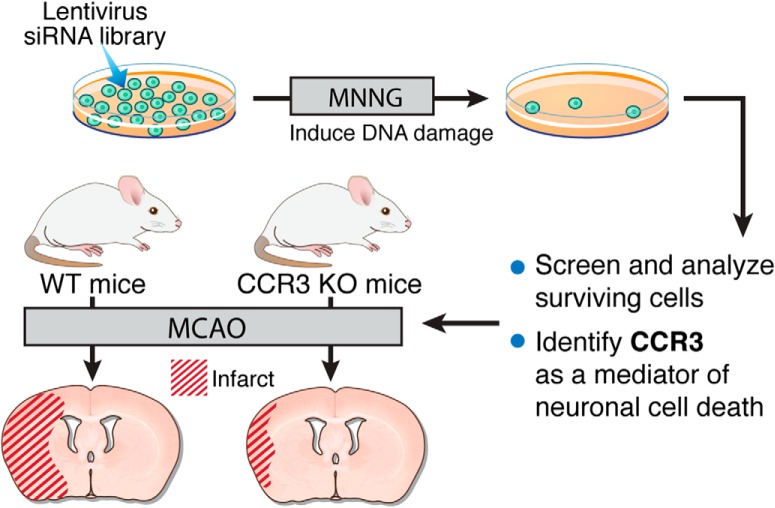
Visual Abstract
Keywords: excitotoxicity, ischemia, library screen, stroke
Abstract
Neuronal loss caused by ischemic injury, trauma, or disease can lead to devastating consequences for the individual. With the goal of limiting neuronal loss, a number of cell death pathways have been studied, but there may be additional contributors to neuronal death that are yet unknown. To identify previously unknown cell death mediators, we performed a high-content genome-wide screening of short, interfering RNA (siRNA) with an siRNA library in murine neural stem cells after exposure to N-methyl-N-nitroso-N′-nitroguanidine (MNNG), which leads to DNA damage and cell death. Eighty genes were identified as key mediators for cell death. Among them, 14 are known cell death mediators and 66 have not previously been linked to cell death pathways. Using an integrated approach with functional and bioinformatics analysis, we provide possible molecular networks, interconnected pathways, and/or protein complexes that may participate in cell death. Of the 66 genes, we selected CCR3 for further evaluation and found that CCR3 is a mediator of neuronal injury. CCR3 inhibition or deletion protects murine cortical cultures from oxygen-glucose deprivation–induced cell death, and CCR3 deletion in mice provides protection from ischemia in vivo. Taken together, our findings suggest that CCR3 is a previously unknown mediator of cell death. Future identification of the neural cell death network in which CCR3 participates will enhance our understanding of the molecular mechanisms of neural cell death.
Significance Statement
A high-content siRNA genome-wide screen identified 80 mediators of cell death in neural stem cells in response to DNA damage injury. These cell death mediators possibly form a molecular network containing several partially overlapping and interconnected pathways or protein complexes. Through in vitro and in vivo experiments, we confirmed that one of the identified molecules, CCR3, is a mediator of neuronal cell death. Further investigation of the mechanisms of CCR3 action in neuronal injury is warranted. Additionally, the other 65 genes may provide new opportunities to understand and prevent neuronal cell death.
Introduction
Neuronal cell death plays an important role in the proper development of the immature nervous system, but in the adult, neuronal cell death underpins neuronal dysfunction caused by disease, trauma, or ischemic injury. Although neuronal cell death in the context of nervous system development is prominently triggered by the absence of cell survival factors and induction of apoptosis, death of mature neurons in the context of neurological disease is induced through the activation of a variety of cell death mediators and their signaling networks (Becker and Bonni, 2004). Although many genetic screens are conducted in smaller organisms such as Drosophila or Caenorhabditis elegans, recently RNA-mediated interference (RNAi) loss-of-function experiments have been applied to genome-wide screens in mammalian cells to identify genes that are required for a wide variety of specific phenotypes (Kittler et al., 2004; Paddison et al., 2004; Moffat et al., 2006; Hitomi et al., 2008). Because DNA damage is a contributing factor in neuronal cell death after stroke or in neurodegenerative diseases, to identify currently unknown mediators of cell death, we performed a genome-wide RNAi screen in neural stem cells after exposure to the DNA alkylating agent, N-methyl-N-nitroso-N′-nitroguanidine (MNNG). MNNG promotes nicks and breaks and is known to activate the enzymatic activity of poly(ADP-ribose) polymerase, resulting in a more necrotic-like rather than apoptotic cell death which has been designated parthanatos. (D’Amours et al., 1999; Zong et al., 2004).
In this study, we performed a genome-wide screen using the mouse GeneNet Mouse 40K FIV lentiviral small, interfering RNA (siRNA) library. We identified 80 key mediators for DNA damage-induced neural cell death. Among these 80 genes, 66 have not been previously been linked to DNA damage response pathways or cell death cascades. After functional analysis and bioinformatics analysis of the gene set, we selected the C-C chemokine receptor 3 (CCR3) for further characterization. CCR3 is a seven-transmembrane G protein–linked receptor that has been extensively studied in the setting of inflammation and allergic reaction in peripheral tissues (Willems and Ijzerman, 2010). In addition to expression in bone marrow–derived cells including basophils, eosinophils, and microglia, CCR3 is expressed in astrocytes and neurons in both the developing and adult brain (van der Meer et al., 2000). Through loss-of-function experiments, we confirm that CCR3 functions in cell death signaling in cortical cultures exposed to oxygen-glucose deprivation (OGD) and contributes to ischemic brain injury in vivo. Understanding how CCR3 contributes to neuronal loss and further investigation into the potential neuroprotective actions of CCR3 inhibition may reveal new therapeutic opportunities for treatment of ischemic injury or neurodegenerative diseases.
Methods
Cell culture and lentiviral siRNA infection
Mouse neural stem cells were cultured and infected with mouse 40K genome-wide lentiviral siRNA provided by System Biosciences (Palo Alto, CA) according to the manufacturer’s manual. Briefly, mouse neural stem cells (ANSO4E) were plated in twelve 10-cm plates at a density of about 5 × 105 cells per plate 24 h before viral infection. On the second day, 1 × 107 infection units of mouse 40K genome-wide lentivirus siRNA was thawed and diluted in 30 ml complete neural cell culture medium (20 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor in Dulbecco’s modified Eagle’s medium/F-12 medium) with 5 μg/ml polybrene. Ten plates of cultured mouse neural cells were infected with 3-ml viral dilutions (ratio of virus and cell of about 1:5). For the mock transduction control, 3 ml Dulbecco’s modified Eagle’s medium with polybrene was added. All cells were incubated at 37°C with 5% CO2. For each plate, the culture medium was removed and replaced with 10 ml fresh complete medium (without polybrene) 12 h after infection. At d 4, the cells were split 1:3 and returned to culture for another 24 h. On d 5 (72 h after transduction), neural stem cells were treated with 500 μm MNNG for 15 min, washed two times with medium, and incubated at 37°C with 5% CO2 for 3 d. The dead cells were removed by changing the medium every 3 d, and the surviving cells were grown and treated with 500 μm MNNG for secondary selection. The surviving cells were harvested after growing to 10 million cells.
Recovery of siRNA inserts
When integrated into genomic DNA, the pFIV constructs produced an alternative transcript from that of a fusion of the marker genes (copGFP or Puro) with the siRNA sequence. This alternative transcript was used as a template to amplify the siRNA insert. Recovery of siRNA inserts was performed according to the manufacturer’s instructions. Briefly, total RNA of surviving cells or reference control cells was isolated with TRIzol Reagent (Invitrogen, San Diego, CA) and purified further with RNeasy minicolumns (Qiagen, Valencia, CA). For each sample, 10 μg total RNA in 15 μl RNA-free water was used for cDNA synthesis as recommended by the manufacturer. The primer sequence for reverse transcription was TGCATGTCGCTATGTGTTCTGGGA. Amplification of the inserts from synthesized cDNA required two rounds of PCR. The primers for the first round of PCR were forward, 5′-AATFTCTTTGGATTTGGGAATCTTA-3′, and reverse, 5′-AAAAGGGTGGACTGGGATGAGT-3′. During the second round of PCR with two nested primers (nested forward primer 5′-Bio-ATCGTCAATCACCTTCCTGTCAGA-3′ and nested reverse primer 5′-AATAGAAAGAATGCTTATGGACGCTA-3′), the amplified siRNA targets were labeled with biotin. Biotinylated siRNA targets (15 μg) were hybridized with Affymetrix GeneChip Mouse Genome 430 2.0 Array (#900495; Affymetrix, Santa Clara, CA) to identify surviving cell-associated siRNAs using the manufacturer’s protocols and recommended reagents.
Individual siRNA knockdown and OGD
Lentiviral siRNA vectors for 17 genes were obtained from Invitrogen, and lentiviruses were individually prepared as described previously (Wang et al., 2011). Primary cultured mouse cortical neurons were cultured and infected with lentiviral siRNAs at 7 days in vitro. Lentiviral siRNAs for DsRed were used as control. Seven days after infection, neurons were treated with lethal OGD (90 min) to induce neural cell death. Twenty-four hours after OGD treatment, cell viability was determined by staining with Hoechst 33342 and propidium iodide. Images were captured by charge-coupled device camera under Axiovert 200M fluorescence microscope, and dead and viable cells were counted by a blinded observer using unbiased computer-assisted cell counting software (Axiovision 4.3; Zeiss, Oberkochen, Germany). Cell death rate was determined as the ratio of dead cells to total cells. Three separate individual experiments were performed using three separate wells.
Middle cerebral artery occlusion
CCR3–/+ mice were crossed to generate CCR3–/– mice and wild-type (WT; CCR3+/+) mice in the same litters to ensure the same genetic background. We used only the males for middle cerebral artery occlusion (MCAO) to avoid the possibility of sex-specific sensitivity to cell death. Thirteen male CCR3 KO mice and nine WT littermates were subjected to MCAO as described previously (Andrabi et al., 2011). In brief, mice were anesthetized with 1.5–2% isoflurane and maintained at normothermic temperature. Occlusion was performed by placing a monofilament to the base of the middle cerebral artery. The whole MCAO procedure was monitored by laser-Doppler flowmetry with a probe placed on thinned skull over the lateral parietal cortex. After 60 min of occlusion, the filament was removed and the reperfusion was verified. Twenty-four hours later, animal brains were coronally sectioned into five 2-mm-thick sections in a mouse brain matrix and stained in 2% 2,3,5-triphenyltetrazolium chloride solution before fixation in 10% formalin overnight. The infarction area was imaged with a digital camera and analyzed using ImageJ (National Institutes of Health, Bethesda, MD) by an operator blinded to genotype. The infarction volume was calculated by summing the infarct volumes of sections. Infarct size was calculated and expressed as a percentage by using the following formula: (contralateral volume – ipsilateral undamaged volume) × 100/contralateral volume, to eliminate effects of edema as described previously (Kishimoto et al., 2010). The investigator performing the surgery and analyzing infarct size was unaware of the genotype of the mouse.
Assessment of neurobehavioral activity
Spontaneous motor activities of mice were recorded for 5 min in an animal cage at 24 h, 3 d, and 7 d after MCAO as described previously (Andrabi et al., 2011). Briefly, neurological deficits were assessed on a scale of 0–4 (0, no neurological deficit; 4, severe neurological deficit) according to the criteria: 0 = normal, the mice explored the cage environment and moved around in the cage freely; 1 = the mice could hesitantly move in the cage but did not approach all sides of the cage; 2 = the mice showed postural and movement abnormalities and had difficulty approaching all walls of the cage; 3 = the mice with postural abnormalities tried to move in the cage but did not approach one wall of the cage, and 4 = mice were unable to move in the cage and stayed at the center. The observer was blinded to the genotypes of the animals.
Data analysis
The software provided with the library on the Data Analysis Program and Gene List was used to analyze the hybridization data and create a report file in a format compatible with common spreadsheets (Excel and .txt file), and further analysis was performed in R. The file lists the intensities of signal, which correspond to the abundance level for each of the specific siRNA species in the library. The data for basic bioinformatics was analyzed with Gene Set Enrichment Analysis (GSEA) and the Eukaryotic Subcellular Localization Database coupled with R. Analysis for gene networks and pathways was performed with Cytoscape v3.3.0 software with GeneMANIA (http://www.genemania.org) plugin according to the instruction manual.
Statistical analysis
All experiments were repeated at least three times, and quantitative data are presented as the mean ± SEM as calculated by GraphPad Prism6 software (Instat; GraphPad, San Diego, CA). Statistical significance was assessed by one-way ANOVA. Significant differences were identified by post hoc analysis using the Tukey–Kramer post hoc method for multiple comparisons. Assessments were considered significant with p < 0.05 and nonsignificant with p > 0.05.
Results
Genome-wide screen of mediators for DNA damage–induced neural cell death
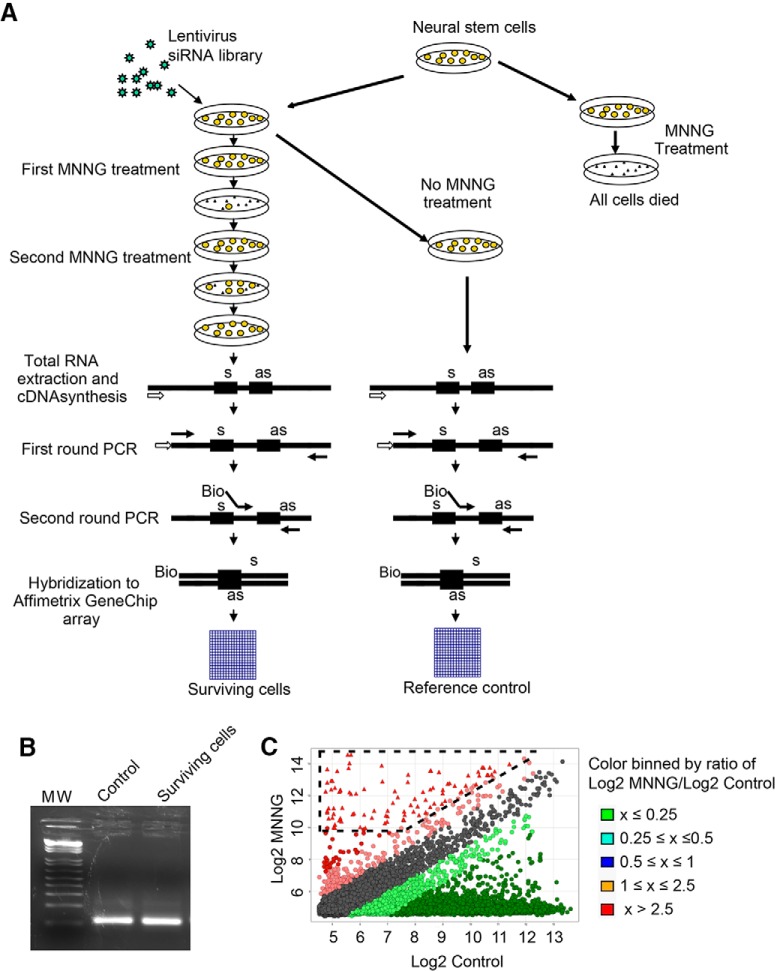
Mouse adult neural stem cells were transduced with the mouse 40K genome-wide FIV lentiviral siRNA library from System Biosciences (Fig. 1A). This lentiviral siRNA library contains 150,000 siRNA sequences that target 39,000 mouse mRNA transcripts with at least four different siRNA sequences for each target mRNA. We used a multiplicity of infection of ∼0.2 to ensure that each cell received a single lentiviral-encoded siRNA. The cell cultures were exposed to 500 μm MNNG treatment for 15 min, which in nontransduced cultures results in 100% lethality 24 h after exposure (data not shown). In FIV lentiviral siRNA–infected cultures, surviving neural stem cells were grown and expanded and subjected to a second treatment with MNNG (Fig. 1A). Surviving neural stem cells were grown and expanded, followed by extraction of mRNA. The siRNA templates were recovered by PCR using primers that recognize common sequences within the siRNA templates. A second PCR using nested primers with a biotin residue at the 5′ end and another containing a 5′-phosphate group were used to identify the siRNA inserts by hybridizing to the GeneChip Mouse Genome 430 2.0 Array (Fig. 1B,C; Huaux et al., 2005). After hybridization, the image of the processed chip was scanned, and data were subjected to quantile normalization and ANOVA. We selected 80 genes with signal intensities of more than 1000 and log2 ratios between surviving cells and reference controls (neural stem cells transduced with the library but not treated with MNNG) of more than 2.5 (Table 1). Of these 80 genes, 14 (17.5%) are known mediators of cell death, whereas the others have not been previously linked to DNA damage–induced cell death or other cell death pathways.
Figure 1.
siRNA screening for genes required for DNA damage–induced neural cell death. A, Schematic diagram of lentiviral siRNA library screening. B, Fragments of siRNA inserts were amplified from lentivirus-infected, MNNG-treated surviving neural stem cells and reference control cells (lentivirus infection but no MNNG treatment) by two rounds PCR. C, Scatterplot analysis of siRNA inserts enriched in neural stem cells treated by MNNG.
Table 1.
Identified genes required for DNA damage–induced neural cell death
| Category and gene symbol | Full gene name | GenBank accession no. | Control | MNNG | Log2 ratio |
|---|---|---|---|---|---|
| Cell death | |||||
| Dedd | Death effector domain-containing | AK006814 | 937 | 9842 | 3.39 |
| Eif2ak2 | Eukaryotic translation initiation factor 2 alpha kinase 2 | BE911144 | 3 | 7879 | 11.35 |
| Fasl | Fas ligand (TNF superfamily, member 6) | NM_010177 | 304 | 3395 | 3.48 |
| Trim39 | Tripartite motif-containing 39 | NM_024468 | 480 | 4511 | 3.23 |
| CCR3 | Chemokine (C-C motif) receptor 3 | NM_009914 | 770 | 4474 | 2.54 |
| NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | AV156640 | 94 | 1710 | 4.19 |
| GAS6 | Growth arrest-specific 6 | NM_019521 | 4 | 7793 | 10.93 |
| VAMP3 | Vesicle-associated membrane protein 3 | BE994144 | 578 | 7037 | 3.61 |
| NCOA1 | Nuclear receptor coactivator 1 | BE996469 | 1242 | 9787 | 2.98 |
| IFI202B | Interferon activated gene 202B | AV229143 | 343 | 12,047 | 5.13 |
| STAT3 | Signal transducer and activator of transcription 3 | AK004083 | 907 | 5156 | 2.51 |
| ABCE1 | ATP-binding cassette, sub-family E, member 1 | BG063303 | 344 | 2480 | 2.85 |
| CAPN2 | Calpain 2, (m/II) large subunit | NM_009794 | 1625 | 13,981 | 3.10 |
| Map3k12 | Mitogen activated protein kinase kinase kinase 12 | BB370469 | 1 | 1069 | 9.92 |
| Signal transduction | |||||
| Sgsm1 | Small G protein signaling modulator 1 | AK010756 | 11 | 5037 | 8.84 |
| Grm7 | Glutamate receptor, metabotropic 7 | BB539404 | 21 | 4723 | 7.81 |
| V1ra5 | Vomeronasal 1 receptor, A5 | NM_053220 | 225 | 3542 | 3.98 |
| Tbc1d8b | TBC1 domain family, member 8B | AK014817 | 1841 | 11,784 | 2.68 |
| Fgf23 | Fibroblast growth factor 23 | AF263536 | 1344 | 14,077 | 3.39 |
| Regulation of transcription | |||||
| Pcgf6 | Polycomb group ring finger 6 | BC016195 | 100 | 1650 | 4.04 |
| Bnc1 | Basonuclin 1 | U88064 | 200 | 2708 | 3.76 |
| Zbtb46 | Zinc finger and BTB domain containing 46 | AK016700 | 91 | 1220 | 3.74 |
| Med30 | Mediator complex subunit 30 | NM_027212 | 312 | 1818 | 2.54 |
| E2f5 | E2F transcription factor 5 | BC003220 | 243 | 2547 | 3.39 |
| Myt1 | Myelin transcription factor 1 | NM_008665 | 661 | 6287 | 3.25 |
| 1300003B13Rik | RIKEN cDNA 1300003B13 gene | AK004870 | 161 | 1427 | 3.15 |
| Snap91 | Synaptosomal-associated protein 91 | BG068132 | 1 | 2092 | 11.3 |
| Ccnl1 | Cyclin L1 | BB543556 | 1 | 2347 | 11.2 |
| Protein kinase activity | |||||
| Stk25 | Serine/threonine kinase 25 (yeast; Stk25) | BG068951 | 1 | 1499 | 10.55 |
| Gucy2e | Guanylate cyclase 2e | NM_008192 | 1211 | 11,563 | 3.26 |
| Tcp10c | T-complex protein-10 complete | AV257292 | 784 | 4904 | 2.65 |
| Stk22a | Serine/threonine kinase 22A | NM_009435 | 1 | 1966 | 10.94 |
| Oxidoreductase activity | |||||
| Mosc2 | MOCO sulfurase C-terminal domain containing 2 | NM_133684 | 11 | 5283 | 8.91 |
| Cyp2c65 | Cytochrome P450, family 2, subfamily c, polypeptide 65 | AK008688 | 283 | 4801 | 4.08 |
| Hydrolase activity | |||||
| Fbp2 | Fructose bisphosphatase 2 | NM_007994 | 3 | 5351 | 10.80 |
| Pgam5 | Phosphoglycerate mutase family member 5 | BC021317 | 4 | 1533 | 8.58 |
| Orc4l | Origin recognition complex, subunit 4-like | BB620704 | 27 | 1877 | 6.12 |
| Mtm1 | X-linked myotubular myopathy gene 1 | NM_019926 | 1395 | 13,912 | 3.32 |
| Ubiquitin-protein ligase activity | |||||
| March1 | Membrane-associated ring finger (C3HC4) 1 | AK013582 | 1 | 4915 | 12.26 |
| Transporter | |||||
| Ubl7 | Ubiquitin-like 7 (bone marrow stromal cell-derived) | BC016456 | 11 | 1513 | 7.1 |
| Sec15l1 | SEC15-like 1 (S. cerevisiae) | BC026859 | 471 | 2855 | 2.6 |
| Clic5 | Chloride intracellular channel 5 | BB028501 | 25 | 1805 | 6.17 |
| Slc8a1 | Solute carrier family 8, member 1 | BM123508 | 548 | 6850 | 3.64 |
| Clcc1 | Chloride channel CLIC-like 1 | BC003247 | 164 | 1391 | 3.08 |
| Protein complex | |||||
| Mrpl17 | Mitochondrial ribosomal protein L17 | BB343967 | 1 | 12,983 | 13.66 |
| Sfrs8 | Splicing factor, arginine/serine-rich 8 | BE688816 | 13 | 2396 | 7.53 |
| Clptm1 | Cleft lip and palate associated transmembrane protein 1 | NM_019649 | 137 | 3821 | 4.8 |
| Metabolism | |||||
| Tssk1 | Testis-specific serine kinase 1 | NM_009435 | 1 | 1966 | 10.94 |
| Ndufb2 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 | NM_026612 | 19 | 24,343 | 10.32 |
| Spopl | Speckle-type POZ protein-like | BM116703 | 436 | 11,419 | 4.71 |
| Mat2a | Methionine adenosyltransferase II, alpha | BB488978 | 1521 | 9954 | 2.71 |
| Unknown | |||||
| Tollip | Toll interacting protein | BB400304 | 10 | 1376 | 7.10 |
| Zmynd19 | Zinc finger, MYND domain containing 19 | NM_026021 | 217 | 2020 | 3.22 |
| ZNF706 | Zinc finger protein 706 | AA165749 | 184 | 7385 | 5.33 |
| Clec14a | C-type lectin domain family 14, member a | NM_025809 | 98 | 1301 | 3.73 |
| Dexi | Dexamethasone-induced transcript | NM_021428 | 6 | 5740 | 9.9 |
| Rg9mtd2 | RNA (guanine-9-) methyltransferase domain containing 2 | BG063557 | 30 | 4464 | 7.22 |
| Rutbc2 | RUN and TBC1 domain containing 2 | AK010756 | 11 | 5037 | 8.84 |
| 4833422M21RIK | RIKEN cDNA 4833422M21 gene | AK014752 | 36 | 1017 | 4.82 |
| 4930455J16RIk | RIKEN cDNA 4930455J16 gene | AK015470 | 43 | 5455 | 6.99 |
| A630057N01RIK | RIKEN cDNA A630057N01 gene | BB224397 | 881 | 6339 | 2.85 |
| AFTPH | Aftiphilin | BG071681 | 12 | 1323 | 6.78 |
| ANKRD13C | Ankyrin repeat domain 13C | AV104707 | 43 | 1760 | 5.36 |
| AU021889 | Expressed sequence AU021889 | BG068079 | 5 | 1846 | 8.53 |
| C2ORF77 | Chromosome 2 open reading frame 77 | BE282424 | 21 | 16,878 | 9.65 |
| D14Abb1e | DNA segment, chromosome 14, Abbott 1 expressed | BM209908 | 58 | 1537 | 4.73 |
| COMMD8 | COMM domain containing 8 | AV365904 | 2687 | 16,102 | 2.58 |
| DLX1AS | Distal-less homeobox 1, antisense | AI325350 | 168 | 1755 | 3.38 |
| FAM19A1 | Family with sequence similarity 19, member A1 | AW121549 | 84 | 7580 | 6.50 |
| Thrap6 | Thyroid hormone receptor associated protein 6 | NM_027212 | 312 | 1818 | 2.54 |
| MSL1 | Male-specific lethal 1 homolog | AW495537 | 8 | 11,950 | 10.54 |
| MTG1 | Mitochondrial GTPase 1 homolog (S. cerevisiae) | BC027306 | 23 | 4386 | 7.58 |
| Nalp9b | NACHT, LRR and PYD containing protein 9b | BG068754 | 6 | 1147 | 7.58 |
| Surf6 | Surfeit gene 6 | NM_009298 | 303 | 1964 | 2.70 |
| TMEM203 | Transmembrane protein 203 | BC022606 | 10 | 4837 | 8.92 |
| TSPYL3 | TSPY-like 3 (pseudogene) | BB308532 | 384 | 2908 | 2.92 |
| Btbd4 | BTB (POZ) domain containing 4 | AK016700 | 91 | 1220 | 3.74 |
| Tssc1 | Tumor suppressing subtransferable candidate 1 | BB082634 | 165 | 2032 | 3.62 |
| Pax6os1 | Pax6 opposite strand transcript 1 | BB255007 | 5 | 1008 | 7.66 |
| Ttc21b | Tetratricopeptide repeat domain 21B | AW554401 | 350 | 12,119 | 5.11 |
Validation of identified genes as mediators of cell death
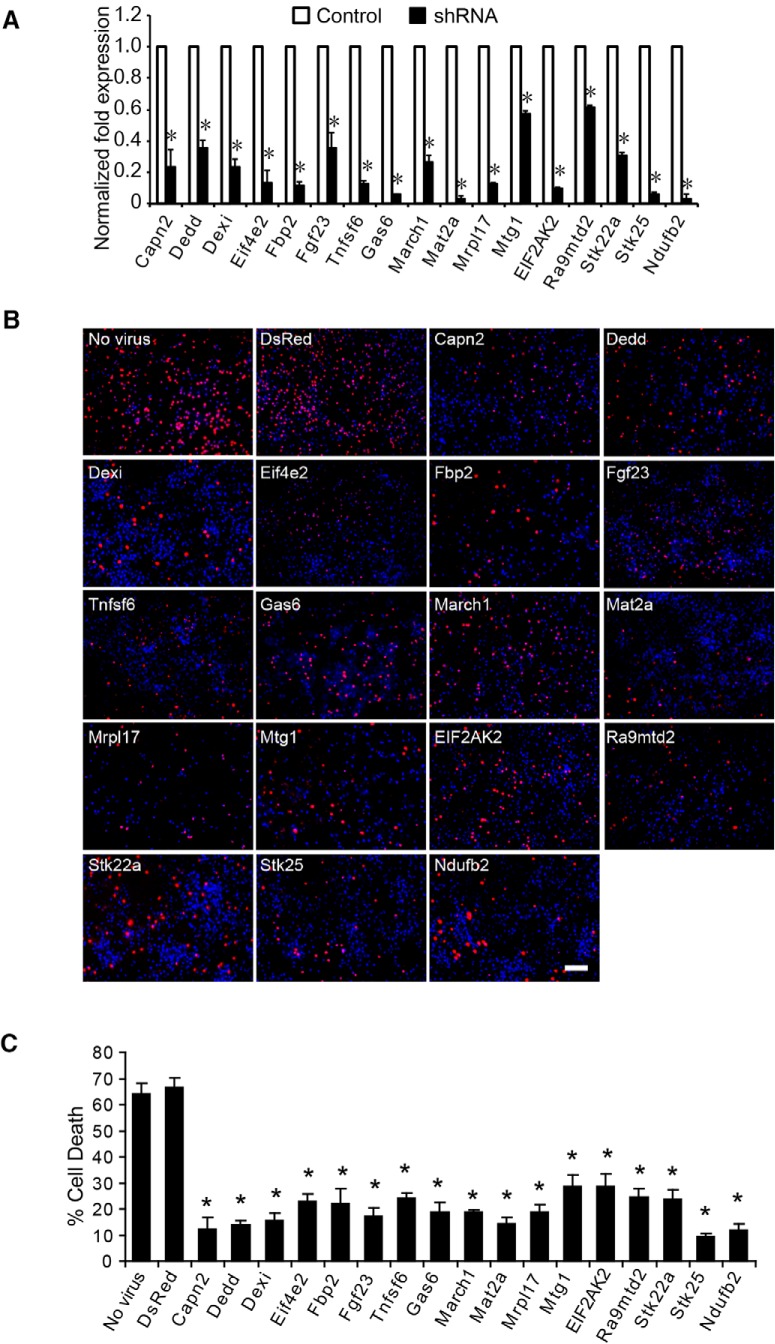
Because the goal of this screen was to identify genes that are unknown cell death effectors that could contribute to neuronal cell death, 17 genes were selected from the 80 identified genes for further analysis in primary mouse cortical neurons exposed to OGD experiments (stroke in a dish). Lentiviral siRNAs for these genes were individually prepared and used to infect primary cultured cortical neurons at 7 days in vitro. Four genes (Capn2, Dedd, FASLG, and EIF2AK2) that have been previously linked to cell death (Zhang and Ghosh, 2002; Scheuner et al., 2006; Lee et al., 2013; de Oliveira et al., 2014) served as positive controls, and an additional 13 genes that have not been previously linked to cell death were selected at random. Confirmation of gene knockdown was confirmed by reverse-transcription (RT)-PCR (Fig. 2A). Nontransduced neurons and neurons transduced with lentiviral siRNA directed against DsRed were used as controls. At day 7 after infection, neurons were subjected to 90 min of OGD, leading to 66.7 ± 3.5% (mean ± SEM) cell death in DsRed siRNA-transduced neurons 24 h after reperfusion with normal media (Fig. 2B,C). Cell death was significantly and substantially decreased in neurons infected by lentiviral siRNAs for the 17 individual genes (Fig. 2B,C). These data, taken together, suggest that the genome-wide siRNA library screen was effective in the identification of cell death signaling molecules.
Figure 2.
Validation of the siRNA library screening. A, Seventeen genes were randomly selected from the 80 identified in the screen, and expression was knocked down in primary cultured cortical neurons (7 days in vitro) by lentiviral siRNAs for the 17 genes. B, Representative images of neurons treated with lethal OGD (90 min) and stained with Hoechst 33342 and propidium iodide (PI). Dead neurons were scored as red cells with condensed or fragmented nuclei. Non-virus-infected neurons were used as viral control, and neurons infected with lentiviral siRNA for DsRed were used as siRNA control. Scale bar =100 µm. C, Quantification of cell death in B. Experiments were performed three times, and data represent the mean ± SEM. Statistical significance from DsRed siRNA control indicated at *p < 0.05; differences between multiple groups were evaluated by one-way ANOVA followed by the Tukey–Kramer post hoc test.
Biological network analysis of identified genes
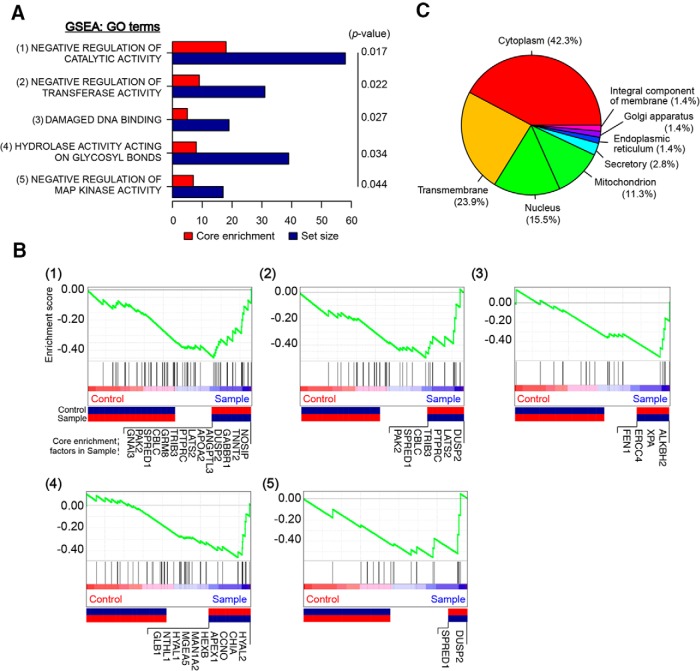
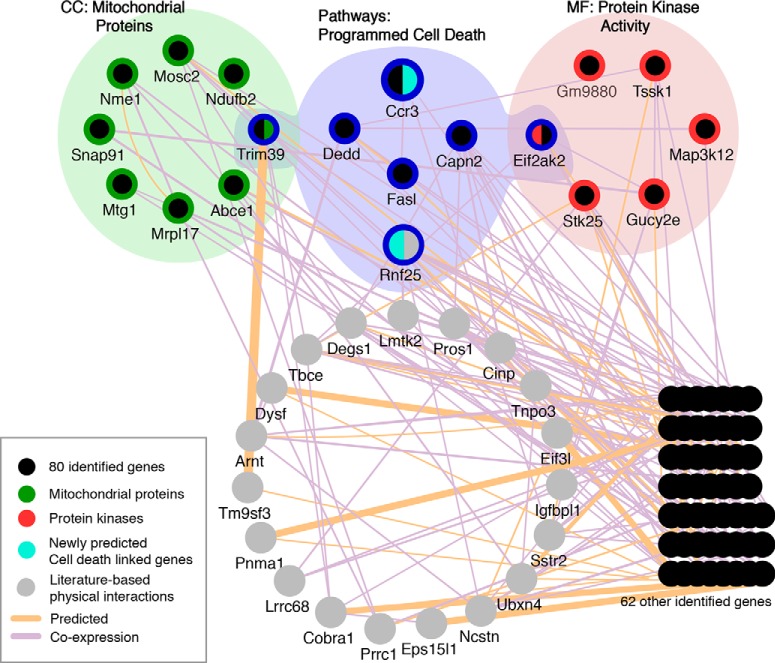
To ascertain which biological functions maybe associated with the gene set identified in the genome-wide siRNA screen, we used GSEA (Mootha et al., 2003; Subramanian et al., 2005). Five significant gene ontology (GO) terms from microarray were isolated to the DNA damage network (Fig. 3A). Of the core enrichment factors in each GO term identified in the siRNA screen, 37 could be classified into GSEA categories as core enrichment factors, and 71 genes from 80 processed signals into subcellular localization categories (Fig. 3B,C). The primary categories enriched in GSEA analysis mainly include the negative regulation of catalytic activity, negative regulation of transferase activity, damaged DNA binding, hydrolase activity acting on glycosyl bonds, and negative regulation of mitogen-activated protein kinase (MAPK) activity (Fig. 3A). We identified 14 core enrichment factors under DNA damage with negative regulation of catalytic activity, seven of which have negative regulation of transferase activity, four with damaged DNA binding, 10 with hydrolase activity acting on glycosyl bonds, and two with negative regulation of MAPK activity (Fig. 3B), suggesting that these biological processes are involved in the process of DNA damage–induced neural cell death. Of 71 subcellular localization categories, eight mediators are mitochondrial proteins (Ndufb2, Mrpl17, Mosc2, Snap91, Trim39, Nme1, Abce1, and Mtg1), reinforcing the known role of mitochondria in response to DNA damage injury and cell death (Fig. 3C).
Figure 3.
GSEA of identified genes required for neural cell death. The expression data are ranked in the order of differential expression. A, Forty-seven core enrichment factors from the siRNA library screen for DNA damage–induced cell death genes classified by GSEA analysis. B, Five individual plots (p-value <0.05) of the running sum for the gene sets in the ranked gene list. C, Subcellular localization was categorized using the top 80 genes from the siRNA library screen. Genes with no annotations were excluded from the analysis.
Canonical pathways in DNA damage–induced neural cell death
In response to DNA damage, a number of proteins called “DNA damage sensors,” such as protein kinases, are first activated and initiate the DNA damage signal networks, such as the activation of a variety of kinase substrates called “effectors,” leading to programmed cell death (Peng and Chen, 2003; Shiotani and Zou, 2009). To gain insights into the network and pathways of genes identified in this siRNA screen, we used Cytoscape v3.3.0 software with GeneMANIA (http://www.genemania.org) plugin (Shannon et al., 2003; Montojo et al., 2010). A search condition specifies criteria for choosing “predicted” and literature (evidence)-based “coexpressed” genes. Network integrated with GO term results was visually rearranged in Cytoscape (cellular components and molecular functions and pathways from GSEA and the Database for Annotation, Visualization, and Integrated Discovery). Screen results for 80 genes from the genome-wide siRNA library screen and 20 evidence-based predicted genes were mapped onto the network, and highly interconnected clusters containing factors identified from the screens were isolated, thus representing the programmed cell death clusters important for DNA damage–induced neural cell death (Fig. 4). From six identified protein kinase activity groups (EIF2AK2, Stk22a, Stk25, Ndufb2, Map3k12, Gucy2e; red colored) and eight mitochondrial proteins (colored green in Fig. 4), Eif2ak2 and Trim39 from each group are associated with five other predicted programmed cell death clusters (Ccr3, Dedd, Capn2, FASLG, and Rnf25; colored blue in Fig. 4). RNF25, a unique gene belonging to the programmed cell death cluster, was generated from the predicted/coexpression network (colored gray; Fig. 4). Apart from four validated genes in Figure 2B, Ccr3 and Trim39 from screening and RNF25 from prediction are key factors identified for programmed cell death pathways.
Figure 4.
Network analysis of identified genes required for DNA damage–induced neural cell death. Screen results for 80 genes from siRNA library screen and 20 evidence-based predicted genes were mapped onto the network, and highly interconnected clusters containing factors identified from the siRNA library screens were isolated, representing the programmed cell death clusters important for DNA damage–induced neural cell death. A network was initially constructed by Cytoscape v3.3.0 software with GeneMANIA plugin using limited database search for predicted and literature (evidence)-based coexpressed genes. The network was then visually rearranged with combination of GO term results [cellular components (CC) molecular functions (MF), and pathways] from the Database for Annotation, Visualization, and Integrated Discovery and GSEA.
CCR3 is a key mediator of neuronal cell death
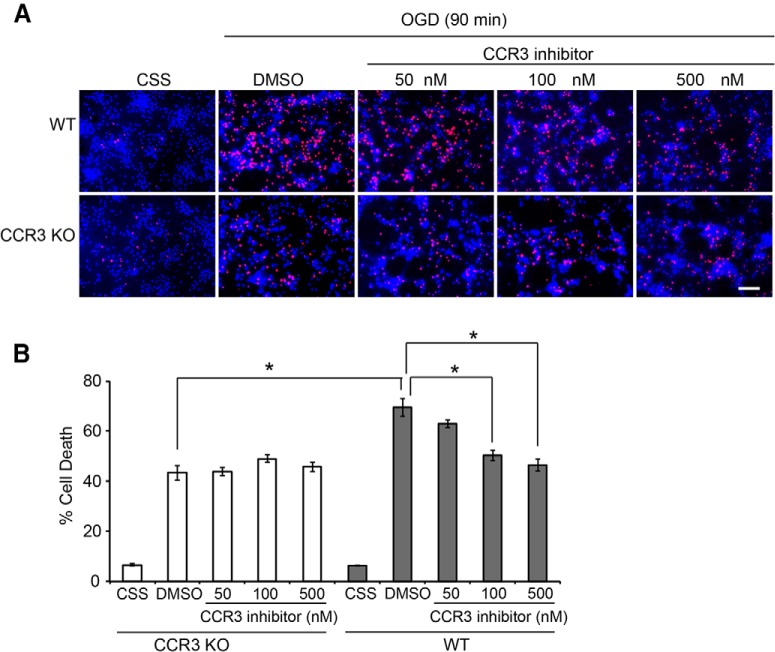
Of the many genes identified, CCR3 is interesting in that it has not been extensively studied in the nervous system. Because of its actions in inflammatory responses and allergic reactions (Willems and Ijzerman, 2010), however, small molecule inhibitors of CCR3 that are relatively selective and specific have been identified, some of which are currently in clinical trials (Bando et al., 2005; Zhang et al., 2005). To test whether CCR3 plays a role in neuronal injury, primary cortical cultures were generated from WT and CCR3 knockout (KO) mice. Cultures were exposed to 90 min of OGD in the presence or absence of the CCR3 inhibitor, SB328437 [N-(1-naphthalenylcarbonyl)-4-nitro-l-phenylalanine methyl ester; van der Meer et al., 2000], and cell viability was assessed 24 h later. WT cultures exhibited 69.56 ± 3.58% cell death that was markedly reduced by the CCR3 inhibitor at the concentrations of 100 nm (50.41 ± 2.05%) and 500 nm (46.49 ± 2.36%; Fig. 5A,B). Knockout of CCR3 also reduced cell death by 37.56%; it was not further reduced by the CCR3 inhibitor, demonstrating the selectivity of the inhibitor (Fig. 5A,B).
Figure 5.
CCR3 deletion or inhibition protects neurons against OGD-induced excitotoxicity. A, Primary cultured cortical neurons were isolated from CCR3 KO and WT embryos at embryonic day 15. Neurons (14 days in vitro) were pretreated with or without CCR3 inhibitor SB328437 at the indicated dose before being subjected to OGD treatment for 90 min. Twenty-four hours after OGD, neurons were stained with Hoechst 33342 and propidium iodide (PI). Dead neurons were scored as red cells with condensed or fragmented nuclei. Scale bar =100 µm. B, Quantification of cell death in different treatments. Experiments were performed at least three times. Data represent the mean ± SEM. * p<0.05, one-way ANOVA with Tukey–Kramer test.
Deletion of CCR3 protects against neuronal injury after stroke
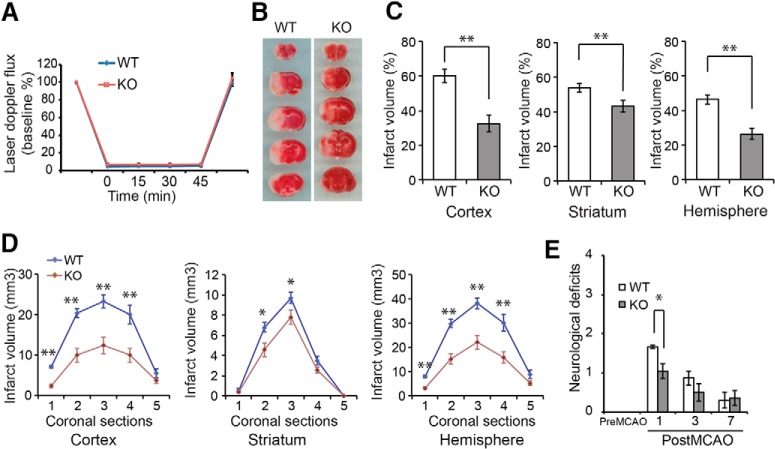
To extend these studies in vivo, WT and CCR3 KO mice were subjected to MCAO. Over the 45-min occlusion, cortical perfusion blood flow monitored by laser-Doppler flowmetry was reduced equivalently in WT (4.88 ± 0.15% of baseline, mean ± SEM) and CCR3 KO (6.74 ± 0.10% of baseline, mean ± SEM) mice. The reduction was stable throughout the occlusion period and recovered to pre-ischemic levels immediately on removal of the filament (Fig. 6A). Despite the similar intensity of the insult, infarct volume was reduced in the cortex and striatum and overall in the infarcted hemisphere by 46.12, 19.56, and 43.43% in CCR3 KO mice compared with WT littermates (Fig. 6B,A). Moreover, the reduction in infarct volume was not skewed to a particular brain region, but was uniformly reduced in CCR3 KO brains (Fig. 6D). Neurobehavioral activity was assessed by spontaneous activity in the open-field assay 24 h after MCAO. Similar to the infarct data, CCR3 KO mice have improved neurobehavioral scores compared with WT mice (Fig. 6E). These results, taken together, demonstrate that CCR3 is a mediator for neuronal cell death after ischemic insult.
Figure 6.
CCR3 deficiency protects against neuronal injury after stroke. A, Laser-Doppler flux measured over the lateral parietal cortex in the core of the ischemic region in CCR3 KO (n = 13) and WT (n = 9) mice. Values are means ± SEM, expressed as a percentage of pre-ischemic baseline values. B, Representative images of 2,3,5-triphenyltetrazolium chloride (TTC) staining of brain slices from CCR3 KO mice and WT littermate controls subjected to 60 min of MCAO. C, Quantification of infarct volumes in the cortex, hippocampus, and whole hemisphere after 60 min of MCAO in CCR3 KO mice and WT mice. Data are expressed as a percentage of the entire ischemic hemisphere and are the means ± SEM. *p < 0.05 from WT by Student’s t test. D, Quantification of infarct volumes among the five coronal levels (level 1 is most anterior) after 60 min of MCAO in CCR3 KO and WT mice. Data represent the mean ± SEM. *p < 0.05 and **p < 0.01 from WT by ANOVA with Tukey–Kramer post hoc test. E, Spontaneous neurobehavioral activity after MCAO was assessed on a scale of 0–4 (0, no neurological deficit; 4, severe neurological deficit) by the criteria described in Methods. Data represent the mean ± SEM, *p < 0.05 from WT by one-way ANOVA with Tukey–Kramer post hoc test.
Discussion
Neuronal cell death after injury or disease significantly impacts quality of life. Although some neuronal cell death pathways have been uncovered, because of the complexity of the brain it is not inconceivable that there remain additional cell death signaling networks to be discovered. DNA damage is an important contributor to the activation and propagation of neural cell death signaling events. DNA damage is a prominent feature in a number of acute and chronic neurological diseases including stroke, Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. In the present study, we identified 80 genes that participate in DNA damage–induced cell death by genome-wide screening using an siRNA library. Bioinformatic analysis suggests that these 80 genes are connected to several partially overlapping and interconnected pathways and protein complexes, including negative regulation of catalytic activity, negative regulation of transferase activity, damaged DNA binding, hydrolase activity acting on glycosyl bonds, and negative regulation of MAPK activity. The results provide new insight into neural cell death signaling pathways. Indeed, many of the genes and pathways identified in this screen have not been previously linked to DNA damage–induced cell death, suggesting that the networks that govern neuronal cell death encompass a broad range of cellular functions. The identification of uncharacterized novel genes in this functional screen will provide valuable cues for investigating their individual functions. Six genes encoding protein kinase activity were identified in this screen, consistent with the noted role of protein kinases as fast transducers for triggering the activation of their substrates and initiating cell death pathways (Peng and Chen, 2003; Shiotani and Zou, 2009). In this screen, eight mitochondrial mediators (Ndufb2, Mrpl17, Mosc2, Snap91, Trim39, Nme1, Abce1, and Mtg1) were identified that contribute to cell death, suggesting that both intrinsic and extrinsic mitochondria-mediated cell death signaling pathways play pivotal roles as well. This is consistent with the concept that mitochondria actively participate in neuronal cell death and are important contributors to the pathogenesis of prominent neurodegenerative diseases and psychiatric disorders (Mattson et al., 2008).
CCR3 is a member of the seven transmembrane G protein-coupled receptors that bind C-C type chemokines. CCR3 is activated by a large number of chemokine ligands with varied selectivity and potency, including CCL5, CCL7, CCL11, CCL13, CCL15, CCL24, CCL26, and CCL28 (Daugherty et al., 1996; Struyf et al., 2001; Willems and Ijzerman, 2010; White et al., 2013; Pease and Horuk, 2014). In the periphery, CCR3 plays significant pathogenic roles in the development of inflammation, allergic reactions, and lung fibrosis (Huaux et al., 2005). In the brain, CCR3 has also been identified in microglia, astrocytes, and neurons (van der Meer et al., 2000; Van Der Meer et al., 2001). Expression of CCR3 is elevated in the setting of human immunodeficiency virus infection (van der Meer et al., 2000; Van Der Meer et al., 2001), and elevated expression of CCR3 is observed in reactive microglia and astrocytes around the amyloid deposits in Alzheimer’s disease (Xia et al., 1998). After MCAO in mice, the expression of CCR3 is induced in neurons around the peri-infarct areas (Tokami et al., 2013). In this study, we found that CCR3 plays a crucial role in neuronal injury. CCR3 deletion or inhibition protects neurons from OGD-induced cytotoxicity in primary cortical cultures. How CCR3 is activated in cortical cultures is not known and requires further study. It may promote activation of a cell death cascade through its activation of the MAPK signaling pathway or phospholipase Cγ signaling (White et al., 2013). CCR3 KO mice also displayed significant reduction of infarct volume in the brain after MCAO. These data are consistent with our findings in cortical cultures for a neuronal role of CCR3. However, in vivo after MCAO, CCR3 will also be directly activated by elevated chemokines because of the inflammatory response (Perez-Alvarez and Wandosell, 2016). Indeed, the CCR3 ligand, CCL5, is rapidly induced in the brain after MCAO and thus would be able to activate CCR3 (Tokami et al., 2013). Interestingly, knockout of CCL5 has been shown to provide protection against MCAO in mice, although the mechanism under study was the regulation of microvascular dysfunction (Terao et al., 2008). It is possible that local production of CCL5 could also be the mechanism of activation of CCR3 in cortical cultures, which comprise both neurons and glia. Although there is strong evidence for inflammatory mediators expressed and acting within the CNS, in CCR3 KO mice we cannot exclude the possibility that protection is also afforded by inhibition of peripheral inflammatory responses. Taken together, our findings imply a role for CCR3 in neuronal cell death and cell death after ischemic injury in murine models. These initial findings support the need for additional investigations into the actions of CCR3 in neuronal demise to determine whether inhibiting CCR3 could provide a new avenue for the future treatment of ischemia injury or neurodegenerative disease.
In summary, siRNA library screening is a powerful and efficient method to globally identify signaling pathways relevant to neural cell death. Identification of the network of neural cell death mediators will greatly enhance our understanding of the molecular mechanisms of neural cell death and in the future may provide new therapeutic targets for injury to or diseases of the nervous system.
Acknowledgments
Acknowledgments: We thank Dr. Hongjun Song (Johns Hopkins University School of Medicine) for providing mouse neural stem cells.
Synthesis
The decision was a result of the Reviewing Editor Jun Chen and the peer reviewers coming together and discussing their recommendations until a consensus was reached. A fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision is listed below. The following reviewer(s) agreed to reveal their identity: Ruth Stetler
Both reviewers identified interesting aspects of the current study. In specific, the technique of high content genome-wide siRNA screen provides a useful tool to screen silenced genes under different conditions, with which the authors have identified novel players in cell death downstream of DNA damage in neurons. Based on the gene screening results, the authors attempted to confirm whether one of these critical proteins (CCR3) mediates neuronal ischemic injury, which is demonstrated to involve DNA damage, through use of both in vitro and in vivo models. The finding that knockdown or knockout out of CCR3 led to protection against OGD or transient focal ischemia helps to support the concept that CCR3 contributes to ischemic injury. Despite of the interesting findings, there are several issues about the study that should be addressed by the authors.
Major points:
1. The expression profiles and responses to the injury of neural stem cells should be different from the neurons. The authors used neural stem cells to screen the mediators for DNA damage-induced neuronal cell death in this paper. So were the identified genes specific mediators for neuronal death or just common mediators for cell death of different kinds of cells?
2. The immune cells including neutrophils and macrophages/microglia express CCR3. So CCR3-/- mice showed smaller infarction and less neurological deficits via reducing the neuronal death or regulating the inflammation after stroke? The authors should show some evidences to support their conclusions.
3. In the ischemic models, the authors use either pharmacological pretreatment to inhibit CCR3, or a global CCR3 knockout model followed by ischemic injury. The outcomes are acute cell survival/death (in vitro) or gross histology (TTC, in vivo). No analyses of DNA damage or specific morphology have been performed. This leads to two issues. First, the role of CCR3 in ischemic injury could be completely independent of its initiation downstream of DNA damage, since the paradigm inhibits CCR3 upstream of DNA damage, and thus decouples the MNNG experiments from the ischemia models. Second, a major issue in cell death protection has been repeatedly described in the literature, wherein inhibition of molecular events simply alters morphology or delays cell death. The authors rely on acute timepoints for histology, and do not perform cellular-based histological assays with the in vivo model.
4. The authors claimed that "CCR3 functions as a death receptor to trigger the neuronal cell death pathway during brain ischemia injury". What ligands of CCR3 would initiate this effect, the regular chemokines ligand of CCR3 or other specific ligands? The author should at least discuss the potential mechanisms underlying the roles of CCR3 in cell death.
Minor points:
1. The authors state in the methods the use of siRNA lentiviral transduction, but the results (p. 9) and figure 2A note shRNA. There is a nuance between the two terms - please amend for consistency or accuracy. Secondly, multiple grammatical errors are present throughout the bulk of the paper that should be corrected.
2. In the validation experiments (Fig 2), did lentiviral shRNA transfection per se affect neuronal viability?
3. Fig 1A, what is "s" or "as"?
References
- Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, West AB, Koehler RC, Poirier GG, Dawson TM, Dawson VL (2011) Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med 17:692–699. 10.1038/nm.2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando Y, Onuki R, Katayama T, Manabe T, Kudo T, Taira K, Tohyama M (2005) Double-strand RNA dependent protein kinase (PKR) is involved in the extrastriatal degeneration in Parkinson’s disease and Huntington’s disease. Neurochem Int 46:11–18. 10.1016/j.neuint.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Becker EB, Bonni A (2004) Cell cycle regulation of neuronal apoptosis in development and disease. Progress in Neurobiology 72:1–25. 10.1016/j.pneurobio.2003.12.005 [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D'Silva I, Poirier GG (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342: (Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Daugherty BL, Siciliano SJ, DeMartino JA, Malkowitz L, Sirotina A, Springer MS (1996) Cloning, expression, and characterization of the human eosinophil eotaxin receptor. J Exp Med 183:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira GP, Maximino JR, Maschietto M, Zanoteli E, Puga RD, Lima L, Carraro DM, Chadi G (2014) Early gene expression changes in skeletal muscle from SOD1(G93A) amyotrophic lateral sclerosis animal model. Cell Mol Neurobiol 34:451–462. 10.1007/s10571-014-0029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J (2008) Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135:1311–1323. 10.1016/j.cell.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaux F, Gharaee-Kermani M, Liu T, Morel V, McGarry B, Ullenbruch M, Kunkel SL, Wang J, Xing Z, Phan SH (2005) Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis. Am J Pathol 167:1485–1496. 10.1016/S0002-9440(10)61235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Li RC, Zhang J, Klaus JA, Kibler KK, Doré S, Koehler RC, Sapirstein A (2010) Cytosolic phospholipase A2 alpha amplifies early cyclooxygenase-2 expression, oxidative stress and MAP kinase phosphorylation after cerebral ischemia in mice. J Neuroinflammation 7:42. 10.1186/1742-2094-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, Yaspo ML, Himmelbauer H, Korn B, Neugebauer K, Pisabarro MT, Buchholz F (2004) An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432:1036–1040. 10.1038/nature03159 [DOI] [PubMed] [Google Scholar]
- Lee EH, Yum MS, Park SJ, Lee BH, Kim GH, Yoo HW, Ko TS (2013) Two cases of X-linked myotubular myopathy with novel MTM1 mutations. J Clin Neurol 9:57–60. 10.3988/jcn.2013.9.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A (2008) Mitochondria in neuroplasticity and neurological disorders. Neuron 60:748–766. 10.1016/j.neuron.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298. 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD (2010) GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26:2927–2928. 10.1093/bioinformatics/btq562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK,. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ (2004) A resource for large-scale RNA-interference-based screens in mammals. Nature 428:427–431. 10.1038/nature02370 [DOI] [PubMed] [Google Scholar]
- Pease JE, Horuk R (2014) Recent progress in the development of antagonists to the chemokine receptors CCR3 and CCR4. Expert Opin Drug Discov 9:467–483. 10.1517/17460441.2014.897324 [DOI] [PubMed] [Google Scholar]
- Peng A, Chen PL (2003) NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J Biol Chem 278:8873–8876. 10.1074/jbc.C300001200 [DOI] [PubMed] [Google Scholar]
- Perez-Alvarez MJ, Wandosell F (2016) Stroke and neuroinflamation: role of sexual hormones. Curr Pharm Des 22:1334–1349. [DOI] [PubMed] [Google Scholar]
- Scheuner D, Patel R, Wang F, Lee K, Kumar K, Wu J, Nilsson A, Karin M, Kaufman RJ (2006) Double-stranded RNA-dependent protein kinase phosphorylation of the alpha-subunit of eukaryotic translation initiation factor 2 mediates apoptosis. J Biol Chem 281:21458–21468. 10.1074/jbc.M603784200 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33:547–558. 10.1016/j.molcel.2009.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyf S, Menten P, Lenaerts JP, Put W, D'Haese A, De Clercq E, Schols D, Proost P, Van Damme J (2001) Diverging binding capacities of natural LD78beta isoforms of macrophage inflammatory protein-1alpha to the CC chemokine receptors 1, 3 and 5 affect their anti-HIV-1 activity and chemotactic potencies for neutrophils and eosinophils. Eur J Immunol 31:2170–2178. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao S, Yilmaz G, Stokes KY, Russell J, Ishikawa M, Kawase T, Granger DN (2008) Blood cell-derived RANTES mediates cerebral microvascular dysfunction, inflammation, and tissue injury after focal ischemia-reperfusion. Stroke 39:2560–2570. 10.1161/STROKEAHA.107.513150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokami H, Ago T, Sugimori H, Kuroda J, Awano H, Suzuki K, Kiyohara Y, Kamouchi M, Kitazono T, REBIOS Investigators (2013) RANTES has a potential to play a neuroprotective role in an autocrine/paracrine manner after ischemic stroke. Brain Res 1517:122–132. 10.1016/j.brainres.2013.04.022 [DOI] [PubMed] [Google Scholar]
- van der Meer P, Ulrich AM, González-Scarano F, Lavi E (2000) Immunohistochemical analysis of CCR2, CCR3, CCR5, and CXCR4 in the human brain: potential mechanisms for HIV dementia. Exp Mol Pathol 69:192–201. 10.1006/exmp.2000.2336 [DOI] [PubMed] [Google Scholar]
- Van Der Meer P, Goldberg SH, Fung KM, Sharer LR, González-Scarano F, Lavi E (2001) Expression pattern of CXCR3, CXCR4, and CCR3 chemokine receptors in the developing human brain. J Neuropathol Exp Neurol 60:25–32. 10.1093/jnen/60.1.25 [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim NS, Haince J-F, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM (2011) Poly (ADP-ribose) (PAR) Binding to Apoptosis-Inducing Factor Is Critical For PAR Polymerase-1-Dependent Cell Death (Parthanatos). Sci. Signal Apr 5;4(167):ra20 10.1126/scisignal.2000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White GE, Iqbal AJ, Greaves DR (2013) CC chemokine receptors and chronic inflammation–therapeutic opportunities and pharmacological challenges. Pharmacol Rev 65:47–89. 10.1124/pr.111.005074 [DOI] [PubMed] [Google Scholar]
- Willems LI, Ijzerman AP (2010) Small molecule antagonists for chemokine CCR3 receptors. Med Res Rev 30:778–817. 10.1002/med.20181 [DOI] [PubMed] [Google Scholar]
- Xia MQ, Qin SX, Wu LJ, Mackay CR, Hyman BT (1998) Immunohistochemical study of the beta-chemokine receptors CCR3 and CCR5 and their ligands in normal and Alzheimer’s disease brains. Am J Pathol 153:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ghosh S (2002) Negative regulation of Toll-like receptor-mediated signaling by Tollip. J Biol Chem 277:7059–7065. 10.1074/jbc.M109537200 [DOI] [PubMed] [Google Scholar]
- Zhang QP, Xie LK, Zhang LJ, Tan JQ (2005) Apoptosis in human germinal centre B cells by means of CC chemokine receptor 3 expression induced by interleukin-2 and interleukin-4. Chin Med J (Engl) 118:665–670. [PubMed] [Google Scholar]
- Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB (2004) Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev 18:1272–1282. 10.1101/gad.1199904 [DOI] [PMC free article] [PubMed] [Google Scholar]