Abstract
AMPD1 is an adenosine monophosphate deaminase that catalyzes the deamination of AMP to IMP. To understand the physiological function of AMPD1, we obtained a strain of Ampd1 mutant mice from KOMP repository, which was generated by a knockout-first strategy. An elevated AMP level and almost complete lack of IMP was detected in the skeletal muscle of E18.5 Ampd1tm1a/tm1a mice. However, Ampd1tm1a/tm1a mice died in 2 days postnatally, which was contradicting to previous reports. After removal of the knockout-first cassette and critical exon, mice homozygous for the Ampd1tm1c/tm1c and Ampd1tm1d/tm1d alleles survived to adulthood. RNA-seq analysis indicated that the expression of two neighboring genes, Man1a2 and Nras, were disrupted in the Ampd1tm1a/tm1a mice, but normal in the Ampd1tm1c/tm1c and Ampd1tm1d/tm1d mice. The neonatal lethality phenotype in the Ampd1tm1a/tm1a mice was consistent with the Man1a2-deficient mice. Our results indicated the knockout-first cassette may cause off-target effect by influence the expression of neighboring genes. This study, together with other reports, strongly suggests that removal of targeting cassette by site-specific recombinases is very important for the accurate phenotypic interpretation on mice generated by target mutations.
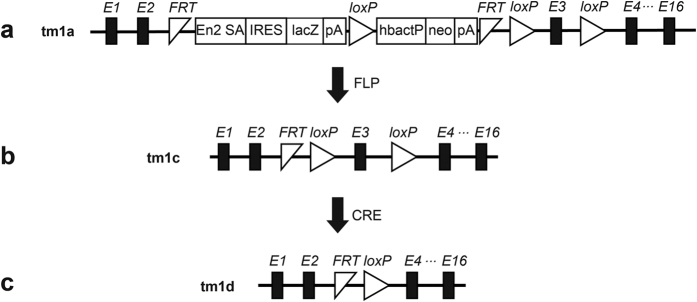
Mice with artificial disruption in a gene have been widely applied to understand the gene function in vivo. With the accomplishment of human and mouse genome projects, a variety of strategies have been employed to generate genome-wide mutagenesis or knockout. Recently, Bradley and colleagues report a knockout-first strategy to generate mutant alleles for thousands of genes in mice. The knockout-first combines the advantage of both a reporter-tagged and a conditional mutation1. As shown in Fig. 1a, a cassette containing mouse En2 splicing acceptor (SA), LacZ, and promoter-driven neomycin resistant gene (Neo) is inserted in introns of target genes. The initial allele (tm1a) is predicted to generate a null allele through splicing to the LacZ trapping element. Conditional alleles (tm1c, Fig. 1b) can be generated by removal of the gene trap cassette using Flp recombinase. The tm1d allele (Fig. 1c) is conventional mutation after crossing tm1c allele with Cre transgenic mice. This multi-purpose strategy is applied to European Conditional Mouse Mutagenesis (EUCOMM) and the National Institutes of Health Knockout Mouse programs (KOMP) to generate knockout mice, which are available for the researchers world-wide.
Figure 1. Schematic diagram of Ampd1tm1(KOMP)Wtsi allele.
(a) The Ampd1 tm1a allele was initially a non-expressive form due to the alternative splicing of Ampd1 exon1-2 to En2 SA in the trapping cassette, disrupting the Ampd1 transcript. (b) Ampd1 tm1c allele was produced by crossing Ampd1 tm1a allele with an Flp transgenic mouse strain, removing the FRT-flanked knockout-first cassette. (c) Ampd1 tm1d allele was generated by mating Ampd1 tm1c allele with a Cre transgenic mouse strain, removing the exon 3 of Ampd1. E1…16, exons of Ampd1; En2 SA, mouse En2 splicing acceptor; IRES, internal ribozyme entry site; lacZ, lair conditionerZ; pA, poly A; hbactP, human beta actin promoter; neo, promoter-driven neomycin resistant gene; FRT, flippase recognition target; Loxp, locus of crossover in p1.
Adenosine monophosphate (AMP) deaminase (AMPD, EC 3.5.4.6) is a group of enzymes that catalyze the deamination of AMP to IMP. AMPD family contains three members, AMPD1-32. AMPD1 is mainly expressed in the skeletal muscle, whereas AMPD2 is widely expressed in non-muscle tissues and AMPD3 is mainly expressed in erythrocytes3,4. AMPD is the initial reaction of the purine nucleotide cycle, which plays a central role in the adenine nucleotides metabolism and energy charge maintenance5. Thus AMPD is thought to play an important role in regulating nucleotide metabolism, as well as maintaining adenylate energy charge. Several studies have reported that AMPD1 deficiency might regulate muscle metabolism6,7. Polymorphism in AMPD1 gene has been reported to be associated with metabolic myopathy, which is also named as AMP deaminase deficiency8,9. The disease is reported an approximate 2–3% incidence in the Caucasian population, as well as a similar prevalence in African-Americans10. In our previous study, a haplotype AMPD1-NRAS-CSDE1 is found to be associated with autism11, and variants in AMPD1 contribute to autism risk in Chinese Han population12.
In this study, we studied a strain of Ampd1 mutant mice generated by a knockout-first strategy. Our data indicated that insertion of knockout-first cassette into Ampd1 gene disrupted the expression of neighboring genes up to 2.4 Mb, which may underlie their neonatal death.
Results
Ampd1 tm1a/tm1a mice died in 2 days postnatally
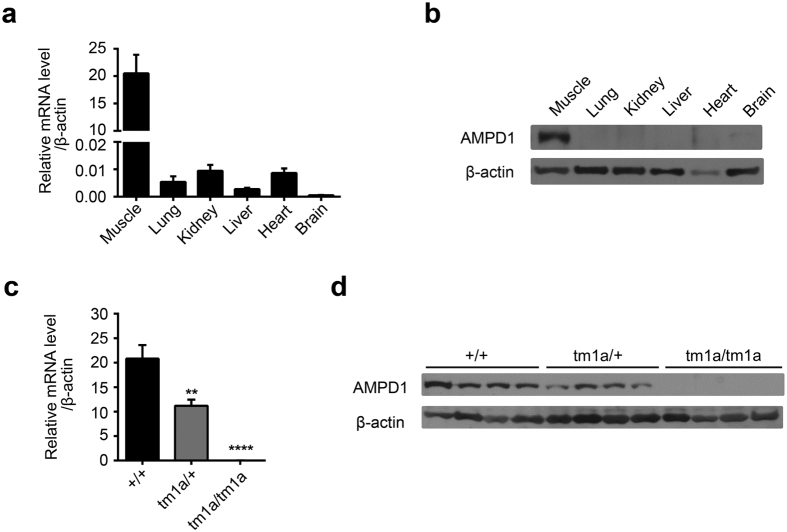
To study the physiological function of AMPD1, we obtained a strain of Ampd1 mutant mice from KOMP repository, which was generated by the knockout-first strategy. The tm1a allele was initially a non-expressive form due to the alternative splicing of the Ampd1 exon1-2 to SA in the trapping cassette, disrupting the Ampd1 transcript. As shown in Fig. 2, Ampd1 was predominantly expressed in the muscle as determined with real-time PCR (RT-PCR) and Western blot (Fig. 2a,b); no Ampd1 mRNA and protein were detected in the hind-leg muscle of E18.5 Ampd1tm1a/tm1a mice (Fig. 2c,d).
Figure 2. The expression of Ampd1 in different tissues and the validation of Ampd1 disruption.
(a,b) The expression of Ampd1 in different tissues. Ampd1 was predominantly expressed in the muscle as determined with real-time PCR (a) and Western blot (b) in E18.5 WT mice. (c,d) The validation of Ampd1 disruption. Ampd1 mRNA (as compared with WT, ****P < 0.0001; **P < 0.001, One-way ANNOVA followed with Bonferroni’s multiple comparisons test) and protein (d) were undetectable in the hind-leg muscle of E18.5 Ampd1tm1a/tm1a mice. Relative mRNA level was normalized to β-actin.
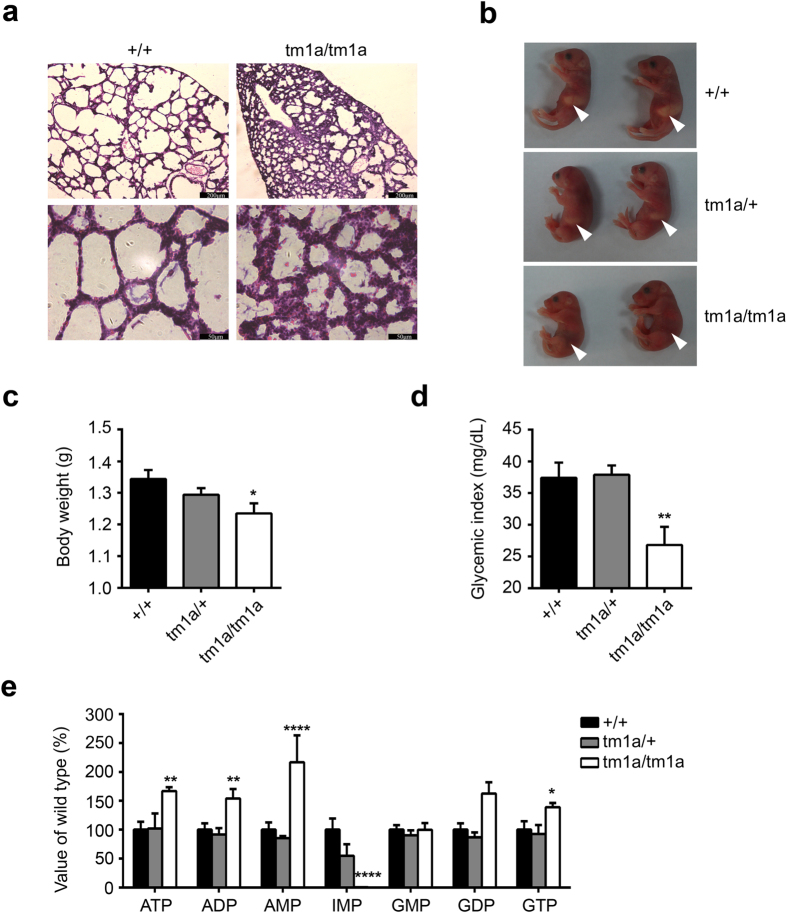
When intercrossing the Ampd1tm1a/+ mice, the progeny at embryonic 18.5 days (E18.5), postnatal 0.5 days (P0.5) and P1 showed a Mendelian distribution, but no Ampd1tm1a/tm1a pups survived more than two days (Table 1), indicating Ampd1tm1a/tm1a mice may die within 24–48 hours postnatally. No apparent histopathological anomaly except lung was found in the P0.5 Ampd1tm1a/tm1a mice. As shown in Fig. 3a, Ampd1tm1a/tm1a mice showed volume reduced pulmonary alveoli and incrassated alveolar septum compared to wide-type (WT) mice.
Table 1. The distribution of progeny from mating of heterozygotes with different Ampd1 alleles.
| Allele | Stage | WT: Heterozygotes: Homozygotes |
|
|---|---|---|---|
| Total Number | Ratio | ||
| tm1a | *E18.5 | 184 | 1:1.98:0.94 |
| #P0.5 | 126 | 1:2.09:0.84 | |
| P1 | 112 | 1:2.07:0.67 | |
| P2 | 89 | 1:2.07:0 | |
| Adult | 407 | 1:1.81:0 | |
| tm1c | Adult | 69 | 1:2.17:0.88 |
| tm1d | Adult | 62 | 1:2.06:0.875 |
*E: embryonic.
#P: postnatal.
Figure 3. Phenotypic analysis of Ampd1tm1a/tm1a mice.
(a) H&E staining of the lung from P0.5 mice. Ampd1tm1a/tm1a mice showed volume reduced pulmonary alveoli and incrassated alveolar septum compared to WT mice. Scale bar: 200 μm (upper), 50 μm (bottom). 7/7 mice showed this phenotype. (b) Representative P0.5 mice. No milk in Ampd1tm1a/tm1a stomachs (arrowheads). (c) Reduced body weight of P0.5 Ampd1tm1a/tm1a pups (WT: 1.343 ± 0.028 g, n = 15; Ampd1tm1a/+: 1.294 ± 0.021 g, n = 35; Ampd1tm1a/tm1a: 1.235 ± 0.031 g, n = 17; mean ± SEM. *P < 0.05, One-way ANNOVA followed with Bonferroni’s multiple comparisons test). (d) Decreased glucose levels in P0.5 Ampd1tm1a/tm1a pups (Ampd1tm1a/+: 37.89 ± 1.48 mg/dL, n = 35; WT: 37.4 ± 2.43 mg/dL, n = 15; Ampd1tm1a/tm1a: 28.76 ± 3.33 mg/dL, n = 17; mean ± SEM. **P < 0.01, One-way ANNOVA followed with Bonferroni’s multiple comparisons test). Glucose was measured using OneTouch UltraMini glucometer (LifeScan, Inc., Milpitas, CA, USA). (e) In the muscle from E18.5 animals, ATP, ADP, AMP, GDP levels were significantly increased 66%, 64%, 117%, 39% respectively and IMP levels were decreased to nearly zero in Ampd1tm1a/tm1a mice (*P < 0.05, **P < 0.01, and ****P < 0.0001, two-way ANNOVA followed with Bonferroni’s multiple comparisons test).
The Ampd1tm1a/tm1a pups could be easily recognized by lack of milk in their stomachs (Fig. 3b). Moreover, the body weight of Ampd1tm1a/tm1a pups at P0.5 was significantly lower than that of WT mice and Ampd1tm1a/+ mice (Fig. 3c). The Ampd1tm1a/tm1a mice at P0.5 also showed significantly decreased blood glucose compared with heterozygous and WT mice (Fig. 3d), implying starvation in neonatal Ampd1tm1a/tm1a mice.
Impaired nucleotide metabolism in the muscle of Ampd1 tm1a/tm1a mice
AMPD1 catalyzes AMP hydrolyzing into IMP and NH3, and plays an important role in the purine nucleotide cycle5. We therefore measured the nucleotide level in the tissues from Ampd1tm1a/tm1a and their WT and Ampd1tm1a/+ controls. A dramatically elevated AMP level (over 200%) and almost complete lack of IMP were detected in the skeletal muscle of E18.5 Ampd1tm1a/tm1a mice (Fig. 3e), indicating that AMPD1 was the major, if not solely, adenosine monophosphate deaminase in the skeletal muscle. Consistent with the restricted expression of Ampd1 in the muscle, there was no genotypic difference in the nucleotide levels of the brain, heart, liver and lung (Supplementary Fig. 1).
Disruption in neighboring genes expression in Ampd1 tm1a/tm1a mice
Another two strains of Ampd1-deficient mice showed similar deficiency in the purine nucleotide cycle as Ampd1tm1a/tm1a mice; however, they were viable and did not show significant difference in life span13,14. We thus speculated an off-target effect in our strain may underlie the phenotypic difference.
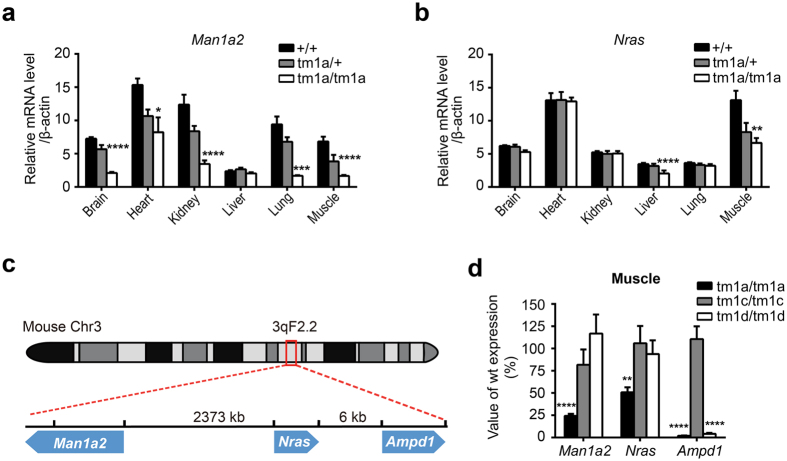
We then analyzed the global gene expression in the skeletal muscle from E18.5 animals by using RNA-seq. In addition to the deficiency in Ampd1, we also identified another two down-regulated genes, Man1a2 and Nras (Supplementary Table 1); RT-PCR analysis indicated that Man1a2 and Nras expression in the muscle of Ampd1tm1a/tm1a mice was reduced to 24% and 51% of WT mice, respectively, The expression of Man1a2 were also dramatically decreased in other tissues, such as the brain, heart, kidney and lung (Fig. 4a), whereas the expression of Nras was slightly reduced in the liver (Fig. 4b). It should be noted that Man1a2-deficient mice have been reported died at postnatal 0.5 day with pulmonary hypoplasia, hypercellular, thicker alveolar septae, and alveolar hemorrhage15.
Figure 4. The knockout-first cassette in Ampd1 tm1a allele influences the neighboring genes expression.
(a) Real time PCR indicated that Man1a2 was significant down-regulated in the brain, heart, kidney, lung and muscle; (b) Nras was slight down-regulated in the liver and the muscle of Ampd1tm1a/tm1a mice (n = 5/each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANNOVA followed with Bonferroni’s multiple comparisons test). (c) Schematic diagram of Man1a2, Nras and Ampd1 on chromosome 3. Nras is 6 kb from Ampd1 gene, whereas Man1a2 is 2.4 Mb away from Ampd1on the other strand. Arrowheads indicate orientation (left, negative strand; right, positive strand). (d) Relative mRNA expression of Ampd1, Man1a2, Nras in the muscle from different mice allele (As compared to WT, n = 5/each genotype, **P < 0.01, ****P < 0.0001, one-way ANNOVA followed with Bonferroni’s multiple comparisons test).
The knockout-first cassette influences neighboring genes expression
Interestingly, Man1a2, Nras and Ampd1 are all located on chromosome 3. Nras is 6 kb from Ampd1 gene, whereas Man1a2 is 2.4 Mb away from Ampd1 (Fig. 4c). Nevertheless, the expression of genes located between Man1a2 and Ampd1 did not change in the muscle and lung (Supplementary Table 1 and Supplementary Fig. 2). To see if down-regulation of Man1a2 and Nras was caused by the insertion of knockout-first cassette or the deficiency of AMPD1 protein per se, we removed the FRT-flanked knockout-first cassette by crossing Ampd1tm1a/+ with a globe Flp transgenic mouse strain to produce tm1c allele. When intercrossing Ampd1tm1c/+ mice, the progeny showed a Mendelian distribution and Ampd1tm1c/tm1c mice were viable in adulthood (Table 1). The Ampd1tm1c/tm1c mice showed normal gene expression for Man1a2, Nras and Ampd1 in the skeletal muscle (Fig. 4d).
We then mated Ampd1tm1c/tm1c with a globe Cre transgenic mouse strain to produce tm1d allele. When intercrossing Ampd1tm1d/+ mice, the progeny showed a Mendelian distribution and Ampd1tm1d/tm1d mice were viable in adulthood (Table 1). Although Ampd1 was disrupted in the Ampd1tm1d/tm1d mice, the expression of Man1a2 and Nras in the skeletal muscle of Ampd1tm1d/tm1d mice was indistinguishable from those of WT controls (Fig. 4d). Our data thus indicated that it was the knockout-first cassette, instead of the deficiency of AMPD1 protein, influenced the expression of Man1a2 and Nras.
Discussion
It is widely accepted that insertion of a selection marker such as PGK-neo in an allele may disrupt neighboring gene expression. Targeted mutation of several genes, including β-globin, Granzyme B, Hox genes, and MRF4, by insertion of PGK-neo selection cassette results in severe reduction of neighboring gene expression up to 100 kb away from the mutation sites16,17,18,19,20,21. Furthermore, FRT/loxP sites used in gene targeting could also influence the expression of neighboring genes22.
Insertion of a knockout-first cassette has also recently been reported to influence neighboring genes expression. Maguire et al. find that homozygous Slc25a21tm1a exhibit orofacial abnormalities, alterations in carpal and rugae structures, hearing impairment and inflammation in the middle ear. However, after removal of the critical exon and knockout-first cassette, Slc25a21 knockout mice homozygous for the Slc25a21tm1c and Slc25a21tm1d alleles are phenotypically indistinguishable from wild-type. RNA expression analysis indicate that the expression of Pax9, located at the 3′ end of Slc25a21 on the opposite strand with 188 bp overlapping at the 3′-UTR regions, is reduced specifically in mice homozygous in tm1a, but not in tm1c and tm1d alleles.
There are several explanations for this phenomenon. One possibility is that a cluster of genes share one regulatory element, and targeting cassette disrupted the interaction between corresponding genes and the upstream regulatory element. For instance, the insertion of PGK-Neo into granzyme B genes, located at the most 5′ of granzyme gene cluster, severely reduces the normal expression of a panel of genes within the locus up to 100 kb away from the mutation17. In contrast, a targeted mutation of the promyelocyte-specific cathepsin G gene, which lies just 3′ to the granzyme genes in the same cluster, has minimal effects on upstream granzyme gene expression. However, our case seems much more complicated. Although insertion of the knockout-first cassette in Ampd1 influences Man1a2 and Nras expression, more than twenty genes localized between Man1a2 and Nras are essentially unaffected.
Another probable explanation is that the targeting cassette disrupted the chromatin conformation-dependent interactions between genes and their distant regulatory elements. A commonly accepted chromatin loop model is that DNA sequences between regulatory elements and promoters form loops to allow the distant regulatory elements to interact directly with the promoters. Chromosome conformation capture (3C) and 3C-derived methods23,24,25,26,27 have demonstrated chromosome three-dimensional conformation in a genome-wide level, which is hosted in the WashU Epigenome Browser28. Interestingly, there are indeed long range interactions between Ampd1 and Man1a229 (Supplementary Fig. 3). We thus speculate that insertion of knockout-first cassette may disrupt some of these interaction, and subsequently downregulate the expression of Man1a2.
The Man1a2 null mice die within 12 h after birth because of respiratory distress and some delay in the lung development15. The Ampd1tm1a/tm1a mice also show deficiency in the lung development, consistent with the downregulation of Man1a2. However, Ampd1tm1a/tm1a mice die within 24–48 hours postnatally, probably due to starvation. Furthermore, Tremblay et al. also report the loss of L-PHA, E-PHA, and DSA reactive glycoproteins in a subpopulation of epithelial cells lining the bronchioles from Man1a2 null mice; however, there is no difference in these lectin-stained glycoproteins in the Ampd1tm1a/tm1a mice (data not shown). We speculate the residual expression of Man1a2 (i.e. 24% in the muscle and 43% in the lung) may underlie the phenotypic difference between Man1a2 null and Ampd1tm1a/tm1a mice. Nevertheless, we cannot exclude the possible contribution of other genes deficiency to the neonatal lethality of Ampd1tm1a/tm1a mice.
In summary, insertion of knockout-first cassette per se in Ampd1 gene results in globally reduced expression of neighboring gene Man1a2, which may partly explain the neonatal lethality phenotype of Ampd1tm1a/tm1a mice. Our results indicate the knockout-first cassette may cause off-target effect by disruption the chromatin conformation-dependent interaction between genes and their distant regulatory elements and influence the neighboring gene expression. We thus strongly suggest users to remove the knockout-first cassette by Flp recombinase, and then generate conditional allele or global allele using specific Cre recombinases. In addition, high throughput assays, such as RNA-seq, may be used to verify if there is any side-effect in the initial allele.
Methods
Ampd1 knockout mice
The Ampd1tm1a/+ mice (#CSD23933) were generated by KOMP. A cassette containing mouse En2 SA, LacZ, Neo, FRT and loxP sites, was inserted in introns around exon3 of Ampd1 gene (Fig. 1a). The tm1a allele was initially an Ampd1 non-expressive form. Ampd1tm1a/+ mice were obtained from the KOMP repository and intercrossed to generate Ampd1tm1a/tm1a. Ampd1tm1c/+ allele was produced by crossing Ampd1tm1a/+ with a globe Flp transgenic mouse strain [Gt(ROSA)26Sortm1(FLP1)Dym/J, The Jackson Laboratory Stock No:003946]. Ampd1tm1d/+ allele was generated by mating Ampd1tm1c/tm1c with a globe Cre transgenic mouse strain [B6.C-Tg(CMV-cre)1Cgn/J, The Jackson Laboratory Stock No: 006054]. PCR primers used for genotyping were listed in Supplementary Table 2. All animal experiments were approved by the ethics committee of State Key Laboratory of Medical Genetics, Central South University of China. All methods were performed in accordance with approved guidelines.
RNA extract and quantitative real-time PCR analysis
Total RNA were extracted from the hindleg muscle of E18.5 mice using Trizol reagent (Life technologies, NY, USA). cDNA was synthesized from total RNA (1 μg) through the RevertAid First Strand cDNA Synthesis Kit (K1622, Thermo Scientific, Inc., Waltham, MA, USA) and oligo dT primers. Quantitative real-time PCR was carried out with a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, lnc. Hercules, CA, USA) using Maxima SYBR Green qPCR Master Mixes (K0251, Thermo Scientific, Inc., Waltham, MA, USA). Data were normalized to β-actin and analyzed by the comparative cycle threshold (CT) method. Specific primers for each gene were as follows: 5′CAGAGCCTCGCTTATCCATC3′ and 5′TCTTGGATCGGAACACATCA3′ for Ampd1; 5′CACATCAGTTGGTGGTCTGG3′ and 5′GCCTCAACAGCATCGTCATA3′ for Man1a2; 5′TGACTTGCCAACAAGGACAG3′ and 5′CTGGCGTATCTCCCTTACCA3′ for Nras; 5′CACGATGGAGGGGCCGGACTCATC3′ and 5′TAAAGACCTCTATGCCAACACAGT3′ for β-actin.
Immunoblotting
Mouse tissues were cut into pieces and added with 2xSDS buffer (63 mM Tris-HCl, 10% Glycerol, 2% SDS, 0.0025% Bromophenol Blue, pH 6.8) supplemented with protease inhibitor cocktails (P8430, Sigma-Aldrich, St. Louis, MO, USA). After ultrasonication, the lysate was centrifuged at 13,000 rpm for 10 min. The protein concentration of the supernatant was measured using the Pierce BCATM protein assay kit (23228, Thermo Scientific, Inc., Waltham, MA, USA). For western blot 40 μg proteins were separated in 10% SDS-PAGE gels and transferred to PVDF membrane, blocked with 5% milk in 0.1% PBS-T (0.1% Triton-X 100 in 1 × PBS), and blotted with primary antibody overnight at 4 °C. The antibodies for AMPD1 (sc-160043, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and β-actin (A2228, Sigma-Aldrich, St. Louis, MO, USA) were purchased. The secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA). ECL plus Western blotting detection system (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) was used to detect immobilized specific antigens, according to manufacturer’s instruction.
Histological examination
Tissues were fixed in 4% paraformaldehyde, following dehydration with sucrose, tissues were frozen immediately in precooling isopentane, and sectioned to 15 μm with Leica CM1950 freezing microtome (Leica Biosystems, Wetzlar, Germany). Sections were stained with Harris’s hematoxylin and eosin (H&E). Images were taken using Leica DM5000B Microscope System (Leica Microsystems, Wetzlar, Germany).
Nucleotide analysis by High Performance Liquid Chromatography (HPLC)
Fresh-frozen tissues were washed once with PBS, and homogenized in 10% Trichloroacetic Acid (T6399, Sigma-Aldrich, St. Louis, MO, USA). After incubating on ice for 10 min, the mixtures were centrifuged at 12,000 g for 15 min at 4 °C, and the nucleotide containing supernatant was washed 4 times with water-saturated diethyl ether. Then the supernatant was froze at −80 °C, and freeze-dried at −20 °C overnight use FreeZone Freeze Dry Systems (LABCONCO, Inc., MO, USA). The extracted nucleotides were dissolved in 100 μL ultrapure water and centrifuged at 8,000 rpm for 5 min. Supernatant was filtered with 0.45 μm filter and then analyzed by HPLC on a Agilent 1260 (Agilent Technologies, Inc., CA, USA) equipped with a Waters SunFireTM C18 column (250 mm × 4.6 mm i.d., 5 μm, Waters, Milford, MA, USA) and detected by UV-VIS absorbance at 260 nm. The nucleotides were eluted with isocratic elution (flow rate: 0.75 ml/min, Column temperature: 25 °C) using a mixture of 10% acetonitrile and 90% water (0. 01 mol/L Tetrabutylammonium bromide, 0.05 mol/L Na2HPO4-NaH2PO4, pH6.4). The amount of each nucleotide was determined by external standard method. The following standards were used: ATP (A2383), ADP (A2754), AMP (01930), IMP (57510), GMP (G8377), GDP(G7127), GTP(G8877) (Sigma-Aldrich, St. Louis, MO, USA).
RNA sequencing (RNA-seq)
RNA sequencing was performed by Novogene Bioinformatics Technology Co. Ltd. The RNA was extracted from the hindleg muscles of embryonic 18.5 day mice (3 WT and 3 Ampd1tm1a/tm1a) by using Trizol reagent (Life technologies, NY, USA). A total of 3 μg RNA per sample was used. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs., MA, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2000 platform and 100 bp single-end reads were generated.
RNA-Seq data analysis
Raw data were firstly processed to obtain clean data by removing reads containing adapter, ploy-N and low quality reads. Index of the reference genome was built using Bowtie v2.0.6 and single-end clean reads were aligned to the reference genome using TopHat v2.0.9. And HTSeq v0.5.4p3 was used to count the reads numbers mapped to each gene. Then RPKM (Reads Per Kilobase of exon model per Million mapped reads) of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis was performed using the DESeq R package (1.10.1). The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by DESeq were assigned as differentially expressed.
Statistical analysis
All statistical analyses were performed using the Prism 6.01 (Graph Pad Software, San Diego, CA). Statistically significant differences between groups were determined by a repeated ANOVA followed by unpaired t test.
Additional Information
How to cite this article: Pan, Y. et al. Insertion of a knockout-first cassette in Ampd1 gene leads to neonatal death by disruption of neighboring genes expression. Sci. Rep. 6, 35970; doi: 10.1038/srep35970 (2016).
Supplementary Material
Acknowledgments
We thank Professors Depei Liu, Xiang Lv, Guangyuan Xiao and other staff from State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & Peking Union Medical College for useful discussions on long range interaction. We thank Professor Shuyun Shi from the School of Chemistry and Chemical Engineering, Central South University for technical assistance in HPLC. This work was supported by the National Basic Research Program of China (2012CB517902 and 2012CB517904), the National Natural Science Foundation of China (81330027, 81525007, 31371187, 81501182 and 31400919), the Lotus Scholar Professorship Funds from Hunan Province Government and a research grant provided by the Central South University of China.
Footnotes
Author Contributions J.-D.L., K.X., J.Z. and Z.H. conceived and designed the experiments; Y.P. and L.Z. performed the experiments; B.T., H.G. and Y.P. helped to analyze the data; Y.P., L.Z. and J.-D.L. wrote the paper; Y.L., Q.L. and H.P. provided experiment assistance.
References
- Skarnes W. C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T., Sabina R. L. & Holmes E. W. Adenylate deaminase. A multigene family in humans and rats. J Biol Chem 265, 11482–11486 (1990). [PubMed] [Google Scholar]
- Fishbein W. N., Sabina R. L., Ogasawara N. & Holmes E. W. Immunologic evidence for three isoforms of AMP deaminase (AMPD) in mature skeletal muscle. Biochim Biophys Acta 1163, 97–104 (1993). [DOI] [PubMed] [Google Scholar]
- Yamada Y., Goto H. & Ogasawara N. A point mutation responsible for human erythrocyte AMP deaminase deficiency. Human molecular genetics 3, 331–334 (1994). [DOI] [PubMed] [Google Scholar]
- Van den Berghe G., Bontemps F., Vincent M. F. & Van den Bergh F. The purine nucleotide cycle and its molecular defects. Prog Neurobiol 39, 547–561 (1992). [DOI] [PubMed] [Google Scholar]
- Sabina R. L. et al. Myoadenylate deaminase deficiency. Functional and metabolic abnormalities associated with disruption of the purine nucleotide cycle. J Clin Invest 73, 720–730 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkeler S. P. et al. AMP deaminase deficiency: study of the human skeletal muscle purine metabolism during ischaemic isometric exercise. Clinical science 72, 475–482 (1987). [DOI] [PubMed] [Google Scholar]
- Morisaki H., Morisaki T., Newby L. K. & Holmes E. W. Alternative splicing: a mechanism for phenotypic rescue of a common inherited defect. J Clin Invest 91, 2275–2280 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki H., Higuchi I., Abe M., Osame M. & Morisaki T. First missense mutations (R388W and R425H) of AMPD1 accompanied with myopathy found in a Japanese patient. Human mutation 16, 467–472 (2000). [DOI] [PubMed] [Google Scholar]
- Yamada Y., Goto H., Murase T. & Ogasawara N. Molecular basis for human erythrocyte AMP deaminase deficiency: screening for the major point mutation and identification of other mutations. Hum Mol Genet 3, 2243–2245 (1994). [DOI] [PubMed] [Google Scholar]
- Xia K. et al. Common genetic variants on 1p13.2 associate with risk of autism. Molecular psychiatry 19, 1212–1219 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. AMPD1 functional variants associated with autism in Han Chinese population. European archives of psychiatry and clinical neuroscience 265, 511–517 (2015). [DOI] [PubMed] [Google Scholar]
- Cheng J. et al. Effect of isolated AMP deaminase deficiency on skeletal muscle function. Molecular Genetics and Metabolism Reports 1, 51–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaideau C. et al. Effects of pharmacological AMP deaminase inhibition and Ampd1 deletion on nucleotide levels and AMPK activation in contracting skeletal muscle. Chemistry & biology 21, 1497–1510 (2014). [DOI] [PubMed] [Google Scholar]
- Tremblay L. O. et al. Respiratory distress and neonatal lethality in mice lacking Golgi alpha1,2-mannosidase IB involved in N-glycan maturation. The Journal of biological chemistry 282, 2558–2566 (2007). [DOI] [PubMed] [Google Scholar]
- Fiering S. et al. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev 9, 2203–2213 (1995). [DOI] [PubMed] [Google Scholar]
- Pham C. T., MacIvor D. M., Hug B. A., Heusel J. W. & Ley T. J. Long-range disruption of gene expression by a selectable marker cassette. Proceedings of the National Academy of Sciences of the United States of America 93, 13090–13095 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijli F. M., Dolle P., Fraulob V., LeMeur M. & Chambon P. Insertion of a targeting construct in a Hoxd-10 allele can influence the control of Hoxd-9 expression. Dev Dyn 201, 366–377 (1994). [DOI] [PubMed] [Google Scholar]
- Horan G. S., Wu K., Wolgemuth D. J. & Behringer R. R. Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc Natl Acad Sci USA 91, 12644–12648 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic D. & Capecchi M. R. Targeted disruptions of the murine Hoxa-4 and Hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech Dev 46, 231–247 (1994). [DOI] [PubMed] [Google Scholar]
- Olson E. N., Arnold H. H., Rigby P. W. & Wold B. J. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85, 1–4 (1996). [DOI] [PubMed] [Google Scholar]
- Meier I. D. et al. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 24, 1714–1724 (2010). [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M. & Kleckner N. Capturing chromosome conformation. Science 295, 1306–1311 (2002). [DOI] [PubMed] [Google Scholar]
- Dostie J. et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Research 16, 1299–1309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38, 1341–1347 (2006). [DOI] [PubMed] [Google Scholar]
- Simonis M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38, 1348–1354 (2006). [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods 10, 375–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.