Abstract
Mitochondrion is an organelle critically responsible for energy production and intracellular signaling in eukaryotic cells and its dysfunction often accompanies and contributes to human disease. Superoxide is the primary reactive oxygen species (ROS) produced in mitochondria. In vivo detection of superoxide has been a challenge in biomedical research. Here we describe the methods used to characterize a circularly permuted yellow fluorescent protein (cpYFP) as a biosensor for mitochondrial superoxide and pH dynamics. In vitro characterization reveals the high selectivity of cpYFP to superoxide over other ROS species and its dual sensitivity to pH. Confocal and two-photon imaging in conjunction with transgenic expression of the biosensor cpYFP to the mitochondrial matrix detects mitochondrial flash events in living cells, perfused intact hearts, and live animals. The mitochondrial flashes are discrete and stochastic single mitochondrial events triggered by transient mitochondrial permeability transition (tMPT) and composed of a bursting superoxide signal and a transient alkalization signal. The real-time monitoring of single mitochondrial flashes provides a unique tool to study the integrated dynamism of mitochondrial respiration, ROS production, pH regulation and tMPT kinetics under diverse physiological and pathophysiological conditions.
Keywords: Reactive oxygen species (ROS), Mitochondria, Mitochondrial flash, Biosensor, In vivo imaging
1. Introduction
Reactive oxygen species (ROS) are a group of oxygen containing molecules and free radicals that regulate diverse physiological processes in the cell [1, 2]. ROS are also critical contributors to cell pathology and death [3, 4]. In most cell types, the mitochondrial electron transport chain (ETC) is the primary site for ROS production, where one electron reduction of an oxygen molecule produces superoxide [5, 6]. Superoxide anions are quickly transformed to other more stable ROS, such as hydrogen peroxide (H2O2) and hydroxyl radicals [7]. Until recently, in vivo and real time determination of intracellular superoxide production remains unsatisfactory largely due to the lack of specific and reversible superoxide indicators that can be targeted into sub-cellular compartments. For example, current methods for ROS detection, including chemiluminescence, electron paramagnetic resonance (EPR) spin trap and small molecule fluorescent indicators, are either non-selective among ROS species or incapable of in situ measurement in living cells [8, 9]. Only in the last few years, genetic fluorescent indicators based on green fluorescent protein (GFP) have begun to be developed for the detection of redox state or ROS, such as the circular permuted yellow fluorescent protein (cpYFP) for superoxide [10], the roGFP for redox [11, 12], and the HyPer for H2O2 [13, 14].
The cpYFP has been targeted to mitochondria (mt-cpYFP) and detected transient and quantal single mitochondrial events, named mitochondrial flashes or mitoflashes, in living cells and live animals [10, 15–18]. A mitochondrial flash is composed of multifaceted signals including a superoxide or ROS signal and an alkalization signal [19]. Mechanistically, mitochondrial flashes are triggered by transient openings of the mitochondrial permeability transition pore (mPTP) and depend on electron flows along the ETC [10, 16, 17, 19, 20]. A number of reports have used mt-cpYFP and other ROS indicators to confirm the existence of transient mitochondrial flash-like events. Collectively, these studies suggest that mitochondrial flashes have important physiological and pathophysiological significances, since flash frequency is closely associated with muscle contraction [16, 17, 21], muscle development [22], cell differentiation [23], neuron development and degeneration [24–27], aging [15], and wound healing [28]. Therefore, real-time monitoring of mitochondrial flashes in living cells, organs and animals provides a unique means for studying the role of integrated mitochondrial functions in health and disease, and represent a significant advance in mitochondrial biomedicine.
Monitoring mitochondrial flash events by using mt-cpYFP and other ROS and pH indicators has been widely applied in biomedical research. Here, we provide detailed methodology for imaging of mitochondrial flashes under physiologically relevant conditions, such as in living cells, intact organs (Langendorff perfused heart), and live animals (skeletal muscles) [10, 16, 29, 30]. This in vivo imaging technique has similarities to methods for measuring other signals such as ROS, pH, and calcium (Ca2+), with their respective fluorescent indicators [11–13, 31–33]. In an effort to reconcile the discrepancy on cpYFP characterization regarding its superoxide sensitivity [10, 34, 35], here we provide a detailed protocol that was used for determining the superoxide and pH sensitivity of cpYFP in vitro.
2. Materials and methods
2.1. Prokaryotic expression and purification of cpYFP protein
The mt-cpYFP expression plasmid (provided by Dr. Robert T. Dirksen, University of Rochester, Rochester, NY) was constructed from mitochondrial targeted ratiometric pericam [36] cloned into pcDNA3 by removing nucleotide sequences encoding calmodulin (886–1323) and M13 (49–126) using the gene splicing by overlap extension (SOE) technique [37]. mt-cpYFP contains the initiation methionine codon followed by 804 nucleotides coding for the mitochondrial matrix targeting sequence (LSLRQSIRFFK), a linker sequence (RSGI) and the 253 amino acid cpYFP sequence (Supplemental Fig 1). The coding region for cpYFP was PCR amplified to add Xho I (5’) and EcoR I (3’) restriction sites and digested by standard molecular biology techniques [38]. The cpYFP was then cloned into pRSET C plasmid (Invitrogen; Carlsbad, CA) between Xho I and EcoR I sites downstream of the 6 × Histidine sequence and transferred into E. coli DH5α cells (Invitrogen) by standard methods [38]. The DH5α cells were plated on Lysogeny broth (LB) agarose plates containing ampicillin (50 µg/mL) and incubated overnight at 37°C. Single colonies were selected and grew overnight in LB with ampicillin. The plasmid DNA was then isolated and checked for the presence of the insert and for the correct orientation by using restriction enzyme digestions and DNA sequencing.
The pRSET-cpYFP plasmid was transformed into BL21(DE3)pLysS E. coli cells (Invitrogen) using standard molecular biology methods [38]. Cells were plated on LB plates containing ampicillin and incubated overnight at 37°C. Single colonies were selected and grew overnight at 37°C in LB containing ampicillin. One liter of LB containing ampicillin was inoculated with aliquots (5 mL) of the overnight cultures and allowed to grow overnight to a density of ~109 cells/mL. Isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.4 mM) was added to induce protein expression for 2–3 h. Cells were harvested by centrifugation (6000g, 10 min). Phenylmethane sulfonyl fluoride (PMSF, 0.5 mM) was added in cell lysis buffer to inhibit proteolysis.
Purification of cpYFP proteins used cobalt affinity chromatography (BD TALON purification kit, BD 635515, BD Biosciences; San Jose, CA). Imidazole was removed from the eluent by using a PD 10 desalting column (GE Healthcare; Piscataway, NJ). Bradford method and SDS-PAGE analysis were used to determine the quantity and purity of cpYFP protein. Two milligrams of cpYFP protein (>80% purity) were obtained from 1 liter cell culture and purified proteins were suspended in protein storage buffer (20 mM HEPES, pH 7.9) at a concentration of 1 mg/ml. Aliquots of purified protein samples were stored at −80°C.
2.2. Protein preparation, fluorescence spectroscopy and redox titration
Unless otherwise stated, all the solutions used for in vitro fluorescence measurement were treated by at least 3 cycles of degassing/evacuation and nitrogen bubbling. All the samples and solutions were prepared and stored in a GloveBag (Sigma; Saint Louis, MO) inflated with nitrogen gas to ensure minimal sample oxidation during the process. All the experimental procedures were carried out at room temperature (20–23°C). To fully reduce cpYFP, the concentrated protein (20 µM) was incubated with 10 mM dithiothreitol (DTT) for more than 3 hr. The reduced protein sample was loaded on a Centri Spin 20 column (Princeton Separations; Freehold, NJ) to remove DTT and diluted to the final concentration (1 µM) in testing buffer (125 mM KCl, 75 mM HEPES, and 1 mM EDTA, pH 8.0). A final volume of 120 µL and a final concentration of cpYFP at 1 µM were used for all fluorescence spectroscopy measurements. Aliquots of the fully reduced cpYFP samples were incubated with aldrithiol (1 mM, Sigma) in aerobic environment for 2 hr to achieve fully oxidation of the protein. For fluorescence spectra scanning, the samples were placed in a sealed quartz cuvette in a spectrofluorimeter (Model: CM1T10I, HORIBA Jobin Yvon; Japan). The cuvette holding chamber was also filled with nitrogen gas and the sample was constantly mixed by a magnetic stirrer during the measurement. Four spectra scans were carried out for each sample:
Emission scan (500–600 nm, 5 nm bandwidth), excitation at 488 nm;
Emission scan (420–600 nm, 5 nm bandwidth), excitation at 405 nm;
Excitation scan (350–505 nm, 1 nm bandwidth), collect emission at 515 nm;
Excitation scan (350–520 nm, 1 nm bandwidth), collect emission at 530 nm.
For each experiment, the testing buffer without protein was measured first to provide background fluorescence, then the fully reduced cpYFP and the fully oxidized cpYFP samples were tested. To determine the redox sensitivity of cpYFP, solutions with different redox potentials (−7.5 to −319 mV) were prepared by mixing reduced DTT with oxidized DTT at different ratios (total DTT concentration remains constant at 10 mM). The exact redox potential was determined by a redox sensitive electrode (Orion 91-80, Thermo Fisher Scientific; Waltham, MA). The pH of these solutions was maintained at 8.0. To do the redox titration, fully reduced cpYFP (after DTT removal) was incubated with the solution of various redox potentials for 1–2 hr before the spectra measurements.
2.3. In vitro characterization of cpYFP sensitivity
To determine the responsiveness of cpYFP to superoxide, fully reduced cpYFP was first incubated with testing buffer saturated with oxygen, then xanthine (2 mM, Sigma) and xanthine oxidase (20 mU, Sigma) were added to produce superoxide. Cu/Zn-superoxide dismutase (SOD, 600 U/ml, Sigma) was added subsequently to remove superoxide. After each addition, the sample was quickly mixed by shaking the cuvette and the fluorescence spectra measured immediately. Fluorescence emission (peak at 515 nm) by 488 nm excitation increased ~2 folds upon oxygen incubation, further increased ~2 folds with superoxide production, and returned to the level of oxygen incubation after SOD addition [10]. In another experiment, fully reduced cpYFP was first incubated with oxygen saturated solution and SOD, then xanthine and xanthine oxidase were added. In the presence of SOD, no fluorescence increase was detected at 488 nm excitation.
Fully reduced cpYFP protein samples were incubated with different ROS species including H2O2 (0.1 and 10 mM) and hydroxyl radical (produced by Fenton reaction: 1 mM H2O2 plus 0.1 mM FeSO4 in the anaerobic environment) and reactive nitrogen species (RNS) including nitric oxide produced by 1 mM 3-morpholinosydnonimine (SIN-1, EMD Chemicals; Gibbstown, NJ) in anaerobic environment and peroxynitrite produced by 1 mM SIN-1 in aerobic environment. Fluorescence measurements were carried out immediately after mixing the ROS and RNS species with cpYFP. To determine the pH response of cpYFP, native cpYFP protein was incubated with air saturated testing buffer at different pH (7, 8, 9 and 10) for 1 hr before the spectra measurements. Fully reduced cpYFP protein was also incubated with other biologically important small molecules and metabolites including Ca2+ (100 nM – 5 mM), ADP (1 mM), ATP (10 mM), NAD+ (10 mM), NADH (1 mM), NADP+ (10 mM) and NADPH (1 mM) and the fluorescence spectra measured [10].
2.4. Detecting mitochondrial flashes in living cells
Adenovirus containing mt-cpYFP was produced by standard molecular biology approach. All the animal procedures used in this study were approved by Internal Review Board of the Institutional Animal Care and Use Committee (IACUC) at the University of Washington. Animals were maintained on rodent diet with water available ad libitum and in a vivarium with a 12 hr light/dark cycle at 22°C. Enzymatically isolated adult rat ventricular myocytes in primary culture were infected with adenovirus carrying the mt-cpYFP coding sequence at a multiplicity of infection of 100 and cultured for 2 to 3 days following previously reported methods [39]. Similar infection conditions were used when expressing mt-cpYFP in other cell types [27].
For live cell imaging, coverslip containing mt-cpYFP expressing cells was transferred to form a chamber on the stage of a confocal laser scanning microscope (LSM510, Carl Zeiss; Germany). The chamber was filled with cell perfusion solution (137 mM NaCl, 4.9 mM KCl, 1 mM CaCl2, 1.2 mM MgSO4, 1.2 mM NaH2PO4, 15 mM glucose and 20 mM HEPES, pH 7.4) at room temperature (20–23°C) throughout confocal imaging process. To verify the expression level and location of mt-cpYFP, two dimensional frame scanning images were taken with a 40 ×, 1.3NA oil immersion objective. The cells were illuminated with 488 nm and 405 nm in sequential, and emission collected at >505 nm.
To monitor mitochondrial flashes real time, dual wavelength and time-lapse frame scanning confocal images were taken at the resolution of 1024 pixels. The actual detecting area was adjusted to contain single cardiomyocyte (rectangle shaped with the long axis set at 1024 pixels) to minimize laser exposure and scanning duration. The laser intensities were set at 1–5% for 488 nm and 5–10% for 405 nm excitation (for the Zeiss LSM 510 confocal, the maximal power of 405 or 488 nm laser was 25 or 30 mW, respectively) and both emissions collected at >505 nm. In the ‘Scan Control’ dialogue box of the confocal scanning software (Zeiss LSM 510, version 3.2, Carl Zeiss, Germany), the ‘Pinhole’ was set at ~200 (which resulted in a pinhole of ~1 airy unit and an optical slice of ~1 µm) and ‘Detection Gain’ at ~600 volts. For adult cardiomyocytes, each frame scan was completed within ~300–400 ms. An interval of 300–700 ms was set in between each frame scan to avoid excessive laser illumination and associated cell damage. A serial scan of 100–200 frames usually took 100–200 seconds (~1 second per frame).
To monitor mitochondrial membrane potential and mitochondrial flash simultaneously, mt-cpYFP expressing cells were loaded with tetramethyl rhodamine methyl ester (TMRM, 20, nM, Invitrogen), a mitochondrial membrane potential indicator, for 20 minutes. The same low concentration of TMRM was always present in the cell perfusion solution to keep the intracellular TMRM level constant. Driven by the negative membrane potential across the inner mitochondrial membrane, TMRM preferentially accumulated in mitochondria matrix, where its fluorescence signal co-localized with that of mt-cpYFP. Tri-wavelength excitation imaging of mtcpYFP and TMRM was done by using “Multi-track Scanning” function of the confocal software. The three tracks were set as (1) 405 nm excitation with emission collected at 500–550 nm, (2) 488 nm excitation with emission collected at 500–550 nm, and (3) 543 nm excitation with emission collected at >560 nm. The three tracks were applied in tandem and switched after each line scan (1024 pixels) to achieve concurrent tri-wavelength imaging.
2.5. Detecting mitochondrial flashes in perfused hearts
The mt-cpYFP transgenic mouse was generated on C57BL/6 background (Charles River) as previously described [16, 29]. The mouse was heparinized (100 U heparin) and euthanized with pentobarbital (150 mg/kg, i. p.). The heart was quickly removed, cannulated via aorta, and perfused in the Langendorff mode using a custom designed perfusion system that allows mounting of the heart onto a chamber on the confocal microscope stage [29]. The heart was perfused with physiological solutions containing 118 mM NaCl, 25 mM NaHCO3, 5.3 mM KCl, 2 mM CaCl2, 1.2 mM MgSO4, 0.5 mM EDTA, and equilibrated with 95% O2 and 5% CO2 (pH 7.4) at 37°C. In addition, metabolic substrates were added to provide glucose only buffer (10 mM glucose, 0.5 mM pyruvate) or mixed substrate buffer (5.5 mM glucose, 0.4 mM mixed long-chain fatty acids (palmitic acid 56.7%, palmitoleic acid 11.7%, stearic acid 1.9%, oleic acid 17.1%, linoleic acid 10.8%, and linolenic acid 1.7%) bound with 1.2% albumin, 1.2 mM lactate, 1.3 mM ketone and 50 µU/mL insulin) [20]. Mitochondrial membrane potential indicator, TMRM (100 nM) was included in the perfusion solution. Blebbistatin (10 µM, Toronto Research Chemicals) was added in the perfusion solution to suppress (but not completely stop) the heartbeat and gentle pressure was applied by using a custom designed apparatus during image acquisition to further prevent motion artifact. About 1 ml of physiological solution was added in the chamber to partially submerge the heart during imaging. Excessive effluent was removed from the chamber by a peristaltic pump. The confocal imaging of perfused heart followed the procedure described above for live cell imaging and was reported previously [10, 20, 29]. The focal plan was carefully adjusted until clear mitochondrial pattern can be revealed in the intact myocardium.
2.6. Detecting mitochondrial flashes in skeletal muscles in vivo
The mt-cpYFP transgenic mouse was anesthetized with pentobarbital (80 mg/kg, i. p.). Hairs on one hindlimb were removed and the skin sterilized with 70% ethanol. An incision was made on the skin along the outer side of the limb to expose the gastrocnemius muscles. An incision was made through the epimysium and the muscle fibers were exposed. The surface of the muscle was immersed in isotonic balanced salt solution containing: 140 mM NaCl, 5 mM KCl, 2.5 mM CaCl2, 2 mM MgCl2 and 10 mM HEPES (pH 7.2). TMRM (500 nM) was also included in the isotonic balanced salt solution and 30 min was allowed for the loading of TMRM before imaging. The mouse was put on its side on the confocal microscope stage and the hindlimb restrained in a position so that the exposed skeletal muscle was facing against the coverslip, which formed the bottom of the chamber. The coverslip was in between the muscle tissue and the inverted objective (40 ×, oil). The leg was pressed down gently by using a custom designed apparatus to make tight contact with the coverslip. Recording of two dimensional confocal images followed the same procedure outlined above for live cell and perfused heart imaging [10, 16, 29].
2.7. Image processing, data analysis and statistics
Initial image processing used the physiological module of LSM510 software (Zeiss LSM 510, version 3.2, Carl Zeiss). Parameters obtained from each serial frame scanning images included the duration of scan, cell size, background fluorescence (no cell area), whole cell fluorescence, and fluorescence of region of Interest (ROI) at both 405 and 488 nm excitations. For quantitative analysis of mitochondrial flashes, we used ROI function of the LSM510 software to identify the size, location and number of mitochondrial flashes in each serial scan. Individual flash was manually detected. When determining whether a fluorescence change (increase) was a flash, we used the following criteria: the fluorescence change was from single mitochondrion, detected only at 488 nm but not 405 nm excitation, can be clearly differentiated from basal fluctuations (noise) or motion artifact, and exhibited a transient time course (e.g., ~5 seconds upstroke and ~10 seconds decay). The time point at which each flash occurred during a scan was also recorded. For comparison purposes, the frequency of mitochondrial flash was defined as the number of flashes within the cell area of 1000 µm2 and during the time period of 100 sec. Further image processing and data analysis used the Interactive Data Language (IDL) software (ITT Visual Information Solutions; Boulder, CO). We developed programs based on a Ca2+ spark detection algorithm [40] to calculate the amplitude and kinetics (upstroke and decay time courses) of individual flashes [10, 41]. Pooled data were used for statistical analysis. Data were presented as mean ± SEM. One way ANOVA, paired and unpaired Student’s T test were used when appropriate to determine statistical significance among groups. A P value less than 0.05 was deemed significant.
3. Results
3.1. In vitro characterization of cpYFP
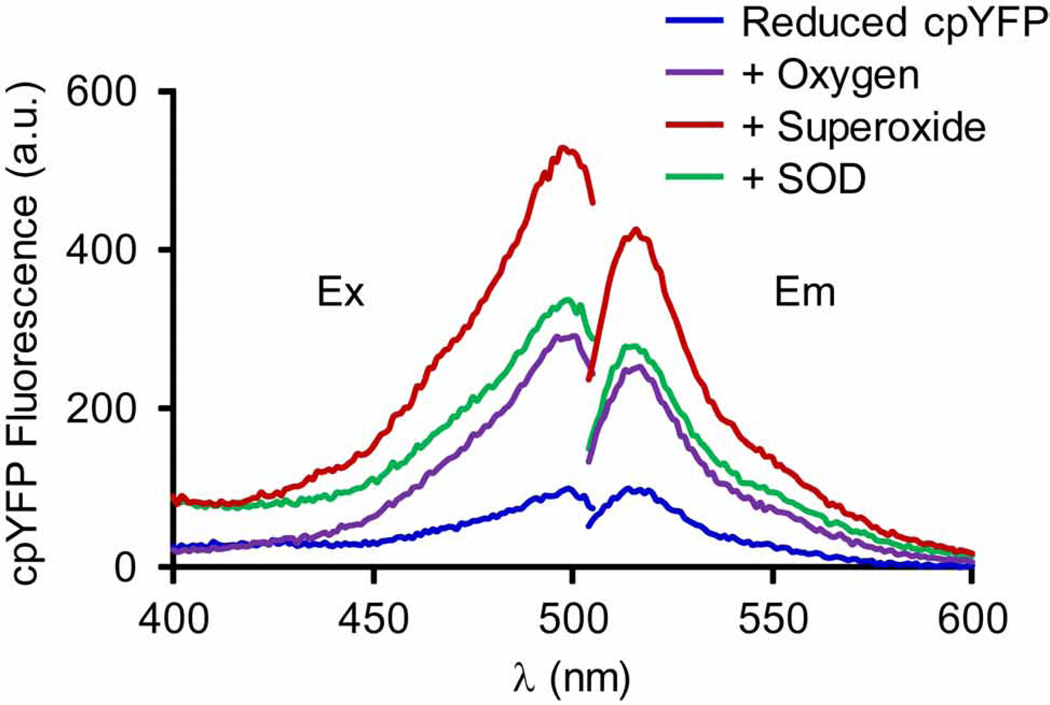
The E. Coli expression and purification of cpYFP follows standard procedures that have been used for many other proteins. Upon induction of cpYFP expression, green fluorescence can be visualized in BL21(DE3) cells under fluorescent/confocal microscope, suggesting functional cpYFP protein exists in prokaryotic cells. Since the purification is done in ambient environment, the purified native cpYFP shows maximal fluorescence at physiological pH and its spectra are identical to the fully oxidized cpYFP. Upon DTT reduction, the fluorescence at 488 nm excitation is significantly decreased for several folds, but is still much higher than the solution background. Fig. 1 shows the emission spectra (488 nm excitation) reported before [10] and the excitation spectra (515 nm emission) during the calibration of superoxide sensitivity. In the oxygen free solutions, adding xanthine or xanthine oxidase does not lead to significant change in cpYFP spectra. The absolute readings of peak emission may vary significantly among different preparations and spectrofluorimeters. Therefore, for each experiment, solution background and fully reduced and fully oxidized cpYFP are always measured to obtain the range of fluorescence change for a particular batch of protein sample. For data analysis, the values of solution background are always subtracted and the peak value of fully reduced protein is set as 100% for normalization and comparison.
Fig. 1.
In vitro characterization of cpYFP fluorescence. Ex: excitation spectra (400 – 505 nm) obtained at 515 nm emission; Em: emission spectra (500 – 600 nm) at 488 nm excitation. Note the two-fold increase in both emission and excitation spectra upon O2 addition, and the additional two-fold increase upon superoxide incubation (provided by 2 mM xanthine plus 20 mU xanthine oxidase). The latter increase in fluorescence signal is reversed by incubation with SOD (600 U/ml). Reproduced from [10].
In a recent brief communication, another group conducted in vitro calibration of purified cpYFP but failed to detect its superoxide response [34]. Detailed methods and the sequence of cpYFP used in that study are not reported [34, 35]. There are several different versions of cpYFP (point mutations) [42], which could have different sensitivity as the one we used (Supplemental Fig. 1). Another potential explanation for this disconnect is the use of another E. Coli strain that may influence the modification, folding or maturation of the protein as we previously presented [35]. In addition, proper control of the oxidative state of the protein is essential. Whether the cpYFP is fully reduced is critical for its response to oxidation and superoxide [34, 35]. The detailed methods described here should help provide a starting point for the reconciliation of this discrepancy.
3.2. Intracellular expression and characterization of mt-cpYFP
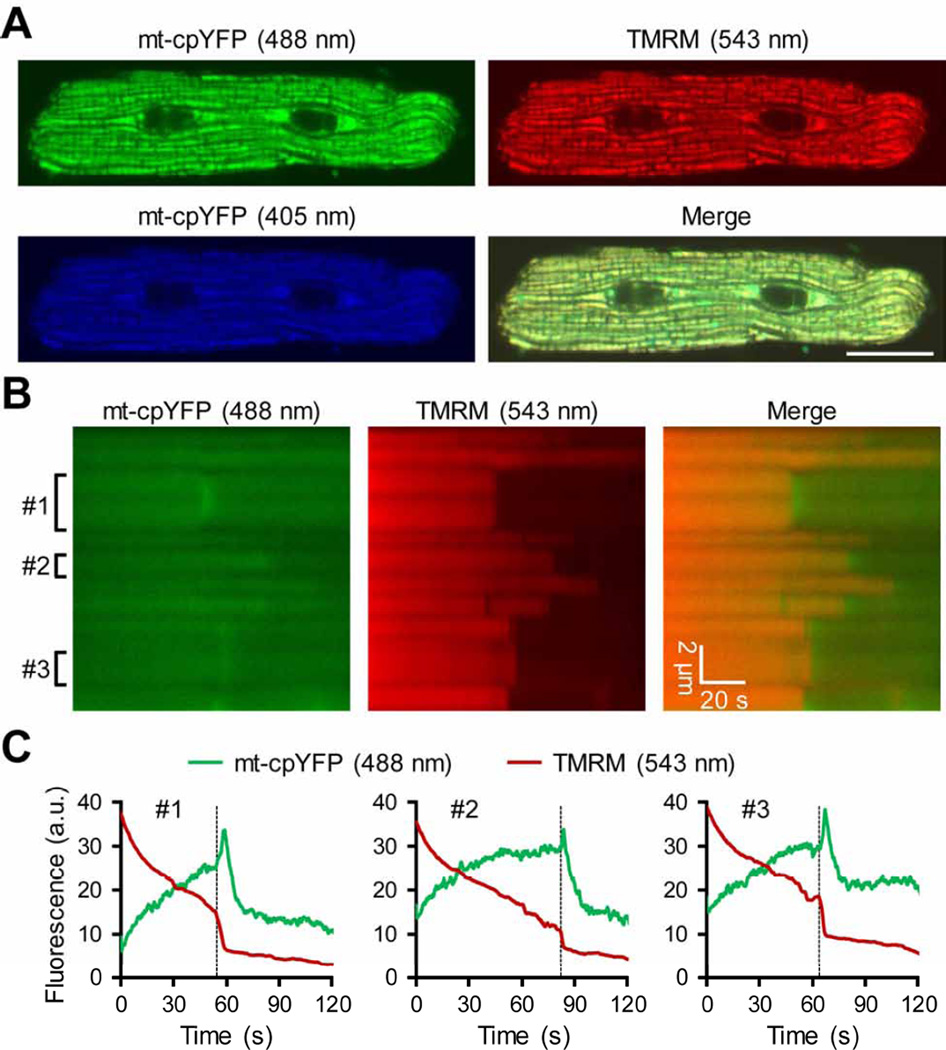
Once expressed in the mammalian cells, mt-cpYFP shows clear mitochondrial localization pattern (Fig. 2A) [10, 15–17, 19, 23, 26–29]. It should be noted that, most cells exhibit autofluorescence at 488 nm excitation, especially from the mitochondria, which is mostly flavin adenine dinucleotide (FAD) autofluorescence. Thus, cells without mt-cpYFP gene transfer are always included as negative control to verify the efficiency of the gene transfer and the expression level of mt-cpYFP. To avoid laser damage of the living cells, laser intensity should be kept as low as possible. For example, we used 1–5% power of the 488 nm laser and 5–10% power of the 405 nm laser on the Zeiss LSM510 confocal. This will also help avoid or minimize photobleaching of mt-cpYFP fluorescence (mainly at 488 nm excitation) and maintain stable basal mt-cpYFP fluorescence during the 100-s time-lapse scan [10]. Since physiological solution used in confocal live cell imaging contains high concentrations of HEPES, fluctuations of extracellular and intracellular pH is mostly suppressed. Usually, live cell imaging is conducted at room temperature. It is possible that the basal fluorescence may change at a high temperature. The mitochondrial flash frequency is increased at a higher temperature (37 °C) [17].
Fig. 2.
Expression pattern of mt-cpYFP in adult ventricular myocyte. (A) Confocal frame scan images showing typical mitochondrial morphology and organization pattern in a rat adult cardiomyocyte two days after adenovirus mediated mt-cpYFP gene transfer. TMRM (20 nM) was used as a mitochondrial marker. Scale bar = 20 µm. (B) Confocal linescan images showing laser-induced mPTP openings (sudden and irreversible loss of TMRM fluorescence) and the accompanying transient increases in mt-cpYFP fluorescence in the very early phase of mPTP opening in each mitochondria. (C) Traces showing time-dependent changes of mt-cpYFP and TMRM fluorescence in the three mitochondria (#1–3) indicated in (B). The broken lines indicate the onset of sustained membrane potential loss and the bursting and transient increase in mtcpYFP fluorescence.
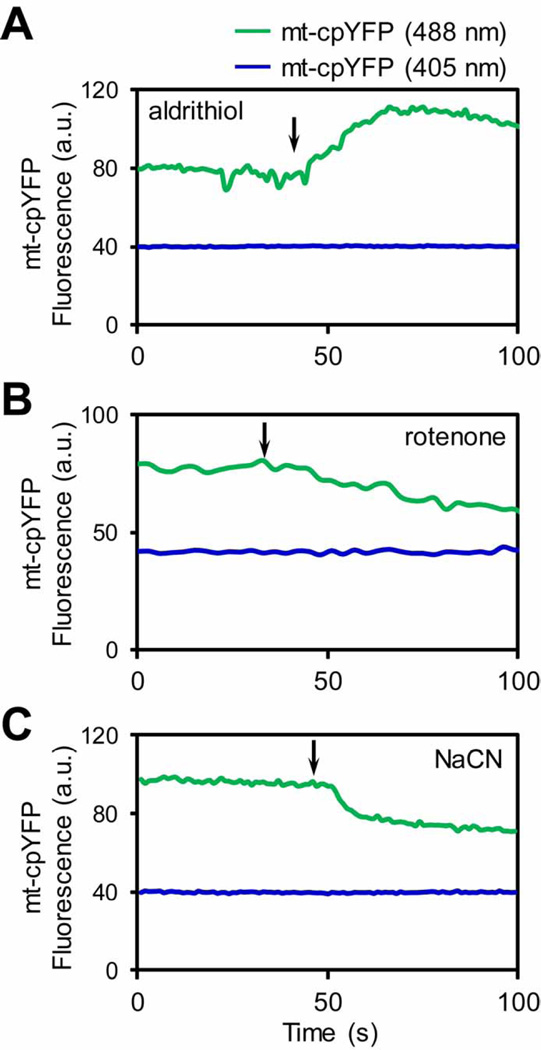
While majority of the studies characterized mt-cpYFP signal during mitochondrial flashes in living cells, some reports also showed that the basal mt-cpYFP fluorescence is responsive to oxidants and antioxidants [10, 28, 43, 44]. Here, we used an approach to induce endogenous ROS production inside mitochondria of living adult cardiomyocytes. The slow laser speed in linescan mode has been shown to induce abrupt loss of membrane potential in individual mitochondria, which is attributed to the permanent opening of the mPTP [45]. Importantly, this triggered mPTP opening is accompanied with local and transient ROS production in that mitochondria [45]. Using the same scanning method, we found that the mt-cpYFP fluorescence is transiently increased when the membrane potential is abruptly dissipated during the linescan (Fig. 2B). There is a one-on-one coupling of the triggered mPTP opening and transient increase in mt-cpYFP fluorescence, which is consistent to the superoxide production accompanying mPTP opening [45]. Furthermore, while exogenous oxidants such as aldrithiol (Fig. 3A) [10], paraquat [28], and menadione [43] significantly increase the basal mt-cpYFP fluorescence, inhibition of ETC electron flow mostly decreases mt-cpYFP fluorescence (Fig. 3B–C) [10].
Fig. 3.
Characterization of mt-cpYFP fluorescence in intact cardiomyocytes. The mt-cpYFP fluorescence (488 nm excitation only) was significantly increased by acute addition of a strong oxidant aldrithiol (0.5 mM) (A), and decreased by ETC inhibitors rotenone (5 µM) or sodium cyanide (NaCN, 5 mM) (B–C). The traces in each panel are averaged fluorescence from 3 individual mitochondria in one cardiomyocyte. Arrows indicate the time of chemical addition.
3.3. In situ and in vivo imaging of mitochondrial flashes
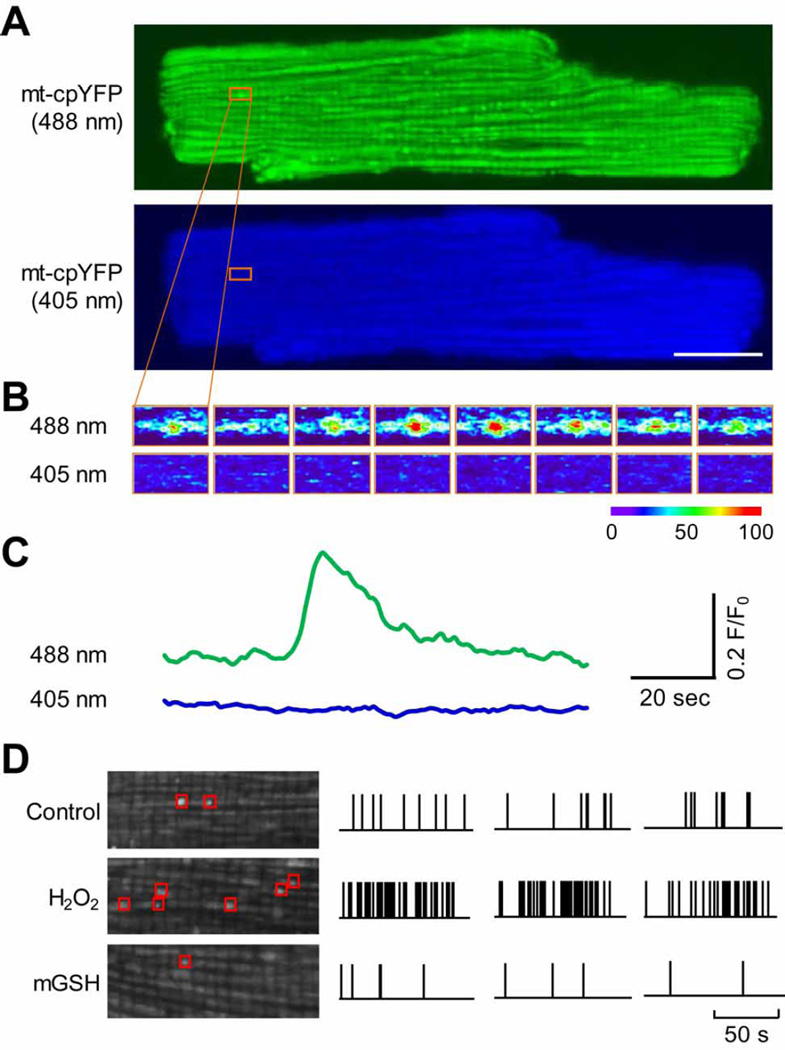
Despite the debate over the superoxide and pH sensitivity of cpYFP, mitochondrial flash events (Fig 4A–C), initially detected by mt-cpYFP [10], has been subsequently detected by other indicators, including ROS indicators MitoSOX Red and 2',7'-dichlorofluorescein (DCF) [19, 46, 47], redox indicator roGFP [24], and pH indicator SypHer [48]. We have further shown that mtcpYFP detected mitochondrial flashes are composed of dual signals, a major superoxide signal and a minor alkalization signal [19]. Several reports also show that mitochondrial flashes are upregulated by oxidants and down regulated by antioxidants (Fig. 4D) [10, 19, 24, 43, 46]. In addition, mitochondrial flashes are always accompanied by loss of membrane potential (Fig. 5) [10, 16, 17, 24, 46, 48].
Fig. 4.
Confocal imaging of single mitochondrial flashes in cultured adult rat cardiomyocyte. (A) Confocal images of a cultured rat cardiomyocyte expressing mt-cpYFP simultaneously excited by 488 and 405 nm laser. The orange boxes indicate the location and onset of one mitochondrial flash. Scale bar = 20 µm. (B) Sequential dual wavelength excitation (488 and 405 nm) images of the area highlighted in (A) showing the time-dependent change of mt-cpYFP fluorescence (indicated by the pseudo-color) during the mitochondrial flash. Time interval between each image is 2 s. (C) Traces showing the time course of the mitochondrial flash in (A) and (B). (D) Left: Enlarged images showing mitochondrial flashes detected during a 100-s scan period in a cardiomyocyte under normal culture condition (Control), after addition of 10 mM H2O2, or after treatment of 50 µM mitochondrial-targeted glutathione (mGSH). Right: Diary plots of mitochondrial flashes onset during a 100-s scan period in typical single cardiomyocytes under the different conditions (Control, H2O2 and mGSH). Note three representative samples are shown for each condition and the vertical bars indicate onset of flashes.
Fig. 5.
Single mitochondrial flashes detected in Langendorff perfused heart. (A) Representative images of the intact myocardium showing the colocalization of mt-cpYFP and TMRM. Scale bar = 20 µm. (B) Image of a single cardiomyocyte in the intact myocardium. The area highlighted by the white boxes is enlarged to show a mitochondrial flash accompanied with decrease in membrane potential (white arrows). Scale bars = 20 and 5 µm for the full size and enlarged images, respectively. (C) Traces showing the fluorescence changes during the single mitochondrial flash highlighted in (B).
The physiological significance of mitochondrial flash is highlighted by the visualization of this event in intact myocardium of Langendorff perfused hearts (Fig. 5), skeletal muscles (Fig. 6), sciatic nerves in anesthetized mice with pan-tissue overexpression of mt-cpYFP [10, 16, 20, 21, 29], and the cortex of roGFP transgenic mice [24]. The evidence so far suggests that single mitochondrial flashes detected in living cells and in vivo are coupled to mitochondrial metabolism/respiration, transient mPTP openings and ROS production [10, 15–17, 19–21, 23–29, 43, 46, 48]. Here, we found that the basal mt-cpYFP fluorescence (488 nm excitation) is also correlated with the frequency of mitochondrial flashes in individual skeletal muscle cells in vivo (Fig. 6). In addition, physiological and pathological perturbations, such as metabolic substrates [16, 20], electrical stimulation [17, 21], Ca2+ [28, 49], ischemic-reperfusion [10, 43], insulin resistance [18], and mitochondrial stresses [20, 48], also modulate mitochondrial flash frequency. Taken together, increasing evidence suggests that mitochondrial flash is a composite biomarker that integrates mitochondrial functions including respiration, ROS production, pH regulation and mPTP opening [50]. Thus, monitoring this single mitochondrial event is a useful tool for studying the role of individual mitochondria in cell signaling, function and dysfunction [50].
Fig. 6.
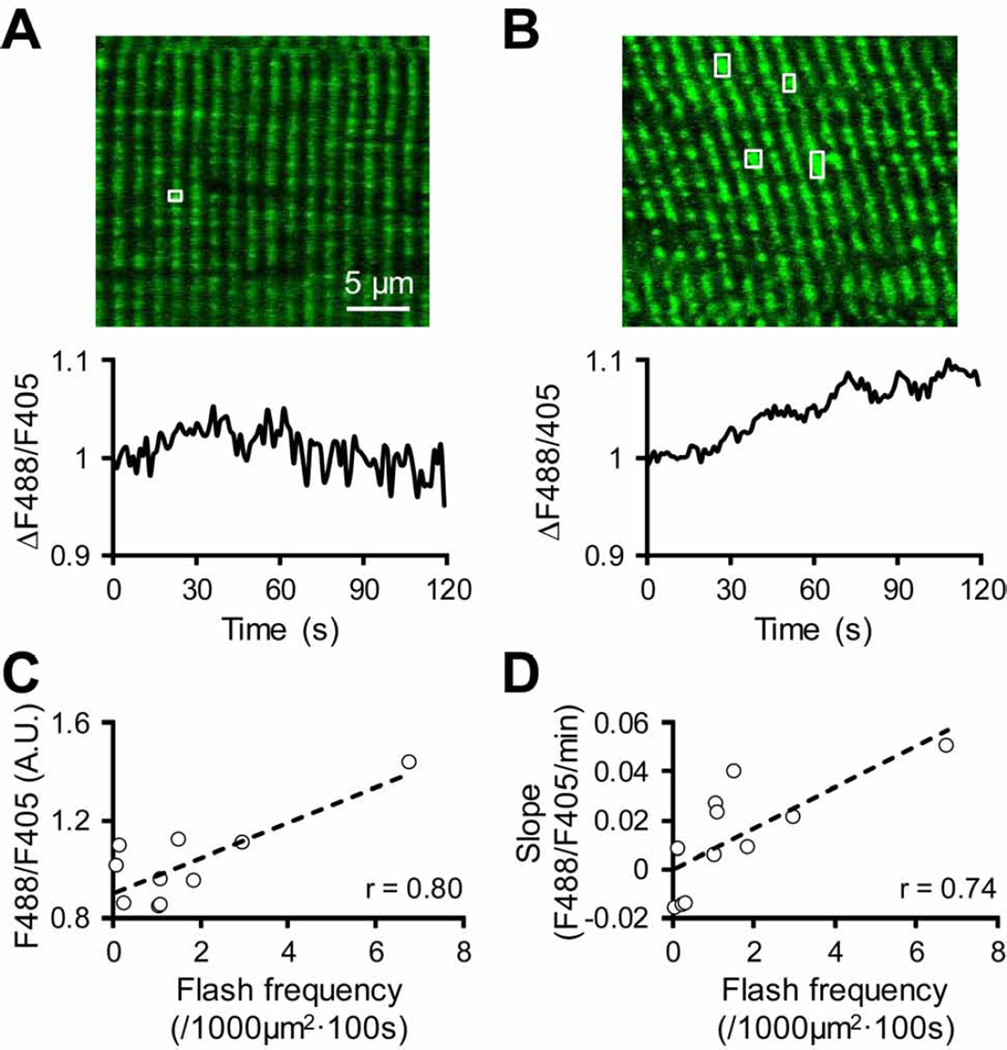
Single mitochondrial flash detected in skeletal muscles in vivo. (A–B) Upper panel: Representative images showing mt-cpYFP fluorescence (488 nm excitation) in two skeletal muscle cells of anesthetized mt-cpYFP TG mice. Lower panel: Representative traces showing time-dependent changes in basal mt-cpYFP fluorescence (ΔF488/F405) in the two skeletal muscle cells during time lapse scanning. The image and trace in (A) are from a skeletal muscle cell that exhibited low basal mt-cpYFP fluorescence, low flash frequency (highlighted by white box), and no increase in basal mt-cpYFP signal during the scan. The example in (B) is from a skeletal muscle cell with high basal mt-cpYFP fluorescence, high flash frequency (highlighted by white boxes) and an obvious increase in basal mt-cpYFP signal during the scan. (C) Correlation between whole cell mt-cpYFP fluorescence and mitochondrial flash frequency in individual skeletal muscle cells in vivo. (D) Correlation between the slope (F488/F405/min) of increased whole cell mt-cpYFP fluorescence and mitochondrial flash frequency in individual skeletal muscle cells in vivo. Data in (C) and (D) are from 10–11 cells/areas in 3 mice and each data point represents an individual muscle cell.
4. Discussion
In this report, we describe the detailed methods for in vitro characterization of the cpYFP indicator and the monitoring of single mitochondrial flash events in living cells and in vivo. Eight years after the first report on cpYFP as a superoxide indicator and the detection of single mitochondrial superoxide flashes [10], many exciting advances have been made and the significance of this single mitochondrial event in cell physiology and pathology has been gradually recognized [50–52]. In more than 40 publications, mitochondrial flash events have been detected in various biological systems ranging from plants, C. elegans, and zebrafish to rodents and human cells [50]. More importantly, when the mechanisms underlying mitochondrial flash generation unfolds, it provides the field with a new view on how mitochondrion actively integrates its functions, including metabolism, ROS, pH, and mPTP. This view is significantly different from the previous view on mitochondrion as simply an energy producer. In addition, real-time monitoring of mitochondrial flashes under pathophysiological conditions has revealed important roles of mitochondria in normal cell function and disease development. One of the examples is dissecting the relationship between muscle contraction and ROS production. We have shown that muscle contraction induced mitochondrial flashes, which provided ROS to further support contractility [17, 21]. Extensive discussion over the pathophysiological significance of mitochondrial flash can be found in a recent review paper [50].
Despite the many exciting advancements regarding the mechanism and pathophysiological roles of mitochondrial flash, a number of issues and questions remain to be fully addressed. Because of the co-presence of ROS and pH signals in a flash, the dual ROS and pH sensitivity of cpYFP has redirected its use as a pure superoxide indicator toward an indicator-of-choice for the detection of mitochondrial flashes. However, the in vitro superoxide and redox manipulation of purified cpYFP protein is yet to be reproduced by others [34] and more importantly, the biochemistry for reversible superoxide-sensing by cpYFP remains elusive and merits further investigation. Therefore, it is necessary that the detailed methods used in the in vitro indicator characterization are presented to facilitate comparison and cross checking by different groups. The protocol detailed here is adopted from the methods used for developing a GFP based redox indicator, in which extra precaution is given to ensure the strict inert environment essential for the successful control of the redox status of the fluorescent protein [11, 12]. The method is also useful in future studies on cpYFP, which should focus on modifying cpYFP to create superoxide-selective reversible indicator for the measurement of superoxide signal independent of the concurrent pH changes during a flash. Recently, a small molecule superoxide indicator has been developed [53]. The indicator, named HKSOX-1, detects superoxide independent of protonation/deprotonation, which makes it pH insensitive. Because it uses nucleophilic property of superoxide anion, it is also not sensitive to redox state. However, with its current reaction kinetics and sensitivity, this probe in its current version is not suitable for detecting fast changes (e.g., 10 seconds) of superoxide burst during a flash (unpublished data).
Many other indicators are able to detect flash events in various cell types and in vivo [19, 24, 46–48]. In addition, mt-cpYFP can be used together or in parallel with other ROS or pH indicators to further differentiate the specific signals detected during the flash [19, 46–48, 50]. We have used mt-cpYFP simultaneously with a pH indicator, SNARF-1, and determined the relative contribution of pH and ROS to the fluorescence change of mt-cpYFP during a mitochondrial flash in adult cardiomyocytes [19]. Similar approaches can be used to differentiate the pH and ROS components of flashes in other cell types. Mitochondrial flash can also be monitored together with other fluorescent indicators, such as indicators for mitochondrial membrane potential and Ca2+, to address the interaction between mitochondrial flash and these intracellular signals [21, 24, 28]. Imaging single mitochondrial events in live animals or intact organs has clear advantage over the traditional methods for mitochondrial study, since it enables real-time and in situ evaluation of mitochondrial function under physiologically relevant conditions [16, 41, 51, 52, 54]. Tissues such as skeletal muscles, subcutaneous structures, and superficial nerves can be imaged by confocal imaging in live animal. Combined with multiphoton microscopy, single mitochondrial flash events can be visualized in internal organs, such as the cortex and spinal axons in live animals [24]. The in vivo imaging also has limitations, such as motion artifact and short-term imaging, which has been discussed before [29]. The recent advent of miniaturized two-photon microscopy that a small animal can carry on its head or other part of the body (unpublished data) can extend the mitochondrial flash imaging to freely moving animals. We hope that the detailed methods presented here will help address the ongoing issue regarding cpYFP sensitivity, and, at the same time, promote the application of in vivo imaging techniques in biomedical research.
Supplementary Material
The full length mt-cpYFP cDNA which was used for the in vitro calibration, mammalian cell transfection (plasmid or adenovirus), and the transgenic animals (mice, zebra fish and C. elegans)
Highlights.
The detailed protocol for in vitro calibration of the cpYFP indicator is provided.
Mitochondrial flash detected by cpYFP is a novel biomarker.
Mitochondrial flash reflects integrated single mitochondrial functions.
Real-time confocal imaging of mitochondrial flashes in intact heart and in vivo.
Acknowledgments
We thank Drs. Huaqiang Fang and Guohua Gong for technical support. This study was partially supported by NIH grant (HL114760) to W.W. and by the National Key Basic Research Program of China (2013CB531200) to H.C..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Droege W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 2.Otani H. Reactive oxygen species as mediators of signal transduction in ischemic preconditioning. Antioxid Redox Signal. 2004;6(2):449–469. doi: 10.1089/152308604322899521. [DOI] [PubMed] [Google Scholar]
- 3.Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu SS. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol. 2004;287(4):C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- 4.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15(22):2922–2933. [PubMed] [Google Scholar]
- 5.Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37(6):755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 6.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552(Pt 2):335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29(3–4):222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 8.Degli Esposti M. Measuring mitochondrial reactive oxygen species. Methods. 2002;26(4):335–340. doi: 10.1016/S1046-2023(02)00039-7. [DOI] [PubMed] [Google Scholar]
- 9.Barja G. The quantitative measurement of H2O2 generation in isolated mitochondria. J Bioenerg Biomembr. 2002;34(3):227–233. doi: 10.1023/a:1016039604958. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Fang H, Groom L, Cheng A, Zhang W, Liu J, Wang X, Li K, Han P, Zheng M, Yin J, Mattson MP, Kao JP, Lakatta EG, Sheu SS, Ouyang K, Chen J, Dirksen RT, Cheng H. Superoxide flashes in single mitochondria. Cell. 2008;134(2):279–290. doi: 10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279(21):22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 12.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279(13):13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 13.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Meth. 2006;3(4):281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Fang H, Huang Z, Shang W, Hou T, Cheng A, Cheng H. Imaging ROS signaling in cells and animals. J Mol Med (Berl) 2013;91(8):917–927. doi: 10.1007/s00109-013-1067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen EZ, Song CQ, Lin Y, Zhang WH, Su PF, Liu WY, Zhang P, Xu J, Lin N, Zhan C, Wang X, Shyr Y, Cheng H, Dong MQ. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature. 2014;508(7494):128–132. doi: 10.1038/nature13012. [DOI] [PubMed] [Google Scholar]
- 16.Fang H, Chen M, Ding Y, Shang W, Xu J, Zhang X, Zhang W, Li K, Xiao Y, Gao F, Shang S, Li J-C, Tian X-L, Wang S-Q, Zhou J, Weisleder N, Ma J, Ouyang K, Chen J, Wang X, Zheng M, Wang W, Zhang X, Cheng H. Imaging superoxide flash and metabolism-coupled mitochondrial permeability transition in living animals. Cell Res. 2011;21(9):1295–1304. doi: 10.1038/cr.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei L, Salahura G, Boncompagni S, Kasischke KA, Protasi F, Sheu S-S, Dirksen RT. Mitochondrial superoxide flashes: metabolic biomarkers of skeletal muscle activity and disease. The FASEB Journal. 2011;25(9):3068–3078. doi: 10.1096/fj.11-187252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Fang H, Shang W, Xiao Y, Sun T, Hou N, Pan L, Sun X, Ma Q, Zhou J, Wang X, Zhang X, Cheng H. Mitoflash altered by metabolic stress in insulin-resistant skeletal muscle. J Mol Med (Berl) 2015;93(10):1119–1130. doi: 10.1007/s00109-015-1278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei-LaPierre L, Gong G, Gerstner BJ, Ducreux S, Yule DI, Pouvreau S, Wang X, Sheu SS, Cheng H, Dirksen RT, Wang W. Respective contribution of mitochondrial superoxide and pH to mitochondria-targeted circularly permuted yellow fluorescent protein (mt-cpYFP) flash activity. J Biol Chem. 2013;288(15):10567–10577. doi: 10.1074/jbc.M113.455709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong G, Liu X, Zhang H, Sheu SS, Wang W. Mitochondrial flash as a novel biomarker of mitochondrial respiration in the heart. Am J Physiol Heart Circ Physiol. 2015;309(7):H1166–H1177. doi: 10.1152/ajpheart.00462.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong G, Liu X, Wang W. Regulation of metabolism in individual mitochondria during excitation-contraction coupling. J Mol Cell Cardiol. 2014;76:235–246. doi: 10.1016/j.yjmcc.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Sun T, Jian C, Lei L, Han P, Lv Q, Yang R, Zhou X, Xu J, Hu Y, Men Y, Huang Y, Zhang C, Zhu X, Wang X, Cheng H, Xiong JW. Remodeling of Mitochondrial Flashes in Muscular Development and Dystrophy in Zebrafish. PLoS One. 2015;10(7):e0132567. doi: 10.1371/journal.pone.0132567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying Z, Chen K, Zheng L, Wu Y, Li L, Wang R, Long Q, Yang L, Guo J, Yao D, Li Y, Bao F, Xiang G, Liu J, Huang Q, Wu Z, Hutchins AP, Pei D, Liu X. Transient Activation of Mitoflashes Modulates Nanog at the Early Phase of Somatic Cell Reprogramming. Cell Metab. 2016;23(1):220–226. doi: 10.1016/j.cmet.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Breckwoldt MO, Pfister FM, Bradley PM, Marinkovic P, Williams PR, Brill MS, Plomer B, Schmalz A, St Clair DK, Naumann R, Griesbeck O, Schwarzlander M, Godinho L, Bareyre FM, Dick TP, Kerschensteiner M, Misgeld T. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat Med. 2014;20(5):555–560. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- 25.Hou Y, Ghosh P, Wan R, Ouyang X, Cheng H, Mattson MP, Cheng A. Permeability transition pore-mediated mitochondrial superoxide flashes mediate an early inhibitory effect of amyloid beta1–42 on neural progenitor cell proliferation. Neurobiol Aging. 2014;35(5):975–989. doi: 10.1016/j.neurobiolaging.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y, Mattson MP, Cheng A. Permeability transition pore-mediated mitochondrial superoxide flashes regulate cortical neural progenitor differentiation. PLoS One. 2013;8(10):e76721. doi: 10.1371/journal.pone.0076721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Xu S, Wang P, Wang W. Transient mitochondrial permeability transition mediates excitotoxicity in glutamate-sensitive NSC34D motor neuron-like cells. Exp Neurol. 2015;271:122–130. doi: 10.1016/j.expneurol.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S, Chisholm AD. C. elegans epidermal wounding induces a mitochondrial ROS burst that promotes wound repair. Dev Cell. 2014;31(1):48–60. doi: 10.1016/j.devcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong G, Wang W. Confocal imaging of single mitochondrial superoxide flashes in intact heart or in vivo. J Vis Exp. 2013;(81):e50818. doi: 10.3791/50818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zong W, Zhao J, Chen X, Lin Y, Ren H, Zhang Y, Fan M, Zhou Z, Cheng H, Sun Y, Chen L. Large-field high-resolution two-photon digital scanned light-sheet microscopy. Cell Res. 2015;25(2):254–257. doi: 10.1038/cr.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochemical Journal. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci U S A. 2006;103(41):15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson BC, Srikun D, Chang CJ. Mitochondrial-targeted fluorescent probes for reactive oxygen species. Curr Opin Chem Biol. 2010;14(1):50–56. doi: 10.1016/j.cbpa.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarzlander M, Wagner S, Ermakova YG, Belousov VV, Radi R, Beckman JS, Buettner GR, Demaurex N, Duchen MR, Forman HJ, Fricker MD, Gems D, Halestrap AP, Halliwell B, Jakob U, Johnston IG, Jones NS, Logan DC, Morgan B, Muller FL, Nicholls DG, Remington SJ, Schumacker PT, Winterbourn CC, Sweetlove LJ, Meyer AJ, Dick TP, Murphy MP. The 'mitoflash' probe cpYFP does not respond to superoxide. Nature. 2014;514(7523):E12–E14. doi: 10.1038/nature13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng H, Wang W, Wang X, Sheu SS, Dirksen RT, Dong MQ, Cheng, et al. Nature. 2014;514(7523):E14–E15. doi: 10.1038/nature13859. reply. [DOI] [PubMed] [Google Scholar]
- 36.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ . Proc Natl Acad Sci U S A. 2001;98(6):3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77(1):61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Second. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Zhou YY, Wang SQ, Zhu WZ, Chruscinski A, Kobilka BK, Ziman B, Wang S, Lakatta EG, Cheng H, Xiao RP. Culture and adenoviral infection of adult mouse cardiac myocytes: methods for cellular genetic physiology. Am J Physiol Heart Circ Physiol. 2000;279(1):H429–H436. doi: 10.1152/ajpheart.2000.279.1.H429. [DOI] [PubMed] [Google Scholar]
- 40.Cheng H, Lederer WJ, Cannell MB. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262(5134):740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 41.Li K, Zhang W, Fang H, Xie W, Liu J, Zheng M, Wang X, Wang W, Tan W, Cheng H. Superoxide flashes reveal novel properties of mitochondrial reactive oxygen species excitability in cardiomyocytes. Biophys J. 2012;102(5):1011–1021. doi: 10.1016/j.bpj.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quatresous E, Legrand C, Pouvreau S. Mitochondria-targeted cpYFP: pH or superoxide sensor? J Gen Physiol. 2012;140(5):567–570. doi: 10.1085/jgp.201210863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Z, Zhang W, Fang H, Zheng M, Wang X, Xu J, Cheng H, Gong G, Wang W, Dirksen RT, Sheu SS. Response to "A critical evaluation of cpYFP as a probe for superoxide". Free Radic Biol Med. 2011;51(10):1937–1940. doi: 10.1016/j.freeradbiomed.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, Li K, Zhu X, Wu D, Shang W, Yuan X, Huang Z, Zheng M, Wang X, Yang D, Liu J, Cheng H. Subsarcolemmal mitochondrial flashes induced by hypochlorite stimulation in cardiac myocytes. Free Radic Res. 2014;48(9):1085–1094. doi: 10.3109/10715762.2014.932114. [DOI] [PubMed] [Google Scholar]
- 45.Zorov DB, Filburn CR, Klotz LO, Zweier JL, Sollott SJ. Reactive oxygen species (ROS)-induced ROS release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J Exp Med. 2000;192(7):1001–1014. doi: 10.1084/jem.192.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pouvreau S. Superoxide flashes in mouse skeletal muscle are produced by discrete arrays of active mitochondria operating coherently. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Huang Z, Hou T, Xu J, Wang Y, Shang W, Ye T, Cheng H, Gao F, Wang X. Superoxide constitutes a major signal of mitochondrial superoxide flash. Life Sci. 2013;93(4):178–186. doi: 10.1016/j.lfs.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Santo-Domingo J, Giacomello M, Poburko D, Scorrano L, Demaurex N. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J. 2013;32(13):1927–1940. doi: 10.1038/emboj.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou T, Zhang X, Xu J, Jian C, Huang Z, Ye T, Hu K, Zheng M, Gao F, Wang X, Cheng H. Synergistic triggering of superoxide flashes by mitochondrial Ca2+ uniport and basal reactive oxygen species elevation. J Biol Chem. 2013;288(7):4602–4612. doi: 10.1074/jbc.M112.398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Gong G, Wang X, Wei-LaPierre L, Cheng H, Dirksen RT, Sheu SS. Mitochondrial Flash: Integrative ROS and pH Signals in Cell and Organelle Biology. Antioxid Redox Signal. 2016 doi: 10.1089/ars.2016.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Jian C, Zhang X, Huang Z, Xu J, Hou T, Shang W, Ding Y, Zhang W, Ouyang M, Wang Y, Yang Z, Zheng M, Cheng H. Superoxide flashes: elemental events of mitochondrial ROS signaling in the heart. J Mol Cell Cardiol. 2012;52(5):940–948. doi: 10.1016/j.yjmcc.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Wei L, Dirksen RT. Perspectives on: SGP symposium on mitochondrial physiology and medicine: mitochondrial superoxide flashes: from discovery to new controversies. J Gen Physiol. 2012;139(6):425–434. doi: 10.1085/jgp.201210790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu JJ, Wong NK, Ye S, Chen X, Lu MY, Zhao AQ, Guo Y, Ma AC, Leung AY, Shen J, Yang D. Fluorescent Probe HKSOX-1 for Imaging and Detection of Endogenous Superoxide in Live Cells and In Vivo. J Am Chem Soc. 2015;137(21):6837–6843. doi: 10.1021/jacs.5b01881. [DOI] [PubMed] [Google Scholar]
- 54.Sheu SS, Wang W, Cheng H, Dirksen RT. Superoxide flashes: illuminating new insights into cardiac ischemia/reperfusion injury. Future Cardiol. 2008;4(6):551–554. doi: 10.2217/14796678.4.6.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The full length mt-cpYFP cDNA which was used for the in vitro calibration, mammalian cell transfection (plasmid or adenovirus), and the transgenic animals (mice, zebra fish and C. elegans)