Abstract
Membrane proteins remain challenging targets for structural biologists, despite recent technical developments regarding sample preparation and structure determination. We review recent progress towards a structural understanding of TRP channels and the techniques used to that end. We discuss available low-resolution structures from electron microscopy (EM), X-ray crystallography and nuclear magnetic resonance (NMR), and review the resulting insights into TRP channel function for various subfamily members. The recent high-resolution structure of TRPV1 is discussed in more detail in Chapter X. We also consider the opportunities and challenges of using the accumulating structural information on TRPs and homologous proteins for deducing full-length structures of different TRP channel subfamilies, such as building homology models. Finally, we close by summarizing the outlook of the “holy grail” of understanding in atomic detail the diverse functions of TRP channels.
Keywords: Ankyrin repeat, coiled coil, α-kinase, EF-hand, Calmodulin
Bottlenecks to TRP channel structure determination
In comparison to soluble proteins, high-resolution structures of membrane proteins are vastly underrepresented, comprising ~1% of structures in the protein database (PDB) versus ~30% of the protein-coding genome. Structure determination of membrane proteins can seem daunting - before the first structures of ion channels were determined not so long ago (Doyle et al., 1998; Dutzler et al., 2002), it had been presumed an almost impossible task (Abbott, 2002).
TRP channel architecture is similar to other ion channels (Cao et al., 2013b; Liao et al., 2013): a six-transmembrane helix topology (S1 through S6) with a reentrant loop between S5 and S6 forming the channel pore is a recurring structural motif (Yu and Catterall, 2004). These channels tetramerize to a 24-helix functional protein complex (Fig. 1). As observed for other ion channels, TRP channel function is strongly influenced by large intracellular domains (Fig. 1), and the responsiveness to functional modulators, e.g. regulation by phosphoinositides (Nilius et al., 2008), or inhibition by quaternary ammonium ions (Jara-Oseguera et al., 2008) and venom toxins (Siemens et al., 2006) is conserved across ion channel families.
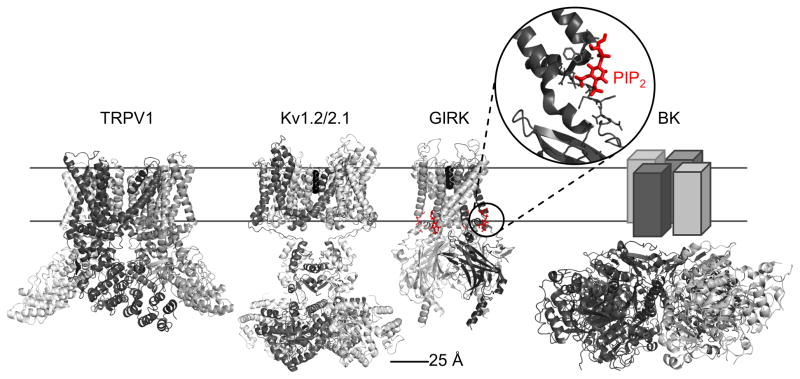
Fig. 1.
Structures of representative ion channels. Structures of TRPV1 (pdb: 3j5p), and other ion channels, such as the Kv1.2/2.1 chimera (pdb: 2r9r) or the GIRK channel in complex with PIP2 (pdb: 3sya) have been determined at high resolution. BK channels (pdb: 3naf), like TRP channels are only moderately voltage dependent and their function is modulated by large cytoplasmic domains. The TRPV1 construct used to determine the structure has been engineered to remove parts of the N- and C-termini (residues 1–109 and 765–838, respectively) and of the extracellular turret (residues 604–626).
Structural studies on TRP channels are challenging, partially due to the difficulty of producing sufficient quantities of protein in available (and financially sustainable) host systems. While structure determination of important membrane proteins has been facilitated through the use of bacterial, archeal or thermophilic homologs, no such homologs have been identified for TRPs. Instead TRP channels are heterologously expressed in yeast, insect or mammalian cells, all more costly and time-consuming than bacterial systems. The fact that neither cell-free nor bacterial expression systems are currently available for full-length TRP channels (except for TRPV3 and TRPM8 (Kol et al., 2013; Zakharian et al., 2010)) shows the importance of carefully selecting the expression system to fulfill the requirements of eukaryotic protein folding and (where applicable) posttranslational modifications. However, TRPs do have many N- and C-terminal cytoplasmic regions for which bacterial systems have yielded soluble fragments for use in X-ray crystallographic and NMR approaches (Fig. 2). Recent technological advances in the field of electron microscopy (EM) make smaller sample amounts amenable for medium to high resolution structure determination (Li et al., 2013), as seen for the 3.4Å TRPV1 structure determined by cryoEM ((Cao et al., 2013b; Liao et al., 2013); see Chapter X for a more detailed description (Hellmich and Gaudet, 2014)).
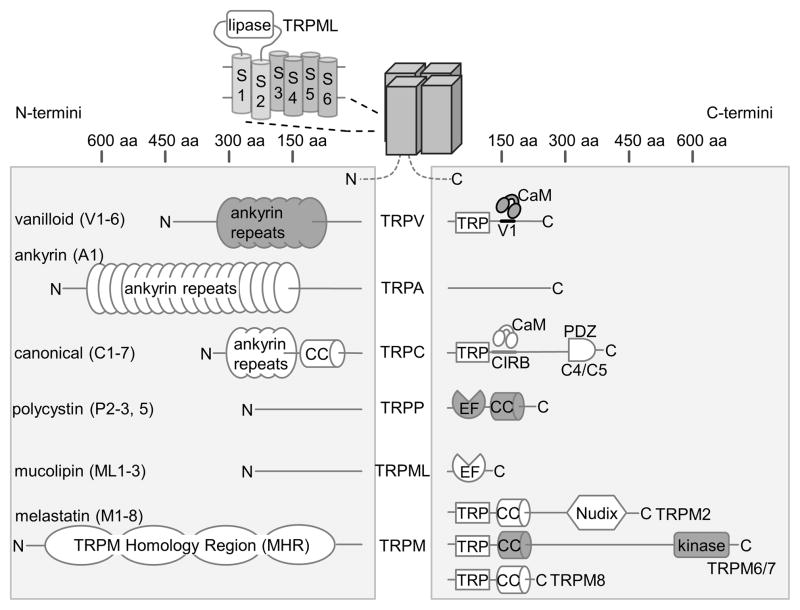
Fig. 2.
Schematic overview of TRP channel domain organization. The domain architecture of N- and C-termini, which differs widely across TRP channel subfamilies, is depicted schematically with approximate size scales. TRPML proteins contain a lipase domain between transmembrane helix S1 and S2 (LaPlante et al., 2011). Domains with available high-resolution structural data are grey.
Membrane protein purification is challenging. It requires stripping the lipid bilayer and replacing it by detergents harsh enough to obtain good yields, but mild enough not to compromise protein integrity. Detergent choice is doubly critical for tetrameric TRP channels, as both the 3D structure of each subunit and the correct oligomeric state need to be retained. Finally, the functional integrity of purified proteins has to be verified, which is particularly challenging for many TRPs as their physiological function and naturally occurring ligands are unknown or coarsely characterized. For membrane transporters, substrate binding is often used as a proxy for activity in detergent. With channels, reconstitution into liposomes or planar bilayers is required to detect ion movement across the membrane with electrophysiology or ion flux assays. Functional reconstitution has been described for TRPA1, TRPM8, TRPV1, TRPV2 and TRPV3 (Cao et al., 2013a; Cvetkov et al., 2011; Huynh et al., 2013; Kol et al., 2013; Moiseenkova-Bell et al., 2008; Zakharian et al., 2010).
Techniques to study TRP channel structures
Once a source of protein has been identified, the next step is to choose an appropriate structure determination technique. While most available protein structures have been obtained using X-ray crystallography, other powerful techniques exist, such as electron microscopy (EM) or nuclear magnetic resonance spectroscopy (NMR). These can provide information not accessible by crystallographic studies, such as dynamics or, in the case of membrane proteins, visualization in lipid bilayers. For TRP channels, current structural information does indeed come from a combination of techniques (Table 1).
Table 1.
Available structures of TRP channels and TRP channel domains.
| Organism | Protein | Fragment | Resolution (Å) | PDB (reference) | |
|---|---|---|---|---|---|
| EM | rat | TRPV1 | full-length (cryoEM) | 19 | (Moiseenkova-Bell et al., 2008) |
| rat | TRPV4 | full-length (cryoEM) | 35 | (Shigematsu et al., 2010) | |
| mouse | TRPA1 | full-length (neg. stain) | 16 | (Cvetkov et al., 2011) | |
| human | TRPM2 | full-length (neg. stain) | 28 | (Maruyama et al., 2007) | |
| mouse | TRPC3 | full-length (cryoEM) | 15 | (Mio et al., 2007a) | |
| rat | TRPV1 | near full-length (cryoEM) | 3.4 | 3j5p (Liao et al., 2013) | |
| rat | TRPV1 | + DkTx & RTX | 3.8 | 3j5q (Cao et al., 2013b) | |
| rat | TRPV1 | + Capsaicin | 4.2 | 3j5r (Cao et al., 2013b) | |
| rat | TRPV2 | full-length (cryoEM) | 13.6 | (Huynh et al., 2013) | |
|
| |||||
| X-ray crystallography | rat | TRPV1 | ARD + ATP | 2.7/3.2 | 2pnn/2nyj (Lishko et al., 2007) |
| human | TRPV2 | ARD | 1.7 | 2f37 (McCleverty et al., 2006) | |
| rat | TRPV2 | ARD | 2.2/1.65/3.1 | 2eta/2etb/2etc (Jin et al., 2006) | |
| mouse | TRPV3 | ARD | 1.95 | 4n5q (Shi et al., 2013) | |
| human | TRPV4 | ARD (+ATP) | 2.85 (2.95) | 4dx1 (4dx2) (Inada et al., 2012) | |
| chicken | TRPV4 | ARD | 2.3/2.8 | 3jxi/3jxj (Landoure et al., 2010) | |
| mouse | TRPV6 | ARD | 1.7 | 2rfa (Phelps et al., 2008) | |
| Gibberella zeae | TRPGz | coiled coil | 1.25 | 3vvi (Ihara et al., 2013) | |
| rat | TRPM7 | coiled coil | 2.01 | 3e7k (Fujiwara and Minor, 2008) | |
| human | TRPP2 | coiled coil | 1.9/1.9 | 3hro/3hrn (Yu et al., 2009) | |
| human | TRPP3 | coiled coil | 2.69 | 3te3 (Molland et al., 2012) | |
| human | TRPP3 | coiled coil | 2.8 | 4gif (Yu et al., 2012) | |
| rat | TRPV1 | C-terminus + CaM | 1.95 | 3sui (Lau et al., 2012) | |
| mouse | TRPM7 | α-kinase (+AMPPNP/ADP·Mg2+) | 2.8 (2.0/2.4) | 1iaj (1ia9/1iah) (Yamaguchi et al., 2001) | |
|
| |||||
| NMR | human | TRPP2 | EF hand | 2kld/2kle (Schumann et al., 2009) | |
| human | TRPP2 | EF hand | 2kq6 (Petri et al., 2010) | ||
| human | TRPP2 | EF hand | 2y4q (unpublished) | ||
| human | PKD1 | PKD domain | 1b4r (Bycroft et al., 1999) | ||
Methods to determine 3D structures each have advantages and limitations (see (Minor, 2007) for a useful primer). Significant differences include resolution, and sample quantity, condition, and preparation. Different levels of resolution are required to answer different questions, and this can guide the choice of method. For example, resolution of 10 Å or worse is adequate to determine approximate molecule shape or observe large domain movements. Detecting secondary structure elements necessitates higher resolution, ~4 Å for β-sheets and ~9 Å for α-helices. Around 3.5 Å, amino acid sidechains are discernible, while ~2.5 Å or better enables the observation of bound water molecules or ions. Below we provide brief overviews of EM, X-ray crystallography and NMR.
Electron Microscopy
Initially, five low-resolution EM structures – of TRPV1, TRPV4, TRPM2, TRPC3 and TRPA1 – were our only experimental glimpse at the overall shapes of full-length TRP channels (Table 1, Fig. 1). Recently, TRPV2 joined this collection (Huynh et al., 2013). The achievable resolution in EM is generally lower than that of X-ray or NMR structures, although the convergence of ideal technical conditions and sample behavior can now yield atomistic structures (Zhou, 2011), including high-resolution cryoEM structures of TRPV1 ((Cao et al., 2013b; Liao et al., 2013); Chapter X (Hellmich and Gaudet, 2014)). EM has no upper size limits on the macromolecular complex under study – in fact the bigger the molecules, the better the signal – and it does not require crystals, although it does require samples of high purity. The protein is applied as a dilute solution to a carbon film surface. It can then be visualized either by negative-staining to yield low resolution shape information, or directly imaged under cryo conditions (cryoEM). Both techniques have been applied to TRP channels.
In cryoEM, the sample is rapidly frozen, i.e. vitrified to avoid ice crystal formation, keeping the protein fully hydrated so that it retains its native conformation. Because the atoms present in biological samples have low electron beam absorption and therefore very low signal-to-noise, cryoEM requires merging tens of thousands of particle images. Under optimal conditions, especially if particle symmetry can be leveraged into further averaging, cryoEM can yield high-resolution structures.
In negative staining an electron-rich dye forms a cast around the protein particles as the sample is dried. The particle’s outline is visualized with good signal-to-noise, but the cast inherently limits the resolution. Staining and drying the sample also leads to loss of the protein’s hydration shell and may consequentially distort it.
The EM 3D reconstruction process consists of merging tens of thousands of 2D pictures of individual protein particles in different orientation into a single 3D representation. Therefore a bias in the protein orientation on the grid, which is particularly common in dried negative-stained samples, can lead to poor averages and loss of resolution and/or distortions of the 3D volume due to undersampling of some orientations.
X-ray crystallography
Most high-resolution structures of TRP channel fragments have been determined by X-ray crystallography (Table 1). We now have an assortment of structures of soluble TRP channel domains that, in combination with the EM structures, begin to delineate larger structural correlations and functional features of various TRP channel subfamilies.
X-ray crystallography requires large quantities of highly pure protein to screen many crystallization conditions as we cannot predict successful conditions. Crystal formation, i.e. the assembly of molecules in an ordered 3D lattice, depends on the chemical and physical properties of the protein and precipitation agents, and the temperature, pH, and other factors such as the presence of ligands and additives (Dumetz et al., 2007; Luft et al., 2011). For membrane proteins, the number of variables increases further, as the nature of the detergent also matters (Bill et al., 2011). Finally, the protein should be conformationally homogenous: highly flexible regions, such as unstructured N- and C-termini and loops are often truncated and dozens of variants may need to be tested to find a candidate that crystallizes successfully. This approach can be used to eliminate low complexity sequence regions in TRP channels and increase the chances to obtain high-resolution structures by either EM (Cao et al., 2013b; Liao et al., 2013) or crystallography. Alternatively, ligands or mutations can be used to restrict protein motion. For example, multiple G-protein coupled receptors crystal structures were obtained after introducing combinations of mutations that significantly increase their melting temperatures (Tate, 2012). A general take-home message is that crystallization is facilitated by taking advantage of accumulated biochemical and structural information on a protein to engineer stable conformational states.
NMR spectroscopy
Only a few structures of TRP channel fragments have been obtained by NMR (Table 1). However, NMR is advantageous for specific questions, especially regarding conformational dynamics. In addition to high concentration (>100 μM) and large sample volumes, NMR also requires isotope labeling to increase the signal from low-abundance nuclei such as 13C and 15N. Efficient labeling protocols exist for bacteria, yeast and even insect cells (Saxena et al., 2012), but production of isotope-labeled membrane proteins in mammalian cells is generally not economically feasible.
Slow tumbling of large particles in solution degrades NMR signals, imposing size limits on proteins in solution NMR, with few examples of protein structures larger than 35 kDa (see (Tugarinov et al., 2005) for one example). This size limitation is particularly acute for membrane proteins because the detergent micelles necessary to maintain a soluble native-like state add significant bulk to the particle size.
Once high-quality NMR spectra with good dispersion of the signals from individual atoms are obtained, determining an NMR structure consists of three steps (Wider, 2000): (i) assign each spectral peak to its corresponding 13C, 15N or 1H atom via through-bond correlations between these peaks, which also yields secondary structure information; (ii) collect through-space correlations linking atom pairs, called the NOEs after the nuclear Overhauser effect allowing nuclei to “see” each other through space to a distance of up to ~5 Å; and (iii) use the resulting constraints to “fold” the protein in silico through iterative molecular dynamics and energy minimization steps, yielding a structure showing atomic details. The NMR assignments also enable further investigations to, for example, probe ligand interactions and protein dynamics. Therefore NMR spectroscopy has a place in “divide and conquer” approaches to understand details of TRP channel structure, function and dynamics.
Solid-state NMR, in contrast to solution NMR, does not have inherent size limitations and can thus be applied to microcrystalline samples or membrane proteins reconstituted into lipid bilayers. However, solid-state NMR structure determination is still far from standardized (Hong et al., 2012; Zhao, 2012). Membrane proteins are challenging because transmembrane regions contain an overabundance of similar hydrophobic residues in a comparable secondary structure environment, making spectrum analyses difficult. The few heroic examples of membrane protein structures determined with solid-state NMR include the G-protein coupled receptor CXCR1, kaliotoxin bound to a KcsA-Kv1.3 chimera, and various bacterial retinylidene proteins (Lange et al., 2006; Park et al., 2012; Wang et al., 2013).
Structural information on TRP channels
In this section, we first discuss the low-resolution electron microscopy (EM) structures of full-length TRP channels, then review the current parts list of high-resolution structures of TRP channel fragments, providing additional context about what we know (and do not know) about how these fragments assemble. The high-resolution cryoEM structures of TRPV1 are discussed separately in Chapter X (Hellmich and Gaudet, 2014).
Overall architecture of a TRP channel – an electron microscopy view
TRP proteins share a common topology, with six membrane-spanning segments flanked by the variable cytoplasmic domains (Fig. 2), and in most cases tetramerize to form channels (TRPP channels however form heteromers; see below). The five low-resolution molecular envelopes obtained by single-particle EM reconstructions (Table 1) gave us first glimpses at the possible overall general shape of full-length TRP channels. Low-resolution structures of TRPC3, TRPV1, TRPV2 and TRPV4 have been determined using cryoEM (Huynh et al., 2013; Mio et al., 2007a; Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010), while negative-stain EM was used for TRPM2 and TRPA1 (Cvetkov et al., 2011; Maruyama et al., 2007). In most of these cases, functional assays (calcium influx or electrophysiology) were used to confirm that the proteins were active in their heterologous expression system. For the low-resolution structures of TRPV1, TRPV2 and TRPA1, the authors also confirmed that the purified proteins retained their ability to be activated by chemical agonists after reconstitution into liposomes (Cvetkov et al., 2011; Huynh et al., 2013; Moiseenkova-Bell et al., 2008).
Although these EM structures vary in resolution and shape details, they provided some useful frameworks for understanding the overall architecture of TRP channels across subfamilies. A common theme is that they all display some signs of a fourfold symmetry axis, however their overall dimensions and surface shapes vary significantly. These differences are in part explained by the differences in molecular mass and intracellular domain composition, although some discrepancies are also apparent, as discussed below.
TRPV proteins have a ~400–450-residue N-terminal domain with six ankyrin repeats and a ~150-residue C-terminus that acts as a platform for interactions with other proteins and ligands (Fig. 2; (Lishko et al., 2007; Numazaki et al., 2003; Prescott and Julius, 2003)). TRPV1 and TRPV4 are ~840 and 870 residues, respectively, and their low-resolution EM structures look similar (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010). TRPV EM structures have a two-domain overall architecture with a larger and a smaller domain connected through four pillars on the four corners of the smaller domain. The overall height of the low-resolution TRPV1 structure is 150 Å. The smaller domain measures ~40 × 60 × 60 Å, while the larger domain, referred to as the “hanging gondola” (a term first coined for the cytoplasmic domains of a voltage-gated potassium channel (Kobertz et al., 2000)), is ~110 × 100 × 100 Å. Such a “hanging gondola” has been observed in other channels including voltage-gated potassium channels (Fig. 1).
Based on superpositions of available crystallographic structures of the N-terminal TRPV1 ankyrin repeat domain (ARD) (Lishko et al., 2007) and the transmembrane domain (TMD) of the Kv1.2 (Long et al., 2005) or MlotiK1 channels (Clayton et al., 2008), the small domain was assigned to the TRPV1 transmembrane region, and the larger domain to the cytoplasmic N- and C-termini. The larger domain can readily accommodate four copies of the TRPV1 ARD, with a good fit obtained when the long axis of the ARD is perpendicular to the membrane plane. Interestingly, evidence of four-fold symmetry was observed in both top and bottom views of the TRPV1 and TRPV4 EM structures (Moiseenkova-Bell et al., 2008; Shigematsu et al., 2010), indicating that both the TMD and the cytoplasmic domains adopt a highly symmetric conformation.
The main difference between the low-resolution TRPV1 and TRPV4 EM structures is that in comparison to TRPV1, the “hanging gondola” is shorter in TRPV4. As a result, the ARD crystal structure had to be tilted diagonally relative to the membrane plane to be accommodated in the volume. It is unclear whether these differences represent two conformations of TRPV channels, structural differences between TRPV1 and TRPV4, or are artifacts of the differences in resolution and structure determination methods. The high-resolution TRPV1 structures do show the ARDs much closer to the transmembrane region (Cao et al., 2013b; Liao et al., 2013). The low-resolution TRPV2 structure is more compact (with an overall height of 115 Å) than the low-resolution TRPV1 and TRPV4 structure (Huynh et al., 2013), and closely resembles that of the high-resolution TRPV1 structures.
The observation that all cytoplasmic domains of the TRPV channels are localized very closely to each other in the “hanging gondola” suggested that their N- and C-termini can interact with each other at least in one conformation. The N-terminal TRPV ARDs do not interact in vitro as isolated domains (Phelps et al., 2010), but N- and C-terminal interactions have been observed for TRPV5 and TRPV6 (Chang et al., 2004; Erler et al., 2004), and the N- and C-termini of TRPV1 indeed interact in the high-resolution structures (Cao et al., 2013b; Liao et al., 2013).
A TRPA1 EM structure also shows a two-domain architecture resembling that of TRPV structures with an overall height of 195 Å and four-fold symmetry (Cvetkov et al., 2011). The larger and taller TRPA1 “hanging gondola” measures ~130 × 120 × 120 Å, and was accordingly assigned to the larger N-terminal region (~800 residues). TRPA1’s smaller domain, designated as the transmembrane region, measures ~70 × 100 × 100 Å, and is therefore larger than that of TRPVs. The authors speculate that this could be due to the amphipatic polymer used in place of detergents to stabilize TRPA1 (Cvetkov et al., 2011).
While the low-resolution TRPV1, TRPV2, TRPV4 and TRPA1 have a similar overall architecture, the EM structures of TRPM2 and TRPC3 look markedly different from the aforementioned and each other. The negative-stain EM structure TRPM2 (Maruyama et al., 2007) shows an overall bell-shape with few features except a bulb-like extension on its widest end that would correspond to the bell clapper. The calculated molecular mass for this structure is about one third heavier than expected. The authors attributed this ~200 kDa difference to attached lipids and detergent molecules. However, the protein was purified in dodecylmaltoside, with an isolated micelle size of ~55 kDa, the same detergent used for TRPV4 where no such large discrepancy between expected and observed size was observed (Shigematsu et al., 2010).
To identify the TRPM2 cytoplasmic region, a gold-conjugated antibody fragment against a C-terminal FLAG-tag was used and the TMD thus assigned to the apex of the large bell hull structure. However, the “bell clapper” of the TRPM2 structure is tantalizingly similar in size and shape to the small domain assigned to the TMD of the TRPV EM structures.
A cryoEM TRPC3 structure consisting of an intricate mesh of thin rods was reported (Mio et al., 2007a) with a ~200 × 200 × 240 Å volume, extraordinarily large for an expected tetramer mass similar to that of a TRPV1 tetramer. The authors observed smaller particles in high-salt conditions, which they attribute to a tetramer-to-monomer transition (Mio et al., 2007b). Alternatively, the mesh-like structure could be an artifact of trying to contour a volume corresponding to a tetramer when they are instead observing a larger aggregate, with high-salt conditions dissociating the aggregate into isolated tetramers. It is also possible that technical issues with a relatively new automated particle picking algorithm may have led to a distorted reconstruction (Moiseenkova-Bell and Wensel, 2009).
In summary, the low-resolution TRPV1, TRPV2, TRPV4 and TRPA1 EM structures are relatively consistent with expectations from our knowledge of distantly related ion channels (Fig. 1). It is noteworthy that only one conformation, with soluble domains forming the hanging gondola, has thus far been observed in low-resolution EM structures. Significant conformational changes in TRPV4 in response to changing levels of phosphoinositides in the cell membrane have been inferred using Förster resonance energy transfer (FRET) between C-termini (Garcia-Elias et al., 2013). In contrast, there was no evidence of conformational changes in TRPV1 in response to capsaicin, heat, or calcium, using a similar experimental strategy (De-la-Rosa et al., 2013). This suggests that either their FRET sensors lack the necessary spatial resolution, and/or that there are only small conformational changes upon TRPV1 activation. In agreement with the latter, no large conformational changes were observed when comparing the high-resolution TRPV1 structures in the absence or presence of agonists (Cao et al., 2013b; Liao et al., 2013).
Current high-resolution parts list: Structures of TRP channel domains
The modularity of ion channels such as TRP channels makes them amenable to a “divide and conquer” approach where high-resolution structures of individual soluble domains are easier or faster to obtain than full-length protein structures (Gaudet, 2009). Below, we describe the available “parts list”, and discuss how the different parts may assemble in the context of full-length tetrameric channels. Because these parts stem from a large number of TRP channels, they allow important insights into the peculiarities and commonalities of various subfamilies that a single structure cannot achieve.
The TRPM7 α-kinase domain
TRPM6 and TRPM7 are involved in Mg2+ homeostasis in mammals and their C-terminal α-kinase domain, unique within the TRP superfamily, is implicated in Mg2+ sensing (Demeuse et al., 2006; Schmitz et al., 2003). The crystal structure of the TRPM7 α-kinase domain was the first high-resolution structure of a TRP channel domain, and coincidentally the first structure of an α-kinase (Yamaguchi et al., 2001). α-Kinases predominantly target residues in α-helical regions, hence their name (Ryazanov et al., 1999). The TRPM7 α-kinase is a Ser/Thr kinase that can be expressed as a soluble protein and retains its activity (Runnels et al., 2001), indicating that it is an independently folding domain.
The TRPM7 α-kinase structure shows a domain-swapped dimer: a short N-terminal α-helix of one kinase subunit reaches into the structural core of the other subunit via an extended peptide region (Fig. 3A; (Yamaguchi et al., 2001)). As we discuss below, this dimerization mechanism suggests a dimer of dimer arrangement in a tetrameric TRPM7 channel.
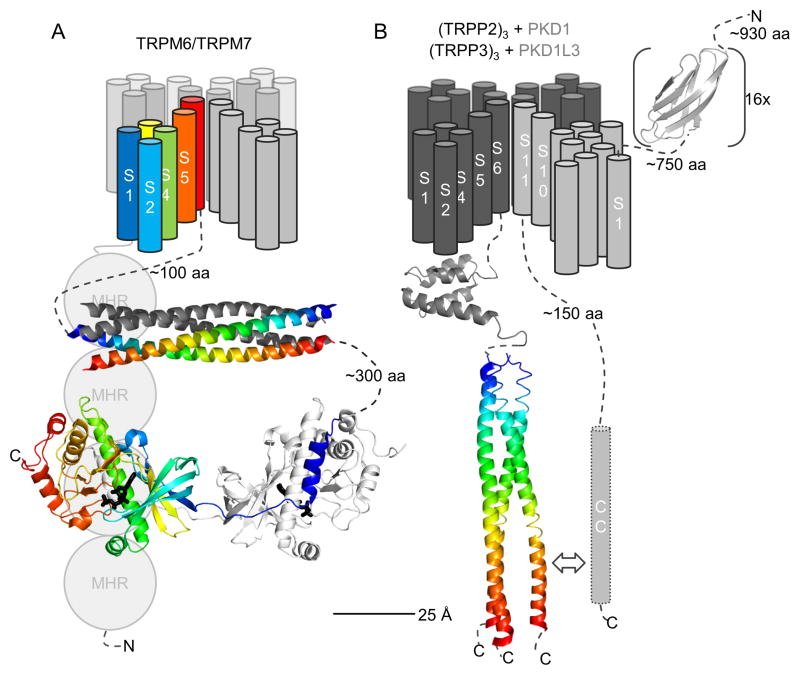
Fig. 3.
Overview of spatial organization of TRPM6/M7 channels and TRPP/PKD complexes. (A) For TRPM6/M7, transmembrane helices S1–S6 are drawn indicating possible 3D topology of the tetramer in the membrane (based on Kv channel topology). For cytoplasmic domains, the antiparallel coiled coil (pdb: 3e7k) and α-kinase domain (pdb: 1ia9) of TRPM7 are shown; no structure is available for the TRPM homology region (MHR). The coiled coil crystallized as a tetramer, and one antiparallel pair is colored. Only one dimer of the α-kinase is shown for clarity. (B) For TRPP/PKC complexes, the three TRPP2/P3 subunits are depicted in dark grey and the 11-transmembrane helix PKD1/PKD1L3 subunit in light grey. One extracellular PKD repeat of PKD1 is depicted (pdb: 1b4r). For TRPP2, the intracellular EF hand (pdb: 2kq6) as well as the parallel coiled coil trimer (pdb: 2hrn) are shown.
The TRPM7 α-kinase structure revealed an unexpected structural similarity to other eukaryotic protein kinases (Yamaguchi et al., 2001), despite the fact that α-kinases do not share all typical sequence motifs of classical kinases (Bates-Withers et al., 2011; Ryazanov et al., 1999). Each individual TRPM7 α-kinase consists of N- and C-terminal lobes connected by a flexible linker region. Similar to classic kinases, the nucleotide-binding pocket is nested in between the two lobes. The TRPM7 α-kinase structure also features a structural zinc-binding site within the C-terminal lobe, which is unique to and conserved in α-kinases.
Within the α-kinase active site, the P-loop (required for nucleotide binding and hydrolysis) and a conserved glycine-rich α-kinase motif are in strikingly similar orientation to the P-loop and activation loop (acting as a structural scaffold for interaction with the substrate) of classical kinases. The glycine-rich α-kinase motif was therefore hypothesized to be the substrate interaction platform (Yamaguchi et al., 2001). Interestingly, point mutations that abrogated TRPM7 activity (Runnels et al., 2001), were not located in the catalytic center of the kinase but rather in this postulated substrate peptide recognition site (Yamaguchi et al., 2001). However, the nature of the substrate(s) cannot be readily inferred from the kinase structure.
The TRPM7 coiled coil
The C-terminal cytoplasmic region of TRPM7 contains a ~50-residue coiled coil assembly domain that precedes and is separated from the α-kinase domain by ~300 residues (Fig. 3A). This coiled coil is found in all TRPM channels, many of which require it for correct oligomerization and channel activation (Tsuruda et al., 2006). The crystal structure of the isolated TRPM7 coiled coil shows a tetrameric antiparallel bundle (Fujiwara and Minor, 2008).
Before describing the structural details of the TRPM7 coiled coil, it is useful to review the salient features of coiled coils (see (Grigoryan and Keating, 2008; Moutevelis and Woolfson, 2009) for in depth reviews). Coiled coils are found in a large number of otherwise functionally and structurally unrelated protein families and consist of two or more α-helices that wind around each other to form a superstructure with helical properties, or supercoil. Coiled coils can be homo- or heteromeric, and the subunits can interact in parallel or antiparallel fashion with regard to the relative orientation of their N- and C-termini. This allows for a broad variety of architectures despite the seemingly simple design of a coiled coil, and hence a variety of functions as oligomerization or protein-protein interaction motifs.
Coiled coils have seven-residue or “heptad” repeats, with individual amino acids designated a through g. Positions a and d typically feature hydrophobic residues that project into the core of the assembled superstructure. This occurs in a “knob-in-holes” fashion, where an a or d residue is a “knob” that protrudes into a “hole” formed by sidechains of the other helices. This makes for a snug helix-helix interaction surface. In contrast, residues at e and g positions are usually hydrophilic and on the outside of the coiled coil, forming inter-helical interactions that determine the number and orientation (parallel or antiparallel) of α-helices in the coiled coil. Residues at positions b, c, and f are at the surface, away from inter-helical contacts, and thus have little influence on coiled coil assembly. In other words, positions a and d provide most of the affinity, whereas positions e and g provide most of the specificity, although predictive rules for coiled coil assemblies have yet to be determined (Grigoryan and Keating, 2008).
The 77-Å long TRPM7 coiled coil is tetrameric, as expected, but surprisingly is antiparallel, with two strands running in opposite direction to the other two. While it is a classical and well ordered a-d coiled coil with knob-in-hole packing geometry at its center, the termini of the coiled coil show distinct splaying and knob-against-knob packing.
Implications for TRPM channel assembly
The available structures of two C-terminal domains of TRPM7 provide some constraints for the assembly of full-length tetrameric TRPM7 channels (Fig. 3A). Because the TRPM7 coiled coil is antiparallel, the four-fold rotational symmetry of the TMD, expected based on homologous voltage-gated potassium channel structures (Long et al., 2005), must be broken in the C-terminal cytoplasmic region. This is in stark contrast to the parallel coiled coil observed in Kv7 channels, which allow for a continuous four-fold rotational symmetry through the entire protein (Howard et al., 2007). However, the TRPM7 TMD is separated by a ~100-residue linker from the coiled coil, which can readily accommodate the divergent symmetry of the TMD and coiled coil.
The antiparallel coiled coil complements the domain-swapped dimer observed for the α-kinase domain (Yamaguchi et al., 2001), suggesting that the four kinase domains in a tetrameric channel assemble as two dimers that may or may not contact each other. It was recently reported that cleavage of the α-kinase domain by caspases leads to increased TRPM7 channel activity in the context of Fas-induced apoptosis (Desai et al., 2012). That the C-terminal kinase domain can influence pore opening implies that the kinase and pore domains communicate, but whether there is direct contact or a long-range network relay remains to be seen. The antiparallel orientation of the TRPM7 coiled coil also raises the question of how this domain is oriented in relation to the rest of the protein (and the membrane plane) and whether its orientation and length remain constant as the TRPM7 channel undergoes activation/deactivation cycles.
Interestingly, sequence alignments of TRPM subfamily members, all of which have a coiled coil sequence motif at their C-terminus, indicate that their coiled coils vary in length and packing properties, hence clustering subfamily members into two groups (Fujiwara and Minor, 2008). This suggests different structural requirements of this domain in different TRPM channels and testable hypotheses of possible hetero-oligomerization partners. Considering that coiled coil assembly can be influenced by minimal sequence changes (Grigoryan and Keating, 2008; Xu and Minor, 2009), it is possible that the antiparallel tetramer assembly of the TRPM7 coiled coil is not a conserved feature of all TRPM channels, and that some may have other arrangements.
In summary, the TRPM7 coiled coil and α-kinase domain structures both suggest that at least some TRPM channels do not maintain four-fold rotational symmetry throughout the whole tetramer. This certainly represents a cautionary note in the processing and interpretation of EM structures: four-fold rotational symmetry averaging should not be blindly enforced, but only after clearly observing symmetry in un-averaged preliminary reconstructions.
Trimeric coiled coils and the assembly of TRPP channels
Putative coiled coils are also found in the N-terminal cytoplasmic region of TRPC proteins and the C-terminal cytoplasmic region of TRPP proteins (Fig. 2). The TRPP subfamily is somewhat unusual as there is emerging consensus that the functional unit is a heteromeric complex (Fig. 3B). Each complex consists of one PKD1 subunit (Polycystic Kidney Disease 1 or polycystin-1; previously known as TRPP1, although it is not a TRP protein) and three TRPP2 subunits (also referred to as polycystin-2, or polycystic kidney disease 2, PKD2) (Yu et al., 2009). A homologous complex is formed by one Polycystic Kidney Disease 1-Like 3 (PKD1L3) subunit and three TRPP3 subunits (also known as polycystic kidney disease 2-Like 1 or PKD2L1) (Yu et al., 2012). The PKD1/TRPP2 complex is important for mechanosensation in the kidney (reviewed in (Qamar et al., 2007)), while the acid-sensing PKD1L3/TRPP3 complex is activated by low pH and implicated in sour taste perception (see (Yu et al., 2012) and references therein).
PKD1, PKD1L3 and their homologs have a large extracellular N-terminal region containing up to 16 PKD domains, for which an NMR structure is available (Bycroft et al., 1999), and other subdomains, followed by 11 predicted transmembrane segments, six of which are homologous to the TMD of ion channels (Li et al., 2003). Mutations in the predicted pore region of PKD1L3 affect ion selectivity of the PKD1L3/TRPP3 complex (Yu et al., 2012). Therefore the PKD1/TRPP2 and PKD1L3/TRPP3 complex likely have a tetrameric channel pore domain analogous to homotetrameric TRP channels (Fig. 3B).
The heterotetrameric 1:3 arrangement raises the question of how these channels are assembled, and structures of TRPP coiled coils are providing valuable insights. X-ray structures of the TRPP2 and TRPP3 C-terminal coiled coils have been determined (Molland et al., 2012; Yu et al., 2009; Yu et al., 2012). Consistent with the heteromeric assemblies described above, the coiled coils of TRPP2 and TRPP3 form stable parallel trimers. Intriguingly, the TRPP2 coiled coil bundle is splayed open at its C-terminus (Yu et al., 2009). Because crystal contacts were observed in this region, the splaying could be a crystal-packing artifact, or suggest a physiological mechanism for partial or complete exchange of coiled coil strands as explained below.
PKD1 contains a short cytoplasmic C-terminus with a predicted coiled coil that interacts with the TRPP2 coiled coil in vitro (Qian et al., 1997; Tsiokas et al., 1997). A heteromeric four-helix bundle of a PKD1 and TRPP2 coiled coil has not yet been observed. However a computational docking study, combined with in vitro crosslinking and mutagenesis, supports a model in which the PKD1 coiled coil replaces one strand of the TRPP2 trimeric coiled coil in the splayed out C-terminal region (Fig. 3B) (Zhu et al., 2011). Furthermore, disruption of these residues prevents assembly of the PKD1/TRPP2 complex in cells (Zhu et al., 2011). Although dynamic exchanges of coiled coil strands may seem unusual, there is ample precedent for coiled coils as dynamic structures. For example, the long stalk of the dynein motor protein is a two-stranded coiled coil that slides to different helical registries separated by half a heptad, as dynein walks on microtubules (Kon et al., 2009).
PKD1L3, in contrast to PKD1, has a very short C-terminus and no predicted coiled coil. TRPP3 thus interacts with PKD1L3 mainly through interactions in the TMD (Ishimaru et al., 2010). However, disruption of TRPP3 coiled coil trimerization impairs surface expression of TRPP3/PKD1L3 (Yu et al., 2012). Coiled coil trimerization is therefore still essential for complex formation, supporting the idea that a TRPP trimer platform is required for a fourth (PKD) subunit to attach itself to form a mature complex.
In summary, the accumulated biochemical and structural data on the TRPP subfamily point to a key role for the coiled coil in the assembly of a functional channel that includes three TRPP subunits and one subunit of a PKD1 family member. The coiled coil structures also enable studies of the assembly mechanisms of this unusual family of ion channels with important physiological roles including kidney function and sour taste perception.
Broader implications of coiled coils on TRP channel assembly
Recently, the crystal structure of a C-terminal coiled coil of a fungal TRP channel, TRPGz from Gibberella zeae, has been determined (Ihara et al., 2013). TRPGz is homologous to the Saccharomyces cerevisiae TRPY1, but has a more polymodal activation profile. When compared to vertebrate TRP channels, it shows the highest similarity to members of the TRPC subfamily. TRPGz contains a coiled coil in its C-terminus, deletion of which disrupts responses to hyperosmotic or temperature shock. In contrast to other TRP channels, deletion of this coiled coil did not lead to oligomerization defects, as TRPGz still formed stable tetramers and retained its responses to voltage. Only the coiled coil shows significant secondary structure within the otherwise unstructured TRPGz C-terminus, as observed with NMR (Ihara et al., 2013). The TRPGz coiled coil was crystallized as a tetramer, with the typical a and d position hydrophobic knobs. Interestingly, the TRPGz coiled coil does not have exact four-fold rotational symmetry: the four coils come together at a significantly steeper angle than in canonical coiled coils, and the knobs-in-hole packing is only maintained for three out of four helices. In solution, this helical bundle is in equilibrium between monomer, dimer and tetramer. The precise role of this packing mode is not clear, and it is worth noting that the peptide used for crystallization is very short (20 amino acid residues).
In addition to the TRPM, TRPP and fungal TRP channels, TRPC subfamily channels also have a potential coiled coil between the ARD and first transmembrane segment (Fig. 2). No high-resolution structures are available for TRPC channels. Biochemical data for TRPC4 suggest that the N-terminal region forms tetramers, although the coiled coil was not directly implicated in this assembly (Lepage et al., 2009). Therefore, it remains to be determined whether the coiled coil sequence motifs actually form coiled coils and if so, whether these are heteromeric in assembly and what their stoichiometry is.
As illustrated above, structural studies of isolated coiled coils provide a wealth of information and constraints to better understand TRP channel assembly and function. However, a cautionary note is in order: coiled coils can be inherently dynamic and/or heavily influenced by their environment in both physiologically relevant and artifactual situations. For example, while the full isolated coiled coil of the Kv7.1 channel yielded the expected parallel tetrameric structure (Howard et al., 2007; Wiener et al., 2008), a shorter construct assembled as a non-physiological stable trimer (Xu and Minor, 2009). It is therefore possible that, in the context of a full-length channel, other oligomerization states of coiled coils are possible other than those observed with short fragments.
TRP channel ankyrin repeats
The highest number of structures of TRP channel domains to date comes from the ankyrin repeat domains (ARDs) of TRPVs. Ankyrin repeats are one of the most prevalent protein sequence motifs, present in a multitude of unrelated protein families where they are mostly associated with protein-protein interaction functions (Sedgwick and Smerdon, 1999). The motif name stems from the protein Ankyrin 1, which contains 24 such repeats (Lux et al., 1990). The basic architecture for this ~33–34 amino-acid stretch consists of short tandem antiparallel α-helices (termed inner and outer helix in regard to their location within the ARD concave or “palm” surface, see below), followed a β-hairpin loop. Individual ankyrin repeats stack against each other with a slight counterclockwise twist from one to the next. This gives them the appearance of a cupped hand: the loops forming the fingers attached to a hand with an open concave palm, the backside accordingly forms a convex surface (Jacobs and Harrison, 1998). The surface residues of the palm and fingers are not particularly conserved, making this region malleable to evolutionary diversification for various interaction partners (Sedgwick and Smerdon, 1999).
Ankyrin repeats are found in the cytoplasmic N-terminal region of TRPA, TRPV, TRPC and TRPN subfamily members (Fig. 2). TRP channel ARDs vary dramatically in size: TRPA1 and TRPN1 have 17 and 29 predicted repeats, respectively, that overall closely match the ankyrin repeat consensus sequence, while TRPC proteins have only four predicted repeats, which, like the six repeats found in TRPV proteins, more loosely follow the consensus sequence motif. So far, ARD structures are only available from the TRPV subfamily.
The ARD structures from TRPV1, TRPV2, TRPV3, TRPV4 and TRPV6 all show six highly conserved repeats (Inada et al., 2012; Jin et al., 2006; Landoure et al., 2010; Lishko et al., 2007; McCleverty et al., 2006; Phelps et al., 2008; Shi et al., 2013). Repeats 5 and 6 have an unusually large angular twist compared to repeats 1–4. Finally, the TRPV ARD finger loops are highly elongated, enhancing the resemblance to a hand (Gaudet, 2008a), although it should be noted that finger lengths varies within TRPV channels (Phelps et al., 2008). Variance in finger sequence and length, and of the palm surface residues, may indicate distinct roles for ARDs in TRPV channels, including interactions with different ligands. In TRPV1, Finger 3 of the ARD interacts with a β-sheet formed by both the proceeding linker region and the C-terminus of an adjacent subunit (Cao et al., 2013b; Liao et al., 2013).
Recent interesting developments include the structures of the TRPV4 ARD and their implications in understanding the molecular basis of TRPV4-linked genetic diseases. A large number of dominant missense mutations in TRPV4 have emerged that lead to very different channelopathies broadly segregating into two groups, skeletal disorders and degenerative neuropathies (see (Nilius and Voets, 2013) for a recent review). We currently do not understand the underlying structural implications of the mutants described to date, and even the functional data are limited somewhat contradictory: both loss- or gain-of-function phenotypes have been described for the same mutant (Auer-Grumbach et al., 2010; Landoure et al., 2010). A different approach is to investigate whether neuronal vs. skeletal manifestations of TRPV4-linked diseases may be the consequence of distinct structural or biochemical differences. However, no correlation was observed between disease phenotype and biochemical properties regarding TRPV4 ARD stability and/or ATP binding (Inada et al., 2012).
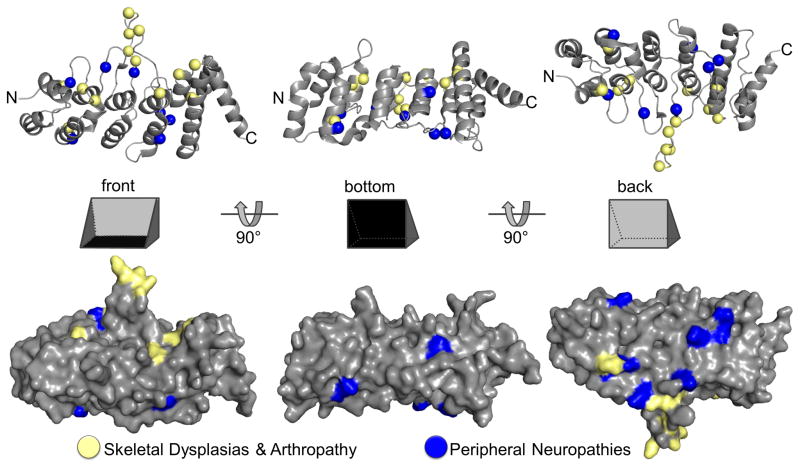
Mapping known disease-causing TRPV4 mutations (Nilius and Voets, 2013) onto the primary and tertiary structure of human TRPV4 allows us to appreciate their relative localization in a new light. While skeletal dysplasia mutations are widely distributed throughout the protein, neuropathy mutations localize primarily, although not exclusively, to the ARD. Furthermore, the skeletal dysplasia mutations located within the ARD map predominantly on its palm surface, whereas the mutants linked to neuropathies are preferentially located to the surface corresponding to the back of the hand (Fig. 4; (Inada et al., 2012; Landoure et al., 2012; Landoure et al., 2010)). Interestingly, the back face of the ARD is much more conserved in TRPV4 than it is in TRPV1 (Phelps et al., 2007; Zimon et al., 2010). This suggests that the physical segregation of TRPV4 channelopathy mutations may underlie their different phenotypic outcome, perhaps by differentially affecting intramolecular contacts or intermolecular interactions with cellular partners.
Fig. 4.
Crystal structure of the human TRPV4 ARD shown in cartoon and surface representations from three angles (pdb: 4dx1). Locations of disease-causing mutations are mapped onto the structure (spheres). Residues described in patients suffering from skeletal dysplasia and arthropathy are predominantly found on the “front” or “palm” surface, and those causing peripheral neuropathies localize preferentially to the “back”.
TRPV ARD structures have already been extensively reviewed (Gaudet, 2008a, b, 2009), and a few themes are emerging. First, the isolated TRPV ARDs show no evidence of self-oligomerization (Phelps et al., 2008; Phelps et al., 2010). The ARDs instead influence TRPV channel assembly and function through intramolecular contacts with other regions of the TRPV proteins, as illustrated in the TRPV1 structure (Liao et al., 2013). Second, engineered mutations of TRPV ARD surface residues tend to increase the sensitivity and/or basal activity of TRPV channels (Lishko et al., 2007; Phelps et al., 2010), suggesting an overall inhibitory role for this domain on TRPV channel function. Third, there is thus far no evidence of large-scale conformational changes within the ARD itself, even upon ATP ligand binding (Inada et al., 2012), although the finger loops do show inherent flexibility (Inada et al., 2012; Jin et al., 2006). Therefore how the ARDs influence TRPV channel behavior remains unclear – perhaps a movement of the entire domain relative to the rest of the channel rather than a conformational change within its boundaries is ultimately responsible for its influence on channel function, sensitization or desensitization. More studies in the context of full-length channels are required to test these hypotheses.
Structures of TRP channel motifs involved in calcium regulation
Many TRP channels are permeable to calcium, and their activation leads to changes in intracellular calcium concentrations. In turn, many of these channels are in turn regulated directly or indirectly by calcium-dependent signaling pathways. Structural biology has begun to yield mechanistic insights into two such calcium regulation mechanisms: (i) calcium-calmodulin (CaM) binding to the C-terminal cytoplasmic region of TRPV1, and (ii) calcium binding to a paired EF-hand motif that precedes the coiled coil in the TRPP C-terminus.
EF hands are the most common Ca2+-binding motifs in proteins, and typically occur in pairs. The basic architecture is a helix-loop-helix motif with the two helices oriented in a V-shape between the thumb and index finger (Fig. 5). Upon calcium binding in the cleft formed by the loop region – by the palm and remaining fingers, using the hand analogy – the thumb moves further away from the index finger, changing the V to an L-shape. This exposes otherwise buried residues that can then interact with downstream target sequences thereby enabling Ca2+-dependent signal propagation (Lewit-Bentley and Rety, 2000). The most famous EF-hand-containing protein is CaM, a ubiquitous 17 kDa protein with flexibly connected N- and C-terminal lobes comprised of two EF-hands that each bind calcium in the μM range (Persechini et al., 1989; Vetter and Leclerc, 2003). CaM regulates many ion channels and signaling proteins, including several TRP channels (Zhu, 2005). Of note, CaM does not necessarily have to be fully calcified to interact with its targets (Delmas and Brown, 2005). This variety in Ca2+-occupancy and structural flexibility make CaM a highly versatile regulatory partner.
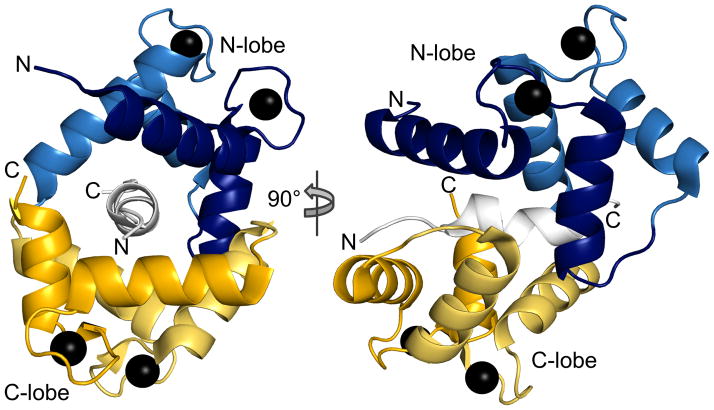
Fig. 5.
Crystal structure of the C-terminal fragment of TRPV1 in complex with Ca2+-calmodulin (pdb: 3sui). The fully Ca2+-loaded CaM tightly winds around the TRPV1 peptide, forming an antiparallel complex where the CaM N-lobe interacts with the TRPV1 peptide’s C-terminus and the C-lobe with the N-terminus. Each EF hand is in a different shade.
Ca2+ influx through TRPV1 is stimulated by noxious heat, pH and capsaicin, but prolonged exposure leads to Ca2+-dependent desensitization (Caterina et al., 1997). Understanding the molecular details of desensitization is a major focus of elucidating TRPV1 function. The TRPV1 C-terminus is important for desensitization and thought to mediate both protein and lipid interactions (Cao et al., 2013a; Nieto-Posadas et al., 2012; Numazaki et al., 2003; Prescott and Julius, 2003). Recently, the first high-resolution view of such an interaction was obtained through a crystal structure of Ca2+-CaM bound to a TRPV1 C-terminal peptide (Fig. 5; (Lau et al., 2012)). This region is missing from the high-resolution TRPV1 structure (Liao et al., 2013).
A 35-amino acid C-terminal TRPV1 fragment (residues 767–801 in rat) binds CaM in a Ca2+-dependent manner with high affinity (KD = 50 nM), and the fully Ca2+-loaded CaM winds itself around a 10-residue α-helix flanked by extended peptide regions in a 1:1 complex (Fig. 5). The complex is antiparallel: the CaM N-lobe interacts with the TRPV1 peptide’s C-terminus and the CaM C-lobe with the peptide’s N-terminus. Binding of CaM to the TRPV1 C-terminus follows the classic 1–10 CaM-binding motif organization (Rhoads and Friedberg, 1997) where two bulky hydrophobic residues at positions 1 and 10 are the main determinants of the CaM interaction.
Intriguingly, TRPV1 ARD also binds to CaM in a Ca2+-dependent manner (Lishko et al., 2007). It had been suggested that CaM may bridge the ARD and C-terminus and thus lock the channel in a ternary complex (Lishko et al., 2007), similar to what has been observed for SK channels and proposed for CaV1 channels (Dick et al., 2008; Schumacher et al., 2001). However, the CaM C-lobe dominates interactions with both the TRPV1 ARD and C-terminus. Accordingly, TRPV1 ARD, Ca2+-CaM and C-terminus do not form a ternary complex, at least not outside the context of the full-length channel (Lau et al., 2012). While mutation of the Ca2+-CaM binding site on TRPV1 ARD essentially abolished desensitization in response to capsaicin, mutation of the Ca2+-CaM binding site in the TRPV1C-terminus led to slower and reduced TRPV1 desensitization, supporting the idea that Ca2+-CaM binding to the two sites differentially regulates TRPV1 (Lau et al., 2012).
In contrast to CaM-mediated regulation, TRPP channel function is directly regulated by calcium through a C-terminal EF-hand motif that precedes the coiled coil (Fig. 2 and 3; (Petri et al., 2010)). Two groups have independently determined the NMR structure of the TRPP2 EF-hand motifs (Petri et al., 2010; Schumann et al., 2009). Schumann and colleagues used a 117-amino acid fragment, which formed a dimer that dissociated upon Ca2+ binding. In each subunit they find a pair of helix-loop-helix motifs assigned as a paired EF-hand motif that binds two Ca2+ with KDs in the 50–200 μM range, although only one of these sites was canonical (Schumann et al., 2009).
Petri and colleagues also find a pair of helix-loop-helix motifs, but only observe Ca2+-binding to one, and did not observe dimerization of their (40-amino acid shorter) construct (Fig. 3; (Petri et al., 2010)). However, the structures themselves are different, and Petri and colleagues raise concerns regarding the validity of the Schumann structure, which instead of the typical L-shape shows the two helices almost perfectly in line, thereby distorting the calcium coordination site. If the paired EF-hand motif indeed dimerizes, it begs the question of how this assembly fits to the observed trimeric TRPP2 coiled coil (Yu et al., 2009).
In summary, structural data on the interaction of TRP channel regions with Ca2+ or Ca2+-dependent regulators are starting to trickle in. While the available structures do not allow a complete view of how TRP channel activity is modulated by these interactions, they provide both tools and constraints for future experiments addressing the molecular mechanisms of Ca2+-mediated TRP channel regulation.
Homology models to understand TRP channel structure and function
Before a high-resolution 3D structure of a full-length TRP channel was available, considerable efforts were invested by several groups to generate homology models of TRP channels. Valuable lessons can be drawn from these approaches and readily applied to homology models based on the new TRPV1 structures and future TRP channel structures. Structural models can be generally divided into two groups – de novo, or template-free, models which rely on general physical properties of proteins; and template-based, or homology models. Template-based models rely on identifying a template structure through sequence similarity with a protein or protein fragment of known structure.
There have been a lot of developments in both types of modeling. The state of the field is assessed every two years through the Critical Assessment of Techniques for Protein Structure Prediction (CASP). The ninth contest, CASP9, in 2011, evaluated over 60,000 template-based models submitted (Mariani et al., 2011). Recent statistical analyses of homology models built using metaservers indicate that sequence identity of >30% between a target sequence and a template provides a very good chance of a model that is accurate enough for such things as site-directed mutagenesis plans (Gront et al., 2012). Sequence identity levels in the 20–30% range yield less accurate models (and the frequency of wrong models increase), whereas identity levels below 20% are generally too low to generate a useable homology model.
Sequence alignments can therefore be used to “thread” the sequence to be modeled onto a known homologous protein structure. One danger for large multidomain proteins such as TRP channels is that a full homology model requires both good templates for each domain to be modeled and untemplated modeling of the connections (either covalent or non-covalent contacts). One must therefore keep in mind that different parts of a homology model warrant different confidence assessments.
Additionally, information from other ion channels should be included in functional and structural assessments. For example the the structure of a GIRK channel bound to PIP2 (Fig. 1; (Whorton and MacKinnon, 2011)) can provide ideas to explore how TRP channels are regulated by phosphoinositides, although its sequence similarity to TRP channels may be too low to be a useful template for homology modeling in the absence of additional constraints.
Considering one of the published TRP channel homology models can serve to illustrate the current challenges. The full-length structure of TRPM8 has been modeled, using Kv2.2/2.1 chimera as a model for the TMD and importin as a model for the N-terminal TRPM homology region (Pedretti et al., 2009), although again the 18% sequence identity is below the thresholds described above. The final model contains more than 16% of residues within the disallowed regions of the Ramachandran plot (Pedretti et al., 2009) compared to less than 1% for typical crystal structures, and does not include a C-terminal coiled coil structure for which there is strong biochemical evidence (Tsuruda et al., 2006)). Finally, there is little experimental data on interdomain contacts in TRPM channels to constrain the relative orientation of each domain within the tetrameric assembly.
How can we improve the quality of TRP channel structural models? We need to use the best modeling tools available, but also generate more and better structural templates and accumulate as many other low- or high-resolution constraints such as knowledge of proximities from crosslinking or FRET studies, and volumes from EM reconstructions.
Conclusion – the way forward in TRP channel structural biology
Although there is continuous progress in TRP channel structural biology, we are still far from the holy grail of seeing the molecular details of a TRP channel in action. In the meantime, many pieces of the structural puzzle have not yet been solved. For example, while we know a lot about the structures of TRPV ARDs, we do not yet have structural insights into the ARDs of TRPC, TRPA and TRPN channels. Also, a number of TRP domains are unique to their subfamily, such as the N-terminal cytoplasmic TRPM homology region common to all TRPM channels, or the C-terminal Nudix domain of TRPM2. Similarly, structures of TRP channels or their fragments in complex with regulatory partners can provide some insights into the functional consequences of their binding.
It is clear that there are vast differences between TRP channel subfamilies, and therefore a need for vast amounts of structural data. While crystal structures will likely be an important component, single-particle electron microscopy techniques are improving and as recently seen, can attain a resolution that rival that of X-ray crystallography. NMR can be particularly useful to study questions of dynamics. Finally, techniques for building homology models are also improving, and the availability of scaffolds is rapidly increasing – although it is important to keep in mind that a homology model is simply a model, even when based on a closely related protein, and needs to be thoroughly validated and tested.
Structures of full-length TRP channel will lead to testable hypotheses for biochemical experiments to answer which residues bestow cation selectivity, how the pore is organized, which regions act as sensors or gates, and why certain mutations lead to channelopathies. However, it should be emphasized that a structure never tells the whole story. Ultimately, to completely understand how TRP channels function, we will need structures of at least one representative of each subfamily, as well as experimental evidence of the multiple conformations associated with different functional states to construct a movie of a TRP channel at work.
Acknowledgments
We thank members of the Gaudet lab, especially Christina Zimanyi, Sriram Srikant and Jeffrey MacArthur, for feedback and discussions on the manuscript. This work was supported by the National Institutes of Health (Grant R01 GM081340 to R.G.), and U.A.H. is the recipient of an EMBO Long-Term Fellowship.
References
- Abbott A. They said it couldn’t be done. Nature. 2002;418:268–269. doi: 10.1038/418268a. [DOI] [PubMed] [Google Scholar]
- Auer-Grumbach M, Olschewski A, Papic L, Kremer H, McEntagart ME, Uhrig S, Fischer C, Frohlich E, Balint Z, Tang B, et al. Alterations in the ankyrin domain of TRPV4 cause congenital distal SMA, scapuloperoneal SMA and HMSN2C. Nature genetics. 2010;42:160–164. doi: 10.1038/ng.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates-Withers C, Sah R, Clapham DE. TRPM7, the Mg(2+) inhibited channel and kinase. Advances in experimental medicine and biology. 2011;704:173–183. doi: 10.1007/978-94-007-0265-3_9. [DOI] [PubMed] [Google Scholar]
- Bill RM, Henderson PJ, Iwata S, Kunji ER, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Overcoming barriers to membrane protein structure determination. Nature biotechnology. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- Bycroft M, Bateman A, Clarke J, Hamill SJ, Sandford R, Thomas RL, Chothia C. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. The EMBO journal. 1999;18:297–305. doi: 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Cordero-Morales JF, Liu B, Qin F, Julius D. TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron. 2013a;77:667–679. doi: 10.1016/j.neuron.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013b;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Chang Q, Gyftogianni E, van de Graaf SF, Hoefs S, Weidema FA, Bindels RJ, Hoenderop JG. Molecular determinants in TRPV5 channel assembly. The Journal of biological chemistry. 2004;279:54304–54311. doi: 10.1074/jbc.M406222200. [DOI] [PubMed] [Google Scholar]
- Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1511–1515. doi: 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkov TL, Huynh KW, Cohen MR, Moiseenkova-Bell VY. Molecular architecture and subunit organization of TRPA1 ion channel revealed by electron microscopy. The Journal of biological chemistry. 2011;286:38168–38176. doi: 10.1074/jbc.M111.288993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Rosa V, Rangel-Yescas GE, Ladron-de-Guevara E, Rosenbaum T, Islas LD. Coarse architecture of the Transient Receptor Potential Vanilloid 1 (TRPV1) ion channel determined by Fluorescence Resonance Energy Transfer (FRET) The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.479618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nature reviews Neuroscience. 2005;6:850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- Demeuse P, Penner R, Fleig A. TRPM7 channel is regulated by magnesium nucleotides via its kinase domain. The Journal of general physiology. 2006;127:421–434. doi: 10.1085/jgp.200509410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai BN, Krapivinsky G, Navarro B, Krapivinsky L, Carter BC, Febvay S, Delling M, Penumaka A, Ramsey IS, Manasian Y, et al. Cleavage of TRPM7 releases the kinase domain from the ion channel and regulates its participation in Fas-induced apoptosis. Developmental cell. 2012;22:1149–1162. doi: 10.1016/j.devcel.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick IE, Tadross MR, Liang H, Tay LH, Yang W, Yue DT. A modular switch for spatial Ca2+ selectivity in the calmodulin regulation of CaV channels. Nature. 2008;451:830–834. doi: 10.1038/nature06529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Dumetz AC, Snellinger-O’brien AM, Kaler EW, Lenhoff AM. Patterns of protein protein interactions in salt solutions and implications for protein crystallization. Protein science: a publication of the Protein Society. 2007;16:1867–1877. doi: 10.1110/ps.072957907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- Erler I, Hirnet D, Wissenbach U, Flockerzi V, Niemeyer BA. Ca2+-selective transient receptor potential V channel architecture and function require a specific ankyrin repeat. The Journal of biological chemistry. 2004;279:34456–34463. doi: 10.1074/jbc.M404778200. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Minor DL., Jr X-ray crystal structure of a TRPM assembly domain reveals an antiparallel four-stranded coiled-coil. Journal of molecular biology. 2008;383:854–870. doi: 10.1016/j.jmb.2008.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Elias A, Mrkonjic S, Pardo-Pastor C, Inada H, Hellmich UA, Rubio-Moscardo F, Plata C, Gaudet R, Vicente R, Valverde MA. Phosphatidylinositol-4,5-biphosphate-dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9553–9558. doi: 10.1073/pnas.1220231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Molecular bioSystems. 2008a;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. TRP channels entering the structural era. The Journal of physiology. 2008b;586:3565–3575. doi: 10.1113/jphysiol.2008.155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. Divide and conquer: high resolution structural information on TRP channel fragments. The Journal of general physiology. 2009;133:231–237. doi: 10.1085/jgp.200810137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Current opinion in structural biology. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gront D, Grabowski M, Zimmerman MD, Raynor J, Tkaczuk KL, Minor W. Assessing the accuracy of template-based structure prediction metaservers by comparison with structural genomics structures. Journal of structural and functional genomics. 2012;13:213–225. doi: 10.1007/s10969-012-9146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmich UA, Gaudet R. High-resolution views of TRPV1 and their implications for the TRP channel superfamily. In: Nilius B, Flockerzi V, editors. Mammalian Transient Receptor Potential (TRP) cation channels. Heidelberg, Germany: Springer DE; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annual review of physical chemistry. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard RJ, Clark KA, Holton JM, Minor DL., Jr Structural insight into KCNQ (Kv7) channel assembly and channelopathy. Neuron. 2007;53:663–675. doi: 10.1016/j.neuron.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KW, Cohen MR, Chakrapani S, Holdaway HA, Stewart PL, Moiseenkova-Bell VY. Structural Insight into the Assembly of TRPV Channels. Structure. 2013 doi: 10.1016/j.str.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Hamamoto S, Miyanoiri Y, Takeda M, Kainosho M, Yabe I, Uozumi N, Yamashita A. Molecular bases of multimodal regulation of a fungal transient receptor potential (TRP) channel. The Journal of biological chemistry. 2013;288:15303–15317. doi: 10.1074/jbc.M112.434795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada H, Procko E, Sotomayor M, Gaudet R. Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry. 2012;51:6195–6206. doi: 10.1021/bi300279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Katano Y, Yamamoto K, Akiba M, Misaka T, Roberts RW, Asakura T, Matsunami H, Abe K. Interaction between PKD1L3 and PKD2L1 through their transmembrane domains is required for localization of PKD2L1 at taste pores in taste cells of circumvallate and foliate papillae. Faseb J. 2010;24:4058–4067. doi: 10.1096/fj.10-162925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- Jara-Oseguera A, Llorente I, Rosenbaum T, Islas LD. Properties of the inner pore region of TRPV1 channels revealed by block with quaternary ammoniums. The Journal of general physiology. 2008;132:547–562. doi: 10.1085/jgp.200810051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Touhey J, Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. The Journal of biological chemistry. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]
- Kobertz WR, Williams C, Miller C. Hanging gondola structure of the T1 domain in a voltage-gated K(+) channel. Biochemistry. 2000;39:10347–10352. doi: 10.1021/bi001292j. [DOI] [PubMed] [Google Scholar]
- Kol S, Braun C, Thiel G, Doyle DA, Sundstrom M, Gourdon P, Nissen P. Heterologous expression and purification of an active human TRPV3 ion channel. The FEBS journal. 2013 doi: 10.1111/febs.12520. [DOI] [PubMed] [Google Scholar]
- Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, Gibbons IR, Burgess SA, Sutoh K. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nature structural & molecular biology. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landoure G, Sullivan JM, Johnson JO, Munns CH, Shi Y, Diallo O, Gibbs JR, Gaudet R, Ludlow CL, Fischbeck KH, et al. Exome sequencing identifies a novel TRPV4 mutation in a CMT2C family. Neurology. 2012;79:192–194. doi: 10.1212/WNL.0b013e31825f04b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landoure G, Zdebik AA, Martinez TL, Burnett BG, Stanescu HC, Inada H, Shi Y, Taye AA, Kong L, Munns CH, et al. Mutations in TRPV4 cause Charcot-Marie-Tooth disease type 2C. Nature genetics. 2010;42:170–174. doi: 10.1038/ng.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M. Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature. 2006;440:959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- LaPlante JM, Falardeau JL, Brown EM, Slaugenhaupt SA, Vassilev PM. The cation channel mucolipin-1 is a bifunctional protein that facilitates membrane remodeling via its serine lipase domain. Experimental cell research. 2011;317:691–705. doi: 10.1016/j.yexcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SY, Procko E, Gaudet R. Distinct properties of Ca2+-calmodulin binding to N- and C-terminal regulatory regions of the TRPV1 channel. Journal of General Physiology. 2012;140:541–555. doi: 10.1085/jgp.201210810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage PK, Lussier MP, McDuff FO, Lavigne P, Boulay G. The self-association of two N-terminal interaction domains plays an important role in the tetramerization of TRPC4. Cell calcium. 2009;45:251–259. doi: 10.1016/j.ceca.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Lewit-Bentley A, Rety S. EF-hand calcium-binding proteins. Current opinion in structural biology. 2000;10:637–643. doi: 10.1016/s0959-440x(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Li A, Tian X, Sung SW, Somlo S. Identification of two novel polycystic kidney disease-1-like genes in human and mouse genomes. Genomics. 2003;81:596–608. doi: 10.1016/s0888-7543(03)00048-x. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Luft JR, Snell EH, Detitta GT. Lessons from high-throughput protein crystallization screening: 10 years of practical experience. Expert opinion on drug discovery. 2011;6:465–480. doi: 10.1517/17460441.2011.566857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):37–58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- Maruyama Y, Ogura T, Mio K, Kiyonaka S, Kato K, Mori Y, Sato C. Three-dimensional reconstruction using transmission electron microscopy reveals a swollen, bell-shaped structure of transient receptor potential melastatin type 2 cation channel. The Journal of biological chemistry. 2007;282:36961–36970. doi: 10.1074/jbc.M705694200. [DOI] [PubMed] [Google Scholar]
- McCleverty CJ, Koesema E, Patapoutian A, Lesley SA, Kreusch A. Crystal structure of the human TRPV2 channel ankyrin repeat domain. Protein science: a publication of the Protein Society. 2006;15:2201–2206. doi: 10.1110/ps.062357206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL., Jr The neurobiologist’s guide to structural biology: a primer on why macromolecular structure matters and how to evaluate structural data. Neuron. 2007;54:511–533. doi: 10.1016/j.neuron.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio K, Ogura T, Kiyonaka S, Hiroaki Y, Tanimura Y, Fujiyoshi Y, Mori Y, Sato C. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. Journal of molecular biology. 2007a;367:373–383. doi: 10.1016/j.jmb.2006.12.043. [DOI] [PubMed] [Google Scholar]
- Mio K, Ogura T, Kiyonaka S, Mori Y, Sato C. Subunit dissociation of TRPC3 ion channel under high-salt condition. Journal of electron microscopy. 2007b;56:111–117. doi: 10.1093/jmicro/dfm012. [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Wensel TG. Hot on the trail of TRP channel structure. The Journal of general physiology. 2009;133:239–244. doi: 10.1085/jgp.200810123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molland KL, Paul LN, Yernool DA. Crystal structure and characterization of coiled-coil domain of the transient receptor potential channel PKD2L1. Biochimica et biophysica acta. 2012;1824:413–421. doi: 10.1016/j.bbapap.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Moutevelis E, Woolfson DN. A periodic table of coiled-coil protein structures. Journal of molecular biology. 2009;385:726–732. doi: 10.1016/j.jmb.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Nieto-Posadas A, Picazo-Juarez G, Llorente I, Jara-Oseguera A, Morales-Lazaro S, Escalante-Alcalde D, Islas LD, Rosenbaum T. Lysophosphatidic acid directly activates TRPV1 through a C-terminal binding site. Nature chemical biology. 2012;8:78–85. doi: 10.1038/nchembio.712. [DOI] [PubMed] [Google Scholar]
- Nilius B, Owsianik G, Voets T. Transient receptor potential channels meet phosphoinositides. The EMBO journal. 2008;27:2809–2816. doi: 10.1038/emboj.2008.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B, Voets T. The puzzle of TRPV4 channelopathies. EMBO reports. 2013;14:152–163. doi: 10.1038/embor.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numazaki M, Tominaga T, Takeuchi K, Murayama N, Toyooka H, Tominaga M. Structural determinant of TRPV1 desensitization interacts with calmodulin. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8002–8006. doi: 10.1073/pnas.1337252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedretti A, Marconi C, Bettinelli I, Vistoli G. Comparative modeling of the quaternary structure for the human TRPM8 channel and analysis of its binding features. Biochimica et biophysica acta. 2009;1788:973–982. doi: 10.1016/j.bbamem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Persechini A, Moncrief ND, Kretsinger RH. The EF-hand family of calcium-modulated proteins. Trends in neurosciences. 1989;12:462–467. doi: 10.1016/0166-2236(89)90097-0. [DOI] [PubMed] [Google Scholar]
- Petri ET, Celic A, Kennedy SD, Ehrlich BE, Boggon TJ, Hodsdon ME. Structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9176–9181. doi: 10.1073/pnas.0912295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Huang RJ, Lishko PV, Wang RR, Gaudet R. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 2008;47:2476–2484. doi: 10.1021/bi702109w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Procko E, Lishko PV, Wang RR, Gaudet R. Insights into the roles of conserved and divergent residues in the ankyrin repeats of TRPV ion channels. Channels (Austin) 2007;1:148–151. doi: 10.4161/chan.4716. [DOI] [PubMed] [Google Scholar]
- Phelps CB, Wang RR, Choo SS, Gaudet R. Differential regulation of TRPV1, TRPV3, and TRPV4 sensitivity through a conserved binding site on the ankyrin repeat domain. The Journal of biological chemistry. 2010;285:731–740. doi: 10.1074/jbc.M109.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott ED, Julius D. A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science. 2003;300:1284–1288. doi: 10.1126/science.1083646. [DOI] [PubMed] [Google Scholar]
- Qamar S, Vadivelu M, Sandford R. TRP channels and kidney disease: lessons from polycystic kidney disease. Biochemical Society transactions. 2007;35:124–128. doi: 10.1042/BST0350124. [DOI] [PubMed] [Google Scholar]
- Qian F, Germino FJ, Cai Y, Zhang X, Somlo S, Germino GG. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nature genetics. 1997;16:179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. Faseb J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- Runnels LW, Yue L, Clapham DE. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science. 2001;291:1043–1047. doi: 10.1126/science.1058519. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG, Pavur KS, Dorovkov MV. Alpha-kinases: a new class of protein kinases with a novel catalytic domain. Current biology: CB. 1999;9:R43–45. doi: 10.1016/s0960-9822(99)80006-2. [DOI] [PubMed] [Google Scholar]
- Saxena K, Dutta A, Klein-Seetharaman J, Schwalbe H. Isotope labeling in insect cells. Methods Mol Biol. 2012;831:37–54. doi: 10.1007/978-1-61779-480-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C, Perraud AL, Johnson CO, Inabe K, Smith MK, Penner R, Kurosaki T, Fleig A, Scharenberg AM. Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7. Cell. 2003;114:191–200. doi: 10.1016/s0092-8674(03)00556-7. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- Schumann F, Hoffmeister H, Bader R, Schmidt M, Witzgall R, Kalbitzer HR. Ca2+-dependent conformational changes in a C-terminal cytosolic domain of polycystin-2. The Journal of biological chemistry. 2009;284:24372–24383. doi: 10.1074/jbc.M109.025635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends in biochemical sciences. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- Shi DJ, Ye S, Cao X, Zhang R, Wang K. Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function. Protein & cell. 2013;4:942–950. doi: 10.1007/s13238-013-3091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Sokabe T, Danev R, Tominaga M, Nagayama K. A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. The Journal of biological chemistry. 2010;285:11210–11218. doi: 10.1074/jbc.M109.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J, Zhou S, Piskorowski R, Nikai T, Lumpkin EA, Basbaum AI, King D, Julius D. Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature. 2006;444:208–212. doi: 10.1038/nature05285. [DOI] [PubMed] [Google Scholar]
- Tate CG. A crystal clear solution for determining G-protein-coupled receptor structures. Trends in biochemical sciences. 2012;37:343–352. doi: 10.1016/j.tibs.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Tsiokas L, Kim E, Arnould T, Sukhatme VP, Walz G. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruda PR, Julius D, Minor DL., Jr Coiled coils direct assembly of a cold-activated TRP channel. Neuron. 2006;51:201–212. doi: 10.1016/j.neuron.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Choy WY, Orekhov VY, Kay LE. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:622–627. doi: 10.1073/pnas.0407792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter SW, Leclerc E. Novel aspects of calmodulin target recognition and activation. European journal of biochemistry/FEBS. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Munro RA, Shi L, Kawamura I, Okitsu T, Wada A, Kim SY, Jung KH, Brown LS, Ladizhansky V. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nature methods. 2013;10:1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wider G. Structure determination of biological macromolecules in solution using nuclear magnetic resonance spectroscopy. BioTechniques. 2000;29:1278–1282. 1284–1290. doi: 10.2144/00296ra01. 1292 passim. [DOI] [PubMed] [Google Scholar]
- Wiener R, Haitin Y, Shamgar L, Fernandez-Alonso MC, Martos A, Chomsky-Hecht O, Rivas G, Attali B, Hirsch JA. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. The Journal of biological chemistry. 2008;283:5815–5830. doi: 10.1074/jbc.M707541200. [DOI] [PubMed] [Google Scholar]
- Xu Q, Minor DL., Jr Crystal structure of a trimeric form of the K(V)7.1 (KCNQ1) A-domain tail coiled-coil reveals structural plasticity and context dependent changes in a putative coiled-coil trimerization motif. Protein science: a publication of the Protein Society. 2009;18:2100–2114. doi: 10.1002/pro.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Molecular cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Science’s STKE: signal transduction knowledge environment. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ulbrich MH, Li MH, Buraei Z, Chen XZ, Ong AC, Tong L, Isacoff EY, Yang J. Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11558–11563. doi: 10.1073/pnas.0903684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ulbrich MH, Li MH, Dobbins S, Zhang WK, Tong L, Isacoff EY, Yang J. Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex. Nature communications. 2012;3:1252. doi: 10.1038/ncomms2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharian E, Cao C, Rohacs T. Gating of transient receptor potential melastatin 8 (TRPM8) channels activated by cold and chemical agonists in planar lipid bilayers. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:12526–12534. doi: 10.1523/JNEUROSCI.3189-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Protein structure determination by solid-state NMR. Topics in current chemistry. 2012;326:187–213. doi: 10.1007/128_2011_287. [DOI] [PubMed] [Google Scholar]
- Zhou ZH. Atomic resolution cryo electron microscopy of macromolecular complexes. Advances in protein chemistry and structural biology. 2011;82:1–35. doi: 10.1016/B978-0-12-386507-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yu Y, Ulbrich MH, Li MH, Isacoff EY, Honig B, Yang J. Structural model of the TRPP2/PKD1 C-terminal coiled-coil complex produced by a combined computational and experimental approach. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10133–10138. doi: 10.1073/pnas.1017669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu MX. Multiple roles of calmodulin and other Ca(2+)-binding proteins in the functional regulation of TRP channels. Pflugers Archiv: European journal of physiology. 2005;451:105–115. doi: 10.1007/s00424-005-1427-1. [DOI] [PubMed] [Google Scholar]
- Zimon M, Baets J, Auer-Grumbach M, Berciano J, Garcia A, Lopez-Laso E, Merlini L, Hilton-Jones D, McEntagart M, Crosby AH, et al. Dominant mutations in the cation channel gene transient receptor potential vanilloid 4 cause an unusual spectrum of neuropathies. Brain: a journal of neurology. 2010;133:1798–1809. doi: 10.1093/brain/awq109. [DOI] [PMC free article] [PubMed] [Google Scholar]