Abstract
A plasmid carrying the colistin resistance gene mcr-1 was isolated from a pig slurry sample in Estonia. The gene was present on a 33,311-bp plasmid of the IncX4 group. mcr-1 is the only antibiotic resistance gene on the plasmid, with the other genes mainly coding for proteins involved in conjugative DNA transfer (taxA, taxB, taxC, trbM, and the pilX operon). The plasmid pESTMCR was present in three phylogenetically very different Escherichia coli strains, suggesting that it has high potential for horizontal transfer.
TEXT
A plasmid containing the mcr-1 gene causing colistin resistance was originally described in Escherichia coli strains from animals, animal products, and human samples in South China (1). This plasmid could cause serious problems when transferred into strains for which colistin is the last treatment option. Since the first report, sequence information from several antibiotic resistance programs has been screened for the presence of the mcr-1 gene (2–25). The gene was found in several samples from around the world. The mcr-1 genes described are present in several plasmid backbones. In addition to mcr-1, two new mcr gene variants were detected (14, 20). Although by now we are aware that mcr-1 is not restricted to China, we need more information for a better understanding of how the gene has spread and how big a concern it might be in different countries. The existing collections of strains and sequences are a good source of information that can be retrieved rapidly.
A survey of the spread of antibiotic resistance in Estonia gathered samples during the years 2011 to 2014. It included 347 E. coli strains: 144 strains from humans, 88 strains from animals, and 115 strains from the environment. Of the 237 Pseudomonas aeruginosa strains, 147 strains were collected from humans, 64 strains were from animals, and 26 strains were from the environment. The collection of E. coli strains was specifically targeted at extended-spectrum β-lactamase (ESBL) producers. All strains were characterized by Illumina HiSeq 2500 sequencing of the DNA (Nextera XT libraries, paired-end 150-bp reads), followed by de novo assembly of the reads into contigs (26). We searched our data set with BLAST against all three mcr sequence variants. The mcr-1 gene was found in three E. coli strains isolated from a single pig slurry sample originating from a farm in November 2013. The farm has a breeding herd of 150 sows and is located in North Estonia. Animals from this farm were not sampled. The strains had been isolated as follows. About 5 g of slurry sample diluted with 0.9% NaCl solution containing ampicillin (10 μg/liter) was incubated overnight at 37°C. E. coli selection was made according to ISO 9308-3 standard. As a second selection step, the samples from positive wells were plated as a dilution series onto ESBL agar (Oxoid Brilliance ESBL agar with cefpodoxime). From the plates, isolates having the E. coli phenotypes (blue or pink) were purified.
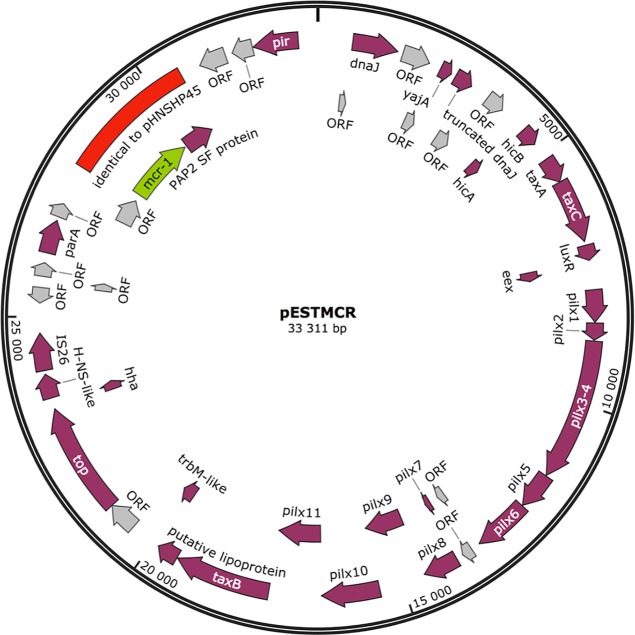
The contigs containing mcr-1 were around 33 kb long. PCR primers were designed to the ends of the contig, and the missing part of the plasmid was amplified and Sanger sequenced. All three plasmids were identical (Fig. 1). The plasmid replication origin groups to IncX4 (27). The plasmid contains 47 open reading frames. These include taxA, taxB taxC, and trbM genes and the pilX operon, which are probably responsible for movement of the plasmid between different bacterial hosts (28–30). mcr-1 is the only antibiotic resistance gene in the plasmid.
FIG 1.
Map of the mcr-1-containing plasmid isolated. The region identical to pHNSHP45 where mcr-1 was originally identified (1) is marked in red. The mcr-1 gene is marked in green. The figure was created with the SnapGene software. ORF, open reading frame.
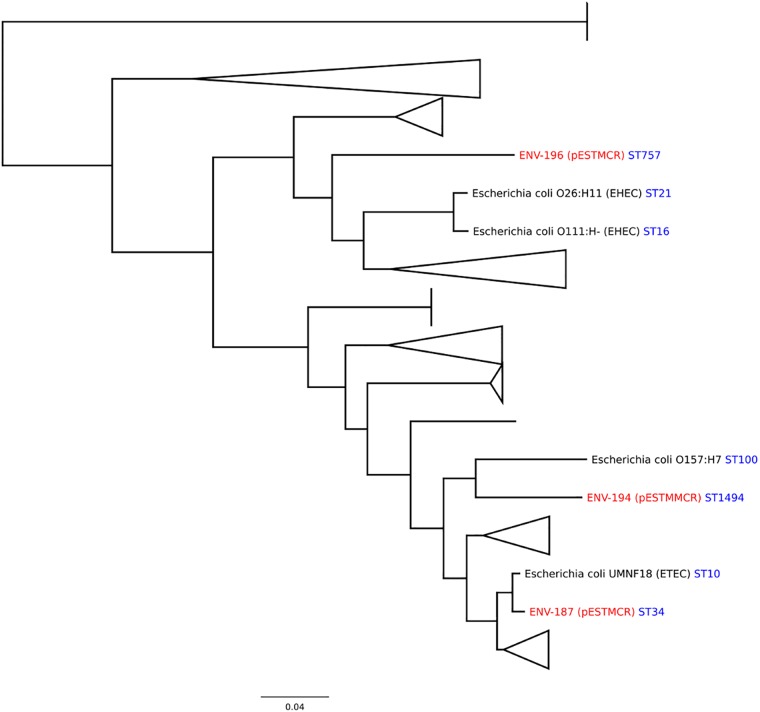
Interestingly, the plasmid is identical to the contigs described in Salmonella isolated from meat samples in France (7) and highly similar to plasmids pMCR1-IncX4 (Klebsiella pneumoniae, China) (17), pAf48 (E. coli, South Africa) (19), and a new mcr variant carried by pMCR1.2-IT (K. pneumoniae, Italy) (20). The mcr-1-containing Salmonella strains were all from one serogroup, O:4. This suggested that the colistin resistance provided by the gene might be limited to certain groups of host strains. To investigate the host spectrum, we compared the genomic sequences of our strains with the completed genome sequences available in the NCBI Genomes database (Fig. 2). All three strains cluster into separate branches of the tree, indicating considerable variation.
FIG 2.
All available E. coli complete genomes were downloaded from the NCBI database in September 2015. These genome sequences were compared with the genomes of strains containing mcr-1 (marked in red). The names of the closest relatives of the mcr-1-containing strains are indicated in black. Core genomes were constructed using rapid core genome multialignment (parsnp) (31). The tree was constructed from core alignment using RAxML under the general time-reversible (GTR) Gamma model. The scale bar represents the mean number of nucleotide substitutions per site. Sequence types (ST) for the sequences present in the database (32) are indicated. ST757 is closely related to clonal complex 10.
All three strains were resistant to colistin, with Etest-based MICs of 2 μg/ml (ENV-196) and 4 μg/ml (ENV-187 and ENV-194). These resistance levels are consistent with the original report (1), where it was found that the colistin MICs for the mcr-1 gene carrying E. coli strains are between 2 and 8 μg/ml. The plasmid was isolated and transformed into laboratory model E. coli strain DH5α, where it increased the colistin MIC from 0.25 μg/ml to 4 μg/ml. It has been reported previously that the mcr-1-carrying plasmid could be transformed into Pseudomonas (1). In our case, transfection into the environmental Pseudomonas putida strain PAW85, which could be a recipient of the plasmid after the sludge reaches the receiving field, was also attempted but failed. No increase in the number of colistin-resistant colonies over the background level of spontaneous resistance mutations was observed.
The strains were ESBL producers, and the corresponding β-lactamase genes could be identified in the genomic sequences: CTX-M-1 in ENV-187, CTX-M-1 and TEM-1A in ENV-194, and CTX-M-1 and AmpC in ENV-196. The strains were sensitive to most of the antibiotics tested: meropenem (MIC, <0.016 μg/ml), ciprofloxacin (MIC, 0.008 μg/ml to 0.012 μg/ml), amikacin (MIC, 2 μg/ml to 4 μg/ml), gentamicin (MIC, 1 μg/ml to 1.5 μg/ml), fosfomycin (MIC, 1 to 4 μg/ml), tigecycline (MIC, 0.125 μg/ml to 0.19 μg/ml), and piperacillin-tazobactam (MIC, 1 μg/ml to 2 μg/ml). Resistance to trimethoprim-sulfamethoxazole was observed in one strain (ENV-196; MIC, >32 μg/ml), while two strains were sensitive (MIC, 0.064 μg/ml to 0.094 μg/ml). This general pattern indicates that these particular strains are not likely to be problematic from an antibiotic treatment viewpoint. Still, considering that the three strains hosting the plasmid, isolated from one sample, were genetically very different, it is probable that the plasmid is highly mobile. It could be transferred into strains resistant to a wide range of antibiotics, thereby creating strains that are very hard to treat. Therefore, the epidemiological situation should be closely followed.
Accession number(s).
The accession number for the pESTMCR sequence in GenBank is KU743383.
ACKNOWLEDGMENT
We thank Marta Putrinš for advice on the transformation experiments.
REFERENCES
- 1.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2015. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. [DOI] [PubMed] [Google Scholar]
- 2.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:pii=21331 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21331. [DOI] [PubMed] [Google Scholar]
- 3.Hu Y, Liu F, Lin IYC, Gao GF, Zhu B. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:146–147. [DOI] [PubMed] [Google Scholar]
- 4.Olaitan AO, Chabou S, Okdah L, Morand S, Rolain J-M. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 15:147. [DOI] [PubMed] [Google Scholar]
- 5.Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J, de Jong MD, Schultsz C, COMBAT Consortium. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:147–149. [DOI] [PubMed] [Google Scholar]
- 6.Tse H, Yuen K-Y. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:145–146. [DOI] [PubMed] [Google Scholar]
- 7.Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 2015. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:144–145. [DOI] [PubMed] [Google Scholar]
- 8.Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW. 2016. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis 16:285–286. doi: 10.1016/S1473-3099(16)00010-4. [DOI] [PubMed] [Google Scholar]
- 9.Petrillo M, Angers-Loustau A, Kreysa J. 2016. Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect Dis 16:280. doi: 10.1016/S1473-3099(16)00005-0. [DOI] [PubMed] [Google Scholar]
- 10.Falgenhauer L, Waezsada S-E, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T, RESET Consortium. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 11.Haenni M, Poirel L, Kieffer N, Châtre P, Saras E, Métayer V, Dumoulin R, Nordmann P, Madec J-Y. 2016. Co-occurrence of extended spectrum β-lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis 16:281–282. doi: 10.1016/S1473-3099(16)00007-4. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. 2016. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis 16:284–285. doi: 10.1016/S1473-3099(16)00008-6. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. 2016. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis 16:283–284. doi: 10.1016/S1473-3099(16)00012-8. [DOI] [PubMed] [Google Scholar]
- 14.Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Hoang HTT, Pham NT, Goossens H. 2016. Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis 16:286–287. doi: 10.1016/S1473-3099(16)00014-1. [DOI] [PubMed] [Google Scholar]
- 15.Du H, Chen L, Tang Y-W, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 16.Yao X, Doi Y, Zeng L, Lv L, Liu J-H. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 17.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang Y-W, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. 2016. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetables in Switzerland. Antimicrob Agents Chemother 60:2594–2595. doi: 10.1128/AAC.00066-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, Giani T, Menichetti F, Rossolini GM. 2016. MCR-1.2: a new MCR variant encoded by a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae of sequence type 512. Antimicrob Agents Chemother 60:5612–5615. doi: 10.1128/AAC.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluytmans-van den Bergh MF, Huizinga P, Bonten MJ, Bos M, De Bruyne K, Friedrich AW, Rossen JW, Savelkoul PH, Kluytmans JA. 2016. Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill 21:pii=21396 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21396. [DOI] [PubMed] [Google Scholar]
- 22.Nordmann P, Lienhard R, Kieffer N, Clerc O, Poirel L. 2016. Plasmid-mediated colistin-resistant Escherichia coli in bacteremia in Switzerland. Clin Infect Dis 62:1322–1323. doi: 10.1093/cid/ciw124. [DOI] [PubMed] [Google Scholar]
- 23.Mulvey MR, Mataseje LF, Robertson J, Nash JHE, Boerlin P, Toye B, Irwin R, Melano RG. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:289–290. doi: 10.1016/S1473-3099(16)00067-0. [DOI] [PubMed] [Google Scholar]
- 24.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Deng H, Li L, Chen M-Y, Fang L-X, Yang Q-E, Liu Y-H, Liao X-P. 2015. Complete nucleotide sequence of cfr-carrying IncX4 plasmid pSD11 from Escherichia coli. Antimicrob Agents Chemother 59:738–741. doi: 10.1128/AAC.04388-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pansegrau W, Lanka E. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol 54:197–251. doi: 10.1016/S0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 29.Núñez B, Avila P, de la Cruz F. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol Microbiol 24:1157–1168. doi: 10.1046/j.1365-2958.1997.4111778.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen C-L, Wang C-Y, Chu C, Su L-H, Chiu C-H. 2009. Functional and molecular characterization of pSE34 encoding a type IV secretion system in Salmonella enterica serotype Enteritidis phage type 34. FEMS Immunol Med Microbiol 57:274–283. doi: 10.1111/j.1574-695X.2009.00612.x. [DOI] [PubMed] [Google Scholar]
- 31.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]