The eosinophilic gastrointestinal disorders (EGIDs) include eosinophilic esophagitis (EoE), eosinophilic gastritis (EG), eosinophilic gastroenteritis (EGE), and eosinophilic colitis (EC). Data on EG, EGE, and EC are relatively sparse,1–4 primarily because the conditions are rare.5 Consensus has not been achieved in the diagnosis of EGIDs other than EoE. Few studies have characterized normal levels of eosinophils present in the GI tract,6, 7 histologic features specific to EGIDs are under-studied,7, 8 and the optimal number and location of biopsies needed to establish the diagnosis remain uncertain. We aimed to determine the number and location of biopsies obtained by gastroenterologists (GIs) attempting to diagnose EG, EGE, and EC.
We used an online tool (Qualtrics, Provo, UT) to conduct a survey assessing biopsy practices among expert GIs in the Consortium for Eosinophilic GI Diseases Researchers (CEGIR), a multi-center research cooperation in the NIH-funded Rare Diseases Clinical Research Network. We also collected information on years in practice after completing fellowship training and whether the GI primarily treated adults or children. This study was exempt from ongoing review by the University of North Carolina Institutional Review Board.
Overall, 18 of 20 GIs in CEGIR responded (90%). There were 11 (61%) pediatric GIs and 7 (39%) adult GIs. The mean practice experience was 17 years, with 7 (61%) having ≤10 years of experience. Most GIs (83%) obtained gastric and duodenal biopsies in patients with suspected EoE, regardless of the endoscopic appearance, but this was more common for pediatric than adult GIs (100% vs 57%; p=0.04). All providers obtained gastric and duodenal biopsies if visible endoscopic abnormalities were noted.
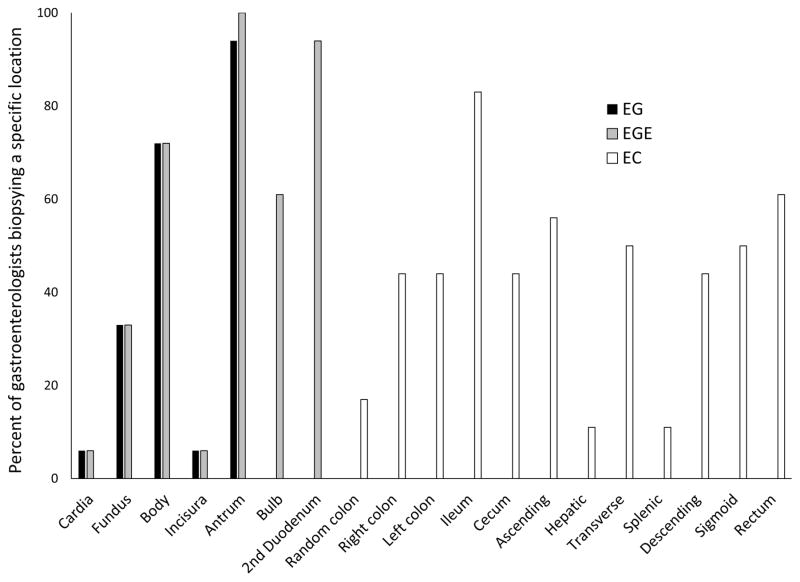
For suspected EG and EGE, biopsies were taken in the gastric body, antrum, and second portion of the duodenum by most GIs, but there was considerable variation by location (Figure 1). The number of biopsy locations throughout the GI tract also varied substantially, ranging from 1–4 for EG, 2–6 for EGE, and 1–10 for EC. Similarly, the number of biopsy fragments obtained varied from 2–13 for EG, 4–18 for EGE, and 4–22 for EC. The median number of fragments obtained was lower for pediatric vs adult GIs for EG (6 vs 11; p=0.03) and EGE (8 vs 14; p=0.005), but not for EC (14 vs 16; p=0.89). Level of practice experience did not explain this variability, with similar median numbers of biopsies for GIs with ≤10 vs >10 years in practice for EG (6 vs 6, p=0.75), EGE (10 vs 10; p=0.89), and EC (12 vs 16 (p=0.41).
Figure 1.
Variability in locations where biopsies are obtained for patients with suspected EGIDs. Specific locations in the GI tract are listed on the x-axis, with the percent of gastroenterologists biopsying at each location noted on the y-axis.
Finally, there was variability in specimen handling. For suspected EG, 67% of GIs placed specimens into one pathology jar, regardless of the gastric anatomic location. For EGE, most GIs (83%) used multiple pathology jars, but there was no standard practice for dividing specimens. For EC, there was a more consistent approach with one jar per colonic location (87%), but the specific colonic locations varied.
Our observations underscore the need for a more consistent approach to diagnosis of these conditions.9 We found that there is substantial variation in the specific anatomic locations that were biopsied, the number of locations that were biopsied, and the number of biopsy fragments that were obtained, regardless of whether EG, EGE, or EC was suspected. Interestingly, some of this variability was explained by whether a GI was an adult or pediatric provider.
For a set of medical conditions lacking diagnostic guidelines, this level of variability is not surprising. Similar variability was noted before the first publication of EoE guidelines, and the variability began to diminish after the guidelines were disseminated.10 Among contemporary EG and EGE case series, there is also notable variation in the approach to diagnosis, not only in the number or locations of biopsies (which is not typically reported), but also in the histologic criteria for diagnosis.2, 3
Our study represents an opportunity for best practices to begin to be developed. For example, most providers took multiple biopsy fragments from multiple anatomic locations, suggesting that such a practice is an important general principle. Some respondents separated samples from different anatomic locations in the upper GI tract. Such a practice is likely to be important since EGIDs are patchy and the distribution may have implications for diagnosis and treatment, though there is an added cost to using multiple specimen jars. Future studies will need to examine the diagnostic yield by the number of biopsies obtained, akin to what was originally done in EoE,11 as well as the optimal sampling locations, to inform diagnostic criteria.
In conclusion, we found substantial variability in the number and location of biopsies obtained in patients with suspected EG, EGE, and EC, even among highly experienced GIs. These results underscore the need for formal diagnostic guidelines for EGIDs that must be informed by additional studies of the correlation between biopsy practice, biopsy findings, and clinical metrics.
Acknowledgments
Grant support: This work was supported in part by U54AI117804 (CEGIR), which is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the Office of Rare Disease Research (ORDR), NCATS, and is funded through collaboration between NIAID, NIDDK, and NCATS, as well as K24DK100303 (GTF) and R01DK101856 (ESD).
Footnotes
Disclosures: There are no potential conflicts of interest for any of the authors pertaining to this study.
Author contributions (all approved final draft):
Dellon: Project conception/design; data collection; data analysis/interpretation; drafting of the article; critical revision
Collins: Project conception/design; data analysis/interpretation; critical revision
Bonis: Data interpretation; critical revision
Capocelli: Data interpretation; critical revision
Dohil: Data interpretation; critical revision
Falk: Data interpretation; critical revision
Furuta: Data interpretation; critical revision
Gupta: Data interpretation; critical revision
Hirano: Data interpretation; critical revision
Hiremath: Data interpretation; critical revision
Kagalwall: Data interpretation; critical revision
Leung: Data interpretation; critical revision
Liacouras: Data interpretation; critical revision
Menard-Katcher: Data interpretation; critical revision
Muir: Data interpretation; critical revision
Mukkada: Data interpretation; critical revision
Putnam: Data interpretation; critical revision
Schoepfer: Data interpretation; critical revision
Straumann: Data interpretation; critical revision
Wershil: Data interpretation; critical revision
Wu: Data interpretation; critical revision
Yang: Data interpretation; critical revision
Rothenberg: Data interpretation; critical revision
Gonsalves: Project conception/design; data analysis/interpretation; critical revision
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Talley NJ, et al. Gut. 1990;31(1):54–8. doi: 10.1136/gut.31.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko HM, et al. Am J Gastroenterol. 2014;109(8):1277–85. doi: 10.1038/ajg.2014.166. Epub 2014/06/25. [DOI] [PubMed] [Google Scholar]
- 3.Reed C, et al. Dig Liver Dis. 2015;47(3):197–201. doi: 10.1016/j.dld.2014.11.009. Epub 2014/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell JM, et al. J Allergy Clin Immunol. 2014;134(5):1114–24. doi: 10.1016/j.jaci.2014.07.026. Epub 2014/09/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen ET, et al. J Pediatr Gastroenterol Nutr. 2016;62:36–42. doi: 10.1097/MPG.0000000000000865. Epub 2015/05/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBrosse CW, et al. Pediatr Dev Pathol. 2006;9(3):210–8. doi: 10.2350/11-05-0130.1. Epub 2006/09/02. [DOI] [PubMed] [Google Scholar]
- 7.Hurrell JM, et al. Adv Anat Pathol. 2011;18(5):335–48. doi: 10.1097/PAP.0b013e318229bfe2. Epub 2011/08/16. [DOI] [PubMed] [Google Scholar]
- 8.Collins MH. Gastroenterol Clin North Am. 2014;43(2):257–68. doi: 10.1016/j.gtc.2014.02.007. Epub 2014/05/13. [DOI] [PubMed] [Google Scholar]
- 9.Bochner BS, et al. J Allergy Clin Immunol. 2012;130(3):587–96. doi: 10.1016/j.jaci.2012.07.024. Epub 2012/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperry SL, et al. Am J Gastroenterol. 2011;106(5):824–32. doi: 10.1038/ajg.2011.10. quiz 33. Epub 2011/02/10. [DOI] [PubMed] [Google Scholar]
- 11.Gonsalves N, et al. Gastrointest Endosc. 2006;64(3):313–9. doi: 10.1016/j.gie.2006.04.037. [DOI] [PubMed] [Google Scholar]