Abstract
Background
Epigenetic alterations of gene or DNA methylation have been highlighted as promising biomarkers for early cervical cancer screening. Herein, we evaluated the diagnostic performance of paired boxed gene 1 (PAX1) and sex determining region Y-box 1 (SOX1) methylation for cervical cancer detection.
Methods
Eligible studies were retrieved by searching the electronic databases. Study quality was assessed according to the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) checklist. The bivariate meta-analysis model was employed to plot the summary receiver operator characteristic (SROC) curve using Stata 12.0 software.
Results
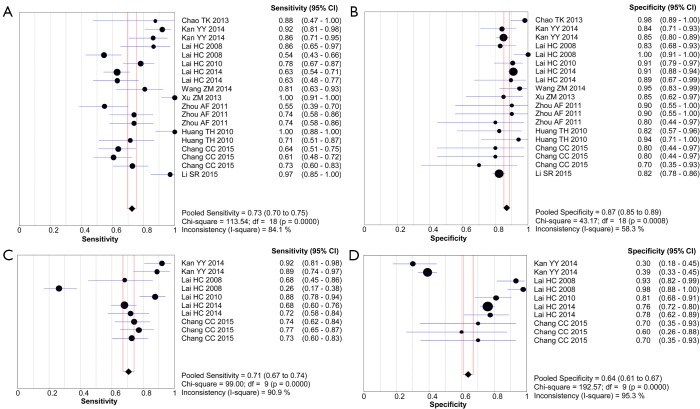
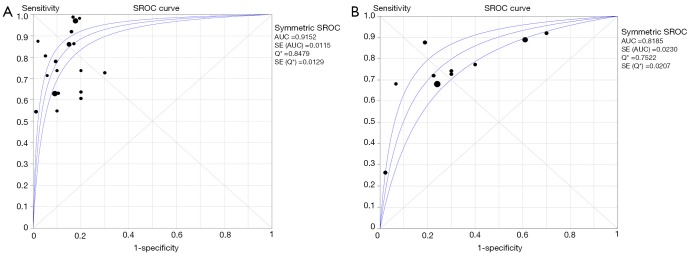
The pooled sensitivity of PAX1 methylation was estimated to be 0.73 [95% confidence interval (CI): 0.70–0.75] in differentiating patients with HSIL (high-grade squamous intraepithelial lesion) or CIN3+ (cervical intraepithelial neoplasia type III/worse) or cervical cancer from normal individuals, corresponding to a specificity of 0.87 (95% CI: 0.85–0.89) and area under the curve (AUC) of 0.91. The SOX1 methylation test yielded an AUC of 0.82, under which, the pooled sensitivity was 0.71 (95% CI: 0.67–0.74) and specificity was 0.64 (95% CI: 0.61–0.67). Notably, the stratified analysis suggested that combing parallel testing of PAX1 methylation and human papillomavirus (HPV) DNA (AUC, sensitivity, and specificity of 0.89, 0.75, and 0.81, respectively) achieved higher accuracy than single HPV DNA testing (AUC, sensitivity, and specificity of 0.77, 0.81, and 0.70, respectively).
Conclusions
PAX1 or SOX1 methylation has a prospect to be an auxiliary biomarker for cervical cancer screening, and parallel testing of PAX1 methylation and HPV DNA in cervical swabs confers an improved diagnostic accuracy than single HPV DNA testing.
Keywords: Paired boxed gene 1 (PAX1), sex determining region Y-box 1 (SOX1), DNA methylation, cervical cancer, human papillomavirus (HPV), meta analysis
Introduction
Cervical cancer occurs frequently in women and every two minutes, a woman dies because of cervical cancer worldwide (1,2). The introduction of cytology screening (Pap smear) has greatly reduced the mortality and morbidity rates for patients with invasive cervical cancers in the past decades. However, it has been demonstrated that the accuracy of Pap smear test ranged from 20% to 80%, and the results varied substantially in areas with different screening infrastructures, which therefore limited its efficacy for cervical cancer diagnosis (3-6). Persistently infected by human papillomavirus (HPV) with high risk type remains the major pathogens of cervical cancer, as a result, HPV DNA testing has been adopted for the triage of patients with atypical squamous cells of undetermined significance (2,4,7-9). Nevertheless, HPV triage yields a poor specificity, which diminishes the overall diagnostic value for cervical cancer screening (10,11). In this respect, discovering and developing new biomarkers which confer high sensitivity and specificity for cervical cancer detection is a matter of great urgency in the clinic. Epigenetic alterations, such as gene or DNA methylation, have been proposed as novel biomarkers for the cervical cancer detection. Among these altered and methylated genes, the paired boxed gene 1 (PAX1) and sex determining region Y-box 1 (SOX1) were both highlighted (12-26). Numerous studies have documented the promise of PAX1 or SOX1 DNA methylation in distinguishing patients with reactive atypia or CIN3+ (cervical intraepithelial neoplasia type III or worse) lesions from normal uterine cervixes (12-23). In order to make a comparison of the accuracy of DNA methylation and HPV DNA testing, we performed a comprehensive meta-analysis and evaluated the diagnostic performance of PAX1 and SOX1 methylation for cervical cancer screening.
Methods
This meta-analysis was conducted in compliance with the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) Statement issued in 2009 (27). The online PubMed and Chinese National Knowledge Infrastructure (CNKI) databases were searched up to August 31st 2015 for all published articles with no language restriction. Relevant publications were identified by searching for combinations of “PAX1 and/or SOX1 methylation”, “HPV” or “human papillomavirus”, “cervical cancer” or “cervical neoplasia” or “cervical carcinoma”, “high-grade squamous intraepithelial lesion” or “cervical intraepithelial neoplasia type III” or “CIN3”, and “diagnosis” or “sensitivity” or “specificity” or “AUC”.
Study selection
The following criteria were used for the literature selection in this meta-analysis: (I) studies evaluated the diagnostic performance of PAX1 and/or SOX1 methylation or HPV DNA testing in cervical neoplasms; and (II) studies explicitly mentioned the sample size, sensitivity, specificity and their 95% confidence intervals (CIs) or other more detailed information. Literature were excluded according to the following criteria: (I) the control group and sample sizes were unclear; (II) studies without complete data including missing information of sensitivity, specificity or area under the curve (AUC) value, etc.; and (III) basic research, animal studies, meta analysis, review articles, letters, commentaries, abstracts presented at conferences, etc.
Data extraction and quality assessment
Two investigators independently judged study eligibility and extracted the relevant data from each article including the name of the first author, year of publication, country, patient size, control sources, sample types, CIN degrees, test method and the diagnostic results. In studies contained both a training and a validation group, data from each group was treated as a single study in the meta-analysis. Study quality was scored using a 14-item quality assessment of diagnostic accuracy studies (QUADAS) checklist and each was assessed as “Yes”, “Unclear” or “No”, corresponding to a score of “1”, “0” and “0”, respectively (28). Any disagreement was resolved by group consensus.
Statistical analysis
Statistical analysis was undertaken using Stata 12.0 (Stata Corporation, College Station, TX, USA), and Meta-disc 1.4 (XI Cochrane Colloquium, Barcelona, Spain) software. Diagnostic performance was assessed using a bivariate random-effects model in case of the existence of heterogeneity among studies (29). Heterogeneity from threshold and non-threshold effects were reflected by the Spearman correlation coefficient, Cochran’s-Q and I2 tests (30), respectively. Influence analysis and meta-regression were performed to trace potential sources of study heterogeneity. The covariates included the following: sample size (<100 or ≥100), cut-off value (PMR % <10 or ≥10), test method (QMSP or MS-HRM), and study quality (QUADAS ≥10 or <10). Deeks’ funnel plot asymmetry test was conducted to evaluate the potential publication bias, and significant level was set at P<0.05.
Results
Study characteristics and quality
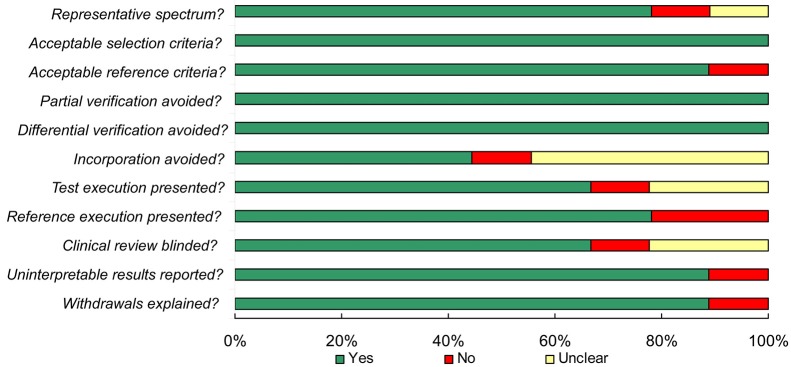
Figure 1 illustrates the articles search procedure: a total of 541 potentially relevant studies were identified from a primary literature search in electronic databases, and 498 studies were excluded due to the status that unrelated to PAX1, SOX1 methylation or cervical cancer diagnosis. The retrieved 43 studies received further evaluation: 2 of them were review articles and 28 were basic research which all failed to meet the inclusion criteria. In one study, the clinical accuracy calculated combined CIN3+ and endometrial complex hyperplasia was further excluded (21). Another study comprises normal and CIN2-3 samples in the paired controls was finally discarded as well (31). Therefore, 11 articles with 19 individual studies for PAX1 methylation, 10 for SOX1 methylation and 10 for HPV DNA testing were included in this meta-analysis. All of the 11 studies were conducted in Asia, including 7 studies in Taiwan and 4 in China mainland. The final diagnoses of all studies were determined by tissue-proven histopathology, and the evaluation method for DNA methylation comprises quantitative methylation-specific polymerase chain reaction (QMSP) and/or methylation-sensitive high-resolution melting (MS-HRM). The main features of each included study were described in Table 1. We evaluated the study quality of each included publications according to the 14 item QUADAS assessment tool (28). As shown in Figure 2, all of the 11 studies got QUADAS scores more than 8 and revealed lower risks of bias, suggesting a relatively high quality of the included studies.
Figure 1.
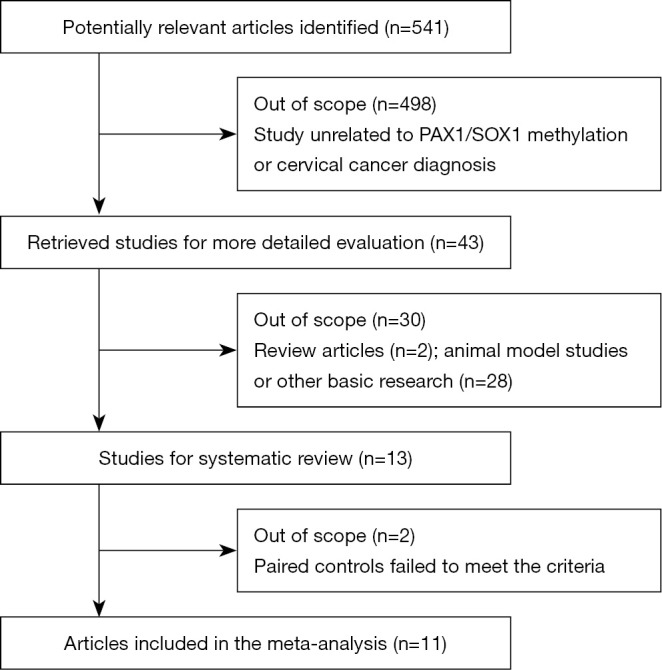
Flowchart of the literature search and study selection procedure.
Table 1. The main features of included studies for PAX1 and SOX1 methylation in diagnosing cervical cancer.
| Author | Year | Study population | Patient size | Control size | Specimen type | HPV type | Method | Cut-off value (PMR %) | |
|---|---|---|---|---|---|---|---|---|---|
| Total | HSIL/CIN3+/cancer | ||||||||
| Chao et al. (16) | 2013 | Taiwanese | 58 | 8 | 50 | Cervical scrapings | High-risk | QMSP | PAX1: 27 |
| Chang et al. (22) | 2015 | Taiwanese | 126 | 66 | 10 | Cervical swabs/tissues | – | QMSP | PAX1: 1.4; SOX1: 15.6 |
| Kan et al. (17) | 2014 | Taiwanese | 172 | 36 | 247 | Cervical scrapings | High-risk | QMSP | PAX1: 11 |
| Lai et al. (12) | 2008 | Taiwanese | 125 | 80 | 45 | Cervical swabs | High-risk | QMSP | Unclear |
| Lai et al. (15) | 2010 | Taiwanese | 132 | 66 | 53 | Cervical scrapings | – | QMSP | PAX1: 10.17; SOX1: 1.8 |
| Lai et al. (14) | 2014 | Taiwanese | 266 | 141 | 410 | Cervical swabs | High-risk type involved 16, 18 | QMSP | PAX1: 4.88 |
| Li et al. (23) | 2015 | Chinese | 34 | 34 | 429 | Cervical scrapings | High-risk | MS-HRM | Unclear |
| Wang et al. (13) | 2014 | Chinese | 91 | 31 | 39 | Cervical scrapings | High-risk | MS-HRM | PAX1: 10 |
| Xu et al. (19) | 2013 | Chinese | 72 | 41 | 20 | Cervical swabs/biopsy tissues | High-risk | QMSP | PAX1: 25 |
| Zhou et al. (18) | 2011 | Chinese | 80 | 42 | 10 | Cervical scrapings | – | QMSP/MS-HRM | Unclear |
| Huang et al. (20) | 2010 | Taiwanese | 56 | 28 | 17 | Cervical scrapings | High-risk | QMSP | PAX1: 4.5 |
PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1; QMSP, quantitative methylation-specific polymerase chain reaction; MS-HRM, methylation-sensitive high-resolution melting; PMR, percentage of methylated reference; HSIL, high-grade squamous intraepithelial lesion; HPV, human papillomavirus.
Figure 2.
Summary of assessment of the included studies analyzed using the QUADAS tool: proportion of studies with low (yes), mediate (unclear), and high risk of bias (no). QUADAS, quality assessment for studies of diagnostic accuracy.
Heterogeneity
Heterogeneity from threshold and non-threshold effects were assessed using Meta-disc 1.4 software. As shown in Table 2, Spearman correlation coefficient in PAX1 methylation test presented a P value of 0.181, indicating that there was no heterogeneity from threshold effect. Additionally, the Cochran’s-Q test yielded a Q value of 30.70 (P>0.01), with I2<50%, suggesting that non-threshold effect is not likely to be a source of heterogeneity. Nevertheless, heterogeneity generated by threshold or non-threshold effects seemed to exist in other pooled analyses (Table S1).
Table 2. Diagnostic indices of PAX1 and SOX1 methylation for cervical cancer screening.
| Analysis | Pooled sensitivity (95% CI) | Pooled specificity (95% CI) | Pooled PLR (95% CI) | Pooled NLR (95% CI) | Pooled DOR (95% CI) | AUC |
|---|---|---|---|---|---|---|
| PAX1 | 0.73 (0.70–0.75) | 0.87 (0.85–0.89) | 5.80 (5.04–6.68) | 0.30 (0.23–0.38) | 27.51 (17.24–43.88) | 0.91 |
| SOX1 | 0.71 (0.67–0.74) | 0.64 (0.61–0.67) | 2.69 (1.77–4.09) | 0.36 (0.23–0.55) | 8.73 (5.75–13.27) | 0.82 |
| HPV | 0.81 (0.77–0.85) | 0.70 (0.67–0.72) | 2.31 (1.78–2.99) | 0.32 (0.20–0.51) | 8.18 (4.15–16.12) | 0.77 |
| PAX1/HPV | 0.75 (0.73–0.78) | 0.81 (0.79–0.82) | 3.93 (3.15–4.91) | 0.28 (0.22–0.35) | 20.49 (14.02–29.93) | 0.89 |
| SOX1/HPV | 0.64 (0.58–0.69) | 0.48 (0.45–0.52) | 2.22 (1.52–3.26) | 0.34 (0.17–0.71) | 11.32 (5.39–23.80) | 0.84 |
| PAX1/SOX1 | 0.72 (0.69–0.74) | 0.77 (0.76–0.79) | 5.41 (3.50–8.36) | 0.29 (0.22–0.39) | 21.41 (13.78–33.28) | 0.89 |
| Outlier excluded for PAX1 | 0.74 (0.71–0.77) | 0.86 (0.83–0.88) | 5.59 (4.82–6.48) | 0.27 (0.19–0.38) | 29.84 (17.72–50.24) | 0.92 |
| Outlier excluded for HPV | 0.84 (0.80–0.88) | 0.70 (0.68–0.73) | 2.36 (1.78–3.12) | 0.29 (0.17–0.50) | 9.30 (4.39–19.69) | 0.79 |
No outlier studies identified in SOX1 methylation test. PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1; CI, confidence interval; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; AUC, area under the curve.
Diagnostic performance
As indicated in Table 2, the pooled accuracies for PAX1 and/or SOX1 methylation, alone, in parallel or in sequential combinations, were determined to assess their usefulness as biomarkers for screening of patients with HSIL and/or cervical carcinoma. The pooled sensitivity, specificity, DOR and AUC for PAX1 methylation test were 0.73 (95% CI: 0.70–0.75), 0.87 (95% CI: 0.85–0.89), 27.51 (95% CI: 17.24–43.88) and 0.91, respectively, versus those of 0.71 (95% CI: 0.67–0.74), 0.64 (95% CI: 0.61–0.67), 8.73 (95% CI: 5.75–13.27) and 0.82 for SOX1 methylation. The forest plots of pooled sensitivity, specificity and summary receiver operator characteristic (SROC) curves for PAX1 and SOX1 methylation are displayed in Figures 3,4. For the HPV DNA testing, it yielded an AUC value of 0.77, with pooled sensitivity of 0.81 (95% CI: 0.77–0.85) and specificity of 0.70 (95% CI: 0.67–0.72). Forest plots of pooled sensitivity, specificity as well as SROC curve for HPV DNA are shown in Figure S1.
Figure 3.
Forest plots of the pooled sensitivity and specificity for PAX1 and SOX1 methylation. (A) Sensitivity for PAX1; (B) specificity for PAX1; (C) sensitivity for SOX1; (D) specificity for SOX1. Only the first author of each study is given. Sensitivity and specificity are given with 95% confidence intervals (CIs). PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1.
Figure 4.
SROC curve of the pooled PAX1 and SOX1 methylation tests. Sample size is indicated by the size of the square. The regression SROC curve indicates the overall diagnostic accuracy. (A) PAX1 methylation; (B) SOX1 methylation. AUC, area under curve; SENS, sensitivity; SPEC, specificity; SROC, summary receiver operator curve; PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1.
Subgroup analyses
A random effect model was applied in the stratified meta-analyses due to the existence of significant heterogeneities among studies. The results for the stratified analyses were listed in Table 2. The paralleled PAX1/HPV, SOX1/HPV and PAX1/SOX1 tests achieved AUC values of 0.89, 0.84 and 0.89, respectively, under which, the pooled sensitivity were 0.75 (95% CI: 0.73–0.78), 0.64 (95% CI: 0.58–0.69) and 0.72 (95% CI: 0.69–0.74), respectively; the pooled specificity were 0.81 (95% CI: 0.79–0.82), 0.48 (95% CI: 0.45–0.52) and 0.77 (95% CI: 0.76–0.79), respectively.
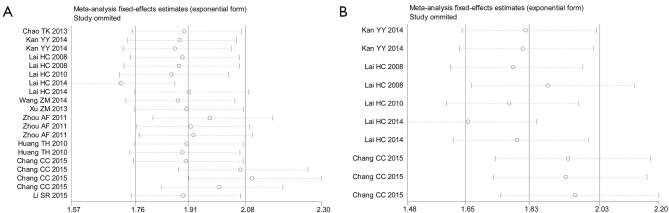
Influence assay and meta-regression
We performed influence analysis based on the platform of Stata 12.0 software. As shown in Figure 5A, influence analysis and outlier detection identified 2 outlier studies for PAX1 methylation test. After adjustment of the outlier studies, the overall pooled sensitivity increased from 0.73 to 0.74, PLR decreased from 5.80 to 5.59, NLR decreased from 0.30 to 0.27, DOR elevated from 27.51 to 29.84. Moreover, the P value of spearman correlation coefficient from threshold effect increased from 0.459 to 0.625, and P value from Cochran’s-Q test was attenuated form 0.03 to 0.05, and I2 declined from 41.4% to 38.3%, hinting that the outlier is likely a source of heterogeneity. For the HPV DNA test, we identified 2 outlier studies as well, and the data revealed that the outliers also contributed to the heterogeneity sources (Figure S2). No outlier studies were identified in SOX1 methylation test (Figure 5B). We further conducted meta-regression analysis based on 5 pre-specified covariates: sample size (<100 or ≥100), cut-off value (PMR % <10 or ≥10), test method (QMSP or MS-HRM), and study quality (QUADAS ≥10 or <10). However, these covariates showed a low likelihood of sources of inter-study heterogeneity (Table 3).
Figure 5.
Influence and outlier detection analyses of PAX1 and SOX1 methylation test. (A) PAX1 methylation test; (B) SOX1 methylation test. PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1.
Table 3. Meta-regression (inverse variance weights) for the potential source of heterogeneity.
| Study characteristic | P value | RDOR | 95% CI |
|---|---|---|---|
| PAX1 methylation | |||
| Patient size (<100 vs. ≥100) | 0.4626 | 1.41 | 0.53–3.79 |
| Control size (<100 vs. ≥100) | 0.8981 | 0.93 | 0.28–3.12 |
| Cut-off value (PMR % <10 vs. ≥10) | 0.0873 | 1.51 | 0.94–2.44 |
| Method (QMSP vs. MS-HRM) | 0.0768 | 0.44 | 0.17–1.10 |
| Study quality (QUADAS ≥10 vs. <10) | 0.4623 | 1.75 | 0.36–8.59 |
| SOX1 methylation | |||
| Patient size (<100 vs. ≥100) | 0.4833 | 0.76 | 0.32–1.80 |
| Control size (<100 vs. ≥100) | 0.9282 | 1.07 | 0.17–6.55 |
| Cut-off value (PMR % <10 vs. ≥10) | 0.1031 | 0.61 | 0.33–1.14 |
| Study quality (QUADAS ≥10 vs. <10) | 0.6732 | 1.2 | 0.45–3.20 |
| HPV DNA testing | |||
| Patient size (<100 vs. ≥100) | 0.2042 | 0.40 | 0.08–2.02 |
| Control size (<100 vs. ≥100) | 0.3471 | 1.83 | 0.45–7.33 |
| Method (QMSP vs. MS-HRM) | 0.0552 | 3.51 | 0.96–12.77 |
| Study quality (QUADAS ≥10 vs. <10) | 0.3654 | 2.06 | 0.34–12.47 |
CI, confidence interval; RDOR, relative diagnostic odds ratio; QUADAS, quality assessment for studies of diagnostic accuracy; QMSP, quantitative methylation-specific polymerase chain reaction; MS-HRM, methylation-sensitive high-resolution melting; HPV, human papillomavirus.
Publication bias
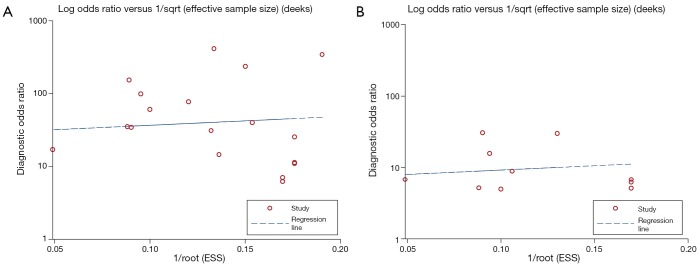
The funnel plots for publication bias showed no asymmetry for the pooled PAX1 and SOX1 methylation analysis, corresponding to a P value of 0.645 and 0.651, respectively, indicating that there was no publication bias in the meta-analysis (Figure 6). Additionally, the P value of the Deeks’ test for HPV DNA was estimated to be 0.869, also showed a low likelihood of publication bias (Figure S3).
Figure 6.
Funnel graph for the assessment of potential publication bias of the included studies. (A) Deeks’ funnel plot asymmetry test for PAX1 methylation, P=0.645; (B) Deeks’ funnel plot asymmetry test for SOX1 methylation, P=0.651. PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1.
Discussion
Infection with oncogenic types of HPV is the most significant risk factor in the etiology of cervical cancer, and is present in nearly all cervical cancers (2,4,8,32,33). Molecular tests detecting HPV DNA are now being available either commercially or in-house developed. The sensitivity of HPV DNA testing is satisfactory, whereas the high prevalence of transient HPV infections had limited the specificity of this approach (10,11). Currently, there are several test methods for HPV DNA assay, such as PCR, hybrid capture II (HC2), strip test (for E6 detection) as well as other novel techniques (5,12-15,34,35), Among these evaluation tools, PCR and HC2 tests remain the most commonly used techniques (34,35). Notwithstanding, single HPV DNA testing using PCR or HC2 method seems to yield inconstant results for the detection of cervical precancerous lesions (CIN2 or worse). For instance, previous studies had documented that the sensitivity of HPV DNA testing using PCR method ranged from 0.66 to 0.89, whereas the specificity was from 0.64 to 0.68 (12,15). However, in a published meta-analysis study, the result showed that PCR test achieved a pooled sensitivity of 0.809 and specificity of 0.947 (5). As reported, HPV DNA test utilizing HC2 method may confer a relatively higher accuracy than using PCR analysis. In the aforementioned meta-analysis, the HC2 method displayed a pooled sensitivity of 0.90 and specificity of 0.865 in primary cervical screening (5). Nevertheless, in Wang’s study, the accuracy of HC2 test in triage of high-grade squamous intraepithelial lesions was unsatisfactory, in which the sensitivity and specificity were only 0.677 and 0.545, respectively (13). Despite the reported inconstant results for these methods, these techniques often lack standardization and comparability (34,35). It appears therefore that single HPV DNA testing is not recommended for screening purposes (10,11).
For early cervical cancer diagnosis, DNA methylation of the PAX1 or SOX1 gene, has been proposed as new biomarkers for the cancer screening against HPV DNA testing (12-26). As the potential diagnostic value of DNA methylation for cervical cancer screening has not yet been well elucidated thus far, this study systematically evaluated and compared the diagnostic accuracy of PAX1 and SOX1 methylation and HPV DNA testing in the diagnosis of HSIL or CIN3+ lesions or cervical cancer. As shown in our data, PAX1 methylation achieved a higher pooled diagnostic accuracy when compared with SOX1 methylation or HPV DNA testing. The pooled sensitivity and specificity of PAX1 methylation was 0.73 and 0.87, respectively. Although the pooled sensitivity appeared not very high, the ROC AUC was 0.91, suggesting an overall high accuracy of this diagnostic test. Diagnostic odds ratio (DOR) is one of the key indicators in assessing the accuracy of one diagnostic test, and that a DOR smaller than 1.0 often suggests a low discriminating value for a diagnostic test (36,37). Importantly, the pooled DOR for PAX1 methylation was 27.51, indicating a better discriminatory test performance of PAX1 methylation for cervical cancer detection. Moreover, the pooled PLR of 5.80 and NLR of 0.30, also suggested that patients with HSIL or CIN3+ or cervical cancer had nearly 6 fold higher chance of being PAX1 methylation test positive than other normal individuals, with a ratio of 30% likely to be false-negative in a negative result of the methylation test, revealing a relatively high power in ruling out high risk of cervical cancer.
As a comparison, SOX1 methylation test revealed an overall accuracy inferior to that of PAX1 methylation: despite a reduction in the sensitivity and specificity, other parameters were lower than PAX1 methylation as well. Yet, the HPV DNA testing harvested the highest combined sensitivity of 0.83 among these assays, but the pooled specificity was estimated to be 0.67. Besides that, other indices like pooled PLR, NLR and DOR, also indicated an overall diagnostic accuracy inferior to PAX1 methylation test. A published research suggested that the methylation test of PAX1 gene harbored higher diagnostic accuracy against HPV DNA testing in detecting cervical cancer (20). Similarly, other studies also documented that quantitative measurement of PAX1 hypermethylation in cervical scrapings is highly accurate than both SOX1 hypermethylation and HPV DNA assay, which the sensitivity was 0.86, and specificity was 0.85 under the ROC curve (12,17). Our results agree well with these data.
In order to verify whether the combing parallel testing of DNA methylation and HPV DNA could benefit the diagnostic accuracy, we further conducted the stratified analyses. As expected, the results displayed that PAX1/HPV paralleled test harbored a high AUC value of 0.89, but both pooled sensitivity and specificity decreased when compared with that of PAX1 methylation alone. However, the overall pooled diagnostic accuracy for PAX1/HPV test was comparable to that of PAX1 methylation alone and better than single HPV DNA testing. For the paralleled SOX1/HPV test, although their AUC and DOR were both elevated as compared to that of SOX1 methylation alone, the pooled sensitivity, specificity as well as other indices was discounted. In addition, we found the parallel testing of PAX1 and SOX1 methylation harvested a sensitivity of 0.72, specificity of 0.77 and DOR of 24.41 with an AUC value of 0.89, suggesting that a combined detection of PAX1 and SOX1 methylation harbored a moderate accuracy for cervical cancer detection. Evidence from a recent study suggested that PAX1 methylation harbored a better diagnostic performance with a specificity of 0.88 when combined with sequential testing of HPV (14). In addition, the paneled assay of PAX1/SOX1 methylation has been previously described as well, which the sensitivity was greater than 0.70, and specificity greater than 0.90 (15). For the combined PAX1 and HPV DNA testing, the published data had documented that when combining with HPV DNA 16/18 typing, the PAX1 testing showed a sensitivity of almost 0.90 and a specificity of 0.83 (17). Consistent with these results, our data showed promising accuracy for the paralleled PAX1/HPV test.
In this study, heterogeneity from threshold and non-threshold effects existed in the pooled studies. For its causes, the uniformed cut-off value setting may contribute to the heterogeneity of threshold effect. On the other hand, the non-threshold effect is another important source of heterogeneity, and parameters of Cochran’s-Q and I2 tests from pooled DOR are good indicators in discussing heterogeneity from nonthreshold effects (36). In this meta-analysis, the forest plots of DOR from each study (except PAX1, SOX1 and SOX1/HPV tests) did not distribute along a straight line (not shown), corresponding to the P values in Cochran’s-Q test less than 0.01 and I2 more than 50%. It is speculated that different measurement methods, or sample types may contribute to the heterogeneity sources. For instance, two different test methods, QMSP and HRM were used in measuring DNA methylation in our study. In addition to the test method, the reference method was also different among studies. In consequence, we further conducted influence and meta-regression analyses and our results revealed that the outlier studies were likely to be a source of heterogeneity.
Conclusions
This meta-analysis demonstrates that PAX1 methylation achieves a promising diagnostic performance for cervical cancer detection, and parallel testing of PAX1 methylation and HPV DNA confers an improved diagnostic accuracy than HPV DNA testing alone. For the SOX1 methylation test, it yields a moderate accuracy in diagnosing cervical neoplasia. Hence, PAX1 or SOX1 methylation could potentially be treated as an auxiliary biomarker for cervical cancer screening. Further high quality studies are still warranted to confirm our analyses.
Acknowledgements
This study was supported by the National Clinical Key Specialty Construction Program of China.
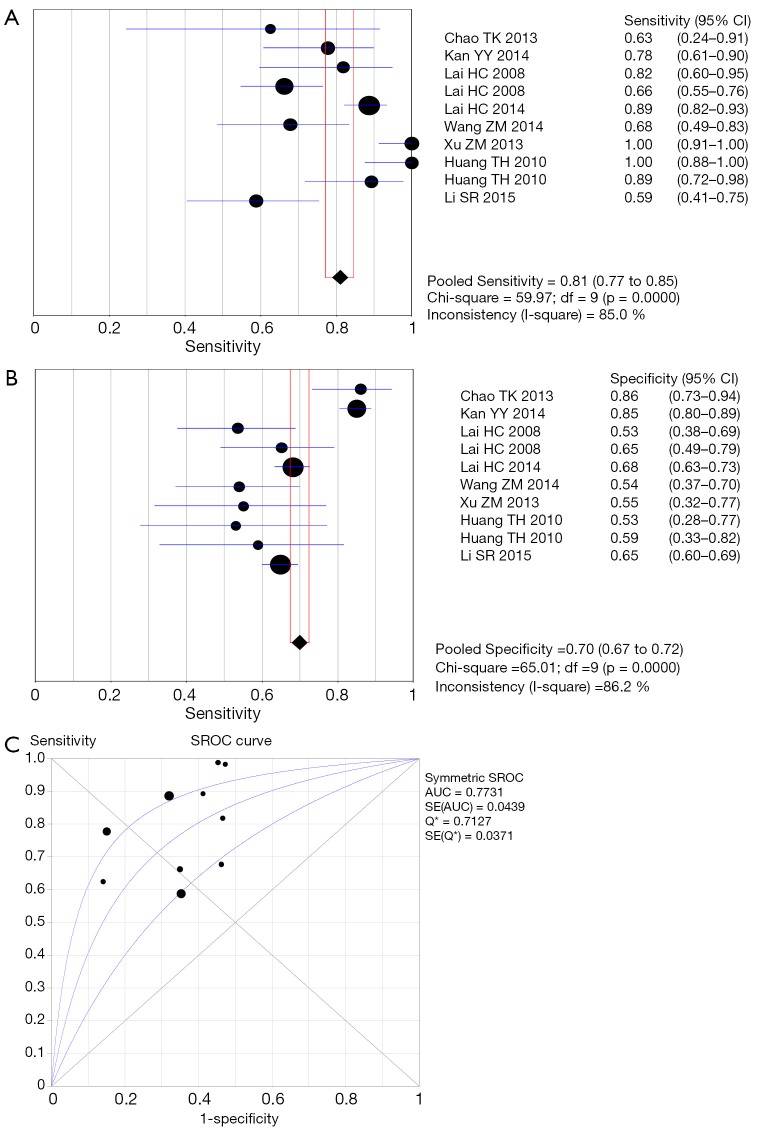
Figure S1.
Forest plots of pooled sensitivity, specificity and SROC curve for HPV DNA test. (A) Sensitivity; (B) specificity; (C) SROC curve. SROC, summary receiver operator curve; HPV, human papillomavirus.
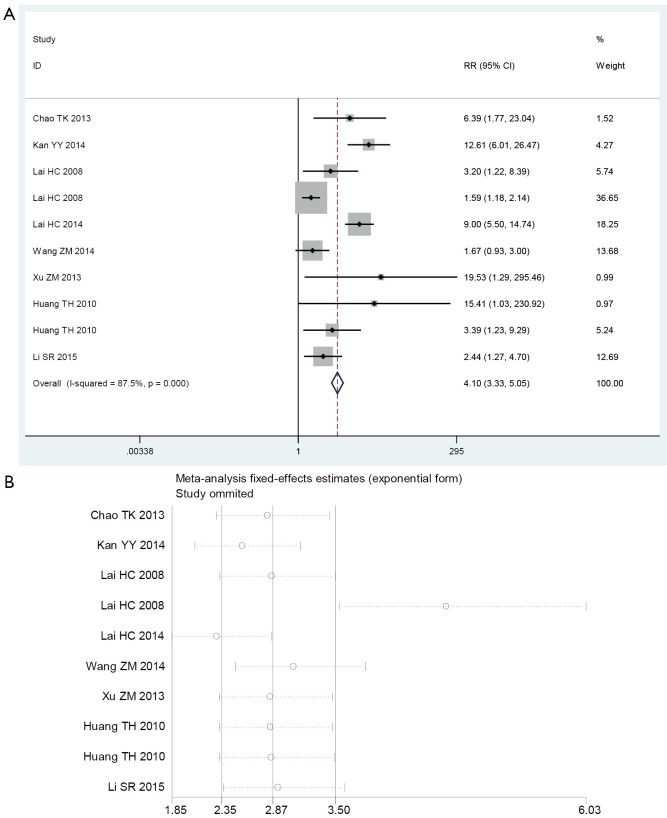
Figure S2.
Influence and outlier detection analyses of HPV DNA test. (A) The intermediate variable of relative risk (RR); (B) outlier detection analysis. HPV, human papillomavirus.
Figure S3.
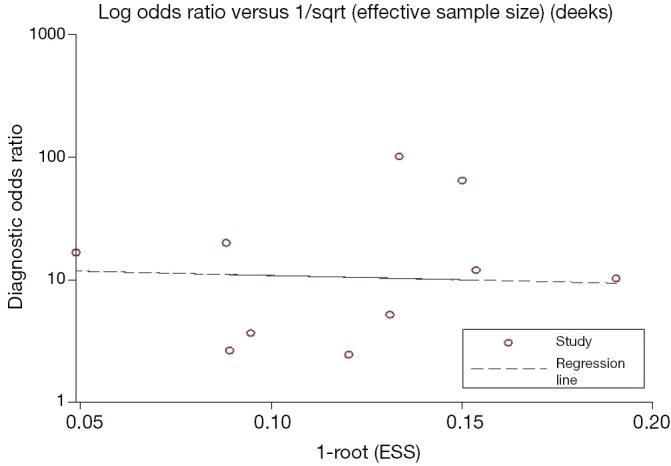
Funnel graph for the assessment of potential publication bias of the HPV DNA testing (P=0.869). HPV, human papillomavirus.
Table S1. Heterogeneity analysis of the pooled studies using meta-disc 1.4 software.
| Analysis | Spearman correlation coefficient | Cochran’s-Q test | I2 test (%) | Heterogeneity | |
|---|---|---|---|---|---|
| Threshold effect | Non-threshold effect | ||||
| PAX1 | 0.181a, P=0.459 | 30.70b, P=0.0311 | 41.4 | No | No |
| SOX1 | 0.766a, P=0.010 | 13.25.00b, P=0.1517 | 32.1 | Yes | No |
| HPV | 0.503a, P=0.138 | 35.57b, P=0.000 | 74.7 | No | Yes |
| PAX1/SOX1 | 0.496a, P=0.019 | 55.10b, P=0.0001 | 61.9 | Yes | Yes |
| PAX1/HPV | 0.593a, P=0.000 | 72.61b, P=0.000 | 58.79 | Yes | Yes |
| SOX1/HPV | 0.964a, P=0.000 | 10.36b, P=0.1103 | 42.1 | Yes | No |
| Outlier excluded PAX1 | 0.124a, P=0.625 | 27.57b, P=0.0502 | 38.3 | No | No |
| Outlier excluded HPV | 0.483a, P=0.187 | 31.57b, P=0.0001 | 74.7 | No | Yes |
a, value of Spearman correlation coefficient; b, Q value. PAX1, paired boxed gene 1; SOX1, sex determining region Y-box 1; HPV, human papillomavirus.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet 2011;378:1461-84. 10.1016/S0140-6736(11)61351-2 [DOI] [PubMed] [Google Scholar]
- 2.Köse FM, Naki MM. Cervical premalignant lesions and their management. J Turk Ger Gynecol Assoc 2014;15:109-21. 10.5152/jtgga.2014.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin RM, Zhao C. Is 58% sensitivity for detection of cervical intraepithelial neoplasia 3 and invasive cervical cancer optimal for cervical screening? Cytojournal 2014;11:14. 10.4103/1742-6413.132997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayrand MH, Duarte-Franco E, Rodrigues I, et al. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med 2007;357:1579-88. 10.1056/NEJMoa071430 [DOI] [PubMed] [Google Scholar]
- 5.Koliopoulos G, Arbyn M, Martin-Hirsch P, et al. Diagnostic accuracy of human papillomavirus testing in primary cervical screening: a systematic review and meta-analysis of non-randomized studies. Gynecol Oncol 2007;104:232-46. 10.1016/j.ygyno.2006.08.053 [DOI] [PubMed] [Google Scholar]
- 6.Nanda K, McCrory DC, Myers ER, et al. Accuracy of the Papanicolaou test in screening for and follow-up of cervical cytologic abnormalities: a systematic review. Ann Intern Med 2000;132:810-9. 10.7326/0003-4819-132-10-200005160-00009 [DOI] [PubMed] [Google Scholar]
- 7.Arbyn M, Buntinx F, Van Ranst M, et al. Virologic versus cytologic triage of women with equivocal Pap smears: a meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia. J Natl Cancer Inst 2004;96:280-93. 10.1093/jnci/djh037 [DOI] [PubMed] [Google Scholar]
- 8.Sankaranarayanan R, Nene BM, Shastri SS, et al. HPV screening for cervical cancer in rural India. N Engl J Med 2009;360:1385-94. 10.1056/NEJMoa0808516 [DOI] [PubMed] [Google Scholar]
- 9.Mandelblatt JS, Lawrence WF, Womack SM, et al. Benefits and costs of using HPV testing to screen for cervical cancer. JAMA 2002;287:2372-81. 10.1001/jama.287.18.2372 [DOI] [PubMed] [Google Scholar]
- 10.Dehn D, Torkko KC, Shroyer KR. Human papillomavirus testing and molecular markers of cervical dysplasia and carcinoma. Cancer 2007;111:1-14. 10.1002/cncr.22425 [DOI] [PubMed] [Google Scholar]
- 11.Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer 2007;7:11-22. 10.1038/nrc2050 [DOI] [PubMed] [Google Scholar]
- 12.Lai HC, Lin YW, Huang TH, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer 2008;123:161-7. 10.1002/ijc.23519 [DOI] [PubMed] [Google Scholar]
- 13.Wang ZM. PAX1 methylation analysis by MS-HRM is useful in triage of high-grade squamous intraepithelial lesions. Asian Pac J Cancer Prev 2014;15:891-4. 10.7314/APJCP.2014.15.2.891 [DOI] [PubMed] [Google Scholar]
- 14.Lai HC, Ou YC, Chen TC, et al. PAX1/SOX1 DNA methylation and cervical neoplasia detection: a Taiwanese Gynecologic Oncology Group (TGOG) study. Cancer Med 2014;3:1062-74. 10.1002/cam4.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai HC, Lin YW, Huang RL, et al. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer 2010;116:4266-74. 10.1002/cncr.25252 [DOI] [PubMed] [Google Scholar]
- 16.Chao TK, Ke FY, Liao YP, et al. Triage of cervical cytological diagnoses of atypical squamous cells by DNA methylation of paired boxed gene 1 (PAX1). Diagn Cytopathol 2013;41:41-6. 10.1002/dc.21758 [DOI] [PubMed] [Google Scholar]
- 17.Kan YY, Liou YL, Wang HJ, et al. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer 2014;24:928-34. 10.1097/IGC.0000000000000155 [DOI] [PubMed] [Google Scholar]
- 18.Zhou AF, Li SR, Wang ZM. The Role of DNA Hypemethylation of PAX1 in early diagnosis of cervical cancer and precursor lesions. Acta Acad Med Weifang 2011;33:439-41. [Google Scholar]
- 19.Xu ZM, Qin SX, Pei F, et al. Quantitative methylation-specific PCR for PAX1 gene and its significance in detection of cervical cancer. Chinese Clinical Oncology 2013;18:10-4. [Google Scholar]
- 20.Huang TH, Lai HC, Liu HW, et al. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer 2010;20:513-9. 10.1111/IGC.0b013e3181c7fe6e [DOI] [PubMed] [Google Scholar]
- 21.Chang CC, Ou YC, Wang KL, et al. Triage of Atypical Glandular Cell by SOX1 and POU4F3 Methylation: A Taiwanese Gynecologic Oncology Group (TGOG) Study. PLoS One 2015;10:e0128705. 10.1371/journal.pone.0128705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CC, Huang RL, Liao YP, et al. Concordance analysis of methylation biomarkers detection in self-collected and physician-collected samples in cervical neoplasm. BMC Cancer 2015;15:418. 10.1186/s12885-015-1411-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li SR, Wang ZM, Wang YH, et al. Value of PAX1 Methylation Analysis by MS-HRM in the Triage of Atypical Squamous Cells of Undetermined Significance. Asian Pac J Cancer Prev 2015;16:5843-6. 10.7314/APJCP.2015.16.14.5843 [DOI] [PubMed] [Google Scholar]
- 24.Lim EH, Ng SL, Li JL, et al. Cervical dysplasia: assessing methylation status (Methylight) of CCNA1, DAPK1, HS3ST2, PAX1 and TFPI2 to improve diagnostic accuracy. Gynecol Oncol 2010;119:225-31. 10.1016/j.ygyno.2010.07.028 [DOI] [PubMed] [Google Scholar]
- 25.Vasiljević N, Scibior-Bentkowska D, Brentnall AR, et al. Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high-risk HPV positive women. Gynecol Oncol 2014;132:709-14. 10.1016/j.ygyno.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CC, Huang RL, Wang HC, et al. High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR, SOX1, and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol Cancer 2014;24:201-9. 10.1097/IGC.0000000000000054 [DOI] [PubMed] [Google Scholar]
- 27.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 2009;62:1006-12. 10.1016/j.jclinepi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 28.Whiting P, Rutjes AW, Reitsma JB, et al. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 2003;3:25. 10.1186/1471-2288-3-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vamvakas EC. Meta-analyses of studies of the diagnostic accuracy of laboratory tests: a review of the concepts and methods. Arch Pathol Lab Med 1998;122:675-86. [PubMed] [Google Scholar]
- 30.Zamora J, Abraira V, Muriel A, et al. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol 2006;6:31. 10.1186/1471-2288-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Xu L, Yang B, et al. Assessing methylation status of PAX1 in cervical scrapings, as a novel diagnostic and predictive biomarker, was closely related to screen cervical cancer. Int J Clin Exp Pathol 2015;8:1674-81. [PMC free article] [PubMed] [Google Scholar]
- 32.Dunne EF, Park IU. HPV and HPV-associated diseases. Infect Dis Clin North Am 2013;27:765-78. 10.1016/j.idc.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 33.Priebe AM. 2012 cervical cancer screening guidelines and the future role of HPV testing. Clin Obstet Gynecol 2013;56:44-50. 10.1097/GRF.0b013e3182836b6a [DOI] [PubMed] [Google Scholar]
- 34.Agorastos T, Sotiriadis A, Chatzigeorgiou K. Can HPV testing replace the pap smear? Ann N Y Acad Sci 2010;1205:51-6. 10.1111/j.1749-6632.2010.05661.x [DOI] [PubMed] [Google Scholar]
- 35.Kroupis C, Vourlidis N. Human papilloma virus (HPV) molecular diagnostics. Clin Chem Lab Med 2011;49:1783-99. 10.1515/cclm.2011.685 [DOI] [PubMed] [Google Scholar]
- 36.Glas AS, Lijmer JG, Prins MH, et al. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 2003;56:1129-35. 10.1016/S0895-4356(03)00177-X [DOI] [PubMed] [Google Scholar]
- 37.Shen Y, Wang T, Yang T, et al. Diagnostic value of circulating microRNAs for lung cancer: a meta-analysis. Genet Test Mol Biomarkers 2013;17:359-66. 10.1089/gtmb.2012.0370 [DOI] [PMC free article] [PubMed] [Google Scholar]