Abstract
Klebsiella pneumoniae is emerging as an important nosocomial pathogen due to its rapidly increasing multidrug resistance, which has led to a renewed interest in polymyxin antibiotics, such as colistin, as antibiotics of last resort. However, heteroresistance (i.e., the presence of a subpopulation of resistant bacteria in an otherwise susceptible culture) may hamper the effectiveness of colistin treatment in patients. In a previous study, we showed that colistin resistance among extended-spectrum-beta-lactamase (ESBL)-producing K. pneumoniae isolates emerged after the introduction of selective digestive tract decontamination (SDD) in an intensive care unit (ICU). In this study, we investigated heteroresistance to colistin among ESBL-producing K. pneumoniae isolates by using population analysis profiles (PAPs). We used whole-genome sequencing (WGS) to identify the mutations that were associated with the emergence of colistin resistance in these K. pneumoniae isolates. We found five heteroresistant subpopulations, with colistin MICs ranging from 8 to 64 mg/liter, which were derived from five clonally related, colistin-susceptible clinical isolates. WGS revealed the presence of mutations in the lpxM, mgrB, phoQ, and yciM genes in colistin-resistant K. pneumoniae isolates. In two strains, mgrB was inactivated by an IS3-like or ISKpn14 insertion sequence element. Complementation in trans with the wild-type mgrB gene resulted in these strains reverting to colistin susceptibility. The MICs for colistin-susceptible strains increased 2- to 4-fold in the presence of the mutated phoQ, lpxM, and yciM alleles. In conclusion, the present study indicates that heteroresistant K. pneumoniae subpopulations may be selected for upon exposure to colistin. Mutations in mgrB and phoQ have previously been associated with colistin resistance, but we provide experimental evidence for roles of mutations in the yciM and lpxM genes in the emergence of colistin resistance in K. pneumoniae.
INTRODUCTION
Klebsiella pneumoniae is emerging as an important nosocomial pathogen due to rapidly increasing resistance to practically all currently available antibiotics, in particular carbapenems (1, 2). This Gram-negative opportunistic pathogen can cause wound and urinary tract infections and other life-threatening, hospital-acquired infections, such as pneumonia, bacteremia, and postoperative meningitis (3, 4). Due to increasing multidrug resistance (MDR) among Gram-negative bacteria, including K. pneumoniae, and the lack of novel antibiotics to treat infections caused by MDR Gram-negative bacteria (5), there is a renewed interest in the antibiotic colistin as a therapy of last resort (6).
Colistin (polymyxin E) is a cationic polypeptide with a lipid tail which targets anionic lipopolysaccharide (LPS) molecules in the outer membranes of Gram-negative bacteria, introducing changes in the permeability of the membrane which lead to leakage of cell contents and, finally, cell death (7, 8). Resistance to colistin among Gram-negative bacteria in clinical isolates was reported recently (9–11). Several strategies are employed by bacteria to gain resistance to colistin, including LPS modifications, particularly modifications of lipid A, the use of efflux pumps, and overexpression of outer membrane proteins (12). Resistance to colistin in clinical isolates may go undetected when traditional in vitro antibiotic susceptibility testing is used, because of heteroresistance, which denotes the presence of subpopulations of bacterial cells with higher levels of antibiotic resistance than those of the rest of the population in the same culture (13). This phenomenon was described recently for Gram-negative organisms including Pseudomonas aeruginosa (14), Acinetobacter baumannii (15, 16), and Enterobacter cloacae (17).
In a previous study (18), we showed that colistin resistance among extended-spectrum beta-lactamase (ESBL)-producing K. pneumoniae isolates emerged after exposure to colistin as part of selective digestive tract decontamination (SDD) in an intensive care unit (ICU), and we postulated that this may be explained by the presence of heteroresistant subpopulations of the colistin-susceptible MDR strains.
In the present study, the existence of colistin-resistant subpopulations among ESBL-producing K. pneumoniae isolates was investigated. Through whole-genome sequencing (WGS) and complementation of mutated alleles in trans, the roles of mutations in resistance to colistin in K. pneumoniae were determined.
MATERIALS AND METHODS
Clinical data and bacterial isolates.
K. pneumoniae isolates were collected during a study on the emergence of colistin resistance in Enterobacteriaceae before and after the introduction of SDD in an ICU (18). Briefly, SDD, a topical mixture of antibiotics, including tobramycin, colistin, and amphotericin B at doses (given four to eight times daily) of 80, 100, and 500 mg, respectively, was introduced in 2002 to control an outbreak of ESBL-producing K. pneumoniae in an ICU. Reexamination of stored isolates from surveillance and clinical cultures from ICU patients before and after the start of SDD revealed that all tested isolates obtained before the start of SDD were colistin susceptible, whereas 71% of isolates from cultures obtained thereafter were resistant. Molecular typing of the isolates revealed that most of them were clonally related (18).
In this study, we included a total of 13 strains: eight genetically related ESBL-producing K. pneumoniae clinical isolates (one isolate per patient) with known colistin MICs and five heteroresistant subpopulations of these isolates. Genetic relatedness was determined by use of the DiversiLab system (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions. The eight clinical isolates included six colistin-susceptible isolates, one of which was obtained before the start of SDD and five thereafter, and two colistin-resistant isolates obtained after the start of SDD. The six colistin-susceptible isolates and one of the two colistin-resistant isolates were genotypically identical based on DiversiLab typing.
K. pneumoniae ATCC 700603 (ATCC, Manassas, VA) was included as a colistin-susceptible reference strain. The identities of all isolates were confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) analysis according to the manufacturer's instructions, and strains were stored at −80°C before the investigations described in this study.
Antibiotic susceptibility testing.
Routine antimicrobial susceptibility testing was performed by use of a Vitek 2 Advanced Expert system and Etest (bioMérieux, Marcy l'Etoile, France), using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (http://www.eucast.org/clinical_breakpoints). The presence of ESBLs was determined with the double-disk synergy test (19). The MICs of colistin against the K. pneumoniae strains were determined by broth microdilution testing (http://www.eucast.org/guidance_documents) using cation-adjusted Mueller-Hinton broth (MHCB). Determination of the colistin MICs of electrotransformed strains was performed with MHCB supplemented with 10 mg/liter tetracycline.
PAPs.
To investigate the presence of colistin heteroresistance, population analysis profiles (PAPs) were determined for two replicates by spiral plating 50-μl aliquots of the starting bacterial cell suspension (corresponding to a 0.5 McFarland standard for K. pneumoniae cultures grown on blood agar plates for 24 h at 37°C; approximately 108 CFU/ml) on Mueller-Hinton agar plates with or without colistin sulfate (0.5, 1, 2, 3, 4, 5, 6, 8, and 10 mg/liter; Sigma-Aldrich, Zwijndrecht, The Netherlands) as described by Li et al. (20). After 24 h of incubation at 37°C, the number of colonies was counted. Colistin heteroresistance was defined as the presence of a colistin-susceptible isolate with a colistin MIC of <2 mg/liter in which detectable colistin-resistant subpopulations were able to grow in the presence of ≥2 mg/liter colistin (20). The detection limit of colistin-resistant subpopulations was 20 CFU/ml.
Genome sequencing and assembly.
Genomic DNAs of K. pneumoniae isolates were isolated from overnight cultures grown in Luria broth at 37°C with shaking at 250 rpm by use of a Wizard Genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. Sequence libraries were prepared with a Nextera XT kit (Illumina, San Diego, CA) used according to the manufacturer's instructions. Libraries were sequenced on an Illumina MiSeq system with a 500-cycle (2 × 250 bp) MiSeq reagent kit v2. High-throughput sequence (HTS) data were analyzed for quality with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and raw 2- by 250-bp paired-end reads were filtered with Nesoni 0.109 (http://github.com/Victorian-Bioinformatics-Consortium/nesoni). De novo genome assembly was performed with SPAdes 2.5.1 (21), with k-mers 25, 35, 45, 57, and 69, using the following cutoffs for the minimum contig/scaffold: a size of 500 bp and average nucleotide coverage (10-fold).
Phylogenetic analysis.
Publicly available WGS sequence data for 25 K. pneumoniae strains were downloaded from the NCBI databases in February 2016. The strains used in phylogenetic analysis were selected to cover all K. pneumoniae clades, as previously determined by Holt et al. (22). For strains for which only raw sequence reads were available, assemblies were generated with SPAdes 2.5.1 (21), as described above. To ensure consistent gene prediction and annotation of all 38 genomes in this study, all genome sequences were reannotated with PROKKA v1.10, using the default settings (23). To identify the core genome of these strains, first an all-against-all protein BLAST sequence similarity search of annotated and translated gene sequences was performed with default settings, except for an E value of 1e−05. Based on the protein BLAST output, orthologous groups were determined and clustered using OrthAgogue v1.0.3 (24) (settings -u and -o 50) and MCL v14-137 (25) (settings -I 1.5), respectively. The nucleotide sequences of orthologous groups containing exactly one representative protein from each of the K. pneumoniae genomes were extracted and then aligned using MUSCLE v3.8.31 (26). Gaps were removed from each alignment by using trimAl v1.6 (27), resulting in alignments of equal length (core genome alignments) which were then concatenated. Subsequently, Parsnp v1.2 (28) (settings -r !, -c, and -C 1000) was used to construct a maximum likelihood phylogenetic tree from the variable positions in these core genome alignments. The tree was midpoint rooted and visualized using FigTree software (v1.4.2; http://tree.bio.ed.ac.uk/software/figtree).
Identification of SNPs and indels.
Mapping of the Nesoni-filtered reads against the complete genome sequence of K. pneumoniae MGH 78578 (NCBI accession number NC_009648) was performed with Bowtie2 v2.2.0 (29) (settings -X 1200, and -a). Genomic repeats were removed from the analyses by filtering out reads that mapped to multiple positions in the K. pneumoniae MGH 78578 genome. To call single nucleotide polymorphisms (SNPs) and insertions and deletions (indels), SAMtools 0.1.18 (30) was used with the following settings: Q score of ≥50, mapping quality of ≥30, mapping depth of ≥10 reads, consensus of ≥75% to support a call, and ≥1 supporting reads in each direction.
Multilocus sequence typing (MLST) and identification of antibiotic resistance genes.
Sequence types of the isolates were determined by submitting the genome assemblies to MLST, version 1.8 (31). Antibiotic resistance genes in the genome assemblies were identified by ResFinder v2.1 (32).
Complementation in trans.
The genes that were mutated in the colistin-resistant K. pneumoniae strains were amplified from both the susceptible and resistant strains by PCR using 2× Phusion HF master mix (Thermo Scientific, Landsmeer, The Netherlands) and the primers listed in Table S1 in the supplemental material. The amplified fragments were cloned into the PCR-Trap cloning system (GenHunter, Nashville, TN), and the resulting plasmids (encoding resistance to tetracycline) were transformed into electrocompetent colistin-susceptible or -resistant K. pneumoniae strains by electroporation. The cloned amplicons were sequenced to ensure the absence of errors introduced during PCR (Macrogen Europe, Amsterdam, The Netherlands). Transformants were selected by overnight incubation at 37°C on Luria agar supplemented with 10 mg/liter of tetracycline. The lacZ gene fragment encoding the LacZ α-peptide was used as a control insert.
Accession number(s).
Sequence data from this study were deposited in NCBI's Short Read Archive (SRA) under accession number SRA354747.
RESULTS
Antimicrobial susceptibility and colistin heteroresistance.
An overview of the 13 K. pneumoniae strains included in this study is listed in Table 1; these strains included eight clinical isolates and five heteroresistant subpopulations. After retesting of antibiotic susceptibilities and confirmation of the presence of the ESBL phenotype by the double-disk synergy test, seven of the eight initially ESBL-producing K. pneumoniae clinical isolates were again found to be ESBL positive (isolate 3-CR lost the ESBL phenotype). None of the strains was resistant to carbapenem antibiotics. The six colistin-susceptible isolates had colistin MICs ranging from 1 to 2 mg/liter, and the two colistin-resistant strains had colistin MICs of 16 and 64 mg/liter.
TABLE 1.
Characteristics of ESBL-producing K. pneumoniae isolatesa
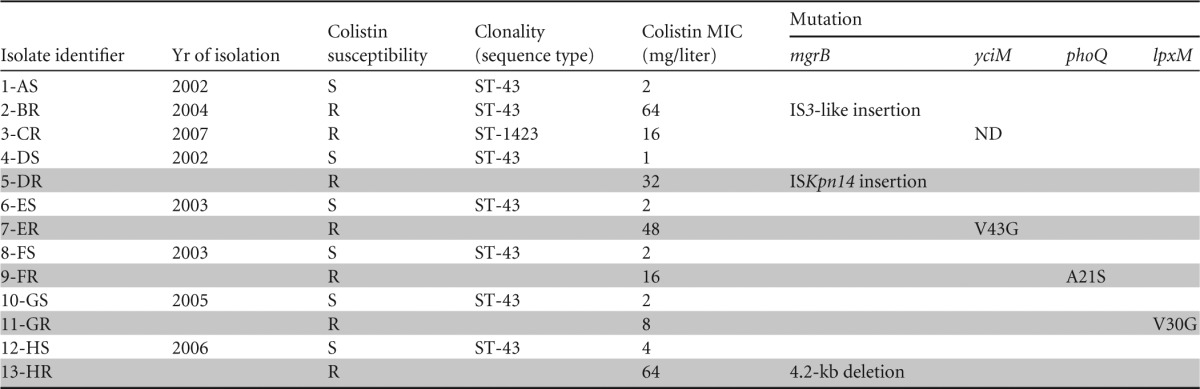
Isolate identifiers consist of unique numbers used in Fig. 1 and 2; letters indicate the code for the patient and whether the strain was susceptible (S) or resistant (R) to colistin. The isolate from patient A was obtained before the introduction of SDD in the ICU, and the remaining seven were obtained thereafter. Clonality was determined by phylogenetic analysis. MICs were determined by the broth microdilution method. The different SNPs, a deletion, and the inactivation of genes due to IS element insertions are indicated in the mutation columns. Heteroresistant strains are indicated with shading. ST, sequence type; ND, not determined.
PAPs revealed the presence of heteroresistance in five clinical isolates (Table 1) (isolates 4-DS, 6-ES, 8-FS, 10-GS, and 12-HS) initially considered colistin susceptible based on MICs ranging from 1 to 2 mg/liter. Subpopulations of these colistin-heteroresistant isolates grew in the presence of colistin at concentrations of 3 to 10 mg/liter (Fig. 1). The MICs for the resistant subpopulations 5-DR, 7-ER, 9-FR, 11-GR, and 13-HR were 32, 48, 16, 8, and 64 mg/liter, respectively (Table 1). The proportion of resistant colonies was on the order of 10−6. The colistin-susceptible reference strain ATCC 700603 survived in the presence of up to 0.5 g/liter colistin sulfate, and no heteroresistant subpopulations were observed.
FIG 1.
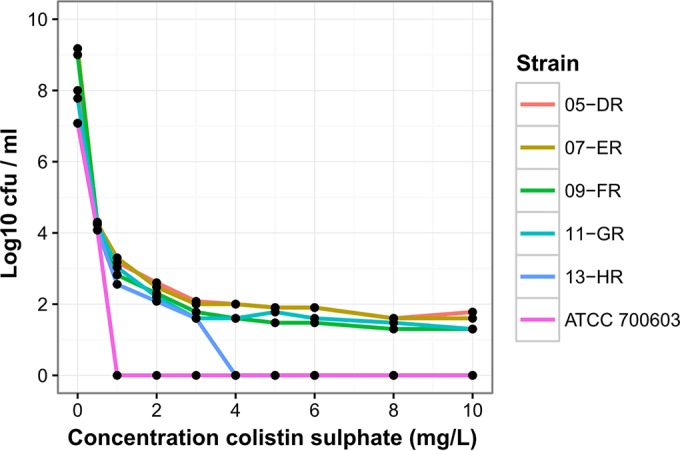
Population analysis profiles indicating colistin heteroresistance. Population analysis profiles are shown for five colistin-susceptible isolates after exposure to colistin sulfate. The y axis indicates the number of colonies on Mueller-Hinton agar plates, and concentrations of colistin sulfate are shown on the x axis.
Phylogenetic analysis of colistin-susceptible and colistin-resistant K. pneumoniae isolates.
A phylogenetic tree (Fig. 2A) for K. pneumoniae was generated based on the core genome sequence of 25 publicly available K. pneumoniae sequences and the 13 sequenced genomes of the K. pneumoniae isolates (Table 1). The core genome consisted of 2,637 orthogroups, with a total alignment length of 2,013,123 bp and 209,626 polymorphic sites. The core genome-based phylogenetic tree recapitulated the previously observed population structure of K. pneumoniae sensu lato, which includes Klebsiella quasipneumoniae (clade KpII) and Klebsiella variicola (clade KpIII) (22, 33, 34). Seven of the eight clinical isolates from the nosocomial outbreak were closely related to each other and clustered in the K. pneumoniae (KpI) clade. A single colistin-resistant isolate (3-CR) was assigned to clade KpIII and therefore appeared to be unrelated to the other isolates from the outbreak. All other strains from the outbreak (seven clinical isolates and five heteroresistant subpopulations) had the same sequence type, i.e., ST-43. These data confirm the previously reported existence of an outbreak with closely related K. pneumoniae isolates in an ICU (18).
FIG 2.
Phylogenetic tree and antibiotic resistance genes of K. pneumoniae strains. (A) The phylogenetic tree represents a concatenated alignment of 2,637 core orthogroups, with a combined length of 2,013,123 bp and 209,626 polymorphic sites, of 38 K. pneumoniae strains. The strains sequenced as part of this study are highlighted in color (orange, colistin-susceptible isolates; and blue, colistin-resistant isolates). (B) Antibiotic resistances detected in the K. pneumoniae strains that were sequenced as part of this study. Classes of antibiotic resistance genes are indicated as follows: A, aminoglycoside resistance genes; B, β-lactam resistance genes; C, chloramphenicol resistance genes; Q, quinolone resistance genes; S, sulfonamide resistance genes; and T, trimethoprim resistance genes.
Although the isolates belonged to the same sequence type, a repertoire of distinctly different antibiotic resistance genes was observed (Fig. 2B). The isolates carried several antibiotic resistance genes, including aminoglycoside resistance genes and β-lactam resistance genes.
Mutations associated with colistin resistance.
SNPs and indels were determined for all paired colistin-susceptible and colistin-resistant isolates. In addition, we determined whether full-length copies of the mgrB and phoQ genes were present in the isolates, as mutations leading to deletion or inactivation of these genes are a common cause of colistin resistance in K. pneumoniae (12, 35–39).
A limited number (1 to 4) of SNPs (see Table S2 in the supplemental material) and indels (see Table S3) distinguished the outbreak isolates. In comparisons of paired colistin-susceptible and colistin-resistant strains originating from the same patients, we identified mutations in mgrB that led to disruption of the gene in three colistin-resistant isolates (2-BR, 5-DR, and 13-HR). The event that led to the inactivation of mgrB was different for each strain. In strain 2-BR, an IS element (IS3-like) was inserted into mgrB. In strain 5-DR, the element ISKpn14 disrupted mgrB. Strain 13-HR had a 4.2-kb deletion including the mgrB gene. In all other colistin-resistant strains, mgrB was not mutated, meaning that other mutations must have led to the colistin resistance phenotypes of 7-ER, 9-FR, and 11-GR.
Interestingly, in these isolates, nonsynonymous SNPs were identified in genes that had predicted roles in outer membrane biosynthesis. Since the outer membrane is the main target of colistin, mutations in genes involving outer membrane biosynthesis may contribute to colistin resistance. In the strains from patient E, the only SNP difference between the colistin-susceptible (6-ES) and colistin-resistant (7-ER) isolates was a nonsynonymous SNP causing a V43G amino acid substitution encoded within the yciM gene. A mutation in phoQ, resulting in an A21S amino acid change, was identified in the colistin-resistant isolate 9-FR. The colistin-susceptible and colistin-resistant strains from patient G (10-GS and 11-GR, respectively) differed from each other by only 2 SNPs. One of these SNPs mapped to the lpxM gene, causing a V30G substitution.
Mutations in mgrB, yciM, phoQ, and lpxM contribute to colistin resistance.
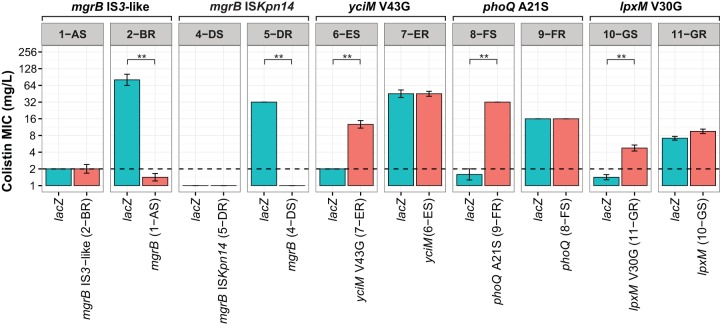
To verify whether the IS element insertions and amino acid substitutions identified by WGS contributed to colistin resistance, the colistin-susceptible and -resistant strain pairs were complemented in trans with the wild-type and mutated genes. Transformation of the strains with a control plasmid containing the lacZ gene did not alter the MIC of colistin for any strain. Complementation in trans with plasmids harboring the parental mgrB gene resulted in a reversal toward colistin susceptibility in isolates 2-BR and 5-DR (Fig. 3). Complementation in trans for the 12-HS and 13-HR pair was not performed due to difficulties in cloning the 4.2-kb deleted region spanning mgrB. Complementation with the mutated forms of phoQ, yciM, and lpxM resulted in decreased susceptibility to colistin. Electrotransformation of the colistin-susceptible strain 6-ES with a plasmid containing the mutated yciM gene resulted in a 3-fold increase in the MIC of colistin. The phoQ and lpxM mutations resulted in a 4-fold increased MIC of colistin for strain 8-FS and a 2-fold increased MIC of colistin for strain 10-GS, respectively. These observations indicate that mutations in yciM, phoQ, and lpxM are dominant when present in trans with the corresponding wild-type alleles, resulting in a colistin resistance phenotype. Because the deletions and inactivations of mgrB are loss-of-function mutations, they are recessive in the presence of the intact mgrB gene.
FIG 3.
Experimental validation of the roles of identified mutations in colistin resistance. Wild-type and mutated mgrB, yciM, phoQ, and lpxM alleles were cloned into corresponding K. pneumoniae strains by use of a PCR-TRAP cloning system (red). The strains from which the corresponding genes originated are indicated in parentheses. A vector containing a gene encoding the LacZ α-peptide of E. coli was used as a vector control (blue). The colistin MICs for the electrotransformed K. pneumoniae strains were determined by reference broth microdilution testing using cation-adjusted Mueller-Hinton broth supplemented with 10 mg/liter tetracycline. The colistin MIC resistance breakpoint (i.e., 2 mg/liter) is indicated with a black dashed line. Significant differences (P < 0.05; Mann-Whitney test) are indicated by double asterisks. The y axis was plotted on a log2 scale.
DISCUSSION
In a previous study, we showed that prolonged use of colistin as part of SDD in an outbreak setting resulted in the emergence of colistin resistance among ESBL-producing K. pneumoniae clinical isolates (18). The main finding of the present follow-up study is that heteroresistance among these apparently susceptible isolates forms a reservoir for the emergence of colistin resistance during treatment.
Heteroresistance has been recognized in both Gram-positive and Gram-negative bacteria and is a phenomenon where subpopulations of seemingly isogenic bacteria exhibit a range of susceptibilities to a particular antibiotic (13). Heteroresistance can be intrinsic or acquired. Intrinsic heteroresistance occurs without preexposure to the antibiotic, but heteroresistance may also be acquired or induced after initial exposure to antibiotics (13). Heteroresistance may have an impact on the outcome of clinical infection, particularly because its detection may be difficult by routine microbiology susceptibility testing (40). The PAP method used in the present study is considered the gold standard for determining heteroresistance (13). Phylogenetic analysis confirmed the clonality of all clinical isolates (excluding 3-CR). However, the absence of overlapping SNPs between clonal colistin-resistant isolates, isolated from different patients over a time span of 4 years, argues in favor of acquired, de novo resistance in individual strains under SDD use, rather than selection of preexisting mutants or transmission of the resistant strains between patients.
Although heteroresistance has previously been described for K. pneumoniae (41, 42), data on the molecular basis of colistin resistance in this species are scarce. Studies have recently shown that mutations in the genes encoding the PhoPQ two-component system and inactivation of the mgrB gene are important pathways by which K. pneumoniae can acquire resistance to colistin (43). In the present study, mutations in phoPQ and mgrB were found in four of the six analyzed isolates.
Mutations in the phoQ gene are a common mechanism by which Gram-negative bacteria, including K. pneumoniae, gain resistance to colistin (12, 38, 39). PhoQ is a sensor histidine kinase which, together with its cognate response regulator, PhoP, forms a two-component system (2CS). PhoPQ is activated under a variety of conditions, including low pH, low concentrations of Mg2+, and the presence of antimicrobial peptides, including colistin. Activation of PhoPQ leads to the expression of genes that modify LPS in a variety of ways, including deacylation of lipid A or modification of lipid A by 4-amino-4-deoxy-l-arabinose, leading to colistin resistance (12). We found that a mutation in phoQ resulting in an amino acid change (A21S) in the sensor domain of PhoQ which leads to colistin resistance was dominant over the nonmutated copy of phoQ. Notably, a mutation in Salmonella phoQ, resulting in a threonine-to-isoleucine change at position 48, in the sensor domain of the PhoQ protein, was also found to be dominant, as it constitutively increased phosphorylation of the response regulator PhoP (44, 45). A similar mechanism may explain why the phoQ mutation of Klebsiella pneumoniae strain 9-FR is dominant. Several studies have recently shown that inactivation of the mgrB gene, which encodes a negative regulator of the 2CS PhoPQ, causes colistin resistance (36–39). The inactivation or deletion of mgrB leads to higher activity of PhoPQ, which in turn activates the pmrHFIJKLM operon, which is responsible for modification of lipid A.
The mgrB and phoPQ genes were not mutated in the remaining two colistin-resistant isolates. In the heteroresistant strain from patient E, a mutation leading to an amino acid substitution encoded within the yciM gene was found. In Escherichia coli, yciM contributes to cell wall integrity by regulating LPS biosynthesis (46, 47), and a deletion in yciM leads to decreased susceptibility to colistin (48). It is possible that the mutation in yciM in K. pneumoniae increases LPS production, leading to higher levels of LPS in the outer membrane, which could titrate out the destabilizing effect of colistin binding to LPS. In the heteroresistant strain from patient G, a nonsynonymous mutation was found in the lpxM gene. LpxM is responsible for the addition of one of the secondary acyl chains to lipid A in Enterobacteriaceae (49, 50). In K. pneumoniae, deletion of lpxM contributes to susceptibility to antimicrobial peptides, including colistin (51). It is possible that the mutation in lpxM alters the acylation of lipid A, thereby making the strain more resistant to colistin. To our knowledge, this is the first time that mutations in yciM and lpxM have been found in K. pneumoniae and linked to reduced susceptibility to colistin. Currently, we cannot mechanistically explain why the mutated alleles of yciM and lpxM are dominant over the wild-type alleles. Conceivably, the presence of these alleles may interfere with the complex regulation of LPS biosynthesis in Klebsiella (52).
The present study shows that heteroresistance to colistin is present in a clonal population of ESBL-producing K. pneumoniae strains that were isolated from ICU patients who had been exposed to colistin. Our study highlights the multiple evolutionary trajectories that can lead to colistin resistance in K. pneumoniae and underscores the importance of monitoring the existence of colistin-resistant subpopulations in diagnostic susceptibility testing of K. pneumoniae.
Supplementary Material
ACKNOWLEDGMENT
W.V.S. was funded through an NWO-Vidi grant (grant 917.13.357).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01344-16.
REFERENCES
- 1.Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 6.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 7.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 8.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giacobbe DR, Del Bono V, Trecarichi EM, De Rosa FG, Giannella M, Bassetti M, Bartoloni A, Losito AR, Corcione S, Bartoletti M, Mantengoli E, Saffioti C, Pagani N, Tedeschi S, Spanu T, Rossolini GM, Marchese A, Ambretti S, Cauda R, Viale P, Viscoli C, Tumbarello M, ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva). 2015. Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin Microbiol Infect 21:1106.e1–1106.e8. doi: 10.1016/j.cmi.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, Veenemans J. 2015. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis 34:1647–1655. doi: 10.1007/s10096-015-2401-2. [DOI] [PubMed] [Google Scholar]
- 11.Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, Network EuSCAPE-Italy, Grundmann H, Pantosti A, Rossolini GM. 2014. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill 19:20939. doi: 10.2807/1560-7917.ES2014.19.42.20939. [DOI] [PubMed] [Google Scholar]
- 12.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermes DM, Pormann Pitt C, Lutz L, Teixeira AB, Ribeiro VB, Netto B, Martins AF, Zavascki AP, Barth AL. 2013. Evaluation of heteroresistance to polymyxin B among carbapenem-susceptible and -resistant Pseudomonas aeruginosa. J Med Microbiol 62:1184–1189. doi: 10.1099/jmm.0.059220-0. [DOI] [PubMed] [Google Scholar]
- 15.Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother 52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez CH, Barberis C, Nastro M, Bombicino K, Granados G, Vay C, Famiglietti A. 2012. Impact of heteroresistance to colistin in meningitis caused by Acinetobacter baumannii. J Infect 64:119–121. doi: 10.1016/j.jinf.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halaby T, Al Naiemi N, Kluytmans J, van der Palen J, Vandenbroucke-Grauls CMJE. 2013. Emergence of colistin resistance in Enterobacteriaceae after the introduction of selective digestive tract decontamination in an intensive care unit. Antimicrob Agents Chemother 57:3224–3229. doi: 10.1128/AAC.02634-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore DM, Brown DF. 2001. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother 48(Suppl 1):S59–S64. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NTK, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 24.Ekseth OK, Kuiper M, Mironov V. 2014. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 30:734–736. doi: 10.1093/bioinformatics/btt582. [DOI] [PubMed] [Google Scholar]
- 25.Van Dongen S. 2008. Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl 30:121–141. doi: 10.1137/040608635. [DOI] [Google Scholar]
- 26.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brisse S, Verhoef J. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol 51:915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 34.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine M-H, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poirel L, Jayol A, Bontron S, Villegas M-V, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 38.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain J-M. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 39.Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charles PGP, Ward PB, Johnson PDR, Howden BP, Grayson ML. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin Infect Dis 38:448–451. doi: 10.1086/381093. [DOI] [PubMed] [Google Scholar]
- 41.Bogdanovich T, Adams-Haduch JM, Tian G-B, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis 53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. 2011. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother 66:946–947. doi: 10.1093/jac/dkr007. [DOI] [PubMed] [Google Scholar]
- 43.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunn JS, Hohmann EL, Miller SI. 1996. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol 178:6369–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baud D, Benyacoub J, Revaz V, Kok M, Ponci F, Bobst M, Curtiss R, De Grandi P, Nardelli-Haefliger D. 2004. Immunogenicity against human papillomavirus type 16 virus-like particles is strongly enhanced by the PhoPc phenotype in Salmonella enterica serovar Typhimurium. Infect Immun 72:750–756. doi: 10.1128/IAI.72.2.750-756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahalakshmi S, Sunayana MR, SaiSree L, Reddy M. 2014. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol Microbiol 91:145–157. doi: 10.1111/mmi.12452. [DOI] [PubMed] [Google Scholar]
- 47.Nicolaes V, El Hajjaji H, Davis RM, Van der Henst C, Depuydt M, Leverrier P, Aertsen A, Haufroid V, Ollagnier de Choudens S, De Bolle X, Ruiz N, Collet J-F. 2014. Insights into the function of YciM, a heat shock membrane protein required to maintain envelope integrity in Escherichia coli. J Bacteriol 196:300–309. doi: 10.1128/JB.00921-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother 54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somerville JE, Cassiano L, Bainbridge B, Cunningham MD, Darveau RP. 1996. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J Clin Invest 97:359–365. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan SA, Everest P, Servos S, Foxwell N, Zähringer U, Brade H, Rietschel ET, Dougan G, Charles IG, Maskell DJ. 1998. A lethal role for lipid A in Salmonella infections. Mol Microbiol 29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 51.Clements A, Tull D, Jenney AW, Farn JL, Kim S-H, Bishop RE, McPhee JB, Hancock REW, Hartland EL, Pearse MJ, Wijburg OLC, Jackson DC, McConville MJ, Strugnell RA. 2007. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J Biol Chem 282:15569–15577. doi: 10.1074/jbc.M701454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, Spence S, Monahan A, Monaghan A, Kissenpfennig A, Ingram RJ, Bengoechea J, Gally DL, Fanning S, Elborn JS, Schneiders T. 2015. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 11:e1004627. doi: 10.1371/journal.ppat.1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.