Abstract
Persister cells are highly tolerant to different antibiotics and are associated with relapsing infections. In order to understand this phenomenon further, we exposed a transposon library to a lethal concentration of ampicillin, and mutants that survived were identified by transposon sequencing (Tn-Seq). We determined that mutations related to carbon metabolism, cell envelope (cell wall generation and membrane proteins), and stress response have a role in persister cell generation.
TEXT
Uropathogenic Escherichia coli (UPEC) is the main etiologic agent of urinary tract infections worldwide, generating approximately 80% of clinical cases every year. Most of these infections are chronic and represent a worldwide public health threat (1). Relapsing infections have been associated with the generation of persister cells (2, 3). This subpopulation is characterized by a transient nonhereditary dormant state that leads to survival in lethal concentrations of different antibiotics. The molecular mechanisms reported to be involved in the generation of persister cells include stochastic processes, toxin-antitoxin (TA) modules, and the stringent response (4–6). However, a deeper understanding of the physiological and genetic regulation of this process is lacking.
Aiming to identify global regulators or metabolic pathways that E. coli might downregulate to enter into a dormant persister state, we used a high-throughput genetic screen to recognize mutants with increased fitness during exposure to a lethal concentration of the cell wall-active antibiotic ampicillin in order to identify novel regulators and/or metabolic pathways involved in the generation of persister cells. This method has previously been used to identify metabolic pathways associated with the generation of persister cells in stationary-phase cultures of E. coli exposed to gentamicin (7).
We used a library of UPEC strain CFT073 containing approximately 360,000 random insertions of the EZ-Tn5 <R6Kγori/KAN-2> transposon (8). A 1:100 dilution of an overnight culture of the library was grown at 37°C in Luria-Bertani (LB) broth with aeration and when the optical density at 600 nm (OD600) reached 0.3, the culture was split in two flasks, and ampicillin was added to a final concentration of 125 μg ml−1 (10 times the MIC). After 6 h of incubation at 37°C with shaking, we enriched the survivors by overnight growth in fresh LB lacking ampicillin (approximately 45 generations of outgrowth).
Total DNA, corresponding to three independent experiments, was extracted from the input and ampicillin-treated output cultures using a QIAamp DNA minikit (Qiagen).
Transposon insertions in mutants showing increased survival in cultures exposed to ampicillin were identified by transposon sequencing (Tn-Seq) using the homopolymer tail-mediated ligation PCR (HTML-PCR) method followed by sequencing on a HiSeq 2500 system (Illumina) (9, 10). Briefly, DNA was fragmented by sonication, and poly(C) tails were added to all 3′ termini using terminal deoxynucleotidyl transferase. Then, a nested PCR strategy was used to amplify the genomic DNA adjacent to transposon insertions, while at the same time the index sequences required for multiplex sequencing were added. Data were analyzed using the Tufts University Core Facility Galaxy server as described previously (11). Mutants were considered positively selected if the following requirements were met: (i) at least three unique insertions were present in each gene in all three input samples, (ii) a Dval genome value (defined as the number of reads of each gene divided by predicted number of reads) of ≥0.01 in each input, which is a criterion used to discard mutants in the library with severely impaired growth, was found in each input, and (iii) an average survival index (Dval genome output/input) of ≥2 was observed.
We detected ∼68,000 mutants in the input samples and ∼55,000 in the output samples. Among those, we identified 50 genes under positive selection in our screen; i.e., transposon insertions in these genes led to increased survival upon ampicillin exposure (Table 1). Among these are genes encoding citric acid cycle enzymes AcnB and FumA, arginine catabolism-related YdjS (12), fructose transport FruA (13), maintenance of NAD+/NADH balance UdhA (14), and gluconeogenic fructose-1,6-bisphosphatase. In addition, we found a putative pyruvate formate-lyase 3-activating enzyme, which might catalyze the conversion of pyruvate into formate in anaerobic respiration (15), and the N-acetylglucosamine repressor NagC, which is necessary for the utilization of N-acetylglucosamine as a carbon source (16). Also, the YfcC (hypothetical protein) has been related to the glyoxylate shunt, which might contribute to the generation of carbon skeletons that can be used as energy in the absence of glucose (17). These results suggest that mutants with impaired metabolism and/or an inability to use different carbon sources might enter into a dormant state that is characteristic of the persister subpopulation (18, 19). These results are supported by recent findings showing that the generation of persister cells is associated with depletion of ATP in Staphylococcus aureus (20).
TABLE 1.
Proteins of mutants under positive selection displaying increased survival when exposed to a lethal concentration of ampicillin
| Locus | Protein | Gene | Mean SIa |
|---|---|---|---|
| c0147 | Aconitate hydratase 2 | acnB | 87.405 |
| c0608 | Hypothetical protein | 5.706 | |
| c0751 | N-Acetylglucosamine repressor | nagC | 6.602 |
| c0764 | Putative pyridoxine phosphate biosynthetic protein | 5.446 | |
| c0861 | Hypothetical protein YbhK | ybhK | 11.288 |
| c0909 | Putative pyruvate formate-lyase 3-activating enzyme | 11.155 | |
| c0943 | Hypothetical protein | 7.865 | |
| c1174 | Hypothetical protein | 2.832 | |
| c1176 | Hypothetical protein | 10.699 | |
| c1179 | 9.273 | ||
| c1254 | Putative glucosyltransferase | iroB | 8.533 |
| c1303 | Hypothetical protein | 16.152 | |
| c1560 | Outer membrane porin protein NmpC precursor | nmpC | 12.465 |
| c1572 | Putative capsid assembly protein of prophage | 8.870 | |
| c1610 | Conserved hypothetical protein | 2.817 | |
| c1822 | Outer membrane protein N precursor | ompN | 7.103 |
| c1884 | Hypothetical protein | 6.538 | |
| c1888 | Conserved hypothetical protein | 5.430 | |
| c1925 | Hypothetical ABC transporter ATP-binding protein YddA | yddA | 3.138 |
| c1927 | Putative sulfatase YdeN precursor | ydeN | 4.039 |
| c1945 | Hypothetical protein YneF | yneF | 3.146 |
| c1956 | Putative outer membrane protein YieC precursor | 4.482 | |
| c2004 | Fumarate hydratase class I, aerobic | fumA | 3.121 |
| c2034 | Transcriptional regulator SlyA | slyA | 15.285 |
| c2144 | Succinylglutamate desuccinylase | ydjS | 3.865 |
| c2199 | Hypothetical protein YeaP | yeaP | 8.016 |
| c2232 | Hypothetical protein YobF | yobF | 62.546 |
| c2377 | Hypothetical protein YedI | yedI | 7.341 |
| c2385 | Protein YedU | yedU | 3.832 |
| c2465 | Hypothetical protein | 12.476 | |
| c2471 | Hypothetical protein | 16.751 | |
| c2475 | Hypothetical protein | 55.491 | |
| c2615 | Hypothetical protein | 3.164 | |
| c2677 | SanA protein | sanA | 3.147 |
| c2702 | PTSb system, fructose-specific IIBC component | fruA | 6.573 |
| c2841 | Hypothetical protein YfcC | yfcC | 2.660 |
| c3028 | Exodeoxyribonuclease VII large subunit | xseA | 4.108 |
| c3174 | Prophage QSR′ DNA packaging protein NU1 homolog | nohB | 5.787 |
| c3255 | Membrane-bound lytic murein transglycosylase B precursor | mltB | 6.485 |
| c3721 | Hypothetical ATP-binding protein YghR | yghR | 5.162 |
| c3888 | PTS system, N-acetylgalactosamine-specific IIB component 2 | agaV | 5.440 |
| c4475 | Guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase | spoT | 7.415 |
| c4704 | Undecaprenyl-phosphate alpha-N-acetylglucosaminyltransferase | rfe | 3.950 |
| c4725 | Adenylate cyclase | cyaA | 76.484 |
| c4804 | Thiol:disulfide interchange protein DsbA precursor | dsbA | 437.265 |
| c4923 | Soluble pyridine nucleotide transhydrogenase | udhA | 52.361 |
| c5177 | Hypothetical protein in ISc | 2.712 | |
| c5207 | Hypothetical protein | 37.124 | |
| c5320 | Hypothetical protein YtfP | ytfP | 2.534 |
| c5329 | Fructose-1,6-bisphosphatase | fbp | 6.908 |
SI, survival index.
PTS, phosphotransferase system.
IS, insertion element.
Moreover, we also identified genes encoding envelope proteins, i.e., peptidoglycan turnover and membrane proteins. Among these, increased survival indices were observed for nmpC, sanA, mltB, agaV, rfe, and dsbA mutants (Table 1). Additionally, transposon insertions in the ybhK gene were identified in our screen. It has been shown that YbhK participates in gluconeogenesis, and a role in the generation of cell wall precursor molecules has been proposed (21). These results indicate that decreasing the turnover of peptidoglycan and/or inhibiting the expression of certain membrane proteins might induce tolerance to ampicillin through envelope stabilization. However, it is also possible that these mutations might in addition or alternatively generate dormancy through some mechanism such as decreasing the rate of multiplication.
Our selection also identified three genes, yedU, xseA, and the transcriptional regulator slyA, which are related to different kinds of stress responses (22–24). We speculate that insertion mutants with an impaired ability to respond to different types of cell damage might, in the presence of ampicillin, exhibit growth arrest, dormancy, and a persister phenotype.
We identified transposon insertions in the noncoding regions upstream of the c1884 and rfe genes, likely in their respective promoters. This observation is consistent with the results of Table 1 showing that insertions in the same genes are positively selected, which indicates that either knocking out or inhibiting the expression of those genes generates increased tolerance to ampicillin. This result was further validated by showing that a clean deletion in c1884 increased survival compared to that of the wild-type strain (Fig. 1).
FIG 1.
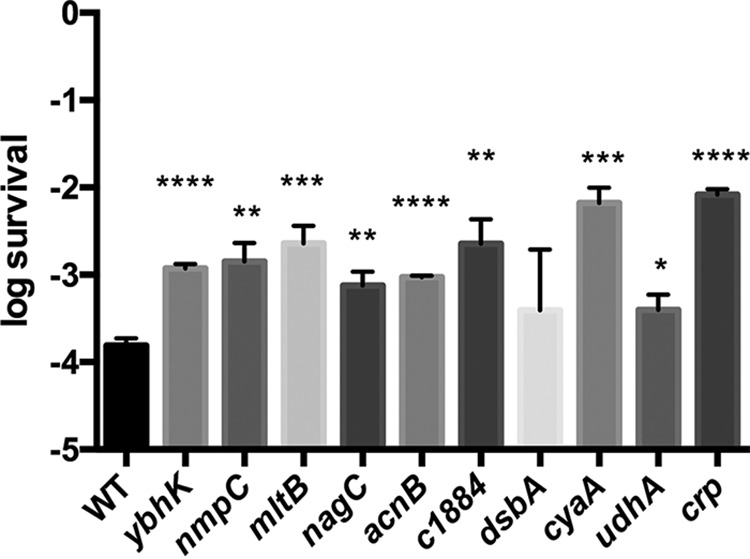
Increased survival of ampicillin exposure by E. coli strains deficient in genes identified by Tn-Seq. The ability to generate persister cells of mutant strains in the background CFT073 was assessed by CFU counting. Error bars denote standard errors corresponding to 3 independent trials. Statistical significance was determined using a two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). WT, wild-type strain.
Consistent with the known role of the stringent response in the persister phenotype, we identified spoT, which encodes the enzyme that synthesizes the stringent response alarmone (p)ppGpp. Based on our results, we cannot exclude the possibility that a secondary mutation(s) in relA and/or another gene(s) that affects the (p)ppGpp level contributes to the observed phenotype, since (i) it has been determined that in E. coli spoT is essential in an relA+ background (25) and (ii) it has been shown that strains deficient in spoT and relA display an impaired ability to generate persister cells (26) Alternatively, the transposon insertions in spoT may have created truncated proteins that retained the essential activity.
Similarly, we identified cyaA and crp promoter mutants in our screen. The effector molecule cyclic AMP (cAMP) synthetized by adenylate cyclase (CyaA) and cAMP receptor protein (CRP) regulate a plethora of biological processes in response to the energy status of the cell (27). It has been previously established that cAMP is involved in the persistence of E. coli through the regulation of indole levels (28). These findings further support our results and the important role of this molecule in the persistence phenomenon.
Most of the genes identified in our selection are hypothetical, which does not allow a deeper interpretation. However, since these results were validated as shown in Fig. 1, a role for those genes in persister generation is proposed.
Finally, insertions in udhA (Table 1 and Fig. 1), which encodes a soluble pyridine nucleotide transhydrogenase, were found to have increased tolerance to ampicillin. We observed that a mutant with this gene deleted grows slower than the wild-type strain (data not shown), and this slow growth might explain the associated increased survival.
To validate our findings we randomly chose and constructed deletions in 10 of the 50 genes identified using the FRT (FLP recombination target)-FLP and lambda Red systems as described previously (29, 30). Each mutation was back-crossed to the wild-type background by generalized transduction using phage φEB49 (31). Finally, the kanamycin resistance cassette was deleted by expression of the FLP resolvase in trans, leaving behind an FRT scar in place of each gene. Tables S1 and S2 in the supplemental material contain the bacterial strains/plasmids and DNA sequences of the primers used to construct these mutations.
The ability to generate persister cells in the deletion strains was evaluated by measuring survival of a lethal dose of ampicillin. Briefly, an overnight culture was diluted 100-fold in fresh LB and incubated at 37°C with aeration; when the OD600 reached ∼0.3 (∼3 × 108 CFU/ml), it was exposed to 125 μg/ml ampicillin for 6 h at 37°C with aeration. For determination of CFU, aliquots were serially diluted in phosphate-buffered saline (PBS) and plated on LB agar plates supplemented with 20 mM MgSO4 and 2 mg/ml sodium pyruvate as previously described (32). Persister cell levels were determined by the ratio of the number of CFU/ml in the culture after 6 h of exposure to ampicillin to the number of CFU/ml before addition of the antibiotic. Increased survival was observed for 9 of 10 mutants tested: ybhK, mltB, nmpC, acnB, c1884, cyaA, udhA, crp, and nagC (Fig. 1). This validates our genetic selection and Tn-Seq analysis. No changes in MICs were observed for any mutants except for cyaA and crp, which showed 2-fold increases compared to that for the wild-type strain. Similarly, no differences in the growth rates were observed for the other mutants evaluated except for the udhA strain as stated above.
This study demonstrates that analysis of a complex pool of mutants under positive selection is a powerful method to study biological phenomena such as persistence, where genetic redundancy is relevant (33).
Taken together, our results suggest that E. coli should be able to enter into a persister state by downregulating pathways involved in central metabolism, catabolism of alternative carbon sources, peptidoglycan turnover, and stress response genes. This adds to our knowledge of how persister cells may be generated and suggests that processes in addition to the stringent response and TA modules may be involved in this important phenotype.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. L. Sonenshein and B. Belitsky for valuable discussions and C. A. Silva for critical reading of the manuscript. We also thank Harry Mobley and Rodney Welch, who kindly provided the transposon library and bacteriophage φEB49.
This work was supported by National Institute of Allergy and Infectious Diseases grant HH4134 from the National Institutes of Health (S.B.L.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01617-16.
REFERENCES
- 1.Bien J, Sokolova O, Bozko P. 2012. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol 2012:681473. doi: 10.1155/2012/681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 6.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 7.Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. 2015. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6:e00078-15. doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HLT. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. doi: 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazinski DW, Camilli A. 2013. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques 54:25–34. doi: 10.2144/000113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonough E, Lazinski DW, Camilli A. 2014. Identification of in vivo regulators of the Vibrio cholerae xds gene using a high-throughput genetic selection. Mol Microbiol 92:302–315. doi: 10.1111/mmi.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider BL, Kiupakis AK, Reitzer LJ. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol 180:4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornberg HL. 2001. Routes for fructose utilization by Escherichia coli. J Mol Microbiol Biotechnol 3:355–359. [PubMed] [Google Scholar]
- 14.Holm AK, Blank LM, Oldiges M, Schmid A, Solem C, Jensen PR, Vemuri GN. 2010. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. J Biol Chem 285:17498–17506. doi: 10.1074/jbc.M109.095570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knappe J, Blaschkowski HP, Gröbner P, Schmitt T. 1974. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem 50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 16.Plumbridge J, Kolb A. 1993. DNA loop formation between Nag repressor molecules bound to its two operator sites is necessary for repression of the nag regulon of Escherichia coli in vivo. Mol Microbiol 10:973–981. doi: 10.1111/j.1365-2958.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Xie Y, Gao P, Zhang S, Tan H, Yang F, Lian R, Tian J, Xu G. 2014. A metabolomics-based method for studying the effect of yfcC gene in Escherichia coli on metabolism. Anal Biochem 451:48–55. doi: 10.1016/j.ab.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 19.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 21.Görke B. 2005. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 151:3777–3791. doi: 10.1099/mic.0.28172-0. [DOI] [PubMed] [Google Scholar]
- 22.Sastry MSR. 2002. Hsp31, the Escherichia coli yedU gene product, is a molecular chaperone whose activity is inhibited by ATP at high temperatures. J Biol Chem 277:46026–46034. doi: 10.1074/jbc.M205800200. [DOI] [PubMed] [Google Scholar]
- 23.Jung H, Liang J, Jung Y, Lim D. 2015. Characterization of cell death in Escherichia coli mediated by XseA, a large subunit of exonuclease VII. J Microbiol 53:820–828. doi: 10.1007/s12275-015-5304-0. [DOI] [PubMed] [Google Scholar]
- 24.Spory A, Bosserhoff A, von Rhein C, Goebel W, Ludwig A. 2002. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J Bacteriol 184:3549–3559. doi: 10.1128/JB.184.13.3549-3559.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 26.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 27.Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol Rev 56:100–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwan BW, Osbourne DO, Hu Y, Benedik MJ, Wood TK. 2015. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol Bioeng 112:588–600. doi: 10.1002/bit.25456. [DOI] [PubMed] [Google Scholar]
- 29.Murphy KC, Campellone KG. 2003. Lambda red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol 4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battaglioli EJ, Baisa GA, Weeks AE, Schroll RA, Hryckowian AJ, Welch RA. 2011. Isolation of generalized transducing bacteriophages for uropathogenic strains of Escherichia coli. Appl Environ Microbiol 77:6630–6635. doi: 10.1128/AEM.05307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


