Abstract
In the human endocrine system many protein hormones including urotensin, glucagon, obestatin, bombesin and secretin, among others, are supplied from amyloidal secretory granules. These granules form part of the so called functional amyloids, which within the whole aggregome appear to be more abundant than formerly believed. Bacterial inclusion bodies (IBs) are non-toxic, nanostructured functional amyloids whose biological fabrication can be tailored to render materials with defined biophysical properties. Since under physiological conditions they steadily release their building block protein in a soluble and functional form, IBs are considered as mimetics of endocrine secretory granules. We have explored here if the in vivo implantation of functional IBs in a given tissue would represent a stable local source of functional protein. Upon intratumoral injection of bacterial IBs formed by a potent protein ligand of CXCR4 we have observed high stability and prevalence of the material in absence of toxicity, accompanied by apoptosis of CXCR4+ cells and tumor ablation. Then, the local immobilization of bacterial amyloids formed by therapeutic proteins in tumors or other tissues might represent a promising strategy for a sustained local delivery of protein drugs by mimicking the functional amyloidal architecture of the mammals’ endocrine system.
As gene therapy is not yet of routine application1, deficiencies in endogenous proteins must be treated by the administration of externally fabricated proteins for protein replacement or as effective drugs2,3, including antibodies, hormones or enzymes that exhibit a desired effect based on signalling, receptor blocking or substrate processing4. These drugs are delivered by systemic administration, but adverse immune reactions and the high cost of treatments linked to the high doses required pose severe obstacles to this common approach. These constraints are exemplified by the treatment of lysosomal disorders, such as Fabry disease. In the treatment of this condition, 1 mg/kg (Fabrazyme®) or 0.2 mg/kg (Replagal®) of protein are administered every two weeks to the patient, with a yearly cost depending on the patient weight but usually representing >200.000 €/year/patient5. This is also commonly associated to undesired immune reactions that minimize the therapeutic effects of the protein drug. Moreover, for systemically administered drugs, only a small portion of the active compound is expected to reach the target6.
In this context, there is a growing interest in the development of new materials as controllable systems for sustained protein release. Ideally, the resulting hybrid materials (scaffold plus functional protein drug) could be immobilized in a given tissue as drug depots for long term local supply of the embedded protein. Then, hydrogels, polymers, and other porous materials and biomaterials are being explored as scaffolds for drug inclusion in vitro and further slow release in vivo7,8,9,10,11,12,13,14,15,16. As in other nanomedical applications, toxicity of the scaffold material is a relevant matter of concern, and the long term stability and interactivity of the scaffold structure itself with body components are critical issues. Then, the use of homogenous delivery platforms in which the protein drug itself is also the immobilizing scaffold would be highly convenient to skip the use of potentially toxic, xenobiotic carrier platforms. In this context, the endocrine system offers some important lessons about sustained protein release from chemically homogenous structures. Many secretory cell types in the human body accumulate functional proteins for long-term storage, in the form of compact protein clusters17,18. These act as secretory granules that are formed by self-assembling polypeptides with a high extent of internal molecular organization. Such protein reservoirs show an amyloidal nature rich in cross-β sheet contacts that provide stable and robust scaffolds to retain protein as in vivo depots. As a response to appropriate stimuli, these functional amyloids release monomeric hormone molecules in a functional form19,20,21,22 in a poorly investigated process probably assisted by molecular chaperones17.
On the other hand, IBs are cross-β sheet rich, functional amyloids formed in recombinant bacteria23 observed as mimetics of the above mentioned secretory granules24,25,26. They combine amyloidal and non-amyloidal protein forms in organized sub-micron particles, in which the amyloidal skeleton offer mechanical stability and retains active, properly folded protein forms through stereospecific interactions8,24. Any therapeutically valuable protein produced in bacteria can be packaged as IBs27, and in addition, bacterial IBs show a high intrinsic and spontaneous penetrability into mammalian cells where they deliver, assisted by molecular chaperones, the functional protein fraction embedded into the amyloidal scaffold28. Noting the structural and functional analogies between secretory granules and IBs, the bacterial products would potentially be ideal homogenous materials for sustained protein delivery in vivo through bioinspired therapeutic approaches. In this context, we have explored here the potential of IBs formed by a model protein with antitumoral activity, in inducing apoptosis upon prolonged local exposition to target tissues.
Results and Discussion
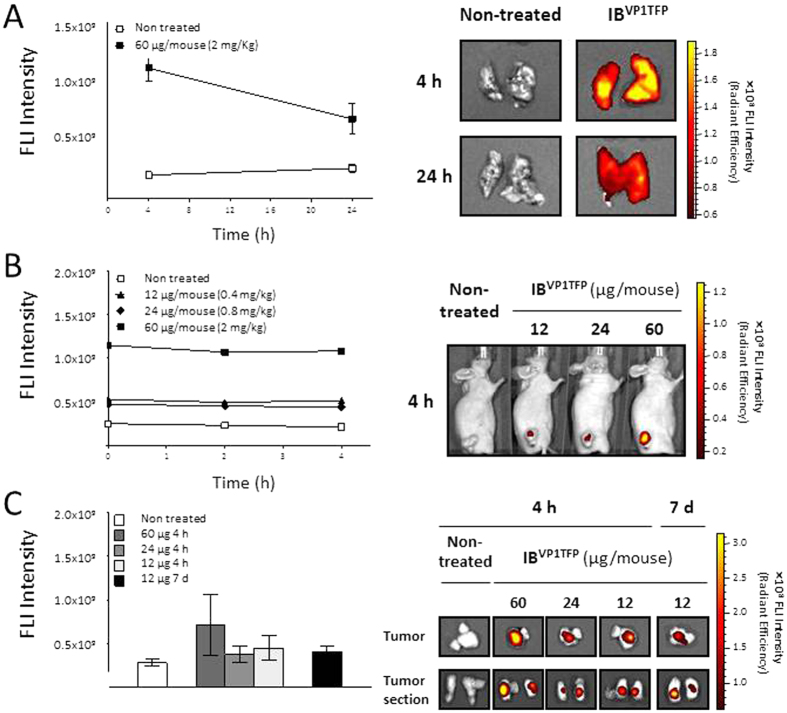
Bacterial IBs are regular shaped soft materials that show a moderate polydispersion in size (Fig. 1)29,30. Their sticky nature31 would prevent them from being used in systemic delivery (as aggregation in lungs would be anticipated) while it would instead favour their retention in tissues if administered locally by injection. These predictions were confirmed by intravenous (iv) tail vein administration of suspended fluorescent IBs formed by VP1TFP (IBVP1TFP) (as model fluorescent materials suitable to easy monitoring) in subcutaneous (sc) tumor-bearing mice. The ex vivo fluorescent imaging (FLI) at early (a few hours) and long-time (1 day) post administration demonstrated that IBVP1TFP got first-pass retained into lung capillaries upon systemic delivery (Fig. 2A), being an indicative of aggregation. Moreover, accumulation of IBVP1TFP into tumor, liver, spleen, kidney, stomach, intestine, heart and skin tissues were not detected at any of the time points studied (4 and 24 h, data not shown). While these data precluded further consideration of IBs as circulating protein drug carrier materials, it was in agreement with possibilities for retention once locally implanted. Solid tumors are highly vascularized tissues, and a poorly stable material would be then easily lost by clearance through the blood stream. In contrast, mechanically stable and firmly attached entities should be predicted as long lasting. Thus, to check the potential of the material as protein depots, IBVP1TFP were intratumorally injected in mice harboring sc tumors of human colon cancer cells. Interestingly, the fluorescent material accumulated in tumor in a dose-dependent way, and the fluorescent light emission signal was stable and steadily localized in the first hours post implantation at each corresponding given dose in vivo (Fig. 2B). Moreover, ex vivo FLI of tumors 7 days post local IBVP1TFP implantation still showed clusters of the material in the local injection site (Fig. 2C), with steady fluorescence intensities along time for the dose of 12 μg per mouse. Even, this fluorescence emission was still within the order of magnitude of the light produced by IBVP1TFP in vitro (Supplementary Figure 1). Since IBVP1TFP signal was not found in other tissues (data not shown) no important fractions of the material appeared to have migrated, at least as major detectable clusters. Importantly, no signs of systemic toxicity were observed such as body weight loss or changes in physical appearance or behavioural patterns (data not shown).
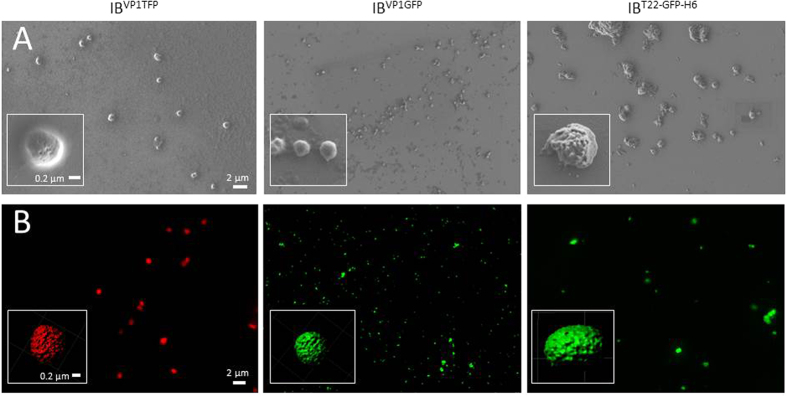
Figure 1. Morphometric analysis of amyloid materials at structural and ultrastructural levels.
(A) Representative FESEM overviews and details (inboxes) of the amyloidal particulate materials used for immobilization, which occurred as rather homogenous populations. Size range of IBVP1TFP was 400–500 nm, 200–300 nm for IBVP1GFP and 500–600 nm for IBT22-GFP-H6 (n > 200). Size bars are common in all images and all inboxes as magnifications are equivalent. (B) Broad field confocal images of the materials and 3D IMARIS reconstructions of representative particles (inboxes). Particle sizes here might be largely overestimated due to fluorescence emission.
Figure 2. Whole-body biodistribution of fluorescent amyloids upon intravenous or intratumoral administration in a HT-29 colorectal cancer model.
(A) Ex vivo lung fluorescence imaging (FLI) at 4 and 24 h post administration of IBVP1TFP protein particles at 60 μg/mouse by intravenous administration route. Ex vivo lung-accumulation of the material was quantified by measuring fluorescent intensity (left plot), while representative ex vivo fluorescent images of the excised lungs are shown (right panel). (B) Non-invasive monitoring of IBVP1TFP tumor-accumulations a long time after intratumoral administration at 12, 24 and 60 μg/mouse of IBVP1TFP. In vivo tumor-accumulation of the material was quantified (left plot), and representative in vivo fluorescent image of tumor-accumulation at 24 h post administration are shown (right panel). (C) Ex vivo tumor FLI at 4 h after intratumoral administration at 12, 24 and or 60 μg/mouse, and 7 days after intratumoral administration of 12 μg/mouse of the material. Ex vivo tumor-accumulation of IBVP1TFP was quantified (left plot), and representative ex vivo fluorescent images of the excised tumor (whole- and sectioned-tumor) are shown (right panel). In all cases, the total tissue-accumulations were quantified by measurements of fluorescent intensity expressed in Radiant Efficiency, MEAN ± SEM. Pseudocolor scale bars were consistent for all images in order to show relative changes for each corresponding images.
These observations, favourable regarding in vivo stability of the material even in irrigated tissues, prompted us to explore further the hypothesis of IBs as functional mimetics of secretory granules. For that, we evaluated the availability of the functional IB protein fraction to the cells in the vicinity of the reservoir implantation site. T22 is a peptidic ligand and a CXCR4 antagonist that acts extracellularly by competing with SDF-1α for binding to this membrane receptor32. High doses of this peptide trigger the inhibition of signal transduction downstream of CXCR4 leading to proliferative block, apoptotic induction and tumor growth inhibition in CXCR4-overexpressing breast33 and brain34 cancer models. The protein fusion T22-GFP-H6 was produced in bacteria in form of fluorescent, T22-containing IBs (IBT22-GFP-H6) for testing the biological effects of the material when implanted in CXCR4+ colorectal cancer models. The coincidence in the same macromolecule of T22 as effector moiety and of GFP as tracking agent renders an unusually convenient system to analyse cell responses with an immediate evidence of protein activity and localization.
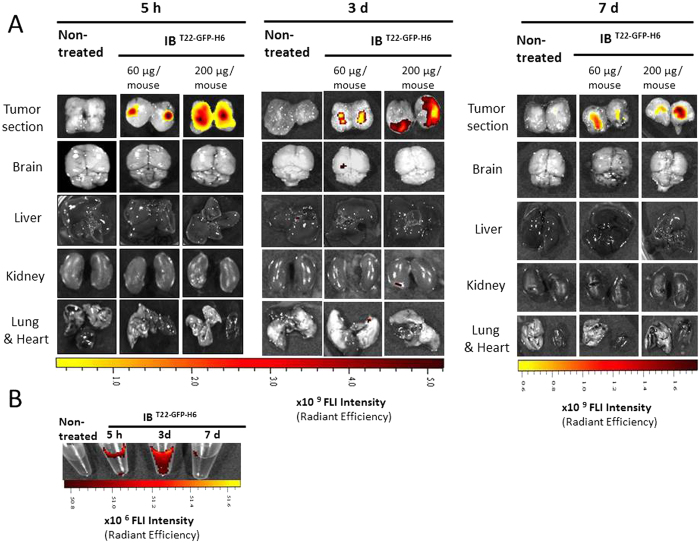
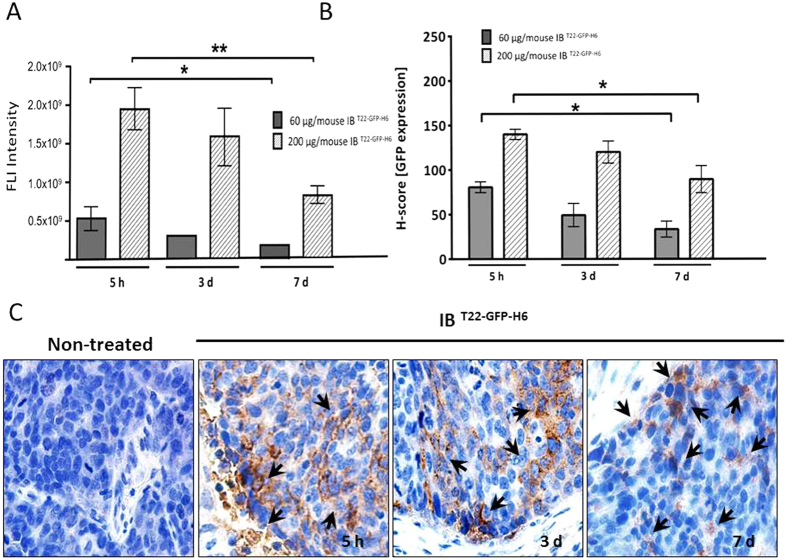
Upon injection in sc tumors, local fluorescence of IBT22-GFP-H6 was clearly dose-dependent and largely stable along time (Fig. 3A). About 75 % of the initial fluorescence emission was still linked to the material in tumor 3 days after local injection and decreased only till 40 % in 7 days postinjection (Fig. 4). No toxic side effects were detected in the animals as indicated by lack of body weight loss or clinical signs (not shown). Again, no significant fluorescence levels were observed in analysed organs other than primary tumor (Fig. 3A). Interestingly, the distribution of the IB through the tumor tissue increased with the dose of injected IBs, clearly covering a wider area and indicating a significant level of diffusion of the material and a residual circulating fraction, probably in soluble form since no lung aggregation was observed (Fig. 3B and Table 1). Fluorescent signals were correlated with anti-GFP immunostaining of tumor (Fig. 4B,C) and GFP was detected both in the extracellular matrix and inside tumor cells at any of the time points studied: 5 h, 3 and 7 days (Fig. 4C).
Figure 3. Whole-body biodistribution of functional fluorescent amyloids in CXCR4+ colorectal murine model.
(A) Representative ex vivo fluorescent images of the excised tumor and organs (brain, liver, kidney and lung and heart) remaining in tumor tissue at 5 h, and 3 or 7 days post administration of IBT22-GFP-H6 protein particles at 60 μg/mouse or 200 μg/mouse using the intratumoral route. (B) Fluorescence emitted by soluble protein species in plasma, which were released by IBT22-GFP-H6 tumor deposits to the bloodstream at 5 h, and 3 or 7 days after injection of a 200 μg/mouse intratumoral dose. Pseudocolor scale bars were consistent among images in order to show relative changes when being compared.
Figure 4. Fluorescent emission by IBT22-GFP-H6 deposits remaining in CXCR4+ colorectal tumors after their administration.
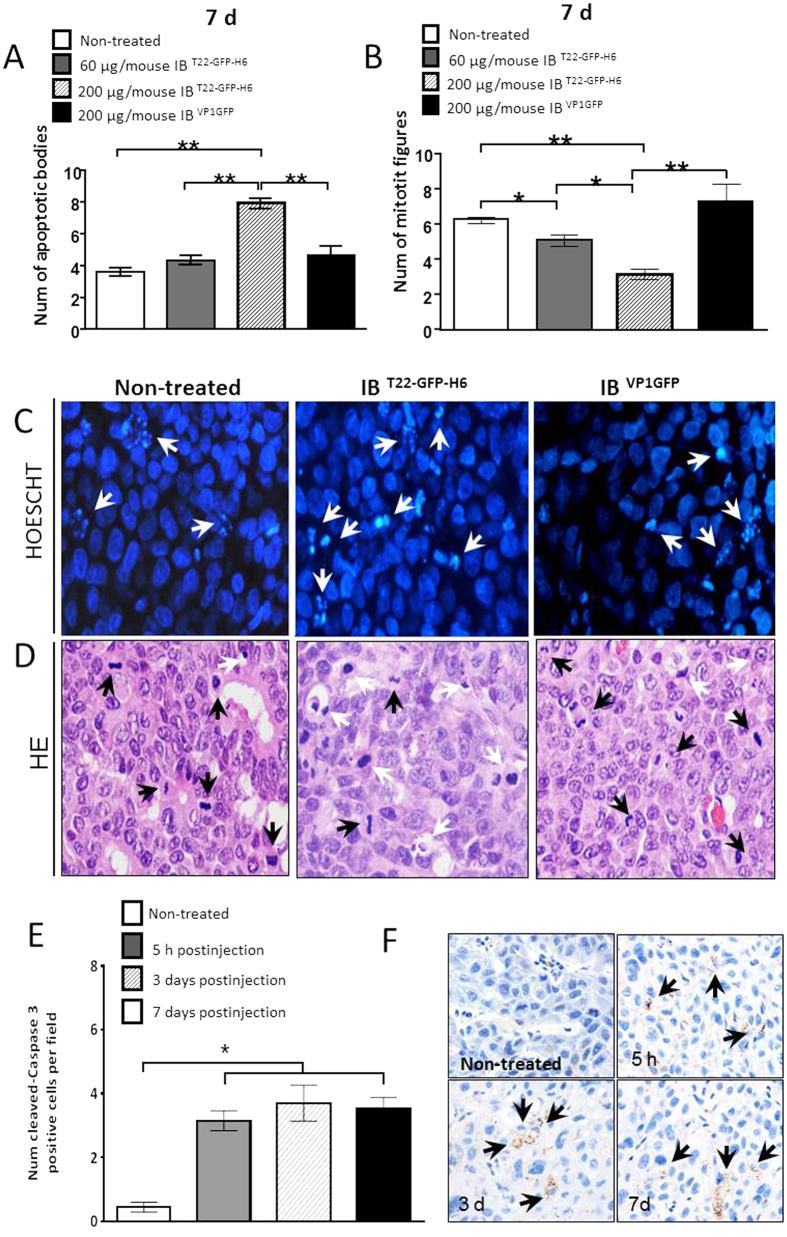
(A) Total IBT22-GFP-H6 protein deposits were quantified measuring fluorescent intensity at 5h, 3 and 7 days after intratumoral injection of 200 μg/mouse. Data were expressed in Radiant Efficiency. (B) Quantitation of total IBT22-GFP-H6 protein deposits plus released soluble proteins in tumor tissue calculated as an H-score for anti-GFP immunostaining (brown colour) at 5 h, and 3 or 7 days post administration. (C) Representative microphotographs of GFP inmunohistochemistry in IBT22-GFP-H6 treated tumors at 5 h, and 3 or 7 days. Note the higher intensity of GFP staining in some tumor areas at 5 h and 3 days (black arrows) and the higher dispersion of protein distribution observed inside the tumor at day 7 post-injection. Quantitative data were expressed as mean ± SE *,**Statistically significant at p < 0.05 or p < 0.01, respectively.
Table 1. Kinetics of IBT22-GFP-H6 protein depots remaining in tumor and soluble species released to plasma, and their antitumor effect in tumor tissue.
| Parameter | Time |
|||
|---|---|---|---|---|
| 0 hours | 5 hours | 3 days | 7 days | |
| IBs in tumor (μg) | 200 | 184.5 | 155.3 | 87.9 |
| IHC GFP H-score | N.d | 140 ± 5 | 120 ± 12 | 89 ± 15 |
| Cleaved-Caspase 3 | N.d | 3.2 ± 0.3 | 3.7 ± 0.6 | 3.5 ± 0.4 |
| Apoptosis | N.d | 3.7 ± 0.2 | 5.9 ± 1 | 8.0 ± 0.2 |
| Mitosis | N.d | 4.9 ± 0.3 | 3.9 ± 0.7 | 3.1 ± 0.1 |
| Plasma (μg/ml) | 0 | 2.9 | 16.4 | 0 |
Samples were analyzed at 0 h, 5 h, 3 and 7 days after intratumoral injection at a 200 μg/mouse dose.
IHC GFP Score: Immunohistochemical quantitation of proteins containing GFP in tumor tissue N.d.: not done.
When exploring the biological impact of the IB-contained effector T22 on surrounding tissues, we observed a dramatic occurrence of apoptotic events in the tumor tissue (Fig. 5A,C), that was not observed for the control, closely related T22-devoid IBVP1GFP. The number of apoptotic bodies was unperceived 5 h after injection but they rose at 3 days and 7 days, in parallel with a significant drop in the number of mitotic cells, not observed when implanting control, non-functional IBs (Fig. 5B,D). The coincidence of inhibited cell proliferation and apoptotic induction was indicative of the fusion protein being available to the surrounding tissues in the course of prolonged time periods. This apoptotic induction was associated with sustained caspase-3 activation along time (Fig. 5E,F).
Figure 5. Apoptotic and mitotic index recorded in CXCR4+ tumors.
Quantitation of apoptotic figures detected by nuclear condensation or nuclear fragmentation after Hoescht staining (A) or mitotic figures (B) after Hematoxylin-eosin staining in SP5 tumors at day 7 post intratumoral administration of IBT22-GFP-H6 protein particles at 60 or 200 μg/mouse. (C,D) Representative microphotographs of apoptotic figures by Hoescht ((C), white arrows) or HE ((D), white arrows) staining or mitotic figures ((D), black arrows) by HE staining after local intratumoral injection of 200 μg/mouse dose of targeted IBT22-GFP-H6 or non-targeted IBVP1GFP (x 400 magnification). (E) Quantitation of the number of positive cells displaying cleaved (active) caspase-3 immunostaining in tumors at 5 h, and 3 or 7 days after intratumoral administration of 200 IBT22-GFP-H6 compared to non-treated tumors. (F) Representative microphotographs of active caspase-3 positive cells (brown stained tumor cells; black arrows) at the studied time point. Data expressed as mean ± SE *,**Statistically significant at p < 0.05 or p < 0.01, respectively.
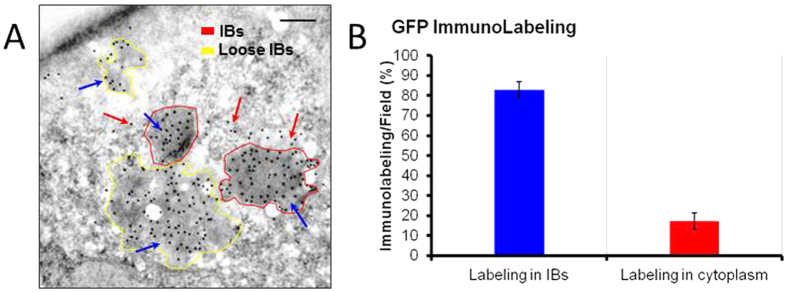
Around 20 % of the IB protein is organized as proteinase K-resistant amyloid fibers35,36, in a pattern similar to that of protein hormones in the human secretory system. Such amyloidal network ensures a stable fibrous scaffold that confers mechanical stability and high porosity to IBs enabling them to be used as functional biomaterials26,37. The remaining IB protein fraction occurs embedded within this network probably by sterospecific interactions35,38, as native-like rich protein versions that are releasable upon internalization by mammalian cells39. In fact, upon in vitro incubation with physiological buffer, an immediate but time-limited amount of IB protein (about 5 %) is released from IBs in the active form40. Such protein loss is probably occurring through an equilibrium between soluble and aggregated protein species, with a kinetics controlled by an aggregation/dissociation constant. However, the fraction of released functional protein appears to be higher upon internalization into mammalian cells, what might be linked to the suggested involvement of cell chaperones in the disintegration of exogenous IBs41,42,43,44 and not subjected to physicochemical equilibrium constants. To confirm such protein release and its time-prolonged nature we determined, by GFP immunostaining, the amount of IB protein still associated to engulfed IBs and the IB protein fraction free from the bulk material, in the cell cytoplasm. As observed (Fig. 6A,B), more than 20% of IB protein is found free in the cell cytoplasm, 24 h after exposure of cultured mammalian cells to IBVP1GFP. Interestingly, some IBs show their natural electrodense nature under transmission electron microscopy (Fig. 6A, red borders) while others are much loser and less dense (Fig. 6A, yellow borders). These images are compatible with those observed upon proteinase K treatment of isolated IBs, in which an important fraction of the protein is degraded and the amyloidal skeleton, free from functional protein, remains36. This amyloidal network represents about 20 % of the total recombinant protein in IBs, but this percentage, as well as the amount of functional protein that can be released from IBs seems to be regulatable by the proper choice of the genetic background of the bacteria where IBs are produced36, and by the production conditions, such as temperature and others45,46,47. The potential to obtain IBs with variable amounts of releasable protein and with different global conformational status48,49 might offer opportunities for a better control of the protein release upon injection, as well as a way to minimize the amount of unfolded protein, what could contribute to undesired immune responses.
Figure 6. Protein release from IBs.
(A) Immunogold labeling of GFP in HeLa cells after 24 h of exposure to VP1-GFP-H6 IBs. Blue arrows indicate labeling in the IB particle while red arrows indicates labeling of released protein to the cell cytoplasm. Highly electrodense IB particles with standard morphometries are shadowed in red while those with loose morphologies and showing lower electrodensity, in yellow. The scale bar represents 200 nm. (B) Quantification of IB-attached and IB-free immunolabeling signals at this incubation time.
In addition to the releasable nature of a fraction of IB protein, the high inherent membrane-activity of bacterial IBs allows efficient adhesion and cell penetration in absence of cytotoxicity, properties that justified the exploration of IBs as both functional topographies in tissue engineering and as protein delivery agents in replacement therapies24. In this context, the absence of intrinsic toxicity of bacterial IBs has been widely demonstrated by conventional procedures on IBs formed by diverse proteins with potential biomedical interest, in therapeutic/prophylactic setting ups. This includes cytoskeleton proteins50, enzymes39, chaperones39 and growth factors42,43,51 among others, and very recently, cytokines in a Zebrafish animal model52.
Beyond the well-known applications of bacterial IBs as vehicles for functional protein in cell culture and oral administration53,54,55,56, we demonstrate here that bacterial IBs act as in vivo depots of functional proteins upon their immobilization by local injection. The availability for interaction and signalling of a CXCR4 peptide ligand that was part of an IB-forming fusion protein and the dramatic impact promoting apoptosis in primary tumor fully confirm the successful mimicry of IBs as implantable homogenous materials for sustained protein release. The fact that the tissue volume in which IBs occur is dose-dependent and expands with increasing IB amounts (Figs 2B,C and 3B,C) opens a possibility to regulate the zone of therapeutic impact, what could be also defined by the selection of multiple injection sites, if required.
Finally, the recent identification of IB formation in food-grade bacteria57, the development of endotoxin-free E. coli strains58 and their adaptation to the production of endotoxin-free IBs59 further expand the opportunities for the in vivo uses of IBs, supporting again the added values of biofabrication versus chemical synthesis in the production of functional materials.
Methods
Bacterial production of IBs
VP1TFP is a far-red fluorescent protein TurboFP635 fused to an aggregation prone viral peptide (from foot-and-mouth disease virus). VP1GFP is an equivalent protein containing the enhanced GFP, while T22-GFP-H6 contains T22 instead of VP1. IB versions of these proteins (Fig. 1) were produced from encoding plasmid vectors by IPTG-mediated induction in E. coli as described39,60,61, purified by a stringent protocol designed to minimize molecular contamination from the producing cells62, and strictly tested by the absence of contaminating, full cell bacteria by described procedures62. The absence of toxicity of IBs on mammalian cell cultures was regularly determined by conventional MTT assays, resulting in viability values always over 90 % (usually over 95 %; not shown). This is coincident with multiple reports showing the lack of intrinsic cytotoxicity of IBVP1GFP and IBTF1GFP produced by optimized protocols28,39,61,63. Protein production and IB purification has been partially performed by the ICTS “NANBIOSIS”, more specifically by the Protein Production Platform of CIBER in Bioengineering, Biomaterials & Nanomedicine (CIBER-BBN)/ IBB, at the UAB SepBioES scientific-technical service.
Field Emission Scanning Electron Microscopy (FESEM)
The ultrastructural details of size, shape and surface aspect were evaluated at nearly native state with a Field Emission Scanning Electron Microscope (FESEM). Purified IBs were diluted in water and microdrops of 10 μl of each sample were deposited over silicon wafers (Ted Pella, Reading, CA, USA) and air dried. Then, the materials were immediately observed without coating in a FESEM Zeiss Merlin (Zeiss, Oberkochen, Germany), operating at 2 kV and equipped with a high resolution in-lens secondary electron detector.
In vivo experiments
All in vivo experiments were performed by the ICTS “NANBIOSIS”, more specifically by the CIBER-BBN’s in vivo Experimental Platform of the Functional Validation & Preclinical Research (FVPR) area (http://www.nanbiosis.es/unit/u20-in-vivo-experimental-platform/) and the Nanotoxicology platform of IIB Sant Pau (http://www.nanbiosis.es/portfolio/u18-nanotoxicology-unit/. Animal care was handled in accordance with the Guide for the Care and Use of Laboratory Animals of the involved institutions and all the in vivo procedures were approved by the Hospital de Sant Pau and Hospital Vall d’Hebron Animal Ethics Committees and performed according to EC directives. Detailed protocols are given in the Supplementary information.
Additional Information
How to cite this article: Céspedes, M. V. et al. Bacterial mimetics of endocrine secretory granules as immobilized in vivo depots for functional protein drugs. Sci. Rep. 6, 35765; doi: 10.1038/srep35765 (2016).
Supplementary Material
Acknowledgments
We are indebted to MINECO (BIO2013-41019-P), AGAUR (2014SGR-132) and CIBER de Bioingeniería, Biomateriales y Nanomedicina (project NANOPROTHER) to AV, Marató de TV3 foundation (TV32013-3930) to EV and to IA and FIS (PI15/00272) to EV and ISCIII (PI15/00378 and PIE15/00028, confinanced by The European Regional Development Fund (FEDER)), Marató TV3 (2013–2030) and AGAUR 2014-PROD0005 to RM for funding our research on protein-based therapeutics. Protein production has been partially performed by the ICTS “NANBIOSIS”, more specifically by the Protein Production Platform of CIBER in Bioengineering, Biomaterials & Nanomedicine (CIBER-BBN)/IBB, at the UAB sepBioEs scientific-technical service. All in vivo experiments were performed by the ICTS “NANBIOSIS”, more specifically by the CIBER-BBN’s in vivo Experimental Platform of the Functional Validation & Preclinical Research (FVPR) area and the Nanotoxicology Unit Platform. We are also indebted to Fran Cortes from the Cell Culture and Cytometry Units of the Servei de Cultius Cel·lulars, Producció d’Anticossos i Citometria (SCAC), and to the Servei de Microscòpia, both at the UAB. We thank Carmen Cabrera for her technical support in the Nanotoxicology Unit Platform. AV received an ICREA ACADEMIA award. MVC is supported by a Miguel Servet contract and UU received a Sara Borrell postdoctoral fellowship, both from ISCIII. JSF is a recipient of a post-doctoral fellowship from Asociación Española Contra el Cáncer (AECC).
Footnotes
Author Contributions M.V.C., Y.F. and P.A. performed most of the in vivo experiments that were designed, coordinated and data finally analysed by I.A., S.S. and Ramon Mangues. Rosa Mendoza, V.T.-R., U.U. and J.S.-F. prepared and characterized the bacterial materials, including molecular cloning, under the supervision of N.F.-M. and E.V. A.S.-C. was in charge of microscopy exploration. J.L.C. and A.V. conceived the whole study, and most of the writing has been done by A.V. All authors have contributed in the final shaping of the manuscript.
References
- Ginn S. L., Alexander I. E., Edelstein M. L., Abedi M. R. & Wixon J. Gene therapy clinical trials worldwide to 2. J. Gene Med. 15, 65–77 (2013). [DOI] [PubMed] [Google Scholar]
- Kaspar A. A. & Reichert J. M. Future directions for peptide therapeutics development. Drug Discov. Today 18, 807–817 (2013). [DOI] [PubMed] [Google Scholar]
- Pavlou A. K. & Reichert J. M. Recombinant protein therapeutics–success rates, market trends and values to 2010. Nat. Biotechnol. 22, 1513–1519 (2004). [DOI] [PubMed] [Google Scholar]
- Leader B., Baca Q. J. & Golan D. E. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 7, 21–39 (2008). [DOI] [PubMed] [Google Scholar]
- Lee K. et al. A biochemical and pharmacological comparison of enzyme replacement therapies for the glycolipid storage disorder Fabry disease. Glycobiology 13, 305–313 (2003). [DOI] [PubMed] [Google Scholar]
- Duncan R. & Gaspar R. Nanomedicine(s) under the microscope. Mol. Pharm. 8, 2101–2141 (2011). [DOI] [PubMed] [Google Scholar]
- Vallet-Regi M., Balas F. & Arcos D. Mesoporous materials for drug delivery. Angew. Chem. Int Ed Engl. 46, 7548–7558 (2007). [DOI] [PubMed] [Google Scholar]
- Seras-Franzoso J. et al. Integrating mechanical and biological control of cell proliferation through bioinspired multieffector materials. Nanomedicine. (Lond) 10, 873–891 (2015). [DOI] [PubMed] [Google Scholar]
- Bhardwaj T. R., Kanwar M., Lal R. & Gupta A. Natural gums and modified natural gums as sustained-release carriers. Drug Dev. Ind. Pharm. 26, 1025–1038 (2000). [DOI] [PubMed] [Google Scholar]
- Lvov Y. M., Shchukin D. G., Mohwald H. & Price R. R. Halloysite clay nanotubes for controlled release of protective agents. ACS Nano. 2, 814–820 (2008). [DOI] [PubMed] [Google Scholar]
- Uhrich K. E., Cannizzaro S. M., Langer R. S. & Shakesheff K. M. Polymeric systems for controlled drug release. Chem. Rev. 99, 3181–3198 (1999). [DOI] [PubMed] [Google Scholar]
- Aarts M. et al. Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science 298, 846–850 (2002). [DOI] [PubMed] [Google Scholar]
- Chew S. Y., Wen J., Yim E. K. & Leong K. W. Sustained release of proteins from electrospun biodegradable fibers. Biomacromolecules. 6, 2017–2024 (2005). [DOI] [PubMed] [Google Scholar]
- Yu L. & Ding J. Injectable hydrogels as unique biomedical materials. Chem. Soc. Rev. 37, 1473–1481 (2008). [DOI] [PubMed] [Google Scholar]
- Liao I. C., Wan A. C., Yim E. K. & Leong K. W. Controlled release from fibers of polyelectrolyte complexes. J. Control Release 104, 347–358 (2005). [DOI] [PubMed] [Google Scholar]
- Prabaharan M. Review paper: chitosan derivatives as promising materials for controlled drug delivery. J. Biomater. Appl. 23, 5–36 (2008). [DOI] [PubMed] [Google Scholar]
- Maji S. K. et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji S. K. et al. Amyloid as a depot for the formulation of long-acting drugs. PLoS Biol. 6, e17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankar S., Anoop A., Sen S. & Maji S. K. Nanomaterials: amyloids reflect their brighter side. Nano Rev. 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badtke M. P., Hammer N. D. & Chapman M. R. Functional amyloids signal their arrival. Sci. Signal. 2, e43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny I. & Gazit E. Amyloids: not only pathological agents but also ordered nanomaterials. Angew. Chem. Int. Ed Engl. 47, 4062–4069 (2008). [DOI] [PubMed] [Google Scholar]
- Iconomidou V. A. & Hamodrakas S. J. Natural protective amyloids. Curr. Protein Pept. Sci. 9, 291–309 (2008). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Montalban N., Garcia-Fruitos E. & Villaverde A. Recombinant protein solubility-does more mean better? Nat. Biotechnol. 25, 718–720 (2007). [DOI] [PubMed] [Google Scholar]
- Villaverde A., Corchero J. L., Seras-Franzoso J. & Garcia-Fruitos E. Functional protein aggregates: just the tip of the iceberg. Nanomedicine. (Lond) 10, 2881–2891 (2015). [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E. et al. Bacterial inclusion bodies: making gold from waste. Trends Biotechnol. 30, 65–70 (2012). [DOI] [PubMed] [Google Scholar]
- Mitraki A. Protein aggregation from inclusion bodies to amyloid and biomaterials. Adv. Protein Chem. Struct. Biol. 79, 89–125 (2010). [DOI] [PubMed] [Google Scholar]
- Villaverde A. et al. Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microb Cell Fact. 11, 76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seras-Franzoso J., Sanchez-Chardi A., Garcia-Fruitos E., Vazquez E. & Villaverde A. Cellular uptake and intracellular fate of protein releasing bacterial amyloids in mammalian cells. Soft. Matter 12, 3451–3460 (2016). [DOI] [PubMed] [Google Scholar]
- Diez-Gil C. et al. The nanoscale properties of bacterial inclusion bodies and their effect on mammalian cell proliferation. Biomaterials 31, 5805–5812 (2010). [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E., Seras-Franzoso J., Vazquez E. & Villaverde A. Tunable geometry of bacterial inclusion bodies as substrate materials for tissue engineering. Nanotechnology 21, 205101 (2010). [DOI] [PubMed] [Google Scholar]
- Seras-Franzoso J. et al. Bioadhesiveness and efficient mechanotransduction stimuli synergistically provided by bacterial inclusion bodies as scaffolds for tissue engineering. Nanomedicine (Lond) 7, 79–93 (2012). [DOI] [PubMed] [Google Scholar]
- Murakami T. et al. Inhibitory mechanism of the CXCR4 antagonist t22 against human immunodeficiency virus type 1 infection. Journal of Virology 73, 7489–7496 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpers H., Huang M. B., Katkoori V., Manne U. & Bond V. Nef-M1, a CXCR4 Peptide Antagonist, Enhances Apoptosis and Inhibits Primary Tumor Growth and Metastasis in Breast Cancer. J. Cancer Ther. 4, 898–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J. B. et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc. Natl. Acad. Sci. USA 100, 13513–13518 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M. et al. Inclusion bodies: specificity in their aggregation process and amyloid-like structure. Biochim. Biophys. Acta 1783, 1815–1825 (2008). [DOI] [PubMed] [Google Scholar]
- Cano-Garrido O. et al. Supramolecular organization of protein-releasing functional amyloids solved in bacterial inclusion bodies. Acta Biomater 9, 6134–6142 (2013). [DOI] [PubMed] [Google Scholar]
- Loo Y. et al. Self-Assembled Proteins and Peptides as Scaffolds for Tissue Regeneration. Adv. Healthc. Mater 4, 2557–2586 (2015). [DOI] [PubMed] [Google Scholar]
- Speed M. A., Wang D. I. & King J. Specific aggregation of partially folded polypeptide chains: the molecular basis of inclusion body composition. Nat. Biotechnol. 14, 1283–1287 (1996). [DOI] [PubMed] [Google Scholar]
- Vazquez E. et al. Functional Inclusion Bodies Produced in Bacteria as Naturally Occurring Nanopills for Advanced Cell Therapies. Adv. Mater 24, 1742–1747 (2012). [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E., Aris A. & Villaverde A. Localization of functional polypeptides in bacterial inclusion bodies. Appl. Environ. Microbiol. 73, 289–294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seras-Franzoso J. et al. A nanostructured bacterial bio-scaffold for the sustained bottom-up delivery of protein drugs. Nanomedicine (Lond) 8, 1587–1599 (2013). [DOI] [PubMed] [Google Scholar]
- Seras-Franzoso J. et al. Functionalization of 3D scaffolds with protein-releasing biomaterials for intracellular delivery. J. Control Release 171, 63–72 (2013). [DOI] [PubMed] [Google Scholar]
- Seras-Franzoso J. et al. Improving protein delivery of fibroblast growth factor-2 from bacterial inclusion bodies used as cell culture substrates. Acta Biomater 10, 1354–1359 (2013). [DOI] [PubMed] [Google Scholar]
- Seras-Franzoso J. et al. A nanostructured bacterial bioscaffold for the sustained bottom-up delivery of protein drugs. Nanomedicine. (Lond) 8, 1587–1599 (2013). [DOI] [PubMed] [Google Scholar]
- Peternel S., Grdadolnik J., Gaberc-Porekar V. & Komel R. Engineering inclusion bodies for non denaturing extraction of functional proteins. Microb. Cell Fact. 7, 34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A. K., Murmu A., Singh A. & Panda A. K. Kinetics of inclusion body formation and its correlation with the characteristics of protein aggregates in Escherichia coli. PLoS One 7, e33951 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Upadhyay V., Upadhyay A. K., Singh S. M. & Panda A. K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 14, 41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura S. & Villaverde A. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 24, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E. et al. Bacterial inclusion bodies: making gold from waste. Trends Biotechnol. 30, 65–70 (2012). [DOI] [PubMed] [Google Scholar]
- Liovic M. et al. Inclusion bodies as potential vehicles for recombinant protein delivery into epithelial cells. Microb Cell Fact. 11, 67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevsevar S. et al. Production of nonclassical inclusion bodies from which correctly folded protein can be extracted. Biotechnol. Prog. 21, 632–639 (2005). [DOI] [PubMed] [Google Scholar]
- Débora Torrealba et al. Nanostructured recombinant cytokines: a highly stable alternative to short-lived prophylactics. Biomaterials 107, 102–114 (2016). [DOI] [PubMed] [Google Scholar]
- Villaverde A. Bacterial inclusion bodies: an emerging platform for drug delivery and cell therapy. Nanomedicine. (Lond) 7, 1277–1279 (2012). [DOI] [PubMed] [Google Scholar]
- Villaverde A. et al. Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microb. Cell Fact. 11, 76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez E. et al. Functional inclusion bodies produced in bacteria as naturally occurring nanopills for advanced cell therapies. Adv. Mater. 24, 1742–1747 (2012). [DOI] [PubMed] [Google Scholar]
- Garcia-Fruitos E. et al. Bacterial inclusion bodies: making gold from waste. Trends Biotechnol. 30, 65–70 (2012). [DOI] [PubMed] [Google Scholar]
- Lu W., Kong J. & Kong W. Construction and application of a food-grade expression system for Lactococcus lactis. Mol. Biotechnol. 54, 170–176 (2013). [DOI] [PubMed] [Google Scholar]
- Mamat U. et al. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb. Cell Fact. 14, 57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda F. et al. Production of functional inclusion bodies in endotoxin-free Escherichia coli. Appl. Microbiol. Biotechnol. 98, 9229–9238 (2014). [DOI] [PubMed] [Google Scholar]
- Cespedes M. V. et al. In vivo Architectonic Stability of Fully de Novo Designed Protein-Only Nanoparticles. ACS Nano. 8, 4166–4176 (2014). [DOI] [PubMed] [Google Scholar]
- García-Fruitós E. et al. Surface Cell Growth Engineering Assisted by a Novel Bacterial Nanomaterial. Advanced Materials 21, 4249–4253 (2009). [Google Scholar]
- Rodriguez-Carmona E., Cano-Garrido O., Seras-Franzoso J., Villaverde A. & Garcia-Fruitos E. Isolation of cell-free bacterial inclusion bodies. Microb. Cell Fact. 9, 71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatkiewicz W. I. et al. Two-dimensional microscale engineering of protein-based nanoparticles for cell guidance. ACS Nano. 7, 4774–4784 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.