Abstract
Objectives
Although antiretroviral therapy (ART) effectively suppresses HIV-1 replication, it does not eradicate the virus and ART interruption consistently results in rebound of viraemia, demonstrating the persistence of a long-lived viral reservoir. Several approaches aimed at reducing virus persistence are being developed, and accurate measurements of the latent reservoir (LR) are necessary to assess the effectiveness of anti-latency interventions. We sought to measure the LR in SIV/SHIV-infected rhesus macaques (RMs) by quantifying integrated SIV-DNA.
Methods
We optimised a repetitive sampling Alu-gag PCR to quantify integrated SIV-DNA ex vivo in ART-naïve and ART-experienced SIV/SHIV-infected RMs.
Results
In ART-naïve RMs, we found the median level of integrated SIV-DNA to be 1660 copies and 866 copies per million PBMC during untreated acute and chronic SHIV infection, respectively. Integrated and total SIV-DNA levels were positively correlated with one another. In ART-treated RMs, integrated SIV-DNA was readily detected in lymph nodes and spleen and levels of total (3319 copies/million cells) and integrated (3160 copies/million cells) SIV-DNA were similar after a median of 404 days of ART. In peripheral blood CD4+ T cells from ART-treated RMs, levels of total (3319 copies/million cells) and integrated (2742 copies/million cells) SIV-DNA were not significantly different and were positively correlated.
Conclusions
The assay described here is validated and can be used in interventional studies testing HIV/SIV cure strategies in RMs. Measurement of integrated SIV-DNA in ART-treated RMs, along with other reservoir analyses, gives an estimate of the size of the LR.
Keywords: SIV, integration, reservoir, latency, rhesus macaques
Introduction
The major obstacle to HIV-1 eradication is a viral reservoir of latently infected cells that persists despite long-term antiretroviral therapy (ART) and causes a rebound of viraemia if treatment is interrupted [1]. This persistent HIV-1 reservoir predominantly comprises a pool of resting memory CD4+ T cells that carry stably integrated, replication competent proviruses and, for the most part, do not produce virus particles [2–5]. As major efforts are underway to develop novel therapeutic approaches aimed at reducing or eliminating the HIV-1 latent reservoir, accurate measurement of this reservoir has become a key priority in contemporary AIDS research.
SIV infection of non-human primates, such as rhesus macaques (RMs), has been utilised for over two decades as an in vivo model for studies of HIV-1 pathogenesis, prevention, and treatment [6–10]. More recently, the RM model of SIV or SHIV infection has been developed and validated for studies of HIV-1 cure as a result of the availability of antiretroviral regimens that suppress plasma viraemia to levels comparable to those in HIV-1-infected patients on ART [11–15]. As such, this animal model enables in vivo evaluation of novel therapeutic strategies that may involve high risk and/or combination interventions that would be difficult to directly test in HIV-1-infected humans. In addition, the SIV/RM model allows for more extensive tissue sampling, including tissues obtained through elective necropsy, thus enabling a more comprehensive anatomical and histological characterisation of the persistent SIV/SHIV reservoir in vivo. For these reasons, the development of assays that accurately measure the latent reservoir (LR) in this non-human primate model of HIV-1 infection is a high priority for HIV/SIV cure research.
Assays to quantify HIV/SIV reservoirs are rapidly evolving. There is currently great interest in identifying simple methods that do not rely on high cell frequencies and complex experimental systems to measure virus persistence and the impact of candidate cure strategies on virus reservoirs. In this setting, a repetitive-sampling Alu-gag PCR assay has been used widely to measure integrated HIV-1 DNA levels. This assay quantifies integrated proviral reservoir including both replication-competent and replication-defective viruses, in HIV-1-infected individuals on ART [16–19]. The current gold standard for measuring replication-competent virus in resting CD4+ T cells from ART-treated, HIV-1-infected patients is the quantitative viral outgrowth assay (or QVOA) [20,21], but this approach requires a large number of cells and is time- and resource-consuming. Results of the integrated HIV-1 DNA assay were strongly positively correlated with those of the QVOA [22], meaning that, while an overestimate, integrated HIV-1 DNA may be helpful as a surrogate measure of virus persistence, particularly when cell numbers available for study are limited. The Alu-gag PCR assay measures integrated HIV-1 DNA by performing a nested PCR, in which the first PCR uses an HIV-1 primer in combination with a specific primer for Alu, the most abundant human short repeat element; and a second PCR that quantifies the provirus. This report describes the development and characterisation of a similar method to quantify integrated SIV-DNA in SIV/SHIV-infected RMs. We used this assay to quantify integrated SIV-DNA levels in 10 RMs during the acute and chronic phases of infection to establish a baseline level. We also quantified total and integrated SIV-DNA in CD4+ T cells from peripheral blood and lymphoid cells from 13 ART-treated RMs and found that these viral forms were positively correlated at each site and that the on-ART total and integrated SIV-DNA levels were not significantly different.
Methods
Animals and samples
We used samples from 10 SHIV-infected untreated Indian rhesus macaques (Macaca mulatta) involved in an ineffective vaccine study (unpublished). All macaques were challenged intrarectally with at least one dose of 10,000 TCID50 SHIVSF162p3. PBMC and plasma samples were collected at regular time points and frozen in liquid nitrogen and at −80°C. Two time points were analysed in this study, one during the early phase of infection (day 10 to 21) and one during the chronic phase of infection (day 126 to 364). We also obtained PBMC and lymphoid tissues from RMs involved in different ART studies. The animals and their treatments are summarised in Table 1. Cells were isolated from lymphoid tissues [23] and frozen in liquid nitrogen until use.
Table 1.
| Animals | Duration of infection (days) | Sample | Virus | Duration on ART (days) | Viral load (RNA copies/mL) | ART regimen |
|---|---|---|---|---|---|---|
| RM1 | 10 | PBMC | SHIVSF162p3 | n/a | 3.80E+05 | n/a |
| 126 | PBMC | SHIVSF162p3 | n/a | 2.11E+05 | n/a | |
| RM2 | 14 | PBMC | SHIVSF162p3 | n/a | 3.78E+05 | n/a |
| 84 | PBMC | SHIVSF162p3 | n/a | 7.80E+04 | n/a | |
| RM3 | 14 | PBMC | SHIVSF162p3 | n/a | 9.20E+07 | n/a |
| 364 | PBMC | SHIVSF162p3 | n/a | 6.21E+05 | n/a | |
| RM4 | 14 | PBMC | SHIVSF162p3 | n/a | 3.76E+07 | n/a |
| 84 | PBMC | SHIVSF162p3 | n/a | 1.83E+05 | n/a | |
| RM5 | 21 | PBMC | SHIVSF162p3 | n/a | 1.54E+06 | n/a |
| 84 | PBMC | SHIVSF162p3 | n/a | 4.41E+02 | n/a | |
| RM6 | 14 | PBMC | SHIVSF162p3 | n/a | 4.85E+06 | n/a |
| 126 | PBMC | SHIVSF162p3 | n/a | 9.47E+05 | n/a | |
| RM7 | 14 | PBMC | SHIVSF162p3 | n/a | 1.63E+06 | n/a |
| 84 | PBMC | SHIVSF162p3 | n/a | 5.20E+03 | n/a | |
| RM8 | 14 | PBMC | SHIVSF162p3 | n/a | 5.37E+06 | n/a |
| 126 | PBMC | SHIVSF162p3 | n/a | 1.37E+05 | n/a | |
| RM9 | 14 | PBMC | SHIVSF162p3 | n/a | 1.55E+06 | n/a |
| 210 | PBMC | SHIVSF162p3 | n/a | 2.99E+05 | n/a | |
| RM10 | 14 | PBMC | SHIVSF162p3 | n/a | 6.62E+05 | n/a |
| 308 | PBMC | SHIVSF162p3 | n/a | 1.00E+02 | n/a | |
| RM13 | 473 | ileac LN | SIVmac251 | 421 | <60 | PMPA-FTC-RAL-DRV/r-MVC |
| RM14 | 456 | ileac LN | SIVmac251 | 404 | <60 | PMPA-FTC-RAL-DRV/r-MVC |
| 456 | colonic LN | SIVmac251 | 404 | <60 | PMPA-FTC-RAL-DRV/r-MVC | |
| RM15 | 477 | ileac lLN | SIVmac251 | 425 | <60 | PMPA-FTC-RAL-DRV/r-MVC |
| 477 | colonic LN | SIVmac251 | 425 | <60 | PMPA-FTC-RAL-DRV/r-MVC | |
| RM16 | 241 | superficial LN | SIVmac251 | 183 | <60 | PMPA-FTC-RAL-DRV/r |
| 241 | spleen | SIVmac251 | 183 | <60 | PMPA-FTC-RAL-DRV/r | |
| RM17 | 261 | mesenteric LN | SIVmac251 | 203 | <60 | PMPA-FTC-RAL-DRV/r |
| 261 | spleen | SIVmac251 | 203 | <60 | PMPA-FTC-RAL-DRV/r | |
| RM18 | 210 | PBMC | SIVmac251 | 144 | <60 | PMPA-FTC-ZDV-LPV/r |
| RM19 | 210 | PBMC | SIVmac251 | 144 | <60 | PMPA-FTC-ZDV-LPV/r |
| RM20 | 210 | PBMC | SIVmac251 | 144 | 7.59E+02 | PMPA-FTC-ZDV-LPV/r |
| RM21 | 210 | PBMC | SIVmac251 | 140 | <60 | PMPA-FTC-ZDV-LPV/r |
| RM22 | 210 | PBMC | SIVmac251 | 140 | 1.07E+02 | PMPA-FTC-ZDV-LPV/r |
| RM23 | 210 | PBMC | SIVmac251 | 144 | <60 | PMPA-FTC-ZDV-LPV/r |
| RM24 | 210 | PBMC | SIVmac251 | 140 | <60 | PMPA-FTC-ZDV-LPV/r |
| RM25 | 210 | PBMC | SIVmac251 | 144 | <60 | PMPA-FTC-ZDV-LPV/r |
PMPA: tenofovir; FTC: emtricitabine; RAL: raltegravir; DRV/r: darunavir/ritonavir; MVC: maraviroc; LPV/r: lopinavir/ritonavir; ZDV: zidovudine.
Plasma SIV RNA quantification
Plasma viral quantification was performed by RT-PCR as described previously [24].
Preparation of a polyclonal integration standard
The strategy used to prepare the polyclonal integration standard was previous described in detail [16,19]. 293T cells were transfected with pSIVmac239Δenv-hygromycin-GFP and pVSV-G plasmids using standard CaPO4 transfection protocol. Supernatants were harvested 48 hours after transfection. RM B-LCL (B lymphoblastoid cells) were infected with the viral supernatant by spinoculation. The cells were infected at a low multiplicity of infection to minimise the chance of multiple transductions within one cell. Briefly, 16×106 B-LCL were mixed with 1 mL of viral stock (VSV-G pseudotyped SIVmac239Δenv-hygromycin-GFP) diluted 1:4 in 10% FBS-RPMI 1640 medium and placed into one well of a flat-bottom six-well tissue culture plate. The plates were centrifuged at 1200g for 2 hours at 25oC. Cells were then maintained in culture for 1 week at 2×106 cells/mL. After hygromycin selection, transduced B-LCL were sorted for GFP expression. Sorted cells were cultured for a further 3 weeks and were again sorted for expression of GFP before DNA extraction. We measured the level of SIV-DNA per cell using quantitative PCR and found ∼1 copy of SIV per cell.
DNA extraction
Genomic DNA was extracted from B-LCL, PBMC or lymphoid tissue cells using the Qiagen Blood and cell culture maxi, midi or mini kit following the manufacturer instructions.
Cell number estimation
The concentrations of RM genomes in the isolated sample DNA were determined by measuring the number of albumin copies by quantitative real-time PCR assay using Invitrogen Platinum Taq DNA polymerase. Two dilutions of the DNA samples were amplified in duplicate wells of a 96-well plate. A standard curve was made using purified DNA from uninfected RM PBMC. The sequences of the primers and probe used were: Forward primer: 5′-TGC ATG AGA AAA CGC CAG TAA-3′; Reverse primer: 5′-ATG GTC GCC TGT TCA CCA A-3′; probe 5′-6-FAM-AGA AAG TCA CCA AAT GCT GCA CGG AAT C-36-TAMSp.
Amplification of integrated SIV-DNA
The amount of integrated SIV-DNA was quantified using a repetitive sampling two-step PCR assay adapted from the method of O’Doherty et al. [19]. The first amplification, targeting a region between SIVgag and the nearest Alu sequence, was performed in 42 replicates on 10,000 cell equivalent per reaction. Simultaneously, 42 replicates of gag-only primer amplification reactions were performed as a control to account for background signals from unintegrated SIV-DNA. The sequences of the first step amplification primers were: Alu Forward: 5′-TCT GCG TCA TCT GGT GCA TTC ACG-3′ and gag Reverse: 5′-TGC CAA CAG GCT CAG AAA ATT-3′. The reactions were carried out in 50 μL containing 1.5 mM MgCl2, 0.21 mM of mixed dNTPs, 100 nM Alu Forward primer and/or 600 nM gag Reverse primer and 5 U/reaction of Platinum Taq DNA polymerase. The thermal cycler was programmed to performed a 2-min hot start at 95oC, followed by 20 cycles of denaturation at 95oC for 15s, annealing at 50oC for 15s and extension at 72oC for 3 min and 30s.
The second-round real-time PCR specific for the SIV LTR was performed using 10 μL of the pre-amplification product diluted 1/8. The sequence of the primers and probes were: Forward 5′-ACG GCT GAG TGA AGG CAG TAA-3′, Reverse 5′-GAC CCG CGC CTT TAT AGG A-3′, Probe: 5′-FAM-CGG CAG GAA CCA ACC ACG ACG-3′-TAMRA. Reactions were carried out in a volume of 20 μL containing 5.5 mM MgCl2, 0.3 mM of mixed dNTPs, 260 nM of primers, 200 nM of probe and 2U of Platinum Taq DNA polymerase. The reactions were performed on a 7500 fast real-time PCR system (Applied Biosystems) with the following thermal program: 20-s hot start at 95oC, followed by 50 cycles of denaturation at 95oC for 3 s and annealing at 59oC for 30 s.
Analysis of integration levels
To generate the standard curve, the integration standard was diluted in PBMC DNA from SIV-negative RMs to obtain samples containing known numbers of integrated SIV-DNA. Dilutions ranging from 1 provirus to 100 proviruses diluted in 10,000 genomes equivalent per reaction were assessed using the two-step PCR assay described above. A student's t-test with a 95% confidence interval was performed to determine if cycle thresholds (Ct) derived from the Alu-gag PCR were significantly lower than those from the gag-only reactions (P<0.1). A standard curve was then generated by plotting the natural log of the average Ct versus the natural log of the number of SIV-DNA copies in order to calculate a regression equation. This standard curve was used to determine the level of integrated SIV-DNA in RM samples based on the average Ct value of the Alu-gag reactions.
Total SIV-DNA quantification
PCR amplifications targeting the LTR region of SIV were performed using the same primers and probe used in the second step of the integration reaction in a single round of 50 cycles. DNA samples were diluted to contain 40,000 cells per well for analysis in triplicate wells of a 96-well plate. Ct values were inserted into a regression line equation obtained from a standard curve from the same reaction using dilutions with known copy numbers of a SIVmac239 full-length plasmid.
Statistical analyses
Quantitative variables were compared using parametric t-tests. Correlations between quantitative variables were estimated by calculating Pearson's rank correlation coefficient. All tests were two-tailed and statistical significance was set at P<0.05. Analyses were performed using GraphPad Prism v4.0.
Results
Generation of a standard curve using an integrated SIV-DNA standard
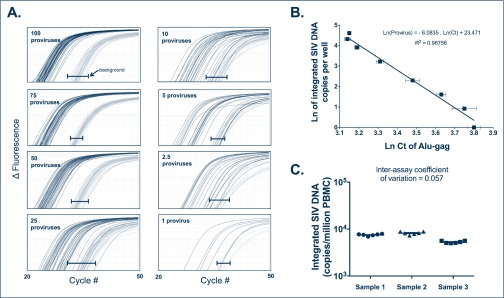
To quantify integrated SIV-DNA levels, peripheral blood mononuclear cells (PBMC) from SIV-infected RMs with unknown levels of SIV integration were compared to a standard containing diverse integration sites to accurately reflect the diversity of integration sites that occur in vivo. A polyclonal integration standard cell line (IS) containing one integration event per cell (not shown) was generated using a strategy similar to that previously described for HIV-1 and detailed here in the Methods. To measure SIV integration events, we used an Alu-PCR assay adapted from the method of O’Doherty and colleagues involving two amplifications and repetitive sampling [16–19]. The first amplification step targets integrated SIV-DNA specifically by amplifying the region between SIVgag and the nearest Alu repeat element of the simian genome. Alu-gag signals are compared to the signals generated using only the gag primer, which approximates the signal expected from unintegrated DNA (Figure 1A). Forty-two replicates of each reaction were assessed to increase the sensitivity of the assay and to compensate for integration specific bias (related to the variable distance between each unique integration site and Alu element) [16]. The nested real-time PCR amplification recognises the R-U5 region of the SIV long terminal repeat. To generate a standard curve, the IS was serially diluted in DNA extracted from uninfected PBMC at 10,000 cell equivalent per reaction in order to obtain samples containing known numbers of integrated SIV-DNA copies. The two-step PCR assay described above was performed for each dilution and detected 1 provirus in 10,000 cells (10% of the time), very similar to the previously described assay [16]. In our hands, the sensitivity of the assay depends on the number of genomes assayed. We robustly detect 1 provirus in 10,000 cells by assaying 10,000 cells per well x 42 wells as shown in Figure 1. As previously reported, a linear relationship was observed between the natural logarithm of the average Alu-gag cycle threshold (Ct) and the natural logarithm of the proviral number: Ln (provirus)=−6.0835 Ln (Ct)+23.471 with r2=0.96756 (Figure 1B), with 1 to 100 proviruses in 10,000 cell equivalents included in the linear range.
Figure 1.
Integration standard curve. The polyclonal integration standard DNA was diluted in uninfected PBMC DNA to a constant amount of 10,000 cell equivalents per reaction. Seven dilutions ranging from 1 to 100 copies per reaction were subjected to our two-step Alu-PCR. (A) The first PCR reaction used primers targeting Alu and gag or only primers to gag. The nested PCR reaction used SIV-specific primers binding to the LTR elements, R and U5. For each dilution sample, Alu-gag and gag-only (background) amplification was measured 42 times. At low proviral copy number, integration is detected at a low frequency as demonstrated by the Alu-gag amplification. For clarity, we did not show the gag-only signals at the lower dilutions, but instead show the bracketed black lines (|—|), which represent where the gag-only signal (background) was detected at each proviral number. (B) The natural log (Ln) of integrated SIV-DNA copy number is plotted on the y-axis against the Ln of the cycle threshold (Ct) of Alu-gag amplification on the x-axis. Each point represents the average Ln(Ct) for 42 replicates and the error bars represent the standard deviation. (C) SIV integration measurements were performed in six independent experiments on PBMC samples from three untreated RM to calculate the inter-assay coefficient of variation of the assay. Horizontal bars show means and standard deviations
Using PBMC samples obtained from three SIV-infected RMs, we determined the reproducibility of the integration assay. We measured the level of integrated SIV-DNA with six independent experiments performed for each of the three RMs (a total of >250 wells per sample). Using untreated RM PBMC samples, the inter-assay coefficient of variation of the SIV Alu-gag integration assay was 0.057 (Figure 1C), confirming its robustness. These results show that the basic principles of the HIV-Alu nested PCR assay to detect HIV-1 integration can be successfully applied to quantify the levels of SIV integration in RM samples.
Excess of unintegrated SIV-DNA is present in SHIV-infected RMs in the absence of ART
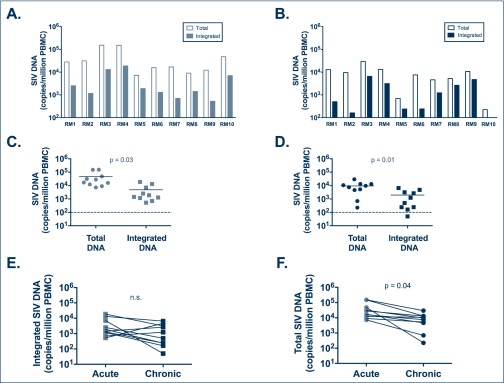
The level of integrated SIV-DNA was determined in the PBMC of 10 SHIV-infected ART-naïve RMs and compared to the level of total SIV-DNA in the same samples (as measured by SIVgag RT-PCR) [24]. PBMC were collected from the same animals during the acute and chronic stages of SHIV infection. Table 1 shows the characteristics of RMs used in this study. Integrated SIV-DNA was detected in 10/10 RMs during the acute phase of infection (Figure 2A) and in 9/10 RMs during the chronic phase of infection (Figure 2B). Of note, the RM for whom integrated SIV-DNA was below the limit of detection was a natural SHIV controller by week 8 post-infection and maintained plasma viral load under 200 copies/mL throughout the study in the absence of ART. Interestingly, the level of integrated SIV-DNA was approximately one log lower than the level of total SIV-DNA; with a median of 1660 copies of integrated SIV-DNA versus 22,500 copies of total SIV-DNA per million PBMC during the acute phase of infection (P=0.03; Figure 2C) and a median of 866 copies of integrated SIV-DNA versus 8580 copies of total SIV-DNA per million PBMC during the chronic phase of infection (P=0.01; Figure 2D). These results suggest an excess of unintegrated SIV-DNA (i.e. 49–96% of the total cell-associated SIV-DNA) as compared to integrated SIV-DNA for all RMs in the absence of ART, as previously reported for HIV-1-infected patients [25,26]. The level of SIV-DNA in CD4+ T cells was also estimated based on the frequency of CD4+ T cells in PBMC determined by flow cytometry and the frequency of infection determined by PCR. The estimated level of integrated SIV-DNA in CD4+ T cells was approximately one log lower than the level of total SIV-DNA during the acute phase of infection; with a median of 15,300 copies of integrated SIV-DNA versus 161,500 copies of total SIV-DNA per million CD4+ T cells (P=0.04; data not shown). When normalised to CD4+ T cell frequencies, the levels of integrated and total SIV-DNA were a median of 9980 copies versus 176,500 copies of SIV-DNA per million CD4+ T cells during the chronic phase of infection (P=n.s.; data not shown).
Figure 2.
Total and integrated SIV-DNA levels in untreated SHIV-infected RMs. The Alu-gag PCR was used to quantify integrated SIV-DNA as compared to total SIV-DNA measured by standard gag RT-PCR in 10 RM. PBMC during the acute phase (A,C) and the chronic phase (B,D) of untreated SIV infection. Levels of integrated (E) and total (F) SIV-DNA were also compared between acute and chronic infection. In (C) and (D) lines are drawn at the median
Frequency of cells harbouring integrated SIV-DNA is similar during the acute and chronic phases of untreated SHIV infection
We next compared the levels of integrated SIV-DNA as determined by the Alu-gag assay in individual animals sampled during the acute and chronic stages of infection. In untreated RMs, there was no significant difference in the level of integrated SIV-DNA between the acute and chronic stages of infection (Figure 2E). By contrast, the level of total SIV-DNA was significantly higher in all RMs during the acute as compared to chronic phase of infection (P=0.04; Figure 2F). However, when normalised to CD4+ T cell frequencies, the levels of total SIV-DNA were not significantly different between acute and chronic phases of infection (data not shown).
Integrated SIV-DNA is positively associated with total SIV-DNA
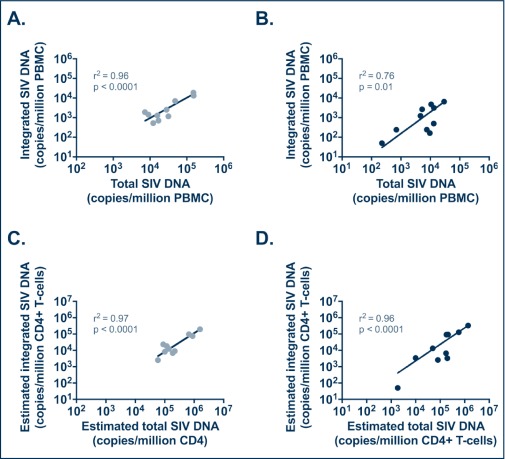
To better understand the relationship between standard virological and immunological assessments in SIV/SHIV-infected RMs and the results of our integrated SIV-DNA assay, we investigated potential correlations between these measures in animals sampled during the acute and chronic stages of infection. Levels of total and integrated SIV-DNA in PBMC were positively correlated during both the acute (r2=0.96, P<0.0001) and chronic (r2=0.76, P=0.01) phases of SIV infection (Figure 3A,B). The same correlations were observed between the estimated levels of total and integrated SIV-DNA in CD4+ T cells during the acute phase (r2=0.97, P<0.0001) and chronic phase (r2=0.96, P<0.0001) of SIV infection (Figure 3C,D). During the chronic phase of infection, integrated SIV-DNA in PBMC was also negatively correlated with the peripheral CD4 absolute count (r2=−0.67, P=0.04, data not shown) and showed a trend towards a positive correlation with the plasma SIV-RNA level (r2=0.61, P=0.07, data not shown).
Figure 3.
Correlations between total and integrated SIV-DNA levels in PBMC and in CD4+ T cells from untreated SHIV-infected RMs. Levels of total and integrated SIV-DNA were not significantly different (A,C) but were positively correlated (B,D). Estimated level of total and integrated SIV-DNA in CD4+ T cells, calculated based on PBMC frequency of infection determined by PCR and the frequency of CD4+ T cells in PBMC determined by flow cytometry, were correlated during both the acute (C) and chronic (D) phases of infection
Use of the integrated SIV-DNA assay to quantify the frequency of latently infected cells in lymphoid tissues and peripheral blood in ART-treated RMs
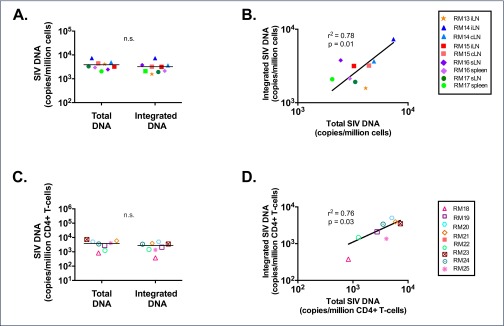
To further validate our assay, levels of total and integrated SIV-DNA were quantified in cells isolated from multiple lymph nodes and the spleen of five ART-treated RMs with plasma viral load <60 copies/mL (the limit of detection of our standard assay). These RMs were all started on ART during the chronic phase of SIV infection and had been treated for a median of 404 days (range 183–425 days) at the time of tissue collection (Table 1). Integrated SIV-DNA was readily detected in all lymphoid compartments tested, demonstrating the persistence of integrated proviruses in the setting of undetectable viraemia in this model. The median frequency of cells harbouring integrated SIV-DNA in lymph nodes and spleen was 3160 per million cells. This value is higher than levels of integrated HIV-1 reported in PBMC or resting CD4+ T cells isolated from the peripheral blood of HIV-1-infected ART-suppressed patients, but similar to total HIV-DNA in CD4+ T cells from gut-associated lymphoid tissue [22,27], reflecting the critical contribution of sites other than peripheral blood to virus persistence. Total SIV-DNA was not significantly different from the frequency of integrated SIV-DNA in ART-treated RMs (Figure 4A), as has been reported for HIV-1-infected patients treated with ART during chronic infection [22].
Figure 4.
Total and integrated SIV-DNA levels in ART-treated SIV-infected RMs. Integrated and total SIV-DNA was measured in lymphoid tissues (A,B) or peripheral CD4+ T cells (C,D) of ART-treated RMs. Levels of integrated and total SIV-DNA were not significantly different (A,C) but were positively correlated (B,C). In (A) and (C) lines are drawn at the median. iLN: ileac lymph node; cLN: colonic lymph node; sLN: superficial lymph node (axillary or inguinal)
Levels of total and integrated SIV-DNA were also quantified in peripheral CD4+ T cells from an additional eight ART-treated RMs. These RMs were all started on ART during the chronic phase of SIV infection and had been treated for a median of 142.5 days (range 140–144 days) at the time of blood collection (Table 1). Integrated SIV-DNA was detected in the peripheral CD4+ T cells of 8/8 RMs. There was no significant difference in the level of integrated SIV-DNA and total SIV-DNA (median of 2742 and 3786 copies per million CD4+ T cells, respectively, Figure 4C). In both cohorts of ART-treated RMs, total and integrated SIV-DNA levels were positively correlated in the lymphoid tissues (r2=0.78, P=0.01; Figure 4B) and in the peripheral blood (r2=0.76, P=0.03; Figure 4D).
Discussion
In this report, we describe the development of an assay quantifying integrated SIV-DNA and its validation using ART-naïve and ART-experienced SIV-infected RM samples. Our assay detects SIV integrated DNA by repetitive sampling Alu-gag PCR. One important feature of our assay is the use of a polyclonal integration standard containing proviruses integrated at different distances from the nearest Alu element, providing a correction factor for the variable efficiency of the of the Alu-gag PCR based on the proximity of the provirus to the Alu repeats in each infected cell. Our integration assay requires a total of 840,000 cell equivalents and can detect down to one provirus. The overall assay including DNA extraction, albumin quantification and quantification of total and integrated SIV-DNA levels can be performed in 2 days. As described previously for HIV [17,28], the limit of detection of the assay can be lowered by increasing the number of genomes per reaction but also by applying a different analysis method, notably by counting the percentage of positive Alu-gag signals after using our repetitive sampling Alu-gag PCR integration assay and quantifying integrated DNA using Poisson statistics. This type of statistical analysis can be readily applied to the assay described here and will be particularly important for studies of RMs on long-term ART and/or rare reservoirs that contain very low levels of viral DNA.
To validate our assay, levels of integrated and total SIV-DNA in ART-naïve animals were measured in both acute and chronic SIV infection. Using untreated RM PBMC samples, the inter-assay coefficient of variation of the SIV Alu-gag integration assay was 0.057; however, this value may differ slightly if samples with lower numbers of integrants are used. As reported in untreated HIV infection, total and integrated SIV-DNA levels in PBMC were positively correlated and the median total SIV-DNA level was significantly higher than the integrated SIV-DNA levels, suggesting the presence of excess unintegrated SIV-DNA forms in ART-naive RMs. Furthermore, in ART-treated RMs, this assay was successfully used to quantify cells harbouring integrated SIV-DNA isolated from peripheral blood, lymph nodes and spleen. During therapy, the median total and integrated SIV-DNA levels did not differ significantly, which presumably reflects the preferential clearance of non-integrated SIV-DNA, as has been reported before [25] and further validates our quantification assays.
We determined that a median of 3160 lymphoid cells per million contain integrated SIV-DNA in RMs on ART for a median of 404 days. Similar levels of integrated SIV-DNA, with a median of 2743 integrated SIV-DNA copies/million CD4+ T cells, were found in peripheral blood in an independent cohort of RMs treated with ART for approximately one-third of this time period (median 142 days) and despite CD4+ T cell enrichment. In line with this result, previous reports in ART-treated RMs and humans showed higher levels of total viral DNA in lymphoid and mucosal tissues as compared to peripheral blood [29–31]. Our results thus confirm in RMs the critical contribution of sites other than peripheral blood to virus persistence.
A limitation of this work is that samples were not available to compare the results of integrated SIV-DNA to SIV QVOA or the more recently described Tat/rev induced limiting dilution assay (or TILDA). In HIV-1-infected patients both the QVOA and TILDA assays positively correlated with frequency of integrated HIV-DNA in PBMC [22,27]. Variations of these assays for the SIV system are in use or in development in our and others’ laboratories and should be employed as desired depending on the question to be addressed in a particular study.
In vivo investigational studies in non-human primates offer a number of important advantages to the HIV cure field. Key amongst these include the ability to control for the timing, dose and route of infection, guaranteed ART adherence, and testing of potentially risky interventions. The integrated SIV-DNA assay described in this report can be added to the armamentarium of evaluations used to measure reservoirs ex vivo from SIV-infected, ART-treated RMs.
Acknowledgements
We thank Rama Amara and Mirko Paiardini for providing samples for this study. This work was partially supported by the amfAR Innovation Grant Program 109353-59-RGRL to MM. AC acknowledges funding from the National Center for Advancing Translational Sciences of the National Institutes of Health under Award UL1TR000454 and Support of the Yerkes National Primate Research Center (P51 OD011132). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Chun TW, Justement JS, Murray D et al. . Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS 2010; 24: 2803– 2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chun TW, Carruth L, Finzi D et al. . Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387: 183– 188. [DOI] [PubMed] [Google Scholar]
- 3. Finzi D, Blankson J, Siliciano JD et al. . Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999; 5: 512– 517. [DOI] [PubMed] [Google Scholar]
- 4. Joos B, Fischer M, Kuster H et al. . HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A 2008; 105: 16725– 16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong JK, Hezareh M, Gunthard HF et al. . Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278: 1291– 1295. [DOI] [PubMed] [Google Scholar]
- 6. Brenchley JM, Paiardini M.. Immunodeficiency lentiviral infections in natural and non-natural hosts. Blood 2011; 118: 847– 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chahroudi A, Bosinger SE, Vanderford TH et al. . Natural SIV hosts: showing AIDS the door. Science 2012; 335: 1188– 1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haase AT. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med 2011; 62: 127– 139. [DOI] [PubMed] [Google Scholar]
- 9. Pandrea IV, Gautam R, Ribeiro RM et al. . Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol 2007; 179: 3035– 3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valentine LE, Watkins DI.. Relevance of studying T cell responses in SIV-infected rhesus macaques. Trends Microbiol 2008; 16: 605– 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Del Prete GQ, Smedley J, Macallister R et al. . Short communication: comparative evaluation of coformulated injectable combination antiretroviral therapy regimens in simian immunodeficiency virus-infected rhesus macaques. AIDS Res Hum Retroviruses 2016; 32: 163– 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mavigner M, Watkins B, Lawson B et al. . Persistence of virus reservoirs in ART-treated SHIV-infected rhesus macaques after autologous hematopoietic stem cell transplant. PLoS Pathog 2014; 10: e1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. North TW, Van Rompay KK, Higgins J et al. . Suppression of virus load by highly active antiretroviral therapy in rhesus macaques infected with a recombinant simian immunodeficiency virus containing reverse transcriptase from human immunodeficiency virus type 1. J Virol 2005; 79: 7349– 7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shytaj IL, Norelli S, Chirullo B et al. . A highly intensified ART regimen induces long-term viral suppression and restriction of the viral reservoir in a simian AIDS model. PLoS Pathog 2012; 8: e1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitney JB, Hill AL, Sanisetty S et al. . Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014; 512: 74– 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agosto LM, Yu JJ, Dai J et al. . HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 2007; 368: 60– 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graf EH, Mexas AM, Yu JJ et al. . Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog 2011; 7: e1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liszewski MK, Yu JJ, O’Doherty U.. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 2009; 47: 254– 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Doherty U, Swiggard WJ, Jeyakumar D et al. . A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol 2002; 76: 10942– 10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laird GM, Rosenbloom DI, Lai J et al. . measuring the frequency of latent HIV-1 in resting CD4(+) T cells using a limiting dilution coculture assay. Methods Mol Biol 2016; 1354: 239– 253. [DOI] [PubMed] [Google Scholar]
- 21. Siliciano JD, Siliciano RF.. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 2005; 304: 3– 15. [DOI] [PubMed] [Google Scholar]
- 22. Eriksson S, Graf EH, Dahl V et al. . Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013; 9: e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chowdhury A, Del Rio Estrada PM, Tharp GK et al. . Decreased T follicular regulatory cell/T follicular helper cell (TFH) in simian immunodeficiency virus-infected rhesus macaques may contribute to accumulation of TFH in chronic infection. J Immunol 2015; 195: 3237– 3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chahroudi A, Cartwright E, Lee ST et al. . Target cell availability, rather than breast milk factors, dictates mother-to-infant transmission of SIV in sooty mangabeys and rhesus macaques. PLoS Pathog 2014; 10: e1003958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koelsch KK, Liu L, Haubrich R et al. . Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis 2008; 197: 411– 419. [DOI] [PubMed] [Google Scholar]
- 26. Yu JJ, Wu TL, Liszewski MK et al. . A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 2008; 379: 78– 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Procopio FA, Fromentin R, Kulpa DA et al. . A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2015; 2: 872– 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Spiegelaere W, Malatinkova E, Lynch L et al. . Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of poisson statistics. Clin Chem 2014; 60: 886– 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kline C, Ndjomou J, Franks T et al. . Persistence of viral reservoirs in multiple tissues after antiretroviral therapy suppression in a macaque RT-SHIV model. PLoS One 2013; 8: e84275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. North TW, Higgins J, Deere JD et al. . Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol 2010; 84: 2913– 2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yukl SA, Gianella S, Sinclair E et al. . Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis 2010; 202: 1553– 1561. [DOI] [PMC free article] [PubMed] [Google Scholar]