Abstract
This report describes the first detection of a blaVEB-2 gene in a Vibrio parahaemolyticus strain isolated from a shrimp sample. The blaVEB-2 gene was carried on a novel Inc-type plasmid that was likely to have originated from aquatic organisms, as indicated by a comparison with other known genetic elements in the GenBank database. However, the plasmid contains resistance elements usually harbored by members of the family Enterobacteriaceae, suggesting that gene transfer events occurred and contributed to the formation of this multidrug resistance-encoding plasmid.
TEXT
Vibrio parahaemolyticus, a halophilic Gram-negative bacterium, is one of the most important seafood-borne pathogens worldwide, especially in areas with high seafood consumption rates. Prescription of antibiotics is necessary for the treatment of life-threatening infections caused by V. parahaemolyticus, with cephalosporins being among the most effective therapeutic agents (1). Resistance to the third-generation cephalosporins in V. parahaemolyticus strains due to carriage of an extended-spectrum beta-lactamase (ESBL) gene, blaPER-1, and the AmpC β-lactamase gene blaCMY-2 has been reported in China (2, 3). Apart from these two resistance elements, however, no other ESBL genes have been reported in V. parahaemolyticus. The emergence of cephalosporinase-encoding genes in V. parahaemolyticus has raised a huge public health concern because cephalosporinase-producing V. parahaemolyticus strains may not only limit the choice of treatment for severe infections caused by these bacteria but also facilitate the transmission of resistance elements to other aquatic pathogens such as V. cholerae. Here, we report the first isolation of a cephalosporin-resistant V. parahaemolyticus strain that carries the blaVEB-2 gene and a characterization of the genetic features of the plasmid that harbors this resistance element.
A cephalosporin-resistant V. parahaemolyticus strain (VPS92) was isolated from a shrimp sample in a supermarket during our foodborne pathogen surveillance in Shenzhen, China in 2015. The genetic identity of this isolate was further confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker), amplification and sequencing of the blaCARB-17 and tlh genes, and API20E strip testing (4). Antimicrobial susceptibility tests were performed by the broth microdilution method according to CLSI guidelines (5), with results showing that VPS92 was resistant to ampicillin, cefotaxime, and sulfamethazine-trimethoprim (Table 1). The isolate was tested for β-lactamase genes by PCR and DNA sequencing as previously described (6). The blaVEB-2 gene was detected for the first time in V. parahaemolyticus. To test the transferability of this gene, a conjugation assay was performed with azide-resistant Escherichia coli J53 as the recipient strain and recovery by screening on LB agar supplemented with 16 μg/ml cefotaxime as previously described (7). Cephalosporin-resistant phenotypes were found to be transferrable to J53. In addition, other phenotypes, such as resistance to chloramphenicol and sulfamethazine-trimethoprim, could also be transferred. S1 pulsed-field gel electrophoresis (S1-PFGE) and Southern hybridization of both the parental strain and the transconjugant were performed as previously described (7). The results showed that the blaVEB-2 gene was located on a plasmid with size of ca. 320 kb.
TABLE 1.
MICs of different antibiotics for blaVEB-2-positive V. parahaemolyticus strain VPS92 and its transconjugant
| Strain | MIC (μg/ml)a of: |
Plasmid size (kb)b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | CTX | AMK | SXT | NAL | CIP | CHL | TET | ||
| J53 | 1 | 4/2 | 0.03 | 0.5 | 0.25/4.75 | 1 | 0.03 | 2 | 0.5 | |
| VPS92 | >64 | 4/2 | >16 | 4 | >8/152 | 4 | 1 | 4 | 2 | 320 |
| VPS92-J53 | >64 | 8/4 | >16 | 0.5 | >8/153 | 16 | 0.25 | 32 | 8 | 320 |
Abbreviations: AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CTX, cefotaxime; AMK, amikacin; SXT, sulfamethoxazole-trimethoprim; NAL, nalidixic acid; CIP, ciprofloxacin; CHL, chloramphenicol; TET, tetracycline.
Determined by S1-PFGE.
To understand the genetic features of the plasmid harboring blaVEB-2, the plasmid recovered from the transconjugant was extracted with the Qiagen plasmid midi kit (Qiagen) and sequenced with the Illumina NextSeq 500 and PacBio RSII single-molecule real-time (SMRT) sequencing platforms (Wuhan Institute of Biotechnology, China). Illumina reads were aligned with PacBio contigs to improve the accuracy of the plasmid sequence data and obtain the complete sequence of the plasmid. The complete plasmid sequence was confirmed by PCR and then annotated with the RAST tool and the NCBI Prokaryotic Genome Annotation Pipeline. Plasmid comparison was performed with the BRIG software (8).
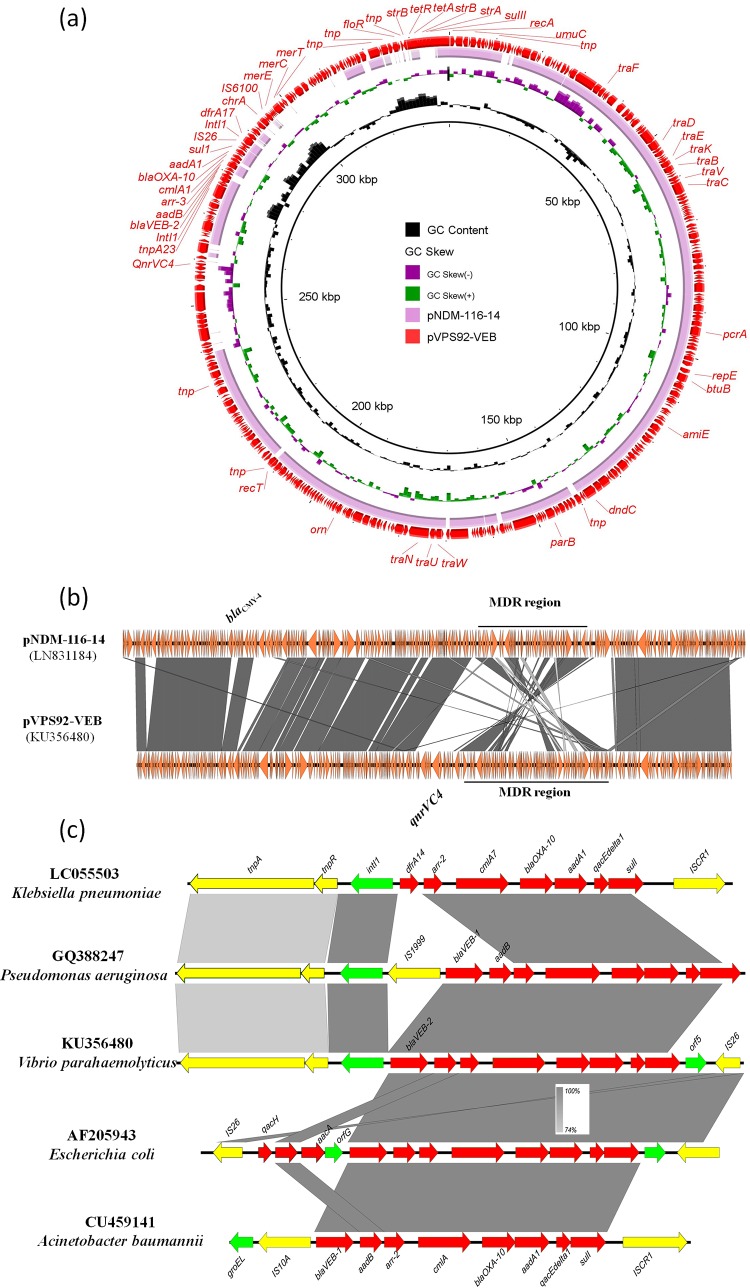
The blaVEB-2-bearing plasmid, designated pVPS92-VEB, was found to be 338,538 bp in length, to contain 390 predicted coding sequences (CDSs), and to exhibit a GC content of 44.3% (Fig. 1). The plasmid harbored different genes, including resistance and mobile elements, conjugative-transfer-related genes, and hypothetical genes. Annotation results showed that blaVEB-2 was located in a class 1 integron with the structure blaVEB-2-aadB-arr2-cmlA-blaOXA-10-aadA1-sul1. After a BLASTN search was conducted, the blaVEB-2 genetic environment was found to be similar to that of the blaVEB-1 gene in integrons of Acinetobacter baumannii (accession no. CU459141), Pseudomonas aeruginosa (GQ388247) and Escherichia coli (AF205943) (9). This indicates that similar integrons harboring blaVEB-2 could mobilize between different plasmids harbored by different species of bacteria and the blaVEB gene could evolve into different alleles. Because blaVEB is always embedded in class 1 integrons, the formation of the blaVEB-2-bearing integron could accelerate the rate of transmission of blaVEB-2 among plasmids and chromosomes (10, 11). A mercury resistance operon downstream of the blaVEB-2-bearing integron, in which the floR, tetA-tetR, and strA-strB genes were clustered into a multidrug resistance (MDR) region, was found to be flanked by mobile elements. This structure may facilitate the transmission of multiple resistance genes in one single mobilization event. In the region upstream of the blaVEB-2-bearing integron, one qnrVC4 gene cassette was found to be inserted into the plasmid backbone, implying that the integron-related qnrVC4 cassette can migrate between integrons and other genetic loci (3, 12). BLASTN analysis showed that the backbone of pVPS92-VEB was similar (99% identity in 78% coverage) to a single plasmid, namely, pNDM-116-14 (LN831184), which was isolated from a V. cholerae strain isolated in India (Fig. 1). After comparison with the plasmid replicon database, the predicted replication initiation gene repE was shown to be novel, with no homology to any of the known replicon genes. The pNDM-116-14 plasmid was known to contain two MDR regions: a class I integron with different gene cassettes, including blaNDM-1 and one gene cassette in which the blaCMY-4 gene was inserted into the backbone of the plasmid (Fig. 1). The major differences between these two plasmids were the mobile elements that carried different antimicrobial resistance genes. In addition, some additional insertion sequences other than those in the MDR region could be seen in pNDM-116-14 (Fig. 1).
FIG 1.
Comparison of plasmids pVPS92-VEB (338,538 bp) and pNDM-116-14 (354,308 bp). (a) Circular map representation of the plasmid comparison results. The red outer circle is a map of plasmid pVPS92-VEB, which was used as the reference for a BLASTN search. The pink inner circle is a homologous alignment of pNDM-116-14 and pVPS92-VEB. Variation was seen mainly in the MDR region of these two plasmids. (b) Linear representation of the plasmid comparison data. Some additional insertion sequences other than those in the MDR region could be seen in pNDM-116-14. (c) Genetic alignment of a class 1 integron harboring blaVEB-2 and other similar integrons from different bacteria. The red arrows represent the resistance gene cassettes embedded in integrons. The yellow arrows denote the mobile elements. The green arrows represent other genes and predicted CDSs. The boxed shading legend shows the percentages of sequence identity found by a BLASTN search.
The dissemination of ESBL genes among pathogens and opportunistic pathogens is a serious public health concern (13). VEB-type β-lactamases can be classified into different subtypes, from VEB-1 through VEB-17, with one to four amino acid substitutions compared to VEB-1 (13–15). The blaVEB genes have been identified mainly in P. aeruginosa and other bacterial species such as Proteus mirabilis and A. baumannii (16, 17). The nature of the novel backbone of pVPS92-VEB and pNDM-116-14 suggested that this type of plasmid may originate from the aquatic environment since it was only reported in Vibrio species twice yet has not been reported in other bacteria, including members of the family Enterobacteriaceae. Although similar plasmids were only reported in Vibrio species, the plasmid could be transferred to E. coli according to the conjugation experiment reported here. In addition, the presence of the qnrVC4 gene cassette in this plasmid is further suggestive of its aquatic origin since the qnrVC gene is typically found in marine organisms. However, it is difficult to draw conclusions about the sources of the integrons bearing blaVEB-2 or blaNDM-1 because of their existence in both Enterobacteriaceae and marine bacteria and their high mobility among different genetic loci.
In conclusion, we report here the first detection of the blaVEB-2 gene in a large conjugative plasmid, pVPS92-VEB, the backbone of which may have originated in the aquatic environment, whereas the integron carrying blaVEB-2 may be obtained through nonaquatic bacteria such as A. baumannii by horizontal gene transfer. The acquisition of various types of ESBL-encoding elements such as blaPER-1, blaVEB, and the AmpC β-lactamase-encoding gene in Vibrio spp. constitutes an increasing public health threat posed by these aquatic pathogens.
Accession number.
The complete plasmid pVPS92-VEB sequence was deposited in the NCBI database under accession number KU356480.
ACKNOWLEDGMENTS
We have no competing interests to declare.
Ethical approval of this study was not required.
REFERENCES
- 1.Liu M, Wong MH, Chen S. 2013. Molecular characterisation of a multidrug resistance conjugative plasmid from Vibrio parahaemolyticus. Int J Antimicrob Agents 42:575–579. doi: 10.1016/j.ijantimicag.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Wong MHY, Liu M, Wan HY, Chen S. 2012. Characterization of extended-spectrum-β-lactamase-producing Vibrio parahaemolyticus. Antimicrob Agents Chemother 56:4026–4028. doi: 10.1128/AAC.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye L, Li R, Lin D, Zhou Y, Fu A, Ding Q, Chan EW, Yao W, Chen S. 2016. Characterization of an IncA/C multidrug resistance plasmid in Vibrio alginolyticus. Antimicrob Agents Chemother 60:3232–3235. doi: 10.1128/AAC.00300-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou J, Li R, Chen S. 2015. CARB-17 family of β-lactamases mediates intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob Agents Chemother 59:3593–3595. doi: 10.1128/AAC.00047-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed CLSI document M45-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 7.Xie M, Lin D, Chen K, Chan EW, Yao W, Chen S. 2016. Molecular characterization of Escherichia coli strains isolated from retail meat that harbor blaCTX-M and fosA3 genes. Antimicrob Agents Chemother 60:2450–2455. doi: 10.1128/AAC.03101-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurya AP, Das Talukdar A, Chanda DD, Chakravarty A, Bhattacharjee A. 2014. Integron-borne transmission of VEB-1 extended-spectrum β-lactamase in Pseudomonas aeruginosa in a tertiary care hospital in India. Antimicrob Agents Chemother 58:6966–6969. doi: 10.1128/AAC.02365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naas T, Aubert D, Lambert T, Nordmann P. 2006. Complex genetic structures with repeated elements, a sul-type class 1 integron, and the blaVEB extended-spectrum beta-lactamase gene. Antimicrob Agents Chemother 50:1745–1752. doi: 10.1128/AAC.50.5.1745-1752.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia R, Guo X, Zhang Y, Xu H. 2010. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob Agents Chemother 54:3471–3474. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zong Z, Partridge SR, Iredell JR. 2009. A blaVEB-1 variant, blaVEB-6, associated with repeated elements in a complex genetic structure. Antimicrob Agents Chemother 53:1693–1697. doi: 10.1128/AAC.01313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Naas T, Guibert M, Chaibi EB, Labia R, Nordmann P. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother 43:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahiri SD, Alm RA. 2016. Identification of novel VEB β-lactamase enzymes and their impact on avibactam inhibition. Antimicrob Agents Chemother 60:3183–3186. doi: 10.1128/AAC.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodford N, Zhang J, Kaufmann ME, Yarde S, Tomas Mdel M, Faris C, Vardhan MS, Dawson S, Cotterill SL, Livermore DM. 2008. Detection of Pseudomonas aeruginosa isolates producing VEB-type extended-spectrum beta-lactamases in the United Kingdom. J Antimicrob Chemother 62:1265–1268. doi: 10.1093/jac/dkn400. [DOI] [PubMed] [Google Scholar]
- 17.Naas T, Bogaerts P, Bauraing C, Degheldre Y, Glupczynski Y, Nordmann P. 2006. Emergence of PER and VEB extended-spectrum beta-lactamases in Acinetobacter baumannii in Belgium. J Antimicrob Chemother 58:178–182. doi: 10.1093/jac/dkl178. [DOI] [PubMed] [Google Scholar]