Abstract
A collection of 74 Enterobacteriaceae isolates found in Bo, Sierra Leone, were tested for quinolone antibiotic susceptibility and resistance mechanisms. The majority of isolates (62%) were resistant to quinolones, and 61% harbored chromosomal gyrA and/or parC mutations. Plasmid-mediated quinolone resistance genes were ubiquitous, with qnrB and aac(6′)-Ib-cr being the most prevalent. Mutated LexA binding sites were found in all qnrB1 genes, and truncated qnrB pseudogenes were found in the majority of Citrobacter isolates.
TEXT
Quinolone and fluoroquinolone compounds are synthetic antibiotics whose antimicrobial activity occurs via concentration-dependent inhibition of type II topoisomerases (1, 2). Fluoroquinolones are currently used to treat a variety of human infections caused by both Gram-positive and Gram-negative bacteria due to their broad-spectrum antimicrobial activity (3–5). Their widespread and often indiscriminate use, however, has resulted in ubiquitous resistance, especially among members of the Enterobacteriaceae (3, 4, 6).
The most common causes for high-level quinolone resistance in Gram-negative bacteria are mutations in quinolone resistance determinant regions (QRDRs) of the type II topoisomerase genes gyrA and parC (5, 7). Another mechanism for quinolone resistance depends on the maintenance of low intracellular drug concentrations via decreased uptake of the drug and/or active efflux using efflux pumps. When upregulated, some of these multidrug efflux pumps confer cross-resistance to multiple classes of antimicrobials (4, 5).
Plasmid-mediated quinolone resistance (PMQR) is conferred by extrinsic resistance determinants that encode efflux pumps (qepA and oqxAB) (8), proteins that protect DNA gyrase and topoisomerase IV through specific binding (qnr genes), or quinolone-inactivating enzymes [aac(6′)-Ib-cr] (9). PMQR genes generally confer low-level resistance, with their MICs falling below Clinical and Laboratory Standards Institute (CLSI) breakpoints for intermediate resistance; therefore, their contribution to quinolone resistance can be masked in strains also harboring QRDR mutations in gyrA and parC. However, their clinical significance stems from the fact that they greatly facilitate the selection of more highly quinolone-resistant strains (10).
An increasing prevalence of quinolone resistance has been reported in West Africa (11–13), with several mechanistic surveys of quinolone nonsusceptibility within this region recently published (13–19). However, no such data are available for Sierra Leone (11). Here, we analyzed 70 Enterobacteriaceae urine sample isolates and four fomite isolates from the indoor environment from a small private hospital located in Bo, Sierra Leone (20). The collection included Citrobacter freundii (n = 22), Enterobacter cloacae (n = 16), Klebsiella pneumoniae (n = 17), Escherichia coli (n = 13), Enterobacter sp./Leclercia sp. (n = 4), Escherichia hermannii (n = 1), and Pantoea dispersa (n = 1). Each isolate in the collection was tested for susceptibility to nalidixic acid, ciprofloxacin, and moxifloxacin using Etest strips (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's recommendations using CLSI interpretative criteria (for nalidixic acid and ciprofloxacin) (21) and FDA recommendations (for moxifloxacin) (22) for phenotype classification.
We sequenced the QRDRs of the chromosomal gyrA and parC genes to identify potential resistance-conferring mutations and screened the isolates by PCR for the following PMQR genes: aac(6′)-Ib-cr, qepA, oqxAB, qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC (see Table S1 in the supplemental material). Detected qnr genes were fully sequenced.
Overall, 62% of the tested isolates were clinically resistant to at least one quinolone antibiotic, while 41% were resistant to all three. The prevalence of resistance was highest in C. freundii and Enterobacter sp./Leclercia sp. and lowest (or absent) in E. cloacae, E. hermannii, and P. dispersa (Table 1). Resistance correlated strongly with the presence of gyrA and parC gene QRDR mutations (5, 7). While a single gyrA mutation was correlated with high-level resistance to nalidixic acid and elevated MICs (not crossing the clinical resistance breakpoint) for ciprofloxacin and moxifloxacin (P < 0.05), additional mutations were correlated with high-level resistance to all three quinolones (P < 0.00005; see Table S2 in the supplemental material).
TABLE 1.
Quinolone resistance, PMQR genes, and QRDR mutation prevalence
| Species | No. of isolates | Quinolone resistance prevalence (%)a |
PMQR gene(s) detected and its prevalence (%)b | QRDR mutation prevalence (%)c |
|||||
|---|---|---|---|---|---|---|---|---|---|
| GyrA |
ParC |
||||||||
| NA | CIP | MOX | Ser83 | Asp87 | Ser80 | Glu84 | |||
| Citrobacter freundii | 22 | 100 | 100 | 95 | aac(6′)-Ib-cr (86), qnrA1 (4), qnrB1a (10), qnrB1d (23), qnrB6 (36), qnrB12 (14), qnrB81 (4), qnrB82 (4), ΔqnrB (86), qnrS1 (50) | 100 | 14 | 59 | 0 |
| Enterobacter sp./Leclercia sp. | 4 | 100 | 100 | 25 | aac(6′)-Ib-cr (100), qnrB1a (100) | 100 | 0 | 100 | 0 |
| Escherichia coli | 13 | 61 | 23 | 23 | qepA1 (23) | 61 | 23 | 31 | 8 |
| Klebsiella pneumoniae | 17 | 53 | 35 | 29 | aac(6′)-Ib-cr (29), qnrB1d (23), oqxAB (100) | 53 | 12 | 53 | 0 |
| Enterobacter cloacae | 16 | 6 | 0 | 19 | aac(6′)-Ib-cr (81), qnrB1a (25), qnrB1i (50), qnrB6a (6), qnrS1 (19), oqxAB (6) | 12 | 0 | 6 | 0 |
| Escherichia hermannii | 1 | 0 | 0 | 0 | aac(6′)-Ib-cr, qnrB1a, qnrS1 | 0 | 0 | 0 | 0 |
| Pantoea dispersa | 1 | 0 | 0 | 0 | None detected | ND | ND | ND | ND |
Prevalence of clinically resistant isolates as measured using Etest assays. NA, nalidixic acid; CIP, ciprofloxacin; MOX, moxifloxacin.
The presence of the following PMQR genes was tested: qnrA, qnrB, qnrC, qnrD, qnrS, qnrVC, aac(6′)-Ib-cr, qepA, and oqxAB. Numbers in parentheses indicate the prevalence (percent) of a particular PMQR gene among tested isolates (when more than one isolate was present).
Presence of mutations was deduced via in silico translation of the obtained DNA sequences. The position numbers are based on E. coli GyrA and ParC protein sequences. ND, not done.
PMQR genes were ubiquitous among the analyzed Enterobacteriaceae (Table 1; for detailed information, see Table S2 in the supplemental material), consistent with previous reports of Enterobacteriaceae in West Africa (13–19). Observed in all species tested except E. coli and P. dispersa, various qnr genes and aac(6′)-Ib-cr were detected in 68% and 58% of the isolates, respectively. Other detected PMQR genes included oqxAB (found in all K. pneumoniae strains and one E. cloacae strain) and qepA (in three E. coli isolates). As indicated above, the phenotypic contributions of the PMQR genes were difficult to determine due to the presence of gyrA and parC QRDR mutations.
Among the PMQR genes, the qnr genes showed the highest diversity and widest distribution. Detected alleles belonged to the qnrA, qnrB, and qnrS families. qnrA and qnrS families were represented by only one allelic variant each (qnrA1 and qnrS1, respectively). In contrast, the qnrB family was represented by three previously described full-length alleles (qnrB1, qnrB6, and qnrB12), two novel variants (qnrB81 and qnrB82), and three variants of 5′ truncated qnrB pseudogenes (ΔqnrB). All of the qnrB gene variants (with the notable exception of qnrB1) and pseudogenes were found only in C. freundii isolates. The majority of the >80 qnrB gene variants described thus far have been found in this species, which is also postulated as the original source of the qnrB gene (23).
The complete DNA sequencing of all detected qnrB1 genes showed that the coding regions were identical but revealed differences in the noncoding sequences upstream of the qnrB1 open reading frame (ORF) within the LexA protein binding site. LexA is a transcriptional repressor that modulates the SOS regulon (24), and the expression of qnrB genes containing LexA binding sequences is induced as a part of the SOS response to a number of environmental stimuli, including quinolone exposure (25, 26). No analogous LexA binding sites are found within other qnr gene families.
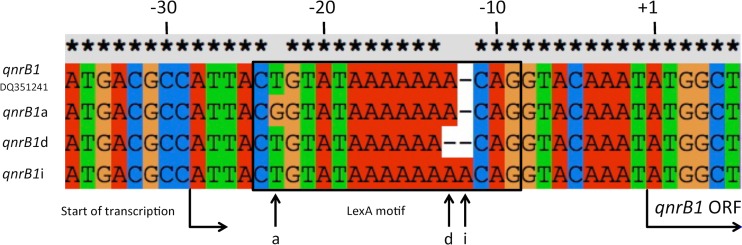
Based on differences within the LexA binding motifs, three qnrB1 subvariants could be distinguished: qnrB1a is identical to the prototype qnrB1 sequence (accession number DQ351241) except for a single T→G substitution at position −23, qnrB1d harbors a single nucleotide deletion, and qnrB1i contains a single nucleotide insertion within the poly(A) sequence located between positions −12 and −18 of the prototype qnrB1 sequence (Fig. 1). Of the remaining qnrB alleles identified in this study, only qnrB6 in E. cloacae harbored mutations within this upstream region; as this mutation was identical to that found in qnrB1a, this allele was thus designated qnrB6a. The mutation in qnrB1a (and qnrB6a) changes the highly conserved 5′ CTGT of the LexA binding palindrome consensus CTGT-N8-ACAG characteristic of Gammaproteobacteria (24), while the LexA binding site mutations in qnrB1i and qnrB1d affect the length of this motif. This is the first report of mutated LexA binding sites identified in qnrB genes from clinical isolates. Although we did not explore the effect of these mutations on LexA binding or phenotype, it has been previously shown that the loss of LexA binding results in increased quinolone MICs (25–27). The qnrB1 subvariants were unevenly distributed among the analyzed species (see Table S2 in the supplemental material).
FIG 1.
Mutations within the LexA binding site directly upstream of the qnrB1 gene. The sequences flanking the 5′ end of the qnrB1 ORF from the three detected qnrB1 subvariants (qnrB1a, qnrB1d, and qnrB1i) were aligned with the analogous region from the prototype qnrB1 gene (GenBank accession number DQ351241). The nucleotide position numbers are based on the prototype qnrB1 gene sequence and denote the distance from the first nucleotide of the translation initiation codon (labeled +1). The sequences enclosed in the black box indicate the LexA binding motif. The vertical arrows indicate the point mutation sites characteristic of the particular qnrB1 gene variants (a, qnrB1a [qnrB6a]; d, qnrB1d; i, qnrB1i).
In addition to the full-length alleles, three truncated qnrB pseudogenes (ΔqnrB) were also identified in the majority (91%) of the C. freundii isolates, and most of the C. freundii isolates contained these pseudogenes together with full-length qnrB genes. To our knowledge, this is the first report in which both full-length qnrB genes and truncated qnrB pseudogenes were found to coexist in the same isolates. While the presence of truncated qnrB pseudogenes in C. freundii has been previously reported, it has never been observed in such a high prevalence as documented in this study (23, 28–30). The three distinct qnrB pseudogene variants (Δ1qnrB, Δ2qnrB, and Δ3qnrB [see Table S2 in the supplemental material]) all encompassed a 283-bp remnant of the ORF that resulted from the deletion of the 360 bp at the 5′ end of the gene.
Flanking region sequences indicated that all of the qnrB pseudogenes and the newly described qnrB variants (qnrB81 and qnrB82) were located within the context of genetic platform GP3 while all other qnrB alleles had flanking sequences consistent with the GP2 environment (30). In addition, the finished genome sequencing of the C. freundii isolate SL151 (T. A. Leski, C. R. Taitt, U. Bangura, R. Ansumana, D. A. Stenger, Z. Wang, and G. Vora, submitted for publication; accession numbers CP016952 [genome], CP017058 [larger plasmid], and CP017059 [smaller plasmid]) revealed that the pseudogene variant Δ2qnrB was located on the chromosome while the other two qnr genes (qnrB6 and qnrS1) as well as aac(6′)-Ib-cr were located on two distinct plasmids. The wide distribution of the qnrB1 allele in five of the seven analyzed species (Table 1) suggests that it is likely located on a transferable plasmid. Overall, the variety of qnrB genes found in the tested C. freundii isolates, the detection of novel variants, and the presence of truncated pseudogenes are consistent with the postulated origin of the qnrB gene family as mobilized chromosomal genes from the C. freundii complex (23, 29).
In summary, the high prevalence and diversity of the PMQR genes make the multidrug-resistant Enterobacteriaceae strains circulating in the population served by Mercy Hospital a potent reservoir of quinolone resistance genes that threaten the continued usefulness of this class of antibiotics in this community.
Accession number(s).
Novel gene sequences obtained in this study were deposited in GenBank under the following accession numbers: new qnrB variants, qnrB81 (SL157) and qnrB82 (SL156), KX372671 and KX372672, respectively; qnrB1 subvariants, qnrB1a (SL166), qnrB1i (SL174), and qnrB1d (SL185), KX372673 to KX372675; and truncated qnrB pseudogenes, Δ1qnrB (SL129), Δ2qnrB (SL151), and Δ3qnrB (SL157), KX372668 to KX372670.
Supplementary Material
Funding Statement
This work was supported in part by the Joint Science and Technology Office (JSTO), Defense Threat Reduction Agency (DTRA), and the Office of Naval Research (ONR) via U.S. Naval Research Laboratory core funds. The funding entities had no role in or any influence over the design of the study, collection, analysis, and interpretation of data, or the final manuscript. One of the funding agencies (DTRA) requested the inclusion of the following distribution statement to this manuscript: “Approved for public release; distribution is unlimited.” This reflects the unclassified nature of the presented data.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01576-16.
REFERENCES
- 1.Hooper DC. 1999. Mode of action of fluoroquinolones. Drugs 58(Suppl 2):S6–S10. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 3.Aldred KJ, Kerns RJ, Osheroff N. 2014. Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 6.Spellberg B, Doi Y. 2015. The rise of fluoroquinolone-resistant Escherichia coli in the community: scarier than we thought. J Infect Dis 212:1853–1855. doi: 10.1093/infdis/jiv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigel LM, Steward CD, Tenover FC. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother 42:2661–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby GA, Strahilevitz J, Hooper DC. 2014. Plasmid-mediated quinolone resistance. Microbiol Spectr 2(5):PLAS-006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robicsek A, Strahilevitz J, Jacoby G, Macielag M, Abbanat D, Park C, Bush K, Hooper D. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 10.Drlica K. 2003. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother 52:11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland: http://www.who.int/drugresistance/documents/surveillancereport/en/. [Google Scholar]
- 12.Lamikanra A, Crowe JL, Lijek RS, Odetoyin BW, Wain J, Aboderin AO, Okeke IN. 2011. Rapid evolution of fluoroquinolone-resistant Escherichia coli in Nigeria is temporally associated with fluoroquinolone use. BMC Infect Dis 11:312. doi: 10.1186/1471-2334-11-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogbolu DO, Daini OA, Ogunledun A, Alli AO, Webber MA. 2011. High levels of multidrug resistance in clinical isolates of Gram-negative pathogens from Nigeria. Int J Antimicrob Agents 37:62–66. doi: 10.1016/j.ijantimicag.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Sumrall ET, Gallo EB, Aboderin AO, Lamikanra A, Okeke IN. 2014. Dissemination of the transmissible quinolone-resistance gene qnrS1 by IncX plasmids in Nigeria. PLoS One 9:e110279. doi: 10.1371/journal.pone.0110279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guessennd N, Bremont S, Gbonon V, Kacou-Ndouba A, Ekaza E, Lambert T, Dosso M, Courvalin P. 2008. Qnr-type quinolone resistance in extended-spectrum beta-lactamase producing enterobacteria in Abidjan, Ivory Coast. Pathol Biol (Paris) 56:439–446. doi: 10.1016/j.patbio.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 16.Namboodiri SS, Opintan JA, Lijek RS, Newman MJ, Okeke IN. 2011. Quinolone resistance in Escherichia coli from Accra, Ghana. BMC Microbiol 11:44. doi: 10.1186/1471-2180-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raufu I, Bortolaia V, Svendsen CA, Ameh JA, Ambali AG, Aarestrup FM, Hendriksen RS. 2013. The first attempt of an active integrated laboratory-based Salmonella surveillance programme in the north-eastern region of Nigeria. J Appl Microbiol 115:1059–1067. doi: 10.1111/jam.12304. [DOI] [PubMed] [Google Scholar]
- 18.Harrois D, Breurec S, Seck A, Delauné A, Le Hello S, Pardos de la Gándara M, Sontag L, Perrier-Gros-Claude JD, Sire JM, Garin B, Weill FX. 2014. Prevalence and characterization of extended-spectrum beta-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin Microbiol Infect 20:O109–O116. doi: 10.1111/1469-0691.12339. [DOI] [PubMed] [Google Scholar]
- 19.Fortini D, Fashae K, García-Fernández A, Villa L, Carattoli A. 2011. Plasmid-mediated quinolone resistance and beta-lactamases in Escherichia coli from healthy animals from Nigeria. J Antimicrob Chemother 66:1269–1272. doi: 10.1093/jac/dkr085. [DOI] [PubMed] [Google Scholar]
- 20.Leski TA, Taitt CR, Bangura U, Stockelman MG, Ansumana R, Cooper WH III, Stenger DA, Vora GJ. 2016. High prevalence of multidrug resistant Enterobacteriaceae isolated from outpatient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infect Dis 16:167. doi: 10.1186/s12879-016-1495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing: 19th informational supplement M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Food and Drug Administration. 2014. Avelox; highlights of prescribing information. Food and Drug Administration, Silver Spring, MD: http://www.accessdata.fda.gov/drugsatfda_docs/label/2014/021085s059,021277s055lbl.pdf Accessed 13 May 2016. [Google Scholar]
- 23.Jacoby GA, Griffin CM, Hooper DC. 2011. Citrobacter spp. as a source of qnrB alleles. Antimicrob Agents Chemother 55:4979–4984. doi: 10.1128/AAC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erill I, Campoy S, Barbé J. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, Jacoby GA, Mills DM, Hooper DC. 2009. SOS regulation of qnrB expression. Antimicrob Agents Chemother 53:821–823. doi: 10.1128/AAC.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Da Re S, Garnier F, Guérin E, Campoy S, Denis F, Ploy MC. 2009. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep 10:929–933. doi: 10.1038/embor.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Wang H, Qi Y, Liang Y, Zhang J, Yu L. 2011. Novel variants of the qnrB gene, qnrB31 and qnrB32, in Klebsiella pneumoniae. J Med Microbiol 60:1849–1852. doi: 10.1099/jmm.0.034272-0. [DOI] [PubMed] [Google Scholar]
- 28.Liao X, Fang L, Li L, Sun J, Li X, Chen M, Deng H, Yang Q, Li X, Liu Y. 2015. Characterization of chromosomal qnrB and ampC alleles in Citrobacter freundii isolates from different origins. Infect Genet Evol 35:214–220. doi: 10.1016/j.meegid.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Saga T, Sabtcheva S, Mitsutake K, Ishii Y, Tateda K, Yamaguchi K, Kaku M. 2013. Characterization of qnrB-like genes in Citrobacter species of the American Type Culture Collection. Antimicrob Agents Chemother 57:2863–2866. doi: 10.1128/AAC.02396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribeiro TG, Novais Â, Branquinho R, Machado E, Peixe L. 2015. Phylogeny and comparative genomics unveil independent diversification trajectories of qnrB and genetic platforms within particular Citrobacter species. Antimicrob Agents Chemother 59:5951–5958. doi: 10.1128/AAC.00027-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.