Abstract
In aged mice peripheral stimulation of the innate immune system with LPS causes exaggerated neuroinflammation and prolonged sickness behavior due in part to microglial dysfunction. Epigenetic changes to DNA may play a role in microglial dysfunction, therefore we sought to determine whether aged microglia displayed DNA hypomethylation of the IL-1β promoter and altered expression of epigenetic regulators. We further examined whether the demethylating agent 5-azacytidine induced IL-1β expression in BV2 and primary microglia similar to microglia from aged mice. Novel findings indicated that aged mice had decreased methylation of the IL-1β gene promoter in primary microglia basally or following systemic LPS that is associated with increased IL-1β mRNA, intracellular IL-1β production, as well as prolonged sickness behavior. Lastly, 5-azacytidine increased IL-1β gene expression and decreased DNA methylation of BV2 and primary microglial cells similar to microglia from aged mice. Taken together, these data indicate that DNA methylation promotes heightened microglial activation in the aged brain.
1. Introduction
In the aged brain it is common for microglia to have an inflammatory gene expression profile and a deramified morphology comparable to the morphology of activated adult microglia (Damani, et al., 2011, Sierra, et al., 2007, Tremblay, et al., 2012). What’s more, microglia in the brain of aged animals are hypersensitive to signals emerging from the peripheral immune system during infection, resulting in an aberrant neuroinflammatory response that is more intense and longer lasting (Henry, et al., 2009, Sierra, et al., 2007). This exaggerated proinflammatory response can be neurotoxic, lead to behavioral pathology (e.g., anorexia, acute cognitive disorders, and delirium), and exacerbate neurodegenerative diseases (e.g., Alzheimer’s disease), greatly enhancing morbidity and mortality in older adults (Franceschi and Campisi, 2014). Why microglia become proinflammatory during aging and why they are hypersensitive to signals from the peripheral immune system is not known.
One way microglial dysfunction may occur is through epigenetic modifications. Epigenetics refers to changes in gene expression that are independent of changes in the DNA sequence; it is thought of as a mediator between gene-environment interactions (Bird, 2007, Jaenisch and Bird, 2003). Epigenetics impact gene regulation though mechanisms such as DNA methylation and post-translational modifications to histone tails, with DNA methylation being the most well studied mechanism. DNA methylation is catalyzed by DNMTs that includes DNMT1, DNMT3a, and DNMT3b (Moore, Le, & Fan, 2013). DNMT1 is a maintenance methyltransferase that maintains DNA methylation patterns during the DNA replication process whereas DNMT3a and DNMT3b are de novo methyltransferases, capable of catalyzing new methylation patterns of previously unmethylated sequences. Another protein associated with DNA methylation is MeCP2, which binds to methylated cytosines and recruits HDACs and other corepressors. This promotes higher-affinity interaction between DNA and histone core, condensing the chromatin, and typically suppresses transcription factor binding and gene expression (Bird, 2002; Strahl & Allis, 2000). Additionally, Gadd45b protein plays an important role in active DNA demethylation (Ma, Guo, Ming, & Song, 2009).
DNA methylation depends on a precise balance of methylation and demethylation reactions that are nicely balanced in mature cells, but with age there is a strong shift to favor DNA hypomethylation (Pogribny and Vanyushin, 2010). Age-related DNA hypomethylation may lead to redistribution of heterochromatin, impair normal gene responsiveness to environmental signals, and increase genomic instability that could compromise proper cell function. Aging could therefore be thought of as a time-dependent, epigenetically mediated loss of phenotypic plasticity (Gravina and Vijg, 2010), suggesting that aberrant DNA methylation patterns may be candidate mechanisms for explaining how microglia from aged mice seem to be “stuck” in a proinflammatory phenotype.
A handful of studies have demonstrated gene expression regulation by DNA methylation in microglia. One using BV2 cells, a murine microglial cell line, reported that DNA methylation influences expression of several genes associated with the pathology of Alzheimer’s disease (Byun, et al., 2012), and another revealed that promoting increased maternal care alters the methylation pattern of the IL-10 gene, leading to increased IL-10 expression in the nucleus accumbens and a reduction in morphine-induced addiction behavior (Schwarz and Bilbo, 2013). Very recently it has also been determined that SIRT1 deficiency in aging microglia is related to increased IL-1β transcription and decreased methylation of CpG sites within the IL-1β proximal promoter (Cho, et al., 2015).
Although increasing evidence suggests microglial phenotype is regulated by epigenetic mechanisms, little is known about DNA methylation of proinflammatory cytokines of healthy microglia from aged animals and the contribution of enzymes that significantly impact the epigenetic landscape. Since it is thought that microglia are the major producers of IL-1β in the brain and one of the most consistently upregulated cytokines with age (Burton, et al., 2016, Chen, et al., 2008), the objectives of this study were to determine whether there are promoter IL-1β DNA methylation modifications as well as altered gene expression of epigenetic regulators present between brain immune cells of young and old mice exposed to an immune challenge. Using the demethylating drug 5-aza, we also tested whether and how IL-1β gene expression is regulated in BV2 and primary microglial cells. We hypothesized that aged mice and those stimulated with an immune challenge would have IL-1β promoter hypomethylation associated with increased IL-1β gene expression, decreased expression of epigenetic regulators that promote DNA methylation, and that BV2 and adult primary microglial cells would have IL-1β promoter hypomethylation similar to an “aging” phenotype with the treatment of 5-aza.
2. Methods
2.1 Animals
Adult (4–6-month-old) C57BL/6 mice were either purchased from Charles River or reared in-house and aged (24–26-month-old) C57BL/6 mice were purchased from the National Institute of Aging. They were individually housed in a temperature-controlled environment with a reversed-phase light/dark cycle (lights on 21:00h). Mice that were purchased were allowed to acclimate to their environment for at least 4 weeks before experimentation. All studies were carried out in accordance with United States National Institutes of Health guidelines and were approved by the University of Illinois Institutional Animal Care and Use Committee.
2.2 Immune Challenge
Escherichia coli LPS (serotype 0127:B8, Sigma, St. Louis, MO, USA) was dissolved in sterile saline before experimentation. Mice from both age groups were given LPS (0.33 mg/kg body weight) or saline IP. This dose of LPS was selected based upon previous studies demonstrating that 0.33 mg/kg LPS produced prolonged sickness behavior in aged compared to young mice (Godbout, et al., 2005). Treatments were administered at 0900 h for all cohorts.
2.3 Behavioral testing
Sickness behavior was assessed by changes in body weight and locomotor activity. Body weight and locomotor behavior were measured at baseline and 4 or 24 h after treatment. Mice were placed in clear plexiglass cages identical to their home cage but devoid of bedding or nesting material. Clear plexiglass lids were placed on top of test cages to prevent escape while facilitating video recording. Locomotor activity was evaluated by virtual division of the cage into four equal quadrants and then tallying the number of line crossings and rearings each mouse displayed during the 5 min test period. Videos were scored by a trained experimenter blinded to treatment.
2.4 Microglia Isolation
Animals were euthanized via CO2 asphyxiation 4 or 24 h after treatment, perfused with sterile ice-cold saline, and brain tissue was collected and used immediately for microglia isolation using a procedure adapted from Nikademova & Watters (Nikodemova and Watters, 2012). To ensure a sufficient number of cells were recovered, tissue samples from two mice were pooled within a given experimental group. Brains were enzymatically digested using the Neural Tissue Dissociation Kit (Miltenyi Biotec, Germany) for 35 min at 37° C. Further processing was performed at 4° C. Tissue debris was removed by passing the cell suspension through a 40 µm cell strainer. After myelin removal using 30% Percoll Plus (GE Healthcare, Princeton, NJ, USA), cells in PBS supplemented with 0.5% BSA and 2 mM EDTA were incubated for 15 minutes with anti-FITC magnetic beads and anti-Cd11b-FITC antibody (BD Biosciences) for flow cytometry or anti-Cd11b magnetic beads (Miltenyi Biotec, Germany) for gene expression and DNA methylation arrays. CD11b+ cells were extensively washed and separated in a magnetic field using MS columns (Miltenyi Biotec, Germany). Cell yields for isolated microglia were around 1 × 106 cells per two brains, and were not different between treatments.
2.5 Extracellular and Intracellular Flow Cytometric Analysis
Flow cytometric analysis of CD11b+ cell surface and intracellular markers was performed based on BD Cytofix/Cytoperm Plus fixation/permeabilization protocol (BD Biosciences, San Jose, CA, USA), as described previously, with a few modifications (Henry, et al., 2009). Isolated cells were incubated in DMEM (Bio-Whittaker, Cambrex, MD, USA) with 10% FBS (Hyclone, Logan, UT), 100 units/ml penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) and brefeldin A (BD Biosciences) at 37° C in a humidified incubator under 5% CO2 for 4 h. After incubation cells were washed and re-suspended in PBS/0.5% BSA/0.01% sodium azide solution (flow buffer) and Fc receptors were blocked with anti-CD16/CD32 antibody (eBioscience, San Diego, CA, USA). Cells were then incubated with anti-CD45-APC, anti-MHC-II-R-PE, and anti-Ly-6C-BV421 (BD Biosciences). Next, cells were fixed and permeabilized with BD Cytofix/CytoPerm™ solution, washed and re-suspended in BD Perm/Wash™ buffer, and incubated with anti-IL-1β-PE (eBioscience) for 30 min. Cells were washed in BD Perm/Wash™ buffer and re-suspended in flow buffer. Expression of surface and intracellular antigens was determined using a Becton-Dickinson LSR II Flow Cytometer (Red Oaks, CA, USA). Ten to twenty thousand events were collected and gating was determined based on fluorescently labeled isotype antibodies for FITC, APC, PE, and BV421 (BD Biosciences) incubated for 15 min with compensation beads from the AbC™ Total Antibody Compensation Bead Kit (Thermo Fisher Scientific, Grand Island, NY, USA). Unstained samples were used as controls. Flow data were analyzed using FCS Express software (De Novo Software, Los Angeles, CA, USA).
2.6 BV2 and primary microglia cell culture and drug treatments
BV2 cells were maintained in 150-cm2 tissue culture flasks (BD Falcon) in DMEM supplemented with 10% FBS and 100 units/ml penicillin/streptomycin at 37° C in a humidified incubator under 5% CO2. When cells reached confluence they were passed by trypsinization, resuspended in serum-free DMEM and seeded in 12-well plates (BD Falcon). Cells were treated one day later with 10µM of 5-aza (Sigma, St. Louis, MO, USA) for 48 h with daily media replacement and stimulated with 100ng/mL LPS 4 or 24 h later. Primary microglia from young adult mice were plated on poly-L-lysine (Sigma) coated 12-well plates in DMEM supplemented with 10% FBS, 100 units/ml penicillin/streptomycin, and 5 ng/mL GM-CSF and maintained at 37° C in a humidified incu bator under 5% CO2. Cells were treated 7–8 days after culturing with 10µM 5-aza for 24 h and then 100ng/mL LPS for 4h. Both media and cells were collected and cells were put directly in Trizol reagent (Invitrogen) for all cell culture experiments. The viability of BV2 and primary cultured microglia cells was evaluated using the Thermo Scientific Pierce Lactate Dehyrogenase Cytotoxicity Assay Kit (Thermo Scientific, Rockford, IL, USA) in accordance with the manufacturer’s instructions and was <15%.
2.7 Gene Expression and DNA Methylation Assays
Total RNA and DNA from microglia were isolated using the Tri Reagent protocol (Invitrogen). Synthesis of cDNA was carried out using a high-capacity RT kit (Applied Biosystems, Grand Island, NY, USA) according to the manufacturer’s instructions. Real-time quantitative RT-PCR was performed on an ABI PRISM 7900HT-sequence detection system (Perkin Elmer, Forest City, CA, USA) to detect changes in mRNA expression of the epigenetic regulators DNMT1 (catalog no. Mm.PT.58.30881142), DNMT3a (Mm.PT.58.13545327), DNMT3b (Mm.PT.58.31955137), HDAC1 (Mm.PT.58.43356830.g), MeCP2 (Mm.PT.58.13934895.g), and Gadd45b (Mm.PT.58.10699383.g), the pro-inflammatory cytokine IL-1β (Mm00434228_m1), and regulators of IL-1β processing and signaling including IL-1RA (Mm00446186_m1), NLRP3 (Mm.PT.58.6779853.gs), and Casp1 (Mm.PT.58.8975671.gs). All genes were analyzed using PrimeTime real-time quantitative RT-PCR Assays (Integrated DNA Technologies, Coralville, IA, USA) and were compared with the housekeeping control gene GAPDH (Mm99999915_g1) using the 2−ΔΔCt calculation method as previously described (Livak and Schmittgen, 2001). Data are expressed as fold change versus controls. To minimize unequal variances, gene expression for BV2 and primary cultured cells were log10 transformed for statistical analyses only (Figure 4–5 and Table 2 still represent the original data before it was log-transformed). Methylation status was assessed via MSP on bisulfite modified DNA (Zymo Research, Irvine, CA, USA). MSP primers were designed using Methprimer software: (http://www.urogene.org/methprimer/) (Li and Dahiya, 2002). Primer sets targeted a methylated and unmethylated CG dinucleotide in DNA associated with the promoter region of IL-1β, or unmethylated β-tubulin-4 as a reference gene as previously published (Gupta, et al., 2010, Levenson, et al., 2006), and are listed in Table 1. Product specificity was determined by melt curve analysis and gel electrophoresis. For the MSP data, a methylation index was calculated by dividing the fold change value for the methylated primer set by the fold change value for the unmethylated primer as previously described (Blaze, et al., 2013, Sui, et al., 2012).
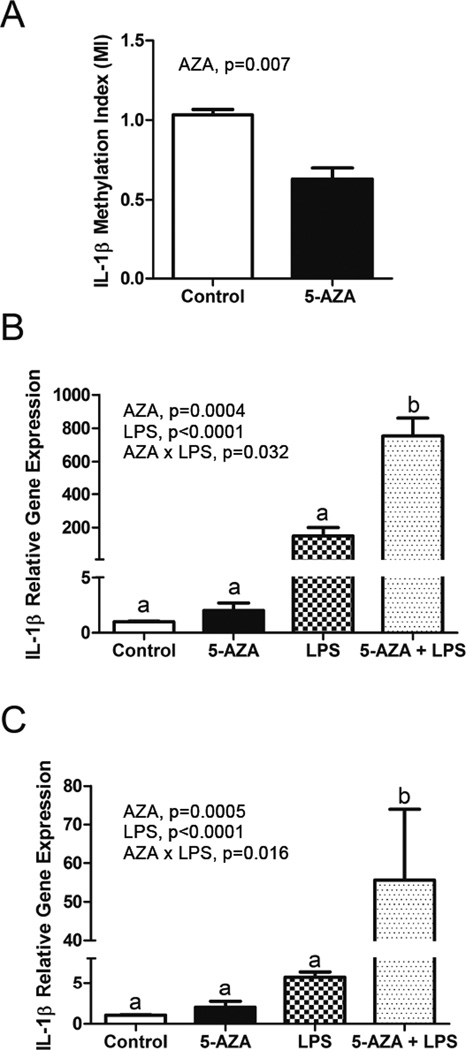
Figure 4. Increased IL-1β gene expression in 5-aza and LPS-treated BV-2 cells coincides with decreased IL-1β promoter DNA methylation.
BV-2 cells were treated with 5-aza for 48 h and/or LPS for 4 or 24 h. Bars represent the mean ± SEM (n = 3–4). A) IL-1β promoter DNA methylation as measured by a Methylation Index (MI). B) lIL-1β mRNA expression after 4h LPS treatment. C) lIL-1β mRNA expression after 24h LPS treatment.
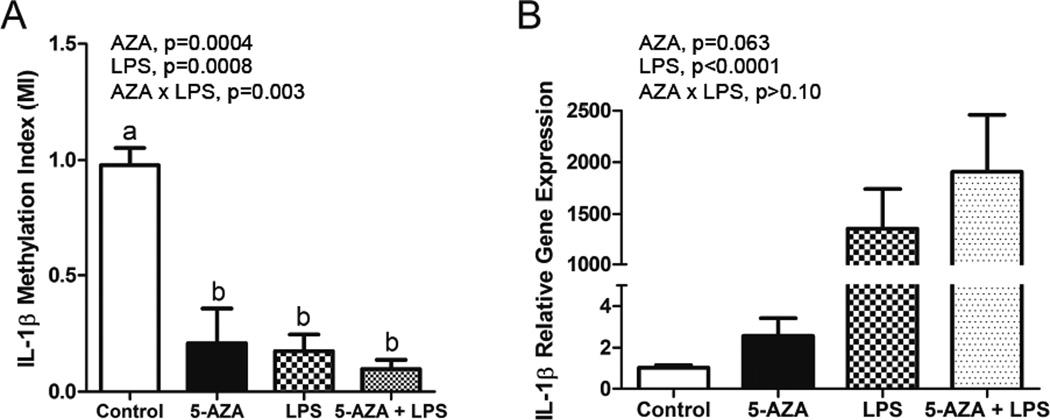
Figure 5. Increased IL-1β gene expression in 5-aza and LPS-treated primary cultured microglia coincides with decreased IL-1β promoter DNA methylation.
Primary cultured microglia were treated with 5-aza for 24 h and/or LPS for 4 h. Bars represent the mean ± SEM (n = 3–4) for A) IL-1β promoter DNA methylation as measured by a Methylation Index (MI) and B) IL-1β mRNA expression.
Table 2.
Expression of IL-1β signaling and processing genes in 5-aza and LPS-treated primary cultured microglia.
| Vehicle | AZA | p-value | |||||
|---|---|---|---|---|---|---|---|
| Gene | Control | LPS | Control | LPS | AZA | LPS | AZA × LPS |
| Casp1 | 1.04±0.12 | 3.09±0.48 | 0.72±0.05 | 2.67±0.17 | 0.074* | <0.0001* | 0.307 |
| IL-1Rn | 1.10±0.18 | 32.73±6.79 | 0.95±0.11 | 26.55±2.86 | 0.632 | <0.0001* | 0.962 |
| NLRP3 | 1.06±0.15 | 42.22±9.60 | 0.68±0.13 | 34.07±2.06 | 0.079* | <0.0001* | 0.285 |
Primary cultured microglia were treated with 5-aza for 24 h and/or LPS for 4 h. Bars represent the mean ± SEM (n = 3–4).
indicates p<0.1.
Table 1.
Primers used in MSP experiments.
| Target Gene | Primer Sequence (5'-3') |
|---|---|
| IL-1β Methylated | TTTTAGTTTAAGTATAAGGAGGCGA |
| ACACATTCGCAAATATATCATCGTA | |
| IL-1β Unmethylated | TTTTAGTTTAAGTATAAGGAGGTGA |
| AACACATTCACAAATATATCATCATA | |
| β –tubulin-4 Unmethylated | GGAGAGTAATATGAATGATTTGGTG |
| CATCTCCAACTTTCCCTAACCTACTTAA |
2.8 Global DNA Methylation Analysis
Global DNA methylation status in primary microglia was performed using the MethylFlash™ Methylated DNA Quantification Kit, an enzyme-linked immunosorbent assay used to detect the total amount of 5mC. The assay was run according to the manufacturer’s instructions (Epigentek, Farmingdale, NY, USA).
2.9 Statistical Analyses
All data were analyzed using Statistical Analysis System (SAS, Cary, NC, USA). Behavior data and data from primary microglia (not cultured) were subjected to two-way analysis of variance for main effects of age and LPS, and all two-way interactions. Data from cell culture experiments were subjected to unpaired t-tests or two-way analysis of variance for main effects of 5-aza and/or LPS, and all interactions. Where analysis of variance revealed a significant interaction (p<0.10 unless noted elsewhere), Fisher’s LSD test was used for post hoc comparisons when appropriate. All data are expressed as means ± SEM.
3. Results
3.1 LPS induced sickness behavior in adult and aged mice
As previously reported in BALB/c mice (Godbout, et al., 2005), aged C57BL/6 mice as well as those given LPS had a decrease in locomotor activity (Supplementary Figure 1). At 4 hours, both age (F (1,35) =11.83, p=0.001) and LPS (F (1,35) =19.51, p<0.0001) produced a significant reduction in the number of line crossings during the locomotor task. At 24 hours, there was still a reduction in locomotor activity with age (F (1,34) =6.24, p=0.018) and LPS (F (1,34) =3.68, p=0.063).
3.2 Primary microglia had increased MHCII expression with age and increased IL-1β expression with age and LPS
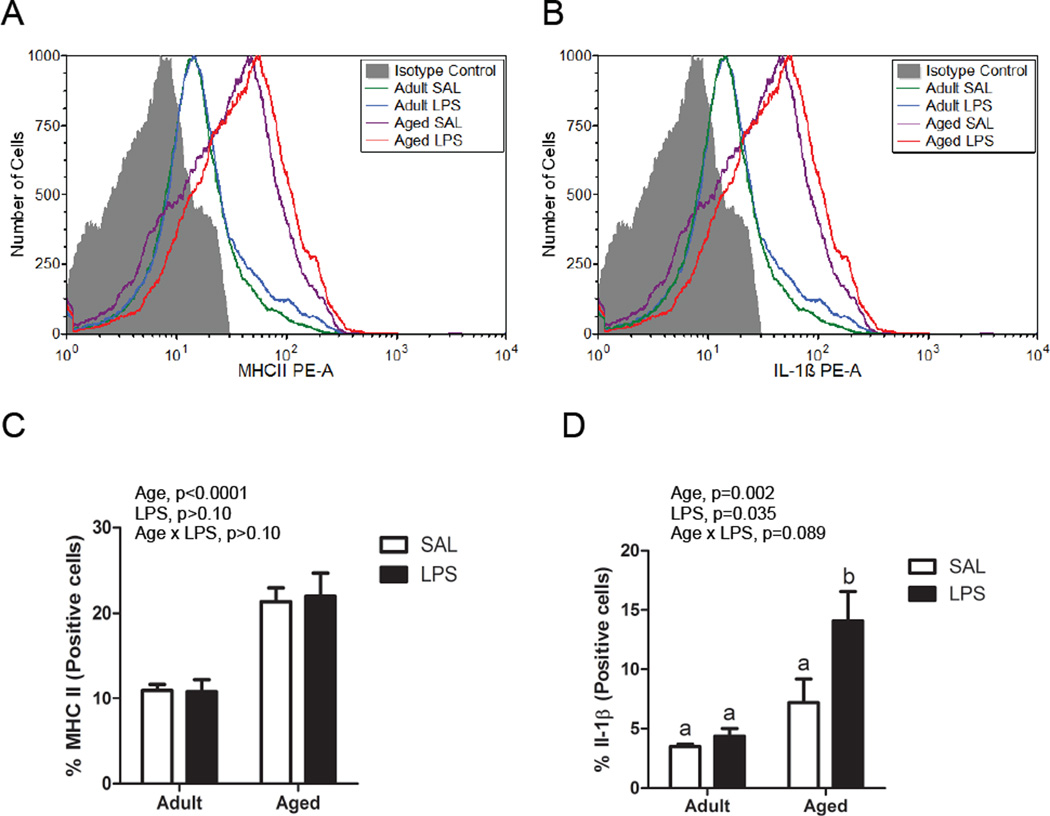
Adult and aged mice were injected with saline or LPS, and brain cells were isolated using Cd11b+ magnetic bead separation. These cells identified as CD11b+, CD45low and Ly6C−, thought to be predominantly microglia, were stained for IL-1β and MHCII and analyzed by flow cytometry. Figure 1A and 1B shows representative histograms and Figure 1C and 1D show the percentages of cells that were IL-1β+ and MHCII+. A greater proportion of microglia from aged mice expressed MHCII compared to adults (F (1,12)=43.19, p<0.0001), while there was no difference between saline and LPS-treated mice, consistent with previous findings (Henry, et al., 2009). Further, both aged animals and those treated with peripheral LPS had an increased percentage of IL-1β+ microglia (F (1,12)=16.82, p=0.002 and F (1,12)=5.66, p=0.035) and there was an interaction in that the LPS-induced increase in IL-1β+ cells was greater in aged mice compared to adults (F (1, 12) = 3.42, p=0.089).
Figure 1. Differential expression of MHCII and IL-1β on microglial cells from saline or LPS-treated adult and aged mice.
Representative histograms of Cd11b+/CD45+/Ly6C- cells incubated with A) anti-MHCII PE and B) anti-IL-1β PE compared with isotype controls. Average percentage of Cd11b+/CD45+/Ly6C- cells that were C) MHCII+ or D) IL-1β+. Bars represent the mean ± SEM (n=4).
3.3 IL-1β promoter DNA methylation decreased in aged and LPS-treated primary microglia
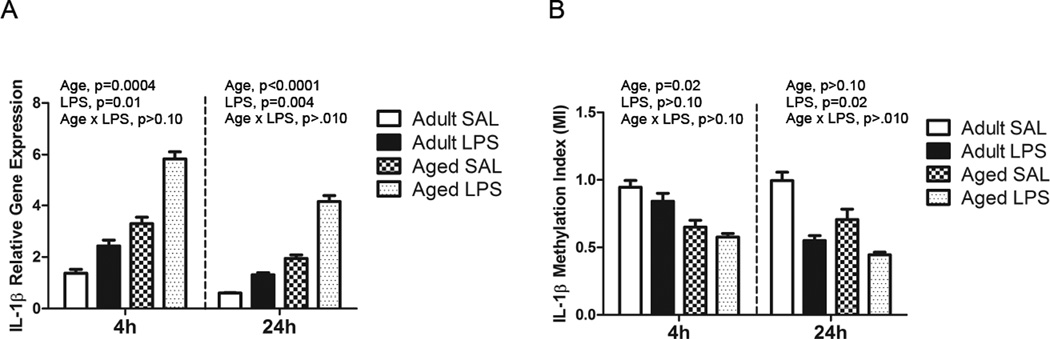
As expected, for IL-1β mRNA there was a main effect of age (F (1, 26) = 16.39, p=0.0004) and LPS (F (1, 26) = 7.51, p=0.01) at 4 hours and main effect of age (F (1, 30) = 20.18, p<0.0001) and LPS (F (1, 30) = 9.9, p=0.004) at 24 hours in that aged mice and mice given LPS had increased IL-1β compared to adult and saline-treated mice (Figure 2A). For IL-1β promoter DNA methylation, there was a main effect of age (F (1, 3) = 6.91, p=0.02) at 4 hours and a main effect of LPS (F (1, 17) = 6.59, p=0.02) at 24 hours in that aged mice and mice given LPS had decreased methylation compared to adult and saline-treated mice (Figure 2B). This coincides with the increase in IL-1β mRNA in aged and LPS-treated mice. DNA from these samples was also used to assess global methylation status, but neither age nor LPS significantly altered 5mC content (data not shown).
Figure 2. Increased microglial IL-1β gene expression in aged and LPS-treated mice coincides with decreased IL-1β promoter DNA methylation.
Adult and aged mice were injected i.p. with LPS or sterile saline and 4 or 24 h later, brain tissue was collected for Cd11b+ cell isolation. Bars represent the mean ± SEM (n = 7–10). A) IL-1β gene expression at 4h and 24h, and B) IL-1β DNA methylation at 4h and 24h.
3.4 Epigenetic regulators are altered by age and LPS in primary microglia
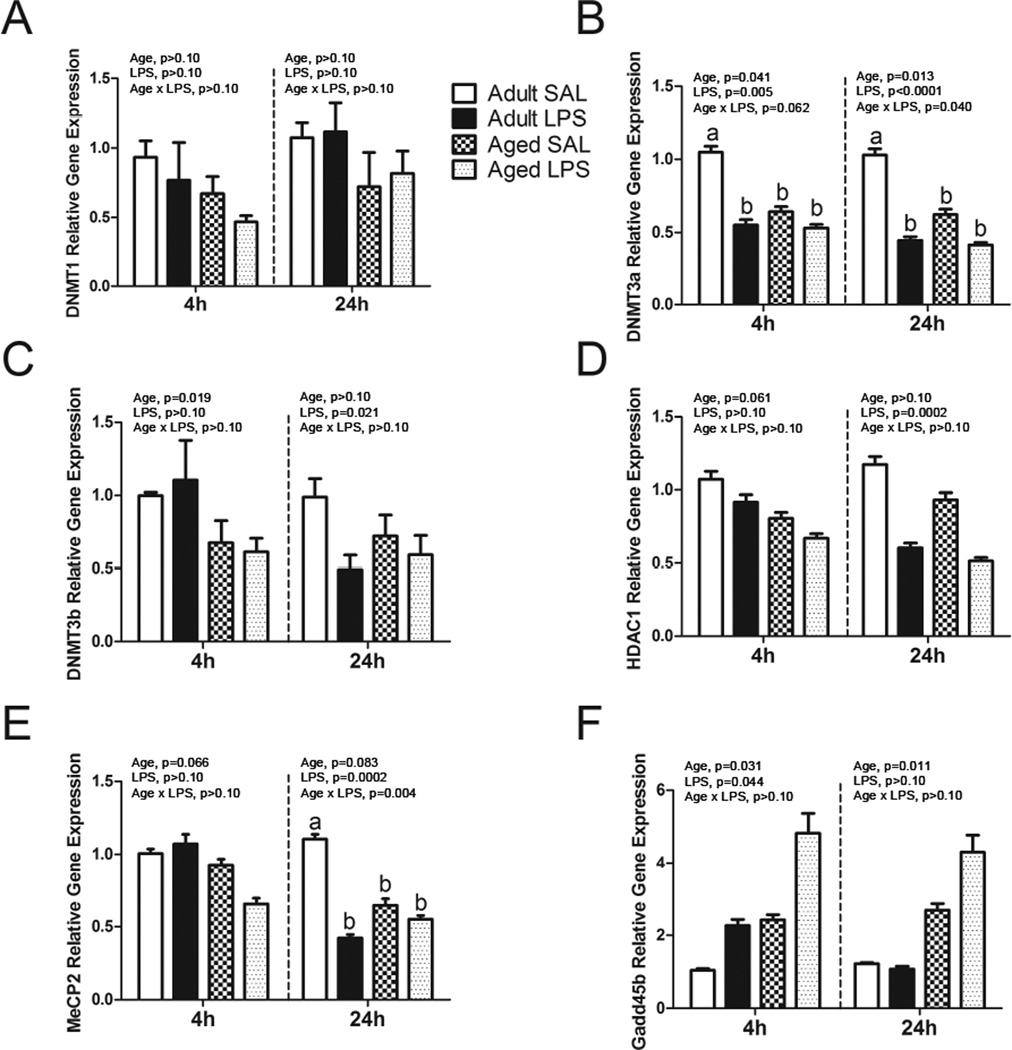
To gain a broader view of the role of epigenetic regulators in neuroinflammation, mRNA for several enzymes that participate in DNA methylation were measured in microglia from adult and aged mice 4 or 24 hours after i.p. injection of LPS (Figure 3). Of note, there were decreases in DNMT3a, DNMT3b, HDAC1, and MeCP2 mRNA with age and/or LPS at 4 and/or 24 hours. Both DNMT3a and MeCP2 had significant age × LPS interactions at 24 hours in that aged saline-treated mice had much lower mRNA levels of DNMT3a and MeCP2 compared to saline-treated adults. For Gadd45b, there were increases in mRNA with age at both 4 and 24 hours and with LPS at 4 hours.
Figure 3. Microglial cell gene expression of epigenetic regulators 4 and 24 h after saline or LPS.
Adult and aged mice were injected i.p. with LPS or sterile saline and 4 or 24 h later, brain tissue was collected for Cd11b+ cell isolation. Data are presented as means ± SEM (n = 7–10) for A) DNMT1, B) DNMT3a, C) DNMT3b, D) HDAC1, E) MeCP2, and F) Gadd45b gene expression.
3.5 Inhibiting DNA methylation increased IL-1β gene expression in BV2 cells and primary microglia
To demonstrate whether or not DNA methylation plays a direct role in primed microglial IL-1β mRNA expression, the BV2 immortalized murine microglial cell line was treated with 5-aza for 48 h and/or LPS for 4 or 24 h. IL-1β promoter DNA methylation was decreased by 5-aza treatment (F (1, 4) = 5.18, p=0.007) (Figure 4A). For IL-1β mRNA expression, there was a main effect of LPS (F (1, 10) = 616.78, p<0.0001), 5-aza (F (1, 10) = 26.73, p=0.0004), and an 5-aza×LPS interaction (F (1, 10) = 6.22, p=0.032) with a 4 hour LPS treatment in that 5-aza and LPS produced a greater increase in IL-1β than 5-aza or LPS alone (Figure 4B). Similarly, for the 24 hour LPS treatment, there was a main effect of LPS (F (1, 10) = 89, p<0.0001), 5-aza (F (1, 10) = 25.7, p=0.0005), and an 5-aza×LPS interaction (F (1, 10) = 8.4, p=0.016) in that 5-aza and LPS produced a greater increase in IL-1β than 5-aza or LPS alone, albeit less than at 4 hours (Figure 4C). Additionally, primary microglial cells were cultured for 7–8 days and treated with 5-aza for 24 h and/or LPS for 4 h. IL-1β promoter DNA methylation was decreased by 5-aza and LPS treatment (F (1, 10) = 26.27, p=0.0004) and F (1, 10) = 22.6, p=0.0008), and there was a 5-aza×LPS interaction (F (1, 10) = 15.08, p=0.003) (Figure 5A). For IL-1β mRNA expression, there was a main effect of LPS (F (1, 9) = 652.96, p<0.0001), and a main effect of 5-aza (F (1, 9) = 4.49, p<0.063) (Figure 5B). The increase in IL-1β mRNA was numerically greatest when LPS and 5-aza were combined, but the 5-aza × LPS interaction was not significant. We also assessed mRNA for genes involved in IL-1β signaling and processing in primary microglia (Table 2). For the 3 genes assessed, there was a main effect of LPS (F (1, 20) = 107.01, p<0.0001, F (1, 20) = 324.97, p<0.0001, and F (1, 20) = 588.09, p<0.0001 for Casp1, IL-1rn, and NLRP3, respectively), and for Casp1 and NLRP3 there was a main effect of 5-aza (F (1, 20) = 3.55, p=0.074 and F (1, 20) = 3.44, p=0.079, respectively). No interactions were detected. These results suggest that several genes involved in IL-1β signaling and processing are affected by DNA methylation, but not in the same way as IL-1β.
4. Discussion
The current study was designed to investigate a link between DNA methylation and the microglial phenotype of aged mice. We clearly demonstrated that age and LPS have the capacity to alter DNA methylation and expression levels of genes important for establishing, maintaining, or reversing DNA methylation in microglial cells. Of note, both aging and stimulation of the immune system with a pro-inflammatory agent (LPS) produces sickness behavior and decreases in methylation of the IL-1β promoter that coincide with increases in IL-1β gene expression and intracellular IL-1β production, suggesting that DNA methylation plays a role in the overexpression of this pro-inflammatory cytokine with age. Although the percentage of IL-1β+ cells do not completely match previous findings (Henry, et al., 2009), it is likely that our cell population is more highly purified now that the magnetic bead separation technique is used instead of the previous isolation method with a Percoll gradient (Burton, et al., 2013, Henry, et al., 2009). Importantly, percentage of Cd11b+/CD45low cells are considerably higher with the new method (Nikodemova and Watters, 2012).
There are a number of possible mechanisms that could be mediating these alterations in DNA methylation with age and LPS. As NF-κB activation is implicated in microglial aging (Zhang, et al., 2013) and sickness behavior (Nadjar, et al., 2005), NF-κB binding activity might play a role in demethylation of the IL-1β promoter. The site of methylation investigated in this study is adjacent to a canonical NF-κB binding site (Lebedeva and Singh, 1997) and recently, the NF-κB subunit RelB at the IL-1β promoter was demonstrated to be required for the attenuated LPS response in LPS-preconditioned microglia (Schaafsma, et al., 2015), thus providing evidence that epigenetic regulation of IL-1β in microglia from aged mice could be regulated by increased NF-κB binding that’s inhibiting proper DNA methylation to be established. Further, it’s been demonstrated that IL-1β induces its own expression in healthy chondrocytes and causes loss of DNA methylation (Hashimoto, et al., 2009). This has the potential to set up a dangerous positive feedback mechanism, and if true in vivo, could partially explain the progression of neuroinflammation in aging. It should be stressed that additional studies are necessary to link these gene methylation and expression changes with histone alterations, subsequent behavioral outcomes, and to explore their reversibility.
Global DNA methylation was unchanged by age or LPS stimulation, suggesting that perhaps DNA hypomethylation is not a global phenomenon as it is in other cell types such as fibroblasts (Wilson and Jones, 1983). As epigenetic regulation of other pro-inflammatory genes such as TNF-α and IL-6 have been demonstrated in other immune cell types such as natural killer cells and peripheral monocytes/macrophages (Eddy, et al., 2014, Sullivan, et al., 2007), it will be pertinent to explore regulation by DNA methylation and other epigenetic modifications in microglia, as well as other signature genes relevant to aging such as those recently found in the microglial sensome (Hickman, et al., 2013) or other recent transcriptomic studies (Holtman, et al., 2015, Orre, et al., 2014).
Gene expression of epigenetic regulators was found to be altered with age and LPS, and support a number of previously established findings. DNMT3a and DNMT3b mRNA decreased with age, and these genes decrease with age in fibroblasts and T cells (Lopatina, et al., 2002, Zhang, et al., 2002). LPS decreased DNMT3a expression, and this is consistent with a previous finding that LPS downregulates DNMT3a in calf peripheral blood mononuclear cells (PBMCs) that occurred concurrently with high expression of pro-inflammatory genes (Doherty, et al., 2013). Further, as differences in DNMT3b expression have the ability to influence adipose tissue macrophage polarization, perhaps the microglial polarization seen in aging is acting through a similar mechanism (Yang, et al., 2014). LPS decreased HDAC1 expression and age also slightly decreased expression at 4 hours (p<0.1), but these results are more difficult to interpret based on existing literature. HDACs in general have been shown to be decreased with aging (Gravina and Vijg, 2010), but evidence has also shown that HDACs are increased with LPS in macrophages (Aung, et al., 2006). It has been suggested that HDACs are important in limiting the inflammatory response as they have been reported to modulate Toll-like receptor (TLR)-mediated signaling and to inhibit NF-κB signaling (Aung, et al., 2006), but inhibiting HDACs has also been demonstrated to suppress innate immune activation in microglia (Kannan, et al., 2013). Additional work will be necessary to delineate these differential effects of HDACs in modulating inflammation, but nonetheless the current findings support the idea that decreased expression of co-repressors like HDAC1 are associated with decreased IL-1β promoter DNA methylation.
The significant decreases in MeCP2 with age are interesting considering a deficiency in MeCP2 has been widely associated with Rett syndrome, a neurodevelopmental disorder causing mental retardation with early onset in childhood, and MeCP2 mutant mice have been shown to exhibit many of the same cognitive deficits and neuroanatomical abnormalities associated with Rett syndrome (Guy, et al., 2001). Intriguingly, selective return of MeCP2 function to microglia arrests Rett syndrome pathology (Derecki, et al., 2013). The mechanism is unclear but may relate to phagocytic ability, as microglia from Mecp2-null mice had severe impairments in phagocytic ability in vitro. The authors proposed that insufficient clearance of debris within the brains of these animals might indeed contribute to the severity of pathology, and it seems that this same phenomena could also be a mechanism involved in aging. Gadd45b was the only gene to increase with age, but expectedly so in that it plays a major role in DNA demethylation. Previous research has demonstrated that LPS, TNF, IL-6 and other inducers of oxidative stress can increase Gadd45b. Further, NF-κB can stimulate the expression of Gadd45 and simultaneously acts as its regulatory target, thus creating a positive feedback loop (Moskalev, et al., 2012).
As DNA hypomethylation is often described in context with gene activation, another important finding was to demonstrate that a demethylating drug like 5-aza has the capacity to decrease IL-1β promoter DNA methylation and subsequently increase IL-1β mRNA in both BV2 and primary microglia, establishing that DNA methylation does indeed play a direct role in IL-1β gene regulation. This also indicates that BV2 and cultured primary microglia have the capacity to display DNA methylation patterns in a manner consistent with what was seen in freshly isolated microglia (Figure 2). This is important because primary microglia and cell lines in culture have been shown to lose their microglial signature (Butovsky, et al., 2014, Gosselin, et al., 2014). The exaggerated increase in IL-1β mRNA with both 5-aza and LPS in BV2 cells suggests that DNA hypomethylation allows LPS to trigger a more pronounced immune response, and is in line with previous findings in other cell lines (Kovacs, et al., 1987, Wessels, et al., 2010).
It is also important to consider other factors that can lead to a heightened proinflammatory response with aging, such as expression of the endogenous receptor antagonist of IL-1β, IL-1ra (Spulber, et al., 2009) or of inflammasomes (Goldberg and Dixit, 2015). As such, mRNA expression of several genes involved in IL-1β signaling and processing were also investigated in primary microglia treated with 5-aza or LPS. All IL-1β-related genes were upregulated with LPS as expected, but 5-aza decreased Casp1 and NLRP3 mRNA expression with or without LPS. This suggests that Casp1 and NLRP3 are additionally affected by DNA methylation, but not in the same way as IL-1β. DNMT inhibitors in general create global demethylation of the genome, but there is some heterogeneity of response of loci (Pandiyan, et al., 2013). Depending on cell type and site of methylation (promoter versus gene body methylation, for example), DNA methylation and transcription can be positively correlated, or variable chromatin accessibility and constitutive DNA hypomethylation can occur (Wagner, et al., 2014). Histone modifications also have a close relationship with DNA methylation, and could be more influential on DNA methylation and transcription of some genes compared to others (Cedar and Bergman, 2009). More analyses of these genes involved in IL-1β signaling and processing will be necessary to understand their effects on microglial IL-1β with aging. However, the observation that 5-aza decreases IL-1β promoter DNA methylation and increases IL-1β mRNA expression (but not mRNA expression of all IL-1β signaling and processing genes) suggests that this is a specific relationship that could be important to understand the changes in IL-1β within the brain and specifically microglia from aged individuals.
Although the increase in IL-1β mRNA with 5-aza and LPS was not significantly exaggerated when combined in primary cultured microglial cells, perhaps they are not capable of the same effects in culture and cannot perform in a similar manner in isolation without the influence of other cell types within the brain. Further, the techniques we used in this study do not allow us to draw any conclusions regarding cytosine methylation verses hydroxymethylation, subregion specificity, or site-specific methylation. In the future it will be useful to use bisulfite pyrosequencing to characterize site-specfic methylation changes of the IL-1β promoter.
These findings highlight the importance of elucidating the interaction of the environment with the epigenome and its role in aging. Manipulating and understanding the epigenome holds promises for preventing and treating age-related diseases, such as restoring the DNA methylation profile of aging cells to that of young or mature cells and preventing significant age-associated demethylation. Specifically, a better understanding of how epigenetic mechanisms like DNA methylation become dysregulated with age in microglia is needed to improve our understanding of neuroinflammatory complications and lead to the development of therapeutic interventions.
Supplementary Material
Highlights.
Immune challenge in aged mice leads to exaggerated neuroinflammation
DNA methylation of IL-1β may play a role in microglial dysfunction with age
5-azacytidine decreases IL-1β methylation in culture similar to microglia from aged mice
Acknowledgments
This work was supported by NIH R01 AG16710. We would like to thank Jalisa Zimmerman for her help in generating animals, behavior coding, and biochemistry.
Abbreviations
- 5-aza
5-azacytidine
- 5-mC
5-methylcytosine
- APC
Allophycocyanin
- Casp1
Caspase 1
- DNMT
DNA methyltransferase
- FITC
Fluorescein isothiocyanate
- Gadd45b
Growth arrest and DNA-damage-inducible beta
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- HDAC
Histone deacetylase
- IL
Interleukin
- IL-1rn
Interleukin-1 receptor antagonist
- IP
Intraperitoneally
- LPS
Lipopolysaccharide
- MeCP2
Methyl-CpG binding protein-2
- MSP
Methylation specific real-time PCR
- NLRP3
NLR family, pyrin domain containing 3
- PBS
Phosphate-buffered saline
- PCR
Polymerase chain reaction
- PE
Phycoerthrin
- RT
Reverse transcriptase
- SIRT
Sirtuin
- TNF
Tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors have no conflicts of interest to disclose.
References
- Aung HT, Schroder K, Himes SR, Brion K, van Zuylen W, Trieu A, Suzuki H, Hayashizaki Y, Hume DA, Sweet MJ, Ravasi T. LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20(9):1315–1327. doi: 10.1096/fj.05-5360com. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Blaze J, Scheuing L, Roth TL. Differential methylation of genes in the medial prefrontal cortex of developing and adult rats following exposure to maltreatment or nurturing care during infancy. Developmental neuroscience. 2013;35(4):306–316. doi: 10.1159/000350716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton MD, Rytych JL, Amin R, Johnson RW. Dietary Luteolin Reduces Proinflammatory Microglia in the Brain of Senescent Mice. Rejuvenation research. 2016 doi: 10.1089/rej.2015.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton MD, Rytych JL, Freund GG, Johnson RW. Central inhibition of interleukin-6 trans-signaling during peripheral infection reduced neuroinflammation and sickness in aged mice. Brain, behavior, and immunity. 2013;30:66–72. doi: 10.1016/j.bbi.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nature neuroscience. 2014;17(1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun CJ, Seo J, Jo SA, Park YJ, Klug M, Rehli M, Park MH, Jo I. DNA methylation of the 5'-untranslated region at +298 and +351 represses BACE1 expression in mouse BV-2 microglial cells. Biochemical and biophysical research communications. 2012;417(1):387–392. doi: 10.1016/j.bbrc.2011.11.123. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chen J, Buchanan JB, Sparkman NL, Godbout JP, Freund GG, Johnson RW. Neuroinflammation and disruption in working memory in aged mice after acute stimulation of the peripheral innate immune system. Brain, behavior, and immunity. 2008;22(3):301–311. doi: 10.1016/j.bbi.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Chen JA, Sayed F, Ward ME, Gao F, Nguyen TA, Krabbe G, Sohn PD, Lo I, Minami S, Devidze N, Zhou Y, Coppola G, Gan L. SIRT1 deficiency in microglia contributes to cognitive decline in aging and neurodegeneration via epigenetic regulation of IL-1beta. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35(2):807–818. doi: 10.1523/JNEUROSCI.2939-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damani MR, Zhao L, Fontainhas AM, Amaral J, Fariss RN, Wong WT. Age-related alterations in the dynamic behavior of microglia. Aging cell. 2011;10(2):263–276. doi: 10.1111/j.1474-9726.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Kipnis J. The role of microglia in brain maintenance: implications for Rett syndrome. Trends in immunology. 2013;34(3):144–150. doi: 10.1016/j.it.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilger RN, Johnson RW. Aging, microglial cell priming, and the discordant central inflammatory response to signals from the peripheral immune system. Journal of leukocyte biology. 2008;84(4):932–939. doi: 10.1189/jlb.0208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty R, O'Farrelly C, Meade KG. Epigenetic regulation of the innate immune response to LPS in bovine peripheral blood mononuclear cells (PBMC) Veterinary immunology and immunopathology. 2013;154(3–4):102–110. doi: 10.1016/j.vetimm.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Eddy JL, Krukowski K, Janusek L, Mathews HL. Glucocorticoids regulate natural killer cell function epigenetically. Cellular immunology. 2014;290(1):120–130. doi: 10.1016/j.cellimm.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. The journals of gerontology Series A, Biological sciences and medical sciences. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- Godbout JP, Chen J, Abraham J, Richwine AF, Berg BM, Kelley KW, Johnson RW. Exaggerated neuroinflammation and sickness behavior in aged mice after activation of the peripheral innate immune system. The FASEB Journal. 2005 doi: 10.1096/fj.05-3776fje. [DOI] [PubMed] [Google Scholar]
- Goldberg EL, Dixit VD. Drivers of age-related inflammation and strategies for healthspan extension. Immunol Rev. 2015;265(1):63–74. doi: 10.1111/imr.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159(6):1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina S, Vijg J. Epigenetic factors in aging and longevity. Pflugers Archiv: European journal of physiology. 2010;459(2):247–258. doi: 10.1007/s00424-009-0730-7. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. HISTONE METHYLATION REGULATES MEMORY FORMATION. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis and rheumatism. 2009;60(11):3303–3313. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CJ, Huang Y, Wynne AM, Godbout JP. Peripheral lipopolysaccharide (LPS) challenge promotes microglial hyperactivity in aged mice that is associated with exaggerated induction of both pro-inflammatory IL-1beta and anti-inflammatory IL-10 cytokines. Brain, behavior, and immunity. 2009;23(3):309–317. doi: 10.1016/j.bbi.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nature neuroscience. 2013;16(12):1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, Wes PD, Moller T, Orre M, Kamphuis W, Hol EM, Boddeke EW, Eggen BJ. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta neuropathologica communications. 2015;3:31. doi: 10.1186/s40478-015-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kannan V, Brouwer N, Hanisch UK, Regen T, Eggen BJ, Boddeke HW. Histone deacetylase inhibitors suppress immune activation in primary mouse microglia. Journal of neuroscience research. 2013;91(9):1133–1142. doi: 10.1002/jnr.23221. [DOI] [PubMed] [Google Scholar]
- Kovacs EJ, Oppenheim JJ, Carter DB, Young HA. Enhanced interleukin-1 production by human monocyte cell lines following treatment with 5-azacytidine. Journal of leukocyte biology. 1987;41(1):40–46. doi: 10.1002/jlb.41.1.40. [DOI] [PubMed] [Google Scholar]
- Lebedeva TV, Singh AK. Constitutive activity of the murine IL-1 beta promoter is regulated by a transcriptional repressor. Biochimica et biophysica acta. 1997;1353(1):32–38. doi: 10.1016/s0167-4781(97)00040-7. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. The Journal of biological chemistry. 2006;281(23):15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics (Oxford, England) 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha S, Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. Journal of cellular biochemistry. 2002;84(2):324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Smit-McBride Z, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Tacutu R, Fraifeld VE. Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing research reviews. 2012;11(1):51–66. doi: 10.1016/j.arr.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjar A, Bluthe RM, May MJ, Dantzer R, Parnet P. Inactivation of the cerebral NFkappaB pathway inhibits interleukin-1beta-induced sickness behavior and c-Fos expression in various brain nuclei. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2005;30(8):1492–1499. doi: 10.1038/sj.npp.1300755. [DOI] [PubMed] [Google Scholar]
- Nikodemova M, Watters J. Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. Journal of Neuroinflammation. 2012;9(1):147. doi: 10.1186/1742-2094-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre M, Kamphuis W, Osborn LM, Melief J, Kooijman L, Huitinga I, Klooster J, Bossers K, Hol EM. Acute isolation and transcriptome characterization of cortical astrocytes and microglia from young and aged mice. Neurobiology of aging. 2014;35(1):1–14. doi: 10.1016/j.neurobiolaging.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Pandiyan K, You JS, Yang X, Dai C, Zhou XJ, Baylin SB, Jones PA, Liang G. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Research. 2013;41(7):3973–3985. doi: 10.1093/nar/gkt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Vanyushin BF. Age-Related Genomic Hypomethylation. 2010:11–27. [Google Scholar]
- Schaafsma W, Zhang X, van Zomeren KC, Jacobs S, Georgieva PB, Wolf SA, Kettenmann H, Janova H, Saiepour N, Hanisch UK, Meerlo P, van den Elsen PJ, Brouwer N, Boddeke HW, Eggen BJ. Long-lasting pro-inflammatory suppression of microglia by LPS-preconditioning is mediated by RelB-dependent epigenetic silencing. Brain, behavior, and immunity. 2015;48:205–221. doi: 10.1016/j.bbi.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Bilbo SD. Adolescent morphine exposure affects long-term microglial function and later-life relapse liability in a model of addiction. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(3):961–971. doi: 10.1523/JNEUROSCI.2516-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A, Gottfried-Blackmore AC, McEwen BS, Bulloch K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia. 2007;55(4):412–424. doi: 10.1002/glia.20468. [DOI] [PubMed] [Google Scholar]
- Spulber S, Bartfai T, Schultzberg M. IL-1/IL-1ra balance in the brain revisited - evidence from transgenic mouse models. Brain, behavior, and immunity. 2009;23(5):573–579. doi: 10.1016/j.bbi.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Sui L, Wang Y, Ju LH, Chen M. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiology of learning and memory. 2012;97(4):425–440. doi: 10.1016/j.nlm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Sullivan KE, Reddy ABM, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, Bhatia M. Epigenetic Regulation of Tumor Necrosis Factor Alpha. Mol Cell Biol. 2007;27(14):5147–5160. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay ME, Zettel ML, Ison JR, Allen PD, Majewska AK. Effects of aging and sensory loss on glial cells in mouse visual and auditory cortices. Glia. 2012;60(4):541–558. doi: 10.1002/glia.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome biology. 2014;15(2):R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels I, Fleischer D, Rink L, Uciechowski P. Changes in chromatin structure and methylation of the human interleukin-1beta gene during monopoiesis. Immunology. 2010;130(3):410–417. doi: 10.1111/j.1365-2567.2009.03243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang X, Liu D, Yu L, Xue B, Shi H. Epigenetic Regulation of Macrophage Polarization by DNA Methyltransferase 3b. Molecular Endocrinology. 2014;28(4):565–574. doi: 10.1210/me.2013-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li J, Purkayastha S, Tang Y, Zhang H, Yin Y, Li B, Liu G, Cai D. Hypothalamic programming of systemic ageing involving IKK-beta, NF-kappaB and GnRH. Nature. 2013;497(7448):211–216. doi: 10.1038/nature12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Deng C, Lu Q, Richardson B. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mechanisms of ageing and development. 2002;123(9):1257–1268. doi: 10.1016/s0047-6374(02)00014-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.