colonies with orange haloes on the chrome azurol S agar, was similarly prevalent among 36 (92%) of the 39 MBL-producing E. coli isolates. The role of iron in the virulence mechanism of human and animal pathogens is well established (48). Siderophore production has been shown to be more frequently associated with extraintestinal pathogenic E. coli (ExPEC).
Whole-genome analysis reveals high resistance and moderate virulence of MBL strains.
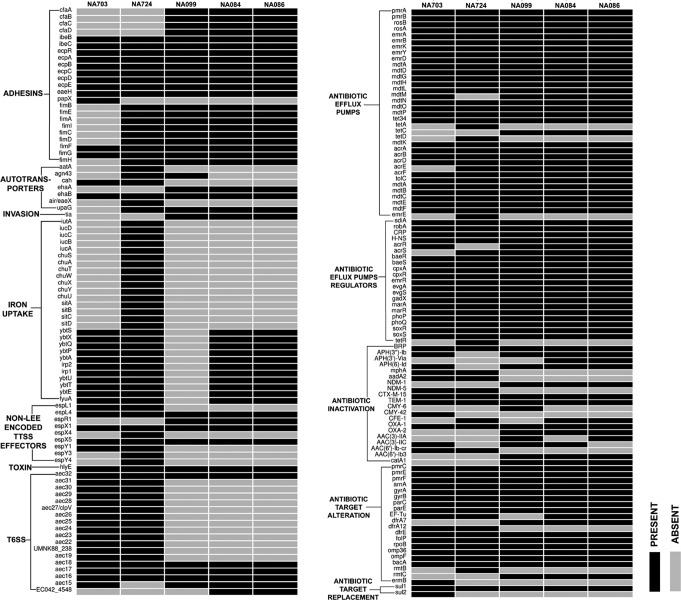
Out of the 39 MBL-positive strains, whole-genome sequencing of five representative strains (three NDM-1 and two NDM-5) was carried out. Their genome characteristics are represented in Table 2. The three NDM-1 strains belonged to ST101 while the two NDM-5 strains were characterized as ST167 and ST405. The whole-genome-based resistome reflected the high prevalence of resistance genes of all categories within all the strains, similar to the phenotypic drug resistance profile (Fig. 4). The virulome of these strains reflected moderate virulence wherein all the strains inherited genes belonging to adhesins, autotransporters, type three secretion systems (TTSS), and type 6 secretion systems (T6SS). NA099 harbors the least number of virulence genes while NA724 carries the maximum number of genes. The details are shown in Fig. 4. All the strains showed different phage contents and variable IS elements. The upstream sequence analysis of these five NDM strains showed insertion elements such as ISAba125 upstream of NDM-1 and IS5 and truncated ISAba125 upstream of NDM-5 sequences.
TABLE 2.
The genome statistics and sequence types of five whole-genome-sequenced NDM-harboring strains comprising three NDM-1 and two NDM-5 strains
| Strain | No. of raw reads | Genome coverage from filtered reads (×) | No. of contigs | ST | Genome size (no. of bases) | G+C content (%) | No. of CDSa | % coding capacity | No. of rRNAs | NDM type | Specimen source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NA084 | 1,741,426 | 73.25 | 109 | 101 | 5,147,001 | 50.59 | 5,000 | 86.8 | 13 | NDM-1 | Urine |
| NA086 | 1,643,674 | 63.13 | 115 | 101 | 5,147,281 | 50.59 | 4,994 | 86.9 | 13 | NDM-1 | Urine |
| NA099 | 2,406,806 | 104.68 | 155 | 101 | 5,197,495 | 50.57 | 5,063 | 85.8 | 17 | NDM-1 | Urine |
| NA703 | 2,401,628 | 92.94 | 211 | 167 | 5,263,381 | 50.66 | 5,186 | 86.0 | 16 | NDM-5 | Pus |
| NA724 | 2,784,910 | 118.36 | 166 | 405 | 5,424,442 | 50.55 | 5,273 | 87.0 | 9 | NDM-5 | Pus |
CDS, coding sequences.
FIG 4.
Whole-genome-based identification of virulence (left) and resistance (right) determinants of five MBL-producing E. coli isolates.
NDM strains reveal high genomic diversity.
PCR-based genotyping was used to analyze the genetic relationships of the organisms. The ERIC-PCR-based dendrogram of these strains (Fig. 5) revealed high diversity among them. However, when strains were grouped at a lower threshold of ∼65% identity, two diffusely clonal clades were observed. As expected, all the strains in this cluster harbored NDM-1 genes. Further, the in silico multilocus sequence typing (MLST) analysis of whole-genome sequences identified all three strains to be ST101. Overall, this cluster might include clonally related ST101 strains which are strongly associated with the NDM genotype. No such clusters were obtained for any other NDM variants. The above observations also support the idea that plasmids are the important means for NDM dissemination.
FIG 5.
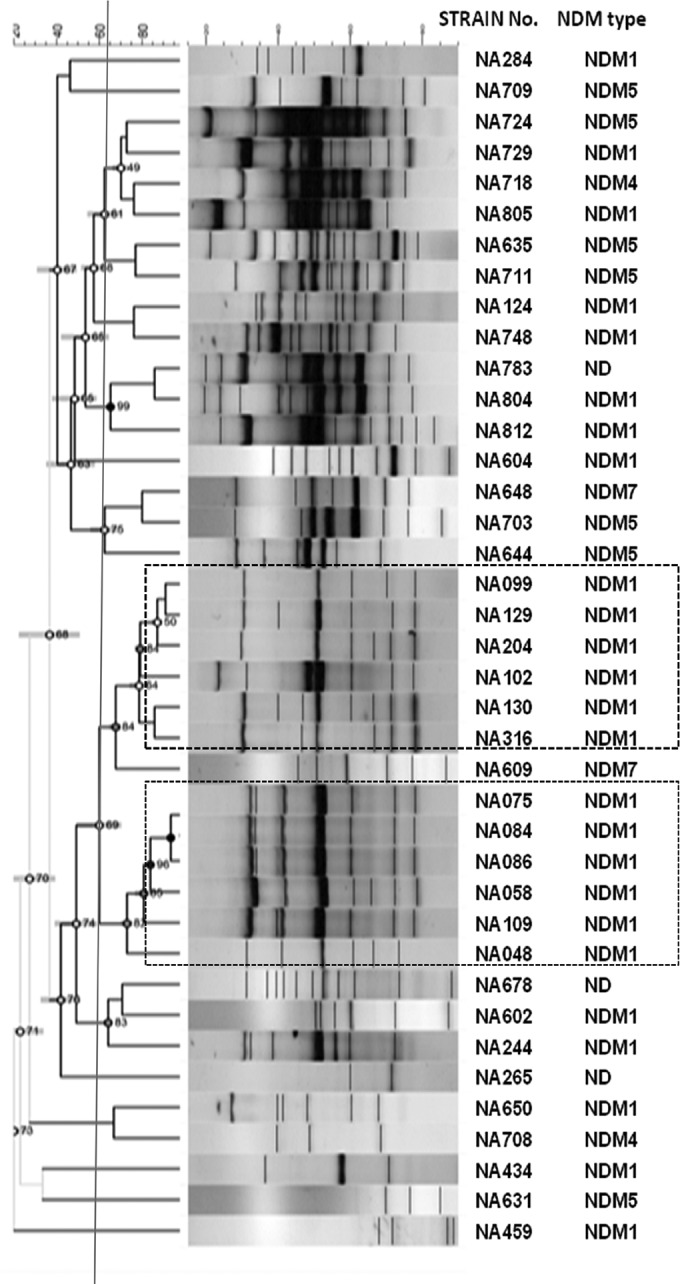
Dendrogram based on the phylogenetic analysis of 39 MBL-producing carbapenem-resistant clinical E. coli strains using ERIC-PCR banding analysis by BioNumerics, version 7.5 (Applied Maths, Belgium). Dotted boxes indicate clusters into which the majority of NDM-1 profiles segregated. ND, not determined.
DISCUSSION
The emerging NDM, an acquired class B carbapenemase from among Enterobacteriaceae, has become a major public health concern worldwide also due to its medical and economic impact (49), and infections with NDM-positive pathogens pose a major health care problem, particularly for high-burden countries such as India. Previous studies from our group have shown an endemic E. coli infection burden and the presence of clonal group ST131 isolates with NDM-1 (23–25, 50). This study was aimed at detection, comprehensive characterization, and whole-genome sequence analysis of MBL-producing MDR organisms that can lead to successful infection. This study would likely be useful in guiding the formulation of control strategies involving antimicrobial stewardship and public health interventions in order to control the escalation of life-threatening MDR bacterial infections involving ExPEC organisms.
In general, production of MBL in Enterobacteriaceae shows an increasing prevalence pattern. However, the prevalence of carbapenem resistance among 510 E. coli isolates analyzed from 2009 to 2014 was found to be 7.6%, which was within the range of previously reported carbapenem resistance from India (51, 52); however, this prevalence might be variable in different geographical locations and different hospital settings. The dissemination of NDM carbapenemases involves several blaNDM gene variants that are associated with different plasmid types among several Gram-negative species. A total of 14 NDM types have been reported globally, which were due to one or more point mutations (http://www.lahey.org/studies). In our study, we identified four NDM variants responsible for MBL production, with NDM-1 being the dominant variant, while other variants comprising NDM-4, NDM-5, and NDM-7 were observed with various frequencies. Previous reports from India have shown the presence of the NDM-6 type in addition to the above-mentioned NDM types (52). These four variants did not demonstrate any significant difference in MIC values for meropenem, nor did they show any specific association with different antimicrobial resistances. The carbapenems are considered the antibiotics of last resort for treatment of MDR bacterial infections; hence, bacteria which are resistant to these agents are often termed superbugs as they possess a resistance phenotype against different classes of broad-spectrum antibiotics. The antibiogram of these 39 isolates (Fig. 1) reflected a similar scenario wherein all MBL-producing strains (100%) revealed pan-drug resistance phenotypes as they were completely resistant to almost all the empirically used antibiotics. Nevertheless, all the strains were susceptible to colistin, and a majority of them were susceptible to fosfomycin. Polymyxins are the antibiotics of last resort to treat infections caused by MBL-producing Enterobacteriaceae (53), and as combination therapies are being advocated for treatment of MDR pathogens (54), the dual combination of colistin and fosfomycin could be the best option for effective treatment of carbapenem-resistant infections, which also minimizes the chances of resistance development.
The spread of antibiotic resistance genes is mediated by horizontal gene transfer (HGT) between bacteria (55). As multidrug-resistant plasmids serve as the most efficient means for dissemination of resistance traits via HGT, we also observed that the NDM genes and other resistances were transferred (via plasmids) to recipient strains, as confirmed genotypically and phenotypically by our conjugation experiments (Fig. 2). ERIC-PCR-based clusters revealed the presence of NDM genes in E. coli isolates of mostly unique genotypes, suggesting that they are clonally unrelated and thereby have similar propensities to acquire the NDM genes (Fig. 5). Recent reports have shown a greater association of the NDM-1 genotype with sequence type 101, which is consistent with our findings as we also observed three NDM-1-bearing E. coli isolates belonging to ST101 (56, 57). We identified NDM-5 to be present in E. coli isolates belonging to ST405 and ST167. These sequence types were also linked to NDM dissemination worldwide (58–60). Further, we found that the NDM-1 sequences were preceded by the ISAba125 insertion element which belongs to the IS30 family while NDM-5 has an IS5 insertion element upstream of its sequence, followed by sequences of ISAba125. ISAba125 is known to be located upstream of NDM in both Acinetobacter and E. coli, similar to our observations. It has also been reported that the IS5 element is followed by a truncated ISAba125 sequence upstream of the NDM gene (61, 62).
The virulence phenotypes such as adhesion to and invasion of epithelial cells play an important role in initiation and establishment of infection. The virulence gene complement as detected at the whole-genome level (Fig. 4) and the adhesion/invasion (Fig. 3a and b) capabilities of the strains toward human urinary bladder epithelial cells demonstrate their pathogenic potentials and associated risks during infection as these strains are resistant to all empirically used antibiotics. Peirano et al. have also shown the ability of CTX-M- and NDM-positive strains to adhere to Caco2 and HepG2 cell lines (63). The ability to resist the bactericidal effect of serum complement as determined by serum resistance assays (Fig. 3c) points toward the enhanced survival advantages of these strains at different extraintestinal infection sites.
In summary, our findings demonstrated that the human extraintestinal pathogenic MBL-producing E. coli strains were associated with promiscuous plasmid types, resulting in a diverse range of clones harboring different variants of the NDM gene. Furthermore, whole-genome sequencing and genetic fingerprinting suggest that blaNDM-1 may also disseminate via dominant clones such as ST101. The genomes newly sequenced herein would be an important addition to the existing uropathogenic E. coli (UPEC)/ExPEC genome archives and would be helpful in future studies involving comparative genomics and molecular epidemiology. Given that the emergence of newer variants of NDM-1 in countries of endemicity and their association with different ExPEC infections represent an alarming problem, large-scale surveillance and molecular genotyping studies are needed in order to develop strategies to circumvent the emergence of dominant clones potentially leading to institutional outbreaks and nosocomial epidemics of the future.
ACKNOWLEDGMENTS
We acknowledge funding received from the Department of Biotechnology, Government of India [BT/HRD/NBA/34/01/2011(ix)], and support from the Indo-German International Research Training Group, Internationales Graduiertenkolleg (GRK1673), Functional Molecular Infection Epidemiology, an initiative of the German Research Foundation and the University of Hyderabad (India), for which N.A. and L.H.W. served as speakers. N.A. is an Adjunct Professor at the Academy of Scientific and Innovative Research, India. A.R. acknowledges the Indian Council of Medical Research for the award of a Junior Research Fellowship.
We thank Ramani Baddam for her help in discussing the data and their interpretation and help with the analysis of plasmid sequences. The Department of Biotechnology and Bioinformatics is supported by DST-FIST and UGC-SAP-DRS (level 1) programs.
REFERENCES
- 1.Bonomo RA. 2011. New Delhi metallo-beta-lactamase and multidrug resistance: a global SOS? Clin Infect Dis 52:485–487. doi: 10.1093/cid/ciq179. [DOI] [PubMed] [Google Scholar]
- 2.Fair RJ, Tor Y. 2014. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem 6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peirano G, Schreckenberger PC, Pitout JD. 2011. Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob Agents Chemother 55:2986–2988. doi: 10.1128/AAC.01763-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. 2013. NDM-8 metallo-beta-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agents Chemother 57:2394–2396. doi: 10.1128/AAC.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watkins RR, Papp-Wallace KM, Drawz SM, Bonomo RA. 2013. Novel β-lactamase inhibitors: a therapeutic hope against the scourge of multidrug resistance. Front Microbiol 4:392. doi: 10.3389/fmicb.2013.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seija V, Medina Presentado JC, Bado I, Papa Ezdra R, Batista N, Gutierrez C, Guirado M, Vidal M, Nin M, Vignoli R. 2015. Sepsis caused by New Delhi metallo-beta-lactamase (blaNDM-1) and qnrD-producing Morganella morganii, treated successfully with fosfomycin and meropenem: case report and literature review. Int J Infect Dis 30:20–26. doi: 10.1016/j.ijid.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Bogaerts P, Bouchahrouf W, de Castro RR, Deplano A, Berhin C, Pierard D, Denis O, Glupczynski Y. 2011. Emergence of NDM-1-producing Enterobacteriaceae in Belgium. Antimicrob Agents Chemother 55:3036–3038. doi: 10.1128/AAC.00049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaheen BW, Nayak R, Boothe DM. 2013. Emergence of a New Delhi metallo-β-lactamase (NDM-1)-encoding gene in clinical Escherichia coli isolates recovered from companion animals in the United States. Antimicrob Agents Chemother 57:2902–2903. doi: 10.1128/AAC.02028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 14.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou D, Huang Y, Zhao X, Liu W, Dong D, Li H, Wang X, Huang S, Wei X, Yan X, Yang Z, Tong Y, Huang L, Yuan J. 2015. A novel New Delhi metallo-β-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob Agents Chemother 59:2450–2453. doi: 10.1128/AAC.05168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L, Revathi G, Bernabeu S, Nordmann P. 2011. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother 55:934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. 2012. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect 18:E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x. [DOI] [PubMed] [Google Scholar]
- 21.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nandanwar N, Janssen T, Kuhl M, Ahmed N, Ewers C, Wieler LH. 2014. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int J Med Microbiol 304:835–842. doi: 10.1016/j.ijmm.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Hussain A, Ranjan A, Nandanwar N, Babbar A, Jadhav S, Ahmed N. 2014. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob Agents Chemother 58:7240–7249. doi: 10.1128/AAC.03320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjan A, Shaik S, Hussain A, Nandanwar N, Semmler T, Jadhav S, Wieler LH, Ahmed N. 2015. Genomic and functional portrait of a highly virulent, CTX-M-15-producing H30-Rx subclone of Escherichia coli sequence type 131. Antimicrob Agents Chemother 59:6087–6095. doi: 10.1128/AAC.01447-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain A, Ewers C, Nandanwar N, Guenther S, Jadhav S, Wieler LH, Ahmed N. 2012. Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum-β-lactamase-producing lineage. Antimicrob Agents Chemother 56:6358–6365. doi: 10.1128/AAC.01099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avasthi TS, Kumar N, Baddam R, Hussain A, Nandanwar N, Jadhav S, Ahmed N. 2011. Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J Bacteriol 193:4272–4273. doi: 10.1128/JB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandanwar N, Hussain A, Ranjan A, Jadhav S, Ahmed N. 2016. Population structure and molecular epidemiology of human clinical multi-drug resistant (MDR) Escherichia coli strains from Pune, India. Int J Infect Dis 45 (Suppl 1):343–344. [Google Scholar]
- 28.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 29.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grobner S, Linke D, Schutz W, Fladerer C, Madlung J, Autenrieth IB, Witte W, Pfeifer Y. 2009. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tubingen, Germany. J Med Microbiol 58:912–922. doi: 10.1099/jmm.0.005850-0. [DOI] [PubMed] [Google Scholar]
- 32.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother 50:2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-lb-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Martinez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 35.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaik S, Kumar N, Lankapalli AK, Tiwari SK, Baddam R, Ahmed N. 2016. Contig-layout-authenticator (CLA): a combinatorial approach to ordering and scaffolding of bacterial contigs for comparative genomics and molecular epidemiology. PLoS One 11:e0155459. doi: 10.1371/journal.pone.0155459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 42.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Q, Wang X, Xu H, Xu Y, Ling J, Zhang D, Gao S, Liu X. 2012. Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiol 12:143. doi: 10.1186/1471-2180-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 50.Jadhav S, Hussain A, Devi S, Kumar A, Parveen S, Gandham N, Wieler LH, Ewers C, Ahmed N. 2011. Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi-urban locality in India. PLoS One 6:e18063. doi: 10.1371/journal.pone.0018063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bora A, Ahmed GU, Hazarika NK, Prasad KN, Shukla SK, Randhawa V, Sarma JB. 2013. Incidence of blaNDM-1 gene in Escherichia coli isolates at a tertiary care referral hospital in Northeast India. Indian J Med Microbiol 31:250–256. doi: 10.4103/0255-0857.115628. [DOI] [PubMed] [Google Scholar]
- 52.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, Singh A, Srivastava AK, Gonzalez-Zorn B. 2014. Prevalence and molecular characterisation of New Delhi metallo-beta-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents 44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Morrill HJ, Pogue JM, Kaye KS, LaPlante KL. 2015. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis 2:ofv050. doi: 10.1093/ofid/ofv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albur MS, Noel A, Bowker K, MacGowan A. 2015. The combination of colistin and fosfomycin is synergistic against NDM-1-producing Enterobacteriaceae in in vitro pharmacokinetic/pharmacodynamic model experiments. Int J Antimicrob Agents 46:560–567. doi: 10.1016/j.ijantimicag.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 55.Dzidic S, Bedekovic V. 2003. Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta Pharmacol Sin 24:519–526. [PubMed] [Google Scholar]
- 56.Toleman MA, Bugert JJ, Nizam SA. 2015. Extensively drug-resistant New Delhi metallo-beta-lactamase-encoding bacteria in the environment, Dhaka, Bangladesh, 2012. Emerg Infect Dis 21:1027–1030. doi: 10.3201/eid2106.141578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo JS, Kim HM, Koo HS, Yang JW, Yoo JI, Kim HS, Park HK, Lee YS. 2013. Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J Antimicrob Chemother 68:2170–2172. doi: 10.1093/jac/dkt126. [DOI] [PubMed] [Google Scholar]
- 58.Zhang LP, Xue WC, Meng DY. 2016. First report of New Delhi metallo-beta-lactamase 5 (NDM-5)-producing Escherichia coli from blood cultures of three leukemia patients. Int J Infect Dis 42:45–46. doi: 10.1016/j.ijid.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 59.Yang P, Xie Y, Feng P, Zong Z. 2014. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother 58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 61.Sole M, Pitart C, Roca I, Fabrega A, Salvador P, Munoz L, Oliveira I, Gascon J, Marco F, Vila J. 2011. First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob Agents Chemother 55:4402–4404. doi: 10.1128/AAC.00642-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krishnaraju M, Kamatchi C, Jha AK, Devasena N, Vennila R, Sumathi G, Vaidyanathan R. 2015. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol 33:30–38. doi: 10.4103/0255-0857.148373. [DOI] [PubMed] [Google Scholar]
- 63.Peirano G, Mulvey GL, Armstrong GD, Pitout JD. 2013. Virulence potential and adherence properties of Escherichia coli that produce CTX-M and NDM beta-lactamases. J Med Microbiol 62:525–530. doi: 10.1099/jmm.0.048983-0. [DOI] [PubMed] [Google Scholar]