Abstract
Acute and chronic alcohol exposure evidently influences epigenetic changes, both transiently and permanently, and these changes in turn influence a variety of cells and organ systems throughout the body. Many of the alcohol-induced epigenetic modifications can contribute to cellular adaptations that ultimately lead to behavioral tolerance and alcohol dependence. The persistence of behavioral changes demonstrates that long-lasting changes in gene expression, within particular regions of the brain, may contribute importantly to the addiction phenotype. The research activities over the past years have demonstrated a crucial role of epigenetic mechanisms in causing long lasting and transient changes in the expression of several genes in diverse tissues, including brain. This has stimulated recent research work that is aimed at characterizing the influence of epigenetic regulatory events in mediating the long lasting and transient effects of alcohol abuse on the brain in humans and animal models of alcohol addiction. In this study, we update our current understanding of the impact of alcohol exposure on epigenetic mechanisms in the brain and refurbish the knowledge of epigenetics in the direction of new drugs development.
Key Words: Alcohol, Epigenetic changes, DNA methylation, Histone modifications
Introduction
The term ‘epigenetics’ has been used to describe events that could not be clarified by genetic principles. Historically, the term ‘epigenetics’ was coined by British biologist Conrad Waddington. According to Waddington, ‘epigenetics’ is the study of interaction between the genome and environment that causes heritable changes in the expression of genes, which occurs without any change in DNA sequence [1]. Epigenetics can also be described as an intermediate link between the genotype and phenotype, a phenomenon that changes the final outcome of a locus or chromosome without changing the underlying DNA sequence. These epigenetic alterations could be passed from the one generation to the other. Although, there is evidence for inheritance of epigenetic changes, there is little data which explain the exact mechanisms of epigenetics inheritance [2,3]. The mechanism of epigenetics is used for information creation and its storage in response to a transient environmental signal. The epigenetic process is also helpful in cell differentiation, regulation of cell type, specific transcriptional programs, which produce remarkable heterogeneity of cellular transcriptome [4].
Alcohol is one of the most commonly used addictive substances, and its persistent use can lead to the manifestation of tolerance, dependence, and finally addiction [5]. Several studies have demonstrated that both genetic and environmental risk factors can play a role in the manifestation of alcohol addiction [6]. Genetic studies in both humans and animal models of alcoholism have identified several genes that may be critical in the pathophysiology of alcoholism [7,8]. From the numerous research findings, it is evident that the exploration of epigenetic mechanisms is a potential area of research for a better understanding of the molecular mechanisms of human diseases including psychiatric and alcohol use disorders (AUDs) [9]. In this study, we highlight the mechanism of different types of epigenetic changes, impact of alcohol on these epigenetic changes in the brain and beyond. In addition, we try delineating the potential epigenetic drug targets.
Epigenetic Mechanisms in the Brain and Impact of Alcohol
The brain is certainly one of the most complex biological organs that makes the organism to do much critical sensory and motor functions. It persistently adapts to environmental stimuli through regulated changes in gene expression. Chronic alcohol exposure leads to extensive changes in brain gene expression in humans and animal models, and there are evidences which indicate that many of these changes mediate the processes of cellular adaptations leading to addiction [10]. Over the years, the role of epigenetic processes in alcohol's effects on the central nervous system (CNS) has been understudied. Conversely, in recent years, there are numerous studies that suggest the important role of epigenetics in alcohol-related molecular and behavioral changes [10]. Different types of epigenetic mechanisms include DNA methylation, histone methylation, phosphorylation, ubiquitylation, sumoylation, and ADP ribosylation. In addition to histone modifications, adenosine triphosphate (ATP), adenine triphosphate-dependent chromatin remodeling, and regulation of gene expression by non-coding RNAs are other epigenetic mechanisms.
DNA Methylation
DNA methylation is a process that transfers a methyl group from S-adenosyl methionine (SAM) to a cytosine residue modifying it to 5-methyl cytosine. The process is catalyzed and maintained by enzymes called DNA methyltransferases (DNMTs). There are 3 main types of DNMTs that regulate DNA methylation. DNMT1 is located at the replication fork and maintains methylation of DNA strands which are newly biosynthesized, thus the enzyme is considered as a primary maintenance DNMT. Another couple of DNMTs, DNMT3A and 3B are mainly responsible for long-term memory formation, causing structural changes in DNA [11]. DNA methylation mainly takes place at CpG sites, nucleotide sequences consisting of a cytosine followed by a guanine. CpG dinucleotides are present in a lesser level throughout the human genome, but are found in high level in the areas called CpG islands which are located in the promoter region of genes near the transcription start site. A CpG island is defined as an area of DNA having at least 200 bp, where the content of GC is more than 60% [12]. DNA methylation promoter region is associated with gene silencing. DNA methylation within regulatory regions (i.e., promoters) of a gene has long been known to attenuate gene expression. However, more recently DNA methylation within the gene also has been shown to regulate tissue- and cell-specific gene expression [13,14].
DNA Methylation and Its Neurological Implications
To date, several studies have produced abundant evidences for the role of DNA methylation in brain physiology and pathophysiology. For instance, a study has reported that DNA methylation results in the difference of the neuronal identity and regional functional specialization [15]. Similarly, DNA methylation (de novo) has been shown to prevent the premature expression of neural stem cellsʼ (NSCs) association and alternate differentiation programs [16]. DNA methylation represses gene expression via the actions of a protein called methyl CpG-binding protein-2 (MeCP2), which selectively binds to methylated DNA, thereby blocking transcription. Mutations in MeCP2 have been linked to a neurodevelopmental disorder, the Rett syndrome [17]. A recent study has further proclaimed the important role of DNA methylation in neuronal health and function, in which the in vivo induction of apoptosis in motor neurons by toxic drugs or peripheral nerve lesions was associated with pre-apoptotic rise in the amounts of DNMT1 and DNMT3A in the cell nucleus [18]. In addition, the role of DNA methylation has been implied in several neurological diseases such as autism spectrum disorders [19], Alzheimer's disease [20,21], schizophrenia, and bipolar disorder [22,23,24].
Impact of Alcohol on DNA Methylation
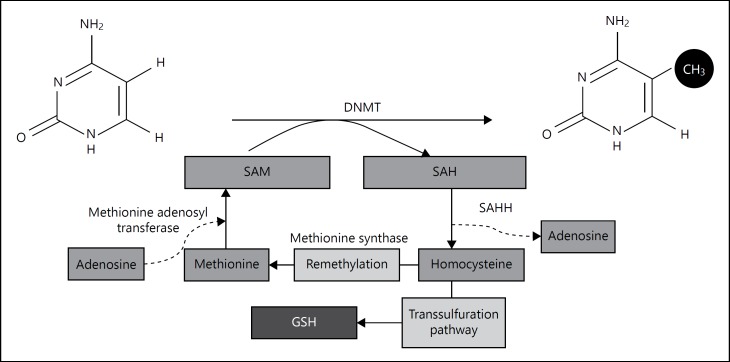
DNA methylation proceeds under the catalysis of DNMTs, which transfer a methyl group from SAM to 5-methyl cytosine and produce S-adenosyl homocysteine (SAH) as an outcome of the reaction. SAH in turn acts as a potent inhibitor of DNMT, which catalyzes the methyl group transfer reaction. The resultant SAH from DNA methylation is then broken down by SAH hydrolase (SAHH) to form homocysteine, which can either enter a set of reactions called the trans-sulfuration pathway to form glutathione (GSH) or be remethylated to form methionine [25]. For remethylation of homocysteine, a methyl group can be transferred either from N5-methyl tetrahydrofolate by methionine synthase, or from betaine by betaine homocysteine methyl transferase (fig. 1). Excessive ROS formation, which can occur during ethanol metabolism, acutely can deplete GSH. This could promote the transsulfuration of homocysteine to generate new GSH and thus divert the reactions from producing methionine and SAM, thereby decreasing DNA methylation. Thus, long-term alcohol consumption leads to substantial DNA hypomethylation as a result of significant reduction in tissue SAM [26].
Fig. 1.
Depiction of DNA methylation process. DNMT transfers the methyl group to 5′ position of cytosine in the DNA sequence. SAM acts as a methyl donor in this process and it is converted to SAH. SAHH converts SAH to homocysteine by the removal of adenosine moiety. Thereafter, homocysteine either undergoes the transsulfuration pathway to form GSH or remethylated through methionine synthase enzyme and form methionine. Subsequently, methionine adenosyl transferase adds adenosine to the methionine and finally forms SAM.
Several emerging evidences support the aggravating effect of alcoholism on DNA methylation. A foremost study reported that dietary changes could affect beverage alcohol consumption in rodents. Especially, diets deficient in vitamin B (folic acid and choline) increased the consumption of solution containing 10% alcohol in rats, whereas vitamin-enriched diets reduced it [26]. It is now well-known that folates and several other B vitamins are vital for one-carbon metabolism and the synthesis of a compound called SAM, which serves as the primary methyl group donor in most transmethylation reactions, including DNA methylation [27]. Hence, it can be legitimate to infer that dietary changes can affect alcohol consumption via changes in DNA methylation and methylation-regulated gene expression. In support of this notion, there are several studies that corroborated the influence of alcohol consumption on vitamin B and folate [27,28]. Besides, alcohol can affect DNA methylation by other mechanisms also. The alcohol metabolite, acetaldehyde can induce inhibition of DNMT, thereby aggravating the normal functioning of DNA methylation. Alcohol-induced DNA damage and the resulting repair mechanisms can lead to demethylation of 5-methylcytosine nucleotides [29]. Both of these mechanisms can lead to reduced levels of methylation throughout the DNA (i.e., global DNA hypomethylation), a chromatin state associated with many pathological conditions, including cancer [30].
In addition, upon the chronic exposure of alcohol, several genes were observed to be affected and found to be aberrant in epigenetic mechanisms involving DNA methylation. For instance, the protein α-synuclein has been implied in alcohol craving, and both protein and mRNA levels of α-synuclein are high with chronic alcohol intake [31]. One of the encoding genes of α-synuclein is Snca. Epigenetic upregulation of α-synuclein, which can result from the decreased cytosine methylation in Snca promoter region, in the substantia nigra, is found to be associated with Parkinson's-like behavior in rats [32]. Methylation also affects transcription of a gene encoding an endogenous opioid called prodynorphin [33]. Prodynorphin is a hormone involved in brain signaling that influences a person's risk of alcoholism. A DNA variation (i.e., single nucleotide polymorphism) of this gene exists and overlaps with a CpG dinucleotide, which is subject to methylation. Methylation of this dinucleotide affects prodynorphin transcription in the prefrontal cortex of alcoholics [33]. Similarly, Htr3a promoter methylation [33,34], methylation process of genes involved in calcium regulation, Alzheimer's disease-associated pathways, muscle contraction processes [35], and genome wide methylation processes are found to be aberrant in chronic alcoholism [36].
Furthermore, studies on the central effects of prenatal alcohol exposure and on gene regulation in cell cultures observed that alcohol shows disparity in its action on different genes and tissues. In a study, prenatal exposure of rats to alcohol shown DNA hypermethylation and a reduced expression of a protein called brain-derived neurotrophic factor (BDNF) in olfactory bulbs of rat pups, along with the loss of neurons in brain region [37]. Similar molecular results were obtained in a separate study, where prenatal alcohol treatment of rats resulted in DNA hypermethylation and a decreased expression of a protein characteristically found in brain cells called astrocytes (i.e., glial fibrillary acidic protein (GFAP)) [38]. In neural cell cultures, alcohol-induced downregulation of cell-cycle genes was paralleled by a raised DNMT activity and hypermethylation of the promoters of those genes [39]. Conversely, a clinical study demonstrated that in alcoholic patients the degree of methylation at the promoter region of the gene encoding the NMDA2B receptor subtype (NR2B) during withdrawal negatively correlated with the severity of alcohol consumption [40]. Moreover, in primary cultured neurons, chronic intermittent ethanol exposure resulted in demethylation of a regulatory region of the NR2B gene, which was correlated with increased NR2B gene expression [41].
Specific DNA methylation patterns differ among tissues and cell types, and these differences contribute to establishing the cell's epigenetic landscape, transcriptional programs, and defining cellular identity [42]. Although alcohol's general effects on DNA methylation may be uniform across various tissues, the specific genes affected by this regulation may differ depending on the cell type. The epigenetic regulation of such proteins as GFAP, which is a marker of astrocytes, and the NR2B subunit, which generally is expressed in neurons, suggests that alcohol-induced epigenetic changes will affect molecular markers of individual cell types to a greater degree than other proteins [42].
Histone Modifications
Histones are the core protein components of chromatin complexes, and they provide the structural backbone around which the DNA wraps at regular intervals generating chromatin. The histone proteins (H2A, H2B, H3, and H4) are each composed of a globular domain and an unstructured tail domain [43]. Histones regulate DNA packaging, with an immense impact on the degree of chromatin compaction, influencing transcriptional activity and transcriptional silencing. Histone proteins are the second major target of epigenetic changes. Histone modifications are post-translational changes on the histone tails, which are flexible stretches of N or C terminal residues extending from the globular histone octamer.
A firm control of histone modifications in the CNS is of critical importance for maintaining neuronal cell viability, function, and connectivity. Histone modifications include methylation of lysine and arginine residues, acetylation of lysine residues, phosphorylation of serine, threonine or tyrosine residues, and ubiquitination of lysine residues present on histone tails, as well as sumoylation and ADP ribosylation.
Histone Methylation and Demethylation
Histone methylation includes addition of methyl groups to lysine and arginine residues of histone tails, while histone demethylation includes removal of methyl groups from lysine and arginine residues of histone tails. Histone methyltransferases (HMTs) and histone demethylases (HDMs) catalyze histone methylation and demethylation processes, respectively (fig. 2a). Based on the target site, methylation can alter histones so that different portions of chromatin are activated or inactivated. In several cases, methylation and demethylation of histones produce dichotomous effect, methylation/demethylation can turn the genes in DNA ‘on’ and ‘off’, respectively, either by relaxing or encompassing their tails, thereby allowing or blocking transcription factors and other proteins to access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different portions of the genome [44]. Histone methylation pattern brings certain stable yet dynamic complexity to the epigenome that can signal vast number of subsequent regulatory marks necessary for the control of transcription at the adult brain during long-term memory formation [4].
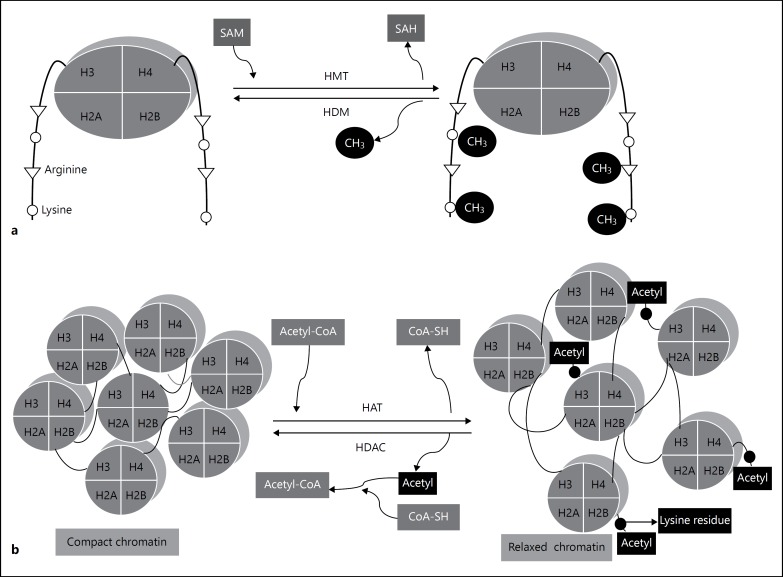
Fig. 2.
Diagrammatic representation of different types of histone modifications. a Histone methylation/demethylation: HMTs and HDMs catalyze histone methylation and demethylation processes, respectively. Similarly, DNA methylation process, SAM acts as a methyl donor in histone methylation at lysine and arginine residues of histone tails. b Histone acetylation/deacetylation: histone acetyltransferase catalyze histone acetylation by adding acetyl group at lysine residues of N-terminal tails of the nucleosome. Acetyl-coenzyme A donates its acetyl group in this process and is changed to coenzyme-SH. Acetyl group neutralizes the positive charge of lysine residue and promotes decondensation of chromatin to initiate the transcriptional process. Conversely, in histone deacetylation process, acetyl group is removed from the lysine residues under the catalysis of HDAC enzyme. Loss of acetyl group from the lysine residues lead to chromatin compaction and transcriptional inactivation. c Histone phosphorylation: protein kinases catalyze the addition of phosphate group to the serine, threonine, and tyrosine residues of histone tails, which generally leads to transcriptional activation. ATP or GTP acts as a donor in the histone phosphorylation process. d Histone ubiquitination: in this process, carboxylic acid group of the last amino acid of ubiquitin, that is, glycine and the epsilon amino group of the histone's lysine get covalently attached through isopeptide bond under the influence of 3 enzymes: (i) ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s).
Histone Methylation and Demethylation: Neurological Implications
Apart from its critical involvement in development, histone methylation remodeling is implied in nervous system also. In neurons, the major role of histone methylation/demethylation is likely to be in the transcriptional regulation. Histone methylation marks at promoters, gene body sequences, or 3′ downstream sequences could affect transcription initiation, elongation, or termination, respectively. Histone methylation is implicated in the development and differentiation processes of nerve cells, including neurons, oligodendrocytes, astrocytes and microglia, Schwann cells, and the myelinating cells of the CNS and PNS. Several studies have corroborated the importance of methylation/demethylation in neurological implications. For an instance, aberrant histone methylation at lysine residues has been found to be associated with several psychiatric disorders such as autism and mental retardation [45]. Similarly, histone methylation at GABAergic gene promoters by a certain HMT may be involved in prefrontal cortex dysfunction during schizophrenia [46]. An additional recent research report suggested the association of histone methylation in the regulation of memory formation [47]. Histone methylation, especially histone H3, has been associated with the expression level of neurotransmitter glutamate, which has been implied in schizophrenia [48]. A recent study in the neonatal mouse brain showed that G9a-mediated histone methylation regulates ethanol-induced neurodegeneration [49]. Similarly, several recent reports suggested the association of histone methylation with neurodegenerative diseases such as Friedreich ataxia, Huntington's disease, and Alzheimer's disease [44]. Taken together, these studies suggest that histone methylation may play an important role in the preservation of neuronal function by regulating the expression of synaptic plasticity-associated genes.
Impact of Alcohol on Histone Methylation
Several studies have shown that histone methylation is substantially affected on alcohol exposure. For instance, chronic alcohol consumption in humans can result in global and gene-specific increases in H3K4 trimethylation in the brain cortex [10,] and either increase or decrease of this modification in promoters of specific genes in the hippocampus [50]. Recent evidence also shows that acute ethanol exposure alters dimethylation levels on lysines 9 and 27 of histone 3, which partially mediate ethanol's teratogenic effects in the brain [51]. The same research group also showed that ethanol exposure increases the levels of this mark in exon 1 of G9a, an HDM involved in alcohol-induced apoptosis [52]. Furthermore, epigenetic marks are emerging as major regulators of gene-by-environment interactions and have been implicated in the etiology of ethanol-induced neurodegeneration. Indeed, G9a-mediated increases of H3K9 and H3K27 dimethylation control proteolytic breakage of histones by caspase-3 and eventual neurodegeneration in the hippocampus and neocortex following acute, low-dose ethanol exposure [49].
Histone methylation was also found to be associated with addiction. In a recent study, intermittent alcohol treatment altered methylation in the promoter region of cFos, Cdk5, FosB, and BDNF genes [53]. These genes were previously found to be associated with behavioral abnormalities characterizing drug addiction [54,55]. Taken together, these studies suggest that histone methylation is a key player in the preservation of neuronal function. Understanding the alcohol impact on histone methylation could provide better insights into drug development for AUDs.
Histone Acetylation/Deacetylation
Histone acetylation is by far the most studied among histone modifications [56]. Histone acetylation includes addition of acetyl groups to lysines present in the N-terminal tails on the surface of the nucleosome, while histone deacetylation includes removal of acetyl groups (fig. 2b) [57]. Generally, histone acetylation promotes decondensation of chromatin and increases gene activity by negating the positive charge of lysine residues in histone tails and increases spacing between nucleosomes, and thus promotes recruiting transcription factors together with other transcription machinery proteins to DNA [58]. Therefore, increasing histone acetylation is usually associated with transcriptional activation. Conversely, histone deacetylation leads to a compact chromatin structure, preventing transcription factor binding [58]. Thus, decreasing histone acetylation is associated with transcriptional repression. Histone acetylation and deacetylation are highly regulated by 2 types of enzymes, histone acetyl transferases (HATs) and histone deacetylases (HDACs), which promote gene activation and gene repression, respectively. Both types of enzymes dynamically interact to regulate the remodeling of the chromatin architecture and gene expression [59].
Histone Acetylation/Deacetylation: Neurological Implications
Numerous studies produced abundant evidence for the role of histone acetylation in neurological functions. For instance, a few studies have indicated that regulation of histone acetylation is important while shaping memories in the hippocampus region of the brain [60]. In addition, aberrant histone methylation was found to be associated with age-dependent memory impairment in mice [61]. The machinery through which histone acetylation and eventual chromatin remodeling can be regulated is via a protein called cyclic-AMP responsive-element binding (CREB) protein. CREB protein helps to modulate the transcription of some genes by binding to a specific sequence on the DNA after it has been stimulated by phosphorylation. Phosphorylated CREB (pCREB) then employs another transcriptional co-factor called CREB-binding protein (CBP), which contains intrinsic HAT activity.
Besides CBP, another molecule called p300, catalyzes the remodeling of the nucleosome by transferring acetyl groups to histones, thereby causing the chromatin to exist in a relaxed conformation that is accessible for transcription machinery [62,63]. CBP is a vital factor for both short- and long-term memory formation and consolidation [64]. A recent study in a mouse model of Alzheimer's disease expressing anomalous CREB function exhibited that ingestion of the CBP gene into the brain increased the levels of BDNF, which is associated with enhanced learning and memory [65]. A recent study further supported the important role of histone acetylation in neurodegeneration. The study examined global histone modifications in immunolabeled inferior temporal gyrus and middle temporal gyrus of post-mortem Alzheimer's disease brains compared with neurologically normal control cases. The study observed that in the Alzheimer's brain, histone H3 and H4 acetylation levels are significantly increased in the inferior temporal gyrus and middle temporal gyrus, but only in proportion to significant increases in total histone H3 and H4 protein levels. These observations were specific to regions of pathology implicated in Alzheimer's disease, as no significant changes were observed in the cerebellum [66]. Apart from this, numerous other studies are corroborating the potential role of histone acetylation in neurological implications.
Impact of Alcohol on Histone Acetylation/Deacetylation
So far the effect of alcoholism on histone acetylation was found to be mediated by modifying the activities of the enzymes that add acetyl groups (i.e., HATs) or remove acetyl groups (HDACs). Numerous studies till now corroborated the definite role of histone acetylation in alcoholism. Even though there are myriad studies explaining alcohol-mediated effects on histone acetylation, the exact mechanisms underlying the influence of alcohol on gene expression are not clearly understood. One such suggested mechanism is the involvement of the transcription factor CREB, to which CBP can bind [65]. CBP has an intrinsic HAT activity and, when recruited by CREB, can promote transcriptional activation by acetylating histones [66]. CREB functioning is found to direct the expression of genes associated with alcohol addiction, such as BDNF, neuropeptide Y and activity-regulated cytoskeleton-associated protein (Arc) [67]. NPY mRNA levels were found to be reduced in the amygdala of alcohol-preferring (P) rats compared to alcohol non-preferring (NP) rats, implying that NPY has a role in anxiety-like and alcohol-drinking behaviors [67]. Direct infusion of NPY into the central region of amygdala (CeA) produced anxiolytic effects and reduced alcohol intake in P rats, which is likely increased by CREB phosphorylation in the CeA region of the brain [68,69]. In addition, alcohol-preferring rats which were never exposed to alcohol showed reduced expression of other CREB target genes, such as BDNF and Arc in the CeA and medial amygdaloid region compared to alcohol NP rats [69]. Also, acute ethanol exposure raised CREB phosphorylation levels and the expression of BDNF and Arc in these amygdaloid structures of alcohol-preferring, but not in alcohol NP rats [70].
Alcohol-induced alterations in histone acetylation are studied in the expression of several genes, including CBP, NPY [65], FosB [53], NR2B [69], pronociceptin, and prodynorphin [70]. Both the amygdaloid pCREB and CBP levels are notably raised by acute ethanol exposure and decreased in rats undergoing withdrawal after chronic ethanol exposure. These findings suggest that changes in CBP levels may be implicated in the dynamic chromatin remodeling in the amygdala caused by acute and chronic ethanol exposures [67]. A key role of histone acetylation was observed in a recent study, in which the hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal in mice was found to be mediated by HDAC [67]. Alcoholism is also suggested to be associated with neurodegeneration; indeed, in a recent study it was found that ethanol-induced acetylation of histone at G9a exon1 leads to neurodegeneration in neonatal mice [52]. Furthermore, in a study, histone acetylation was found to play a crucial role in regulating the transcriptional activity and contribute to the drug-induced alterations in gene expression and behavior. Similarly, a study found that ethanol treatment upregulates the HAT activity in adolescent prefrontal cortex and increases histone (H3 or H4) acetylation of the promoter region of genes cFos, Cdk5, and FosB [53]. Gene expression studies provide further evidence on the role of histone acetylation in alcoholism. The studies focused on brain changes in human alcoholics have shown a general downregulation of genes involved in histone acetylation and upregulation of genes promoting histone deacetylation [67].
Similar to DNA methylation, alcohol's effects on histone acetylation are tissues, brain regions, and cell type specific. For example, alcohol-induced alterations in H3/H4 acetylation were found only in the central and medial, but not in the basolateral nuclei region of the amygdale [67]. In addition, the raised histone acetylation seemed to be specific to neurons [68]. In line with other studies, ethanol-induced behavioral sensitization was found to be associated with striatal HDAC activity decrease in a recent work [69].
Additional factors that can affect alcohol-induced changes in histone acetylation include species, the organism's specific genetic makeup, age, the dose and route of ethanol administration, and duration of exposure. For instance, irregular alcohol exposure showed different effects on histone acetylation in adolescent and adult rats, with juvenile animals generally showing more changes [53,69]. Similarly, alcohol exposure during the early postnatal period in rats resulted in a marked reduction of CBP levels and histone acetylation in the developing cerebellum [70]. In ddY mice species, chronic ethanol vapor treatment showed an increase in both global and gene-specific histone acetylation in the ventral midbrain during withdrawal that hiked around 10 h post-alcohol exposure [71]. Additionally, probable communications among various factors may result in different time courses for alcohol-induced alterations, as histone acetylation measured 24 h after the last repeated alcohol injections was raised in some brain areas (e.g., frontal cortex and nucleus accumbens), decreased in others (e.g., striatum), and unaffected in still other regions of the brain (e.g., hippocampus) [71]. Further activation of different genes is coupled with acetylation of different residues of H3 and H4 at various time intervals [72].
Histone Phosphorylation
Histone phosphorylation is a transcription-activating modification achieved by protein kinases that catalyze the transfer of a phosphate group from ATP or guanosine-5′-triphosphate (GTP) to the serine, threonine or tyrosine residue of histones. All 4 nucleosomal histone tails can be phosphorylated and can be induced by extracellular signals, DNA damage or entry into mitosis. However, the best-known function of histone phosphorylation takes place during cellular response to DNA damage, when phosphorylated histone H2A(X) demarcates large chromatin domains around the site of DNA breakage. However, multiple studies have also shown that histone phosphorylation plays a crucial role in chromatin remodeling linked to other nuclear processes. Phosphorylated residues can have dissimilar effects on chromatin structure based on the situation in which it occurs. Phosphorylation of H3S10 and H3S28 is a fine example of this duality: both phosphorylated residues are involved in chromatin condensation associated with mitosis and meiosis, as well as in chromatin relaxation linked to transcription activation [73].
Histone Phosphorylation: Neurological Implications
Histone phosphorylation is also associated with neurological implications. For instance, a recent study found an increase in histone H3 phosphorylation in neurons in specific brain structures (caudate putamen and medio-lateral axis) in the status epileptic mice (fig. 2c) [74]. Similarly, long-term mental and pain stress on the neurons of the sensory motor cortex and midbrain reticular formation in rats with high excitability threshold found to exhibit higher basal level of H3Ser10 histone phosphorylation in the midbrain reticular formation neurons than rats with low excitability threshold [75]. There is a need for some more studies of histone phosphorylation, to establish its clear role in neurological implications.
Impact of Alcohol on Histone Phosphorylation
Impact of alcohol was also studied on histone phosphorylation. For instance, acute alcohol exposure in rats, was found to result in increased phosphorylation of histone H3 at ser-10 and ser-28, which in turn leads to transient activation of p38 [76]. In another work, acute and rapid intraperitoneal administration of ethanol was found to promote site-specific histone H3 phosphorylation at Ser10 and Ser28 in rat liver in vivo [77]. Similarly, in a more recent study chronic ethanol administration alone caused an increase in histone H3 ser10 and ser28 (H3S10 or S28) phosphorylation, and binge ethanol reduced their levels. This pattern of histone phosphorylation is also associated with differential induction of genes implicated in liver injury [76].
Histone Ubiquitination
Histone ubiquitination (also ubiquitylation or ubiquitylation) is an enzymatic, post-translational modification process in which an ubiquitin protein is covalently attached to histone proteins. This process generally binds the last amino acid of ubiquitin (glycine 76) to a lysine residue on the histone protein. An isopeptide bond is formed between the carboxylic acid group of the ubiquitin's glycine and the epsilon amino group of the histone's lysine. Ubiquitination requires 3 types of enzymes: ubiquitin-activating enzymes, ubiquitin-conjugating enzymes, and ubiquitin ligases, known as E1s, E2s, and E3s, respectively (fig. 2d) [77]. Histone ubiquitination changes the chromatin structure and allows the access of enzymes involved in transcription. Ubiquitin on histones also acts as a binding site for proteins that either activate or inhibit transcription, and also can induce further post-translational modifications of the protein. All these effects can modulate the transcription of genes [78].
Among the histone proteins, histone H2A was the first protein identified to be ubiquitinated [79]. It is now known that H2A and H2B are 2 of the most abundant ubiquitinated proteins in the nucleus. In addition to H2A and H2B, core histones H3, H4, and linker histone H1 have also been reported to be modified by ubiquitin. But the biological function of these modifications has not been well elucidated [80,81]. Ample studies provide insights into the roles of H2A/B ubiquitination in transcriptional regulation [82]. The mechanistic approaches through which histone ubiquitination affects chromatin structure are: it regulates the accessibility of DNA to the transcriptional machinery; ubiquitin moieties constitute signaling molecules which control the staffing of downstream regulators that activate or inhibit transcription; through trans-histone cross talk, histone ubiquitination acts as a prerequisite modification for other histone modifications that alter the structure and function of chromatin. In addition to transcription regulation, monoubiquitinated H2B is required for chromatin function in other ways. H2Bub is required for chromatin boundary integrity, and loss of H2Bub leads to the spread of other histone modifications [83]. Ubiquitination of histone H2B was also found to regulate the chromatin dynamics by enhancing nucleosome stability [82].
Histone Ubiquitination: Neurological Implications
Histone ubiquitination is also associated with neurological implications. Indeed, a recent study found that bre1, a histone h2b ubiquitin ligase regulates the cell cycle and differentiation of neural precursor cells (NPCs) [83]. The study found that knockdown of the gene Bre1a in NPCs of the developing cerebral cortex suppresses proliferation and differentiation, suggesting that Bre1a is a multifunctional molecule that regulates both the cell cycle and differentiation of NSCs [83]. Some other study showed the implication of dysfunction of the ubiquitin proteasome associated with schizophrenia [84]. Further studies are required to know more about histone ubiquitination in neurological implications.
Impact of Alcohol on Ubiquitination
The effect of alcohol on histone ubiquitination has not yet been studied well, but still it could be legitimate to predict that alcohol can produce exacerbating effects even on ubiquitination. In a study of transcription profiling of post-mortem brains from subjects with a history of alcohol abuse or dependence, it showed abnormalities in ubiquitin signaling system [84].
Histone Sumoylation
Sumoylation is another post-translational modification involved in diverse biological processes, including stress response, protein stability, cell cycle progression, apoptosis, nuclear-cytoplasm transport, and transcriptional control. Histone sumoylation can regulate transcriptional repression [85]. A study in this regard reports that histone H4 is modified by small ubiquitin-related modifier (SUMO) family proteins both in vivo and in vitro. H4 binds to the SUMO-conjugating enzyme (E2), UBC9, and can be sumoylated in an E1 (SUMO-activating enzyme) and E2-dependent manner. Also, the study reported that histone sumoylation mediates gene silencing through recruitment of HDAC and heterochromatin protein-1 [85]. Similarly, a study in Saccharomyces cerevisiae found that all 4 core histones are sumoylated, and it was also demonstrated that histone sumoylation sites were involved directly in transcriptional repression [86]. A recent research report also suggested the repressing effect of histone sumoylation. An in vitro study shows that sumoylation of the polycomb group protein L3MBTL2 facilitates the repression of its target genes. Lethal (3) malignant brain tumor like 2 (L3MBTL2) is an integral component of the polycomb repressive complex 1.6 and has been implicated in transcriptional repression and chromatin compaction [86].
Although the repressing effect of sumoylation on transcription is well known, the exact mechanism through which it occurs is yet to be explored. A recent study made some progress in this regard; it showed how sumoylation mediates the gene repression without chromatin compaction. The study showed that sumoylation of H4 forces chromatin to adopt an open structure rather than a compact heterochromatin-like structure, indicating that sumoylation-mediated transcriptional silencing is not a direct result of chromatin condensation [86]. From a recent research findings, it was also proposed that sumoylation by human polycomb group protein 2 (hPc2) regulates the activity of lysine-specific demethylase 5B (KDM5B), which plays a key role in gene repression. hPc2 is a protein which enhances sumoylation [87].
Histone Sumoylation: Neurological Implications
Sumoylation is also implicated in neurological functions. In a recent study, CREB sumoylation by the E3 ligase pias1 was found to enhance spatial memory [87]. In a similar work, it was reported that the protein expression levels of SUMO-modified substrates and several components of the sumoylation machinery are temporally and spatially regulated in the developing rat brain [87]. Similarly, sumoylation dysfunction was also found to be associated with schizophrenia [84]. Likewise, sumoylation has also been implicated in neurodegenerative diseases [85]. Furthermore, in a similar study, it was found that protein sumo/desumoylation equilibrium is unbalanced in a mouse model of Alzheimer's diseases at a very early stage of the pathology, implying the role of sumoylation in the Alzheimer's disease [86].
Impact of Alcohol on Histone Sumoylation
There are very few studies that explored the effect of alcohol on sumoylation. In a recent study, ethanol feeding to mice was found to significantly reduce sumoylation in the liver tissue [87]. Similar result was found in another study, in which alcohol exposure in the normal and heat shock factor 2-deficient mice and cell systems was found to reduce the sumoylation in the brain [88].
miRNA/Non-Coding RNA and Alcoholism
Besides the existence of miRNA, several types of non-coding RNA molecules exist, which have been shown to play critical regulatory roles in cell function [88]. Generally, the non-coding RNA binds to the complimentary sequences on the target mRNAs to repress translation, and thereby silence the gene expression [56]. The role of miRNA in alcoholism was also examined in numerous studies. A study by Pietrzykowski group provides confirmation for the association of non-coding RNA in alcohol-related gene expression. The study demonstrated that alcohol upregulates the expression of non-coding RNA 9 (miRNA9) in rat brain, which culminates in miRNA9-dependant downregulation of big potassium channel variants with high sensitivity to alcohol. This mechanism is supposed to be linked to the progress of cellular tolerance and neuronal adaptation to alcohol [88]. Furthermore, a recent study discovered miR-29b, a novel target of ethanol in the developing cerebellar granule neurons [89]. Normally, the over expression of miR-29b renders neurons protection against ethanol-induced apoptosis. The study found that ethanol exposure suppresses miR-29b and induces neuronal apoptosis [89]. In a more recent study, simultaneous examination of miRNA and mRNA expression profiles in the frontal cortex of ethanol-treated mice and comparison with similar studies conducted in brains of human alcoholics revealed that activated expression of miRNAs correlates with activated expression of mRNAs rather than with mRNA downregulation in experimental mouse model [90]. Over expression of miRNA-30a-5p in the prefrontal cortex was found to control the transition from moderate to excessive alcohol consumption, showing the importance of miRNA in alcoholism [91]. Promising results from the above study and other numerous ongoing studies of miRNA, are paving the way for establishing miRNA as a novel therapeutic target for alcoholism.
Epigenetic Drug Targets
It comes as no surprise that the rapid advancement in neuropsychiatric/neuro-epigenetic research has led to heightened enthusiasm in the development and clinical implementation of pharmacotherapies aimed at targeting epigenetic modifications. While considering epigenetic drug targets, it is important to understand that epigenetic malfunctions, unlike genetic ones, are reversible. With properly identified targets and successful development of compounds against them, aberrant distribution of epi-marks can be corrected or prevented. Obviously, the better understanding of the connection between alcohol consumption, epigenetic modifications, and illness and disease could lead researchers to develop medications or therapies for prevention or treatment. The following is a brief discussion of the current understanding of different epigenetic drug targets.
Epigenetic Drug Targeting DNA Methylation
Common motto of the drugs targeting DNA methylation would be usually to attenuate the DNA hypermethylation either by inhibiting DNMTs or promoting DNA demethylation. Sometimes the drugs targeting DNA methylation could exhibit tissue and gene-specific actions, that is, either the drug may promote DNA methylation or reduce the DNA hypermethylation [92]. DNA methylation is an excellent target for anticancer therapies, as cancer generally associates with highly disrupted methylome. Numerous studies in preclinical and clinical setting are underway, targeting DNA methylation as a drug target for anticancer interventions. Some drugs are available in the market currently. For instance, DNMT inhibitors, 5-azacytidine and its variant 5-aza-2′-deoxycytidine both were approved for the treatment of higher-risk myelodysplastic syndrome [58]. Apart from cancer, there are some studies, which are still in their infancy, which target neurological and other implications. It is even legitimate to predict that preventing DNA hypermethylation could also be beneficial to neurological and other implications. Studies in this regard support this notion. For instance, a recent study in rats examining the effects of olanzapine on genome-wide DNA methylation in the hippocampus and cerebellum showed that widespread and tissue-specific changes in genome-wide methylation could mediate the activity of olanzapine while treating schizophrenia [93]. These examples raise the possibility that the mechanisms of action of certain approved drugs may involve previously uncharacterized epigenetic processes. In addition, there is a clinical study employing a dietary supplement, creatine monohydrate for targeting DNA methylation to treat Rett syndrome [93]. Rett syndrome is a neurodevelopmental disorder that shows anomalies in methyl MeCP2, which is essential for the DNA methylation process, to discharge its effects. DNA methylation seems to be the target in dealing with alcoholism, indeed a recent study supports this perception. The study found that inhibiting the activity of DNMT with systemic administration of the FDA-approved drug, 5-azacitidine prevents excessive alcohol consumption in mice [94].
Drugs Targeting Histone Acetylation/Deacetylation
Owing to the myriad studies, histone acetylation/deacetylation has now become a well-established histone modification to target upon. The main theme of targeting histone acetylation is to increase the acetylation by employing HDAC inhibitors (HDACi). HDACi are now being used routinely in the clinic to treat a variety of human illnesses, including numerous cancers, parasitic infections, and inflammatory diseases. Indeed, 2 HDACi, vorinostat and romidepsin, were approved by the FDA for the treatment of advanced cutaneous T-cell lymphoma. In addition, there is a group of HDACi in different phases of clinical studies for various neurological indications, such as nicotinamide for Friedreich's Ataxia (NCT01589809), sodium valproate for Rubinstein-Taybi Syndrome (NCT01619644), and Vorinostat for Neimann-Pick Disease [95].
Apart from cancer, these HDACi also show strong neuroprotective potential [58]. In a recent study, systemic treatment with several HDACi was found to reduce mice binge-like alcohol drinking [96]. The same result was reproduced even with FDA-approved HDACi vorinostat (SAH) [96]. Furthermore, the activity of HDACi was also found to be specific to alcohol intake, indicating that HDACi can be used to treat harmful effects of alcohol abuse. The applicability of HDACi was further strengthened by a recent study, in which HDACi valproic acid reduced ethanol consumption and ethanol-conditioned place preference in rats [97]. The study implicated HDAC inhibition in the behavioral and reinforcement-related effects of alcohol and raised the question of whether specific drugs that target HDAC could potentially help in dealing with alcoholism in humans.
Drugs Targeting Histone Methylation
The clinical potential of drugs that can intervene with the regulation of histone methylation is largely unexplored. As HDACi, a group of HMT inhibitors is being studied; they are expected to be explored for neurological indications in the near future [98]. From the numerous studies, it is found that histone methylation is crucial for drug addiction [99]. An attractive drug candidate is BIX-01294, which is an inhibitor for the histone H3K9-specific methyltransferases G9a and Glp84. BIX-01294 attenuates neuronal gene expression [100] and, when given directly into the ventral striatum (a vital structure in the brain's addiction circuitry), robustly increases the development of reward behaviors in mice exposed to the stimulant drug cocaine [101]. Furthermore, it needs to be determined whether BIX-01294 elicits learning and memory-enhancing effects outside of the stimulant addiction paradigm. However, it is interesting to note that the drug's mechanism of action could, at least in part, involve the inhibition of G9a- and Glp-mediated repressive chromatin remodeling at the promoters of Bdnf, Cdk5, Arc, and other genes that function as key regulators for spine density and synaptic connectivity in the mouse brain.
Conclusion and Future Perspectives
The field of neuroepigenetics is evolving at a rapid pace. We now understand more about the neural mechanisms that mediate the reversible and bidirectional regulation of DNA methylation and various histone modifications. Now, it is evident that epigenetic changes can play an important and complex role in the neuroplastic changes associated with alcohol and other substance addiction. Presently, DNA methylation, histone acetylation, and methylation are by far the most substantially studied epigenetic mechanisms. Indeed, drugs targeting these epigenetic mechanisms are in different phases of development, preclinical, clinical and in the market. However, there is no such drug approved, which targets epigenetic mechanisms and is indicated for alcohol or other substance addiction. The major drawback of drugs targeting epigenetics is their lack of specificity. As we learn more about the epigenetic mechanisms, we are now beginning to truly appreciate the biological complexities of human psychiatric illness associated with alcohol addiction and are starting to gain a better understanding of how to clinically treat these disorders based on their molecular and biochemical attributes. The developing technologies such as epigenetic editing, can allow us to direct the activity of epigenetic enzymes towards specific genes, and has the potential to bring epigenetic-based interventions to a completely new level in the near future.
Author Contribution
All the authors significantly contributed to the manuscript writing. A.J. and C.S.S. wrote the abstract, ‘DNA methylation’, ‘non-coding RNA and alcoholism’ section in the manuscript. S.P., P.R., B.S. and P.C. wrote the ‘histone modifications’ section in the manuscript. A.J., B.S. and M.L. wrote the ‘epigenetic drug targets’ section in the manuscript. B.K.B. made all the figures and did English corrections in the manuscript. A.J. did referencing of the manuscript.
Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Waddington CH. The Strategy of the Genes, a Discussion of Some Aspects of Theoretical Biology. London: Allen & Unwin; 1957. p. 262. [Google Scholar]
- 2.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 4.Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trudell JR, Messing RO, Mayfield J, Harris RA. Alcohol dependence: molecular and behavioral evidence. Trends Pharmacol Sci. 2014;35:317–323. doi: 10.1016/j.tips.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick DM, Kendler KS. The impact of gene-environment interaction on alcohol use disorders. Alcohol Res. 2012;34:318–324. [PMC free article] [PubMed] [Google Scholar]
- 7.Contet C, Gardon O, Filliol D, Becker JA, Koob GF, Kieffer BL. Identification of genes regulated in the mouse extended amygdala by excessive ethanol drinking associated with dependence. Addict Biol. 2011;16:615–619. doi: 10.1111/j.1369-1600.2010.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botia B, Legastelois R, Alaux-Cantin S, Naassila M. Expression of ethanol-induced behavioral sensitization is associated with alteration of chromatin remodeling in mice. PLoS One. 2012;7:e47527. doi: 10.1371/journal.pone.0047527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 13.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 17.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 18.Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M, Martin LJ. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nardone S, Sams DS, Reuveni E, Getselter D, Oron O, Karpuj M, et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry. 2014;4:e433. doi: 10.1038/tp.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiol Aging. 2014;35:1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 21.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, Zhang C, Cheng L, Reilly JL, Bishop JR, Sweeney JA, et al. Correlation between DNA methylation and gene expression in the brains of patients with bipolar disorder and schizophrenia. Bipolar Disord. 2014;16:790–799. doi: 10.1111/bdi.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong E, Ruzicka WB, Grayson DR, Guidotti A. DNA-methyltransferase1 (DNMT1) binding to CpG rich GABAergic and BDNF promoters is increased in the brain of schizophrenia and bipolar disorder patients. Schizophr Res. 2014;167:35–41. doi: 10.1016/j.schres.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pidsley R, Viana J, Hannon E, Spiers H, Troakes C, Al-Saraj S, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grillo MA, Colombatto S. S-adenosylmethionine and its products. Amino Acids. 2008;34:187–193. doi: 10.1007/s00726-007-0500-9. [DOI] [PubMed] [Google Scholar]
- 26.Williams RJ, Berry LJ, Beerstecher E. Individual metabolic patterns, alcoholism, genetotrophic diseases. Proc Natl Acad Sci U S A. 1949;35:265–271. doi: 10.1073/pnas.35.6.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rush EC, Katre P, Yajnik CS. Vitamin B12: one carbon metabolism, fetal growth and programming for chronic disease. Eur J Clin Nutr. 2014;68:2–7. doi: 10.1038/ejcn.2013.232. [DOI] [PubMed] [Google Scholar]
- 28.Blasco C, Caballería J, Deulofeu R, Lligoña A, Parés A, Lluis JM, Gual A, Rodés J. Prevalence and mechanisms of hyperhomocysteinemia in chronic alcoholics. Alcohol Clin Exp Res. 2005;29:1044–1048. doi: 10.1097/01.alc.0000169265.36440.ee. [DOI] [PubMed] [Google Scholar]
- 29.Chen CH, Pan CH, Chen CC, Huang MC. Increased oxidative DNA damage in patients with alcohol dependence and its correlation with alcohol withdrawal severity. Alcohol Clin Exp Res. 2011;35:338–344. doi: 10.1111/j.1530-0277.2010.01349.x. [DOI] [PubMed] [Google Scholar]
- 30.Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223–230. doi: 10.1016/j.canlet.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, et al. Association of alcohol craving with alpha-synuclein (SNCA) Alcohol Clin Exp Res. 2007;31:537–545. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Li J, Zhang Z, Wang H, Wang Z. Epigenetic upregulation of alpha-synuclein in the rats exposed to methamphetamine. Eur J Pharmacol. 2014;745:243–248. doi: 10.1016/j.ejphar.2014.10.043. [DOI] [PubMed] [Google Scholar]
- 33.Taqi MM, Bazov I, Watanabe H, Sheedy D, Harper C, Alkass K, et al. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol. 2011;16:499–509. doi: 10.1111/j.1369-1600.2011.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker JM, Zhang Y, Wang F, Taylor JR, Zhang H. Ethanol-induced Htr3a promoter methylation changes in mouse blood and brain. Alcohol Clin Exp Res. 2013;37(suppl 1):E101–E107. doi: 10.1111/j.1530-0277.2012.01906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013;37(suppl 1):E108–E115. doi: 10.1111/j.1530-0277.2012.01928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng JT, Wu LS, Lee CS, Hsu PW, Cheng AT. Integrative epigenetic profiling analysis Identifies DNA methylation changes associated with chronic alcohol consumption. Comput Biol Med. 2015;64:299–306. doi: 10.1016/j.compbiomed.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN. Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013;18:392–403. doi: 10.1111/adb.12037. [DOI] [PubMed] [Google Scholar]
- 38.Maier SE, Cramer JA, West JR, Sohrabji F. Alcohol exposure during the first two trimesters equivalent alters granule cell number and neurotrophin expression in the developing rat olfactory bulb. J Neurobiol. 1999;41:414–423. doi: 10.1002/(sici)1097-4695(19991115)41:3<414::aid-neu9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 39.Vallés S, Pitarch J, Renau-Piqueras J, Guerri C. Ethanol exposure affects glial fibrillary acidic protein gene expression and transcription during rat brain development. J Neurochem. 1997;69:2484–2493. doi: 10.1046/j.1471-4159.1997.69062484.x. [DOI] [PubMed] [Google Scholar]
- 40.Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- 41.Biermann T, Reulbach U, Lenz B, Frieling H, Muschler M, Hillemacher T, et al. N-methyl-D-aspartate 2b receptor subtype (NR2B) promoter methylation in patients during alcohol withdrawal. J Neural Transm (Vienna) 2009;116:615–622. doi: 10.1007/s00702-009-0212-2. [DOI] [PubMed] [Google Scholar]
- 42.Qiang M, Denny A, Chen J, Ticku MK, Yan B, Henderson G. The site specific demethylation in the 5ʼ-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS One. 2010;5:e8798. doi: 10.1371/journal.pone.0008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 44.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 45.Pattaroni C, Jacob C. Histone methylation in the nervous system: functions and dysfunctions. Mol Neurobiol. 2013;47:740–756. doi: 10.1007/s12035-012-8376-4. [DOI] [PubMed] [Google Scholar]
- 46.Balemans MC, Huibers MM, Eikelenboom NW, Kuipers AJ, van Summeren RC, Pijpers MM, Tachibana M, Shinkai Y, van Bokhoven H, Van der Zee CE. Reduced exploration, increased anxiety, and altered social behavior: autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behav Brain Res. 2010;208:47–55. doi: 10.1016/j.bbr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stadler F, Kolb G, Rubusch L, Baker SP, Jones EG, Akbarian S. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem. 2005;94:324–336. doi: 10.1111/j.1471-4159.2005.03190.x. [DOI] [PubMed] [Google Scholar]
- 50.Subbanna S, Basavarajappa BS. Pre-administration of G9a/GLP inhibitor during synaptogenesis prevents postnatal ethanol-induced LTP deficits and neurobehavioral abnormalities in adult mice. Exp Neurol. 2014;261:34–43. doi: 10.1016/j.expneurol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A. 2011;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subbanna S, Shivakumar M, Umapathy NS, Saito M, Mohan PS, Kumar A, et al. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subbanna S, Nagre NN, Shivakumar M, Umapathy NS, Psychoyos D, Basavarajappa BS. Ethanol induced acetylation of histone at G9a exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice. Neuroscience. 2014;258:422–432. doi: 10.1016/j.neuroscience.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pascual M, Do Couto BR, Alfonso-Loeches S, Aguilar MA, Rodriguez-Arias M, Guerri C. Changes in histone acetylation in the prefrontal cortex of ethanol-exposed adolescent rats are associated with ethanol-induced place conditioning. Neuropharmacology. 2012;62:2309–2319. doi: 10.1016/j.neuropharm.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 56.Mashayekhi FJ, Rasti M, Rahvar M, Mokarram P, Namavar MR, Owji AA. Expression levels of the BDNF gene and histone modifications around its promoters in the ventral tegmental area and locus ceruleus of rats during forced abstinence from morphine. Neurochem Res. 2012;37:1517–1523. doi: 10.1007/s11064-012-0746-9. [DOI] [PubMed] [Google Scholar]
- 57.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivanov M, Barragan I, Ingelman-Sundberg M. Epigenetic mechanisms of importance for drug treatment. Trends Pharmacol Sci. 2014;35:384–396. doi: 10.1016/j.tips.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 63.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33:1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen G, Zou X, Watanabe H, van Deursen JM, Shen J. CREB binding protein is required for both short-term and long-term memory formation. J Neurosci. 2010;30:13066–13077. doi: 10.1523/JNEUROSCI.2378-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2010;107:22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narayan PJ, Lill C, Faull R, Curtis MA, Dragunow M. Increased acetyl and total histone levels in post-mortem Alzheimer's disease brain. Neurobiol Dis. 2015;74:281–294. doi: 10.1016/j.nbd.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 68.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36:61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascual M, Boix J, Felipo V, Guerri C. Repeated alcohol administration during adolescence causes changes in the mesolimbic dopaminergic and glutamatergic systems and promotes alcohol intake in the adult rat. J Neurochem. 2009;108:920–931. doi: 10.1111/j.1471-4159.2008.05835.x. [DOI] [PubMed] [Google Scholar]
- 71.Guo W, Crossey EL, Zhang L, Zucca S, George OL, Valenzuela CF, et al. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLoS One. 2011;6:e19351. doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shibasaki M, Mizuno K, Kurokawa K, Ohkuma S. Enhancement of histone acetylation in midbrain of mice with ethanol physical dependence and its withdrawal. Synapse. 2011;65:1244–1250. doi: 10.1002/syn.20947. [DOI] [PubMed] [Google Scholar]
- 73.Renthal W, Nestler EJ. Histone acetylation in drug addiction. Semin Cell Dev Biol. 2009;20:387–394. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liokatis S, Stützer A, Elsässer SJ, Theillet FX, Klingberg R, van Rossum B, et al. Phosphorylation of histone H3 Ser10 establishes a hierarchy for subsequent intramolecular modification events. Nat Struct Mol Biol. 2012;19:819–823. doi: 10.1038/nsmb.2310. [DOI] [PubMed] [Google Scholar]
- 75.Mori T, Wakabayashi T, Ogawa H, Hirahara Y, Koike T, Yamada H. Increased histone H3 phosphorylation in neurons in specific brain structures after induction of status epilepticus in mice. PLoS One. 2013;8:e77710. doi: 10.1371/journal.pone.0077710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pavlova MB, Dyuzhikova NA, Shiryaeva NV, Savenko YN, Vaido AI. Effect of long-term stress on H3Ser10 histone phosphorylation in neuronal nuclei of the sensorimotor cortex and midbrain reticular formation in rats with different nervous system excitability. Bull Exp Biol Med. 2013;155:373–375. doi: 10.1007/s10517-013-2157-6. [DOI] [PubMed] [Google Scholar]
- 77.Aroor AR, Restrepo RJ, Kharbanda KK, Shukla SD. Epigenetic histone modifications in a clinically relevant rat model of chronic ethanol-binge-mediated liver injury. Hepatol Int. 2014;8(suppl 2):421–430. doi: 10.1007/s12072-014-9546-4. [DOI] [PubMed] [Google Scholar]
- 78.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 79.Hammond-Martel I, Yu H, Affar El B. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012;24:410–421. doi: 10.1016/j.cellsig.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 80.Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a ‘histone-like’ non-histone chromosomal protein. J Biol Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 81.Jones JM, Bhattacharyya A, Simkus C, Vallieres B, Veenstra TD, Zhou M. The RAG1 V(D)J recombinase/ubiquitin ligase promotes ubiquitylation of acetylated, phosphorylated histone 3.3. Immunol Lett. 2011;136:156–162. doi: 10.1016/j.imlet.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 83.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishino Y, Hayashi Y, Naruse M, Tomita K, Sanbo M, Fuchigami T, et al. Bre1a, a Histone H2B ubiquitin ligase, regulates the cell cycle and differentiation of neural precursor cells. J Neurosci. 2014;34:3067–3078. doi: 10.1523/JNEUROSCI.3832-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rubio MD, Wood K, Haroutunian V, Meador-Woodruff JH. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology. 2013;38:1910–1920. doi: 10.1038/npp.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krumova P, Weishaupt JH. Sumoylation in neurodegenerative diseases. Cell Mol Life Sci. 2013;70:2123–2138. doi: 10.1007/s00018-012-1158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nisticò R, Ferraina C, Marconi V, Blandini F, Negri L, Egebjerg J, et al. Age-related changes of protein SUMOylation balance in the AβPP Tg2576 mouse model of Alzheimer's disease. Front Pharmacol. 2014;5:63. doi: 10.3389/fphar.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu M, Wang F, Li X, Rogers CQ, Liang X, Finck BN, et al. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology. 2012;55:437–446. doi: 10.1002/hep.24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El Fatimy R, Miozzo F, Le Mouël A, Abane R, Schwendimann L, Sabéran-Djoneidi D, et al. Heat shock factor 2 is a stress-responsive mediator of neuronal migration defects in models of fetal alcohol syndrome. EMBO Mol Med. 2014;6:1043–1061. doi: 10.15252/emmm.201303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qi Y, Zhang M, Li H, Frank JA, Dai L, Liu H, Chen G. MicroRNA-29b regulates ethanol-induced neuronal apoptosis in the developing cerebellum through SP1/RAX/PKR cascade. J Biol Chem. 2014;289:10201–10210. doi: 10.1074/jbc.M113.535195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA, et al. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics. 2013;14:725. doi: 10.1186/1471-2164-14-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. 2015;20:1219–1231. doi: 10.1038/mp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melka MG, Laufer BI, McDonald P, Castellani CA, Rajakumar N, O'Reilly R, et al. The effects of olanzapine on genome-wide DNA methylation in the hippocampus and cerebellum. Clin Epigenetics. 2014;6:1. doi: 10.1186/1868-7083-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Medical University of Vienna Effects of Creatine Supplementation in Rett Syndrome: A Randomized, Placebo-controlled Trial, 2009. https://clinicaltrials.gov/ct2/show/record/NCT01147575?term=NCT01147575&rank=1>; [DOI] [PubMed]
- 97.Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling - a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.University Hospital, Bordeaux Rubinstein-Taybi Syndrome: Functional Imaging and Therapeutic, 2014. https://clinicaltrials.gov/ct2/show/NCT01619644?term=NCT01619644&rank=1
- 99.Al Ameri M, Al Mansouri S, Al Maamari A, Bahi A. The histone deacetylase (HDAC) inhibitor valproic acid reduces ethanol consumption and ethanol-conditioned place preference in rats. Brain Res. 2014;1583:122–131. doi: 10.1016/j.brainres.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 100.Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol Med. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker DM, Cates HM, Heller EA, Nestler EJ. Regulation of chromatin states by drugs of abuse. Curr Opin Neurobiol. 2015;30:112–121. doi: 10.1016/j.conb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 103.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arora DS, Nimitvilai S, Teppen TL, McElvain MA, Sakharkar AJ, You C, Pandey SC, Brodie MS. Hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal: reversal by histone deacetylase inhibitors. Neuropsychopharmacology. 2013;38:1674–1684. doi: 10.1038/npp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.D'Addario C, Caputi FF, Ekström TJ, Di Benedetto M, Maccarrone M, Romualdi P, Candeletti S. Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. J Mol Neurosci. 2013;49:312–319. doi: 10.1007/s12031-012-9829-y. [DOI] [PubMed] [Google Scholar]