Abstract
We aimed to investigate polymyxin B (PMB) resistance and its molecular mechanisms in 126 Klebsiella pneumoniae isolates from rectal swabs in Brazil. Ten isolates exhibited PMB resistance with interruption of mgrB gene by insertion sequences or missense mutations. Most of the PMB-resistant isolates harbored blaKPC-2 (n = 8) and belonged to clonal complex 258 (CC258) (n = 7). These results highlight the importance of monitoring the spread of polymyxin-resistant bacteria in hospitals, since few options remain to treat multidrug-resistant isolates.
TEXT
Polymyxin has been widely used to treat infections caused by multidrug-resistant (MDR) Gram-negative bacteria, including Klebsiella pneumoniae. However, reports of polymyxin-resistant K. pneumoniae (PRKP) isolates have increased worldwide, becoming a great public health concern (1).
Most studies on PRKP have focused on patients with infections. However, there have been few reports assessing data on PRKP carriage in patients around the world (2). Some studies have described a remarkable and concerning number of patients who developed infection by PRKP after previous colonization, resulting in elevated mortality rates (3, 4). Colonization by KPC-producing K. pneumoniae and polymyxin therapy are considered important risk factors for PRKP infection (5, 6).
Studies have demonstrated that modifications of PmrA/PmrB and PhoP/PhoQ two-component systems and inactivation of the mgrB gene (a regulator of the PhoP/PhoQ system) led to polymyxin resistance by modification of the lipopolysaccharide target (7). Recently, the plasmid-mediated transferable polymyxin resistance mcr-1 gene, causing resistance by modification of lipid A, was identified in China in Escherichia coli and K. pneumoniae strains (8).
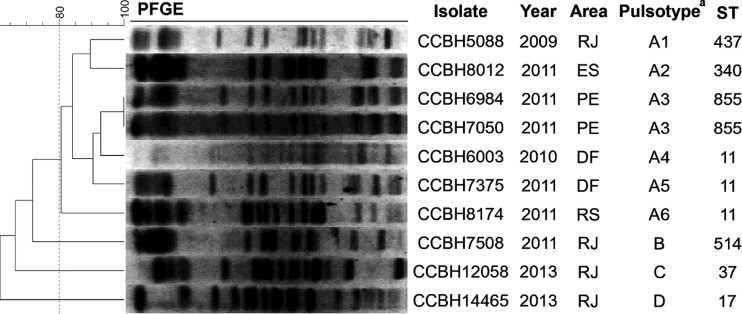
Here, we searched for molecular mechanisms associated with polymyxin resistance in K. pneumoniae isolates from Brazil. A first-step screening for polymyxin B (PMB) resistance was conducted using Etest (bioMérieux, France) in 126 isolates randomly selected from approximately 850 K. pneumoniae isolates with reduced susceptibility to carbapenems recovered from rectal swabs from 11 Brazilian states during 2007 to 2013. The bacterial identification was confirmed by conventional biochemical techniques. Considering the PRKP strains showing a MIC of >2.0 mg/liter (9), 10 (8%) PRKP isolates were observed and included in this study. These 10 PRKP isolates were collected between 2009 and 2013 from five Brazilian states (Fig. 1).
FIG 1.
Epidemiological and molecular typing of polymyxin B-resistant K. pneumoniae isolates analyzed in this work. DF, Distrito Federal; ES, Espírito Santo; PE, Pernambuco; RJ, Rio de Janeiro; RS, Rio Grande do Sul; ST, sequence type. The PFGE pulsotypes and subtypes (indicated by a superscript “a”) were defined as strains with at least 80% and 95% similarities, respectively.
To confirm the resistance phenotype, the PMB MIC was retested in duplicate by microdilution with cation-adjusted Mueller-Hinton broth (10). The isolates showed a MIC50 of 64 mg/liter, a MIC90 of >128 mg/liter, and a MIC range of 16 to >128 mg/liter (Table 1). Concordant Etest and microdilution results were found for only three isolates. The Etest MICs tended to be 1.3-fold to 4.0-fold lower than the microdilution MICs. Discrepancy between the two methodologies demonstrated that the Etest provided a conservative estimate (11). Furthermore, the cation concentration variability of culture media was correlated to the low accuracy and discrepancies in Etest results (12); thus, the use of a dilution method to confirm the PMB susceptibility is recommended (13).
TABLE 1.
Phenotypic and molecular characterization of polymyxin B-resistant K. pneumoniae isolates analyzed in this work
| Isolate | PMB MIC (mg/liter)a |
Modification in proteinb: |
Additional susceptibility profilec |
Resistance genesg | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Etest | Broth dilution | MgrB | PmrB | PmrA | PhoP | PhoQ | Nonsusceptible to: | Susceptible to: | ||
| CCBH5088d | 192 | >128 | Gene disrupted by IS903B | T246Af, R256Ge | WT | A30Sf | WT | FOT, CAZ, ATM, FEP, CTX, TZP, GEN, CIP, SXT, CHL, MEM, ERT, IPM | AMK, TGC | blaKPC-2, blaTEM, blaSHV, blaCTX-M, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH6003d | 64 | 128 | Gene disrupted by IS903B | Gene deletion (570 bp) | WT | WT | WT | CAZ, ATM, FEP, CTX, TZP, CIP, SXT, MEM, ERT, IPM | FOT, AMK, GEN, CHL, TGC | blaKPC-2, blaTEM, blaSHV, qnrS, aadA, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH6984d | 32 | 128 | Gene disrupted by IS903B | T246Af, R256Ge | WT | WT | WT | FOT, CAZ, ATM, FEP, CTX, TZP, AMK, GEN, CIP, SXT, CHL, MEM, ERT, IPM, TGC | blaKPC-2, blaTEM, blaSHV, blaCTX-M, qnrB, aadA, aac(3′)IIa, aac(6′)-Ib | |
| CCBH7050d | 32 | >128 | Gene disrupted by IS903B | T246Af, R256Ge | WT | WT | WT | CAZ, ATM, FEP, CTX, TZP, AMK, GEN, CIP, SXT, CHL, ERT, TGC | FOT, MEM, IPM, | blaTEM, blaSHV, blaCTX-M, qnrB, aadA, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH7375d | 24 | 64 | Gene disrupted by IS10L | T246Af, R256Ge | WT | WT | WT | FOT, CAZ, ATM, FEP, CTX, TZP, AMK, CIP, SXT, MEM, ERT, IPM | GEN, CHL, TGC | blaKPC-2, blaSHV, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH7508 | 16 | 16 | Gene disrupted by IS5 | WT | WT | WT | WT | CAZ, ATM, FEP, CTX, TZP, AMK, GEN, CIP, MEM, ERT, IPM | FOT, SXT, CHL, TGC | blaKPC-2, blaTEM, blaSHV, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH8012d | 64 | 64 | C28Rb | T246Af, R256Ge | WT | WT | WT | CAZ, ATM, FEP, CTX, TZP, AMK, GEN, CIP, SXT, CHL, MEM, ERT, IPM | FOT, TGC | blaKPC-2, blaTEM, blaSHV, blaCTX-M, qnrA, aadA, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH8174d | 48 | 128 | Gene disrupted by ISKpn26 | T246Af, R256Ge | WT | WT | WT | CAZ, ATM, FEP, CTX, TZP, AMK, GEN, CIP, SXT, CHL, MEM, ERT, IPM | FOT, TGC | blaTEM, blaSHV, blaCTX-M, aadA, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH12058 | 12 | 16 | Q30stopb | T246Af, R256Ge | WT | WT | WT | CAZ, ATM, FEP, CTX, TZP, SXT, CHL, MEM, ERT, IPM | FOT, AMK, GEN, CIP, TGC | blaKPC-2, blaTEM, blaSHV, aadB, aac(3′)IIa, aac(6′)-Ib |
| CCBH14465 | 24 | 32 | Gene disrupted by IS102 | T246Af | E57Gf | WT | WT | CAZ, ATM, FEP, CTX, TZP, AMK, CIP, SXT, CHL, MEM, ERT, IPM, TGC | FOT, AMK | blaKPC-2, blaTEM, blaSHV, qnrB, qnrS, aadB, aac(3′)IIa, aac(6′)-Ib |
PMB, polymyxin B.
WT, wild type.
ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; TZP, piperacillin-tazobactam; ERT, ertapenem; GEN, gentamicin; AMK, amikacin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol; FOT, fosfomycin/trometamol; MEM, meropenem; IPM, imipenem; TGC, tigecycline.
Isolates belonging to CC258.
Mutation predicted as deleterious by PROVEAN.
Mutation predicted as neutral by PROVEAN.
The resistance genes searched were related to polymyxin resistance (mcr-1), plasmid-mediated quinolone resistance (qnrA, qnrB, qnrS), aminoglycoside resistance [aadA, aadB, aac(3′)IIa, aac(6′)-Ib], and β-lactam resistance (blaKPC, blaNDM, blaOXA-48, blaIMP, blaVIM, blaSHV, blaTEM, blaCTX-M).
The MICs for meropenem, imipenem, and tigecycline were also determined by Etest, and susceptibility to other antimicrobial agents was determined by agar diffusion (Table 1). Most of the isolates were nonsusceptible to β-lactams (n = 10), ciprofloxacin (n = 9), sulfamethoxazole-trimethoprim (n = 9), gentamicin (n = 7), chloramphenicol (n = 7), and amikacin (n = 6) and remained susceptible to fosfomycin/trometamol (n = 7) and tigecycline (n = 7). Data from SENTRY (2008 to 2010) showed a 3.2% incidence of PRKP isolates in Brazil (14). This rate increased to 6.6% for extended-spectrum-β-lactamase (ESBL)-producing K. pneumoniae isolates from medical centers in Latin America in 2011 (15) and 9.7% for KPC-2-producing K. pneumoniae isolates from Brazil in 2010 (16), showing a clear association between PMB resistance and other acquired resistance mechanisms.
We performed PCR to detect resistance genes to β-lactams, quinolones, and aminoglycosides and performed sequencing when required. Genetic determinants associated with resistance to those classes were observed in all isolates (Table 1), with blaKPC-2 observed in eight strains. The pulsed-field gel electrophoresis (PFGE) (17) and multilocus sequence typing (MLST) (18) analyses (Fig. 1) revealed four pulsotypes (A [A1 to A6], B, C, and D) and seven sequence types (ST), which may indicate independent events of PMB resistance acquisition. A total of seven PRKP isolates belonged to clonal complex 258 (CC258), the most important CC associated with KPC production (19). Corroborating our findings, the expansion of PRKP isolates belonging to ST11 (CC258) and harboring blaKPC-2 was previously reported in Brazil in 2014 (20). The data raise concerns about the surveillance of PRKP spread, since PMB is one of the limited number of treatment options against infections caused by the endemic KPC-2-producing K. pneumoniae strains in Brazil (21).
To investigate the presence of mcr-1 and related variants and mutational events affecting mgrB, pmrA, pmrB, phoP, and phoQ genes, PCR and DNA sequencing were conducted. Mutation analysis of genes involved in polymyxin resistance was performed using Geneious (6.1.8) software and the BLASTN (NCBI) tool. The online platform ISfinder (https://www-is.biotoul.fr) was employed to identify insertion sequences (IS) and the PROVEAN platform (http://provean.jcvi.org/index.php) to predict alterations of biological functions of the proteins by the use of K. pneumoniae MGH 78578 (CP000647.1) as a reference.
Regarding polymyxin resistance, the presence of the mcr-1 gene was not detected. All of the PRKP strains exhibited alterations in the mgrB gene (Table 1), including disruption by IS903B, IS5, IS102, ISKpn26 (IS5 family), and IS10L (IS4 family). Previous studies (2, 22, 23, 24) have already shown the interruption of the mgrB region by IS10-like and IS5-like elements. This mechanism seems to be common in K. pneumoniae, including KPC-producing isolates of CC258 (22). In Brazil, disruption of mgrB by IS903 was already reported in a BKC-1 (Brazilian Klebsiella carbapenemase-1)-producing K. pneumoniae isolate from São Paulo (25). In addition, deleterious mutations (C28R and Q30stop) were observed in mgrB. Mutations at these same amino acid positions were also reported in PRKP isolates from Europe (2, 22, 26).
Alterations in the phoQ gene were not detected. However, a partial deletion of the PmrB-encoding gene was identified in one isolate and a deleterious mutation (R256G) was found in seven isolates (Table 1). This specific mutation was not related to polymyxin resistance as previously reported (27). The T246A, E57G, and A30S mutations detected in PmrB, PmrA, and PhoP, respectively, were not considered deleterious by PROVEAN. Furthermore, we suggest that the specific PmrB (T246A) and PmrA (E57G) mutations found in this study were not capable of producing PMB resistance alone, since these mutations were also found in PMB-susceptible isolates (data not shown).
In this study, the disruption of the mgrB gene was shown to be associated with PMB resistance in K. pneumoniae. It is worrisome that the spread of PRKP in Brazil is associated with KPC-producing strains belonging to the epidemic CC258 variant. The present findings indicate the importance of broad and effective monitoring of PMB-resistant Gram-negative bacteria in order to follow the evolution of PMB resistance in Brazil, as well the importance of screening for PMB resistance in colonized nosocomial patients in order to prevent possible infection by these pathogens.
ACKNOWLEDGMENTS
We thank the PDTIS-IOC DNA Sequencing Platform for DNA sequencing.
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and PAPES/Oswaldo Cruz Institute (IOC-FIOCRUZ).
We declare no conflict of interests.
REFERENCES
- 1.Ah YM, Kim AJ, Lee JY. 2014. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents 44:8–15. doi: 10.1016/j.ijantimicag.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain JM. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Kontopidou F, Plachouras D, Papadomichelakis E, Koukos G, Galani I, Poulakou G, Dimopoulos G, Antoniadou A, Armaganidis A, Giamarellou H. 2011. Colonization and infection by colistin-resistant Gram-negative bacteria in a cohort of critically ill patients. Clin Microbiol Infect 17:E9–E11. doi: 10.1111/j.1469-0691.2011.03649.x. [DOI] [PubMed] [Google Scholar]
- 4.Rodrigues Perez LR, Dias CG. 2016. Emergence of infections due to a polymyxin B-resistant KPC-2-producing Klebsiella pneumoniae in critically ill patients: what is the role of a previous colonization? Infect Control Hosp Epidemiol 37:240–241. doi: 10.1017/ice.2015.294. [DOI] [PubMed] [Google Scholar]
- 5.Giacobbe DR, Del Bono V, Trecarichi EM, De Rosa FG, Giannella M, Bassetti M, Bartoloni A, Losito AR, Corcione S, Bartoletti M, Mantengoli E, Saffioti C, Pagani N, Tedeschi S, Spanu T, Rossolini GM, Marchese A, Ambretti S, Cauda R, Viale P, Viscoli C, Tumbarello M; ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva). 2015. Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin Microbiol Infect 21:1106.e1–1106.e8. [DOI] [PubMed] [Google Scholar]
- 6.Gaspar GG, Bellissimo-Rodrigues F, de Andrade LN, Darini AL, Martinez R. 2015. Induction and nosocomial dissemination of carbapenem and polymyxin-resistant Klebsiella pneumoniae. Rev Soc Bras Med Trop 48:483–487. doi: 10.1590/0037-8682-0041-2015. [DOI] [PubMed] [Google Scholar]
- 7.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 9.Brazilian Committee on Antimicrobial Susceptibility Testing—(BrCAST)—EUCAST. 2016. Tabelas de pontos de corte para interpretação de CIMs e diâmetros de halos. Versão 6.0, 2016. http://brcast.org.br.
- 10.Clinical and Laboratory Standards Institute (CLSI). 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 11.Lat A, Clock SA, Wu F, Whittier S, Della-Latta P, Fauntleroy K, Jenkins SG, Saiman L, Kubin CJ. 2011. Comparison of polymyxin B, tigecycline, cefepime, and meropenem MICs for KPC-producing Klebsiella pneumoniae by broth microdilution, Vitek 2, and Etest. J Clin Microbiol 49:1795–1798. doi: 10.1128/JCM.02534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girardello R, Bispo PJM, Yamanaka TM, Gales AC. 2012. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J Clin Microbiol 50:2414–2418. doi: 10.1128/JCM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez LR. 2015. Evaluation of polymyxin susceptibility profile among KPC-producing Klebsiella pneumoniae using Etest and MicroScan WalkAway automated system. APMIS 123:951–954. doi: 10.1111/apm.12438. [DOI] [PubMed] [Google Scholar]
- 14.Gales AC, Castanheira M, Jones RN, Sader HS. 2012. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010). Diagn Microbiol Infect Dis 73:354–360. doi: 10.1016/j.diagmicrobio.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Jones RN, Guzman-Blanco M, Gales AC, Gallegos B, Castro ALL, Martino MDV, Vega S, Zurita J, Cepparulo M, Castanheira M. 2013. Susceptibility rates in Latin American nations: report from a regional resistance surveillance program (2011). Brazilian J Infect Dis 17:672–681. doi: 10.1016/j.bjid.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira PS, de Araujo CFM, Seki LM, Zahner V, Carvalho-Assef APD, Asensi MD. 2013. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340). J Antimicrob Chemother 68:312–316. doi: 10.1093/jac/dks396. [DOI] [PubMed] [Google Scholar]
- 17.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 18.Diancourt L, Passet V, Verhoef J, Patrick AD, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitout JDD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrade LN, Vitali L, Gaspar GG, Bellissimo-Rodrigues F, Martinez R, Darini ALC. 2014. Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS-1, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J Clin Microbiol 52:2530–2535. doi: 10.1128/JCM.00088-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM; COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 24.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins WMBS, Nicoletti AG, Santos SR, Sampaio JLM, Gales AC. 22 July 2016. Frequency of BKC-1-producing Klebsiella species isolates. Antimicrob Agents Chemother doi: 10.1128/AAC.00470-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Britto XB, Plantinga NL, Lammens C, Poirel L, Nordmann P, Bonten MJ, Goossens H, Malhotra-Kumar S. 2016. Novel mgrB variants identified in colistin-resistant Klebsiella pneumoniae, abstr O594 Abstr 26th Eur Cong Clin Microbiol Infect Dis (ECCMID), 9 to 12 April 2016, Amsterdam, Netherlands. [Google Scholar]
- 27.Cheng YH, Lin TL, Pan YJ, Wang YP, Lin YT, Wang JT. 2015. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother 59:2909–2913. doi: 10.1128/AAC.04763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]