Abstract
Background
Formalin-fixed paraffin-embedded (FFPE) samples can be DNA-extracted and used for human papillomavirus (HPV) genotyping. The xylene-based gold standard for extracting FFPE samples is laborious, suboptimal and involves health hazards for the personnel involved.
Objectives
To compare extraction with the standard xylene method to a xylene-free method used in an HPV LabNet Global Reference Laboratory at the Centers for Disease Control (CDC); based on a commercial method with an extra heating step.
Study design
Fifty FFPE samples were randomly selected from a national audit of all cervical cancer cases diagnosed in Sweden during 10 years. For each case-block, a blank-block was sectioned, as a control for contamination. For xylene extraction, the standard WHO Laboratory Manual protocol was used. For the CDC method, the manufacturers’ protocol was followed except for an extra heating step, 120°C for 20 minutes. Samples were extracted and tested in parallel with β-globin real-time PCR, HPV16 real-time PCR and HPV typing using modified general primers (MGP)-PCR and Luminex assays.
Results
For a valid result the blank-block had to be betaglobin-negative in all tests and the case-block positive for beta-globin. Overall, detection was improved with the heating method and the amount of HPV-positive samples increased from 70% to 86% (p=0.039). For all samples where HPV type concordance could be evaluated, there was 100% type concordance.
Conclusions
A xylene-free and robust extraction method for HPV-DNA typing in FFPE material is currently in great demand. Our proposed standardized protocol appears to be generally useful.
Keywords: Human papillomavirus (HPV), Formalin-Fixed Paraffin-Embedded (FFPE), DNA-extraction, Genotyping, xylene
Background
Human Papillomavirus (HPV) infections are known to be a major cause of cervical cancer (1). More than 200 types of HPV have been fully characterized (2). HPV types are divided in high-risk and low-risk viruses, where the high-risk types, notably HPV16 and HPV18, predominate in HPV-related cancers (1). In light of current developments in HPV-based screening and HPV vaccination, it is important to develop a robust method for HPV genotyping and detection in cervical tissue, to be able to analyze the HPV-type-specific disease burden and to monitor the effectiveness of HPV prevention programs (3).
Formalin-fixed paraffin-embedding (FFPE) of tissues is suitable for long-term storage of cancer tissue samples and has been globally used in pathology for more than a century. FFPE cancer tissue has often been used for analyzing presence and type of HPV in cancers, for example in order to determine the HPV type-specific burden of disease in various regions of the world and over time (4). However, the analysis of FFPE specimens includes several potential problems including DNA cross-linking, DNA fragmentation and the presence of PCR inhibitors (4, 5). The goldstandard method for extracting these samples before HPV testing is xylene-based extraction (6). However, xylene-based extraction is laborious, known to result in suboptimal sensitivity and also involves health hazards for the personnel involved. Recently, a method combining heat treatment and a commercially available DNA extraction kit was evaluated compared to xylene-based extraction, showing higher DNA yield and increased sensitivity for HPV testing (5) We wished to further evaluate and investigate the robustness of this method.
Objectives
The aim of this study was to evaluate a xylene-free method for the extraction of HPV-DNA in FFPE samples. If found robust, this method could become a new standard method for FFPE extraction before HPV genotyping. We thus compared the xylene-based gold standard method to a commercial column-based extraction with an extra heating step (5).
Study design
FFPE samples
Fifty FFPE-samples from patients with cervical cancer were randomly selected from a national case-control audit encompassing all invasive cervical and unspecified uterine cancers diagnosed from 2002 until 2011 in Sweden, the Advancing Cervical Cancer Eradication Strategies, ACCES, study. The diagnostic slides and the formalin-fixed paraffin-embedded (FFPE) tissue blocks were collected from different pathology biobanks throughout Sweden. Here, we selected fifty samples from one biobank, Gävleborg. The diagnostic slides were re-reviewed by an experienced histopathologist and only tissues where presence of cervical cancer could be verified were retained in this study.
Sectioning
All FFPE blocks were sent to a commercial, accredited laboratory (HistoCenter, Inc., Gothenburg, Sweden) for sectioning. In-between each case-block, a paraffin blank-block was sectioned, as a control for contamination. Every case required new gloves and a new knife. The blank-block was sectioned first. Four 5 μm sections were transferred to a 1,5 mL screw-cap Eppendorf tube using a new, clean, toothpick. The case-block was mounted on the microtome and sectioned with the same knife. Each case was sectioned in six 5 μm sections. The first and last section were set aside for histological review and two times two 5 μm sections were put in two tubes, one tube per extraction method, for HPV testing. The slides for histology were stained with hematoxylin-eosin. The tubes were marked with the blocks specific lab number, B (blank-block) and C (case-block). The slides were marked in the same way, with the specific lab number followed by letters F (first) and L (last). After each case the knife was removed and the microtome was cleaned with DNAZap (Applied Biosystems). Specimen size was determined in millimeters, using a ruler, with size in square millimeters being the product of length and width. Although all cases included in the study had been verified to contain tumor tissue, sections from HPV-negative tumors were re-reviewed a second time and still found to contain tumor tissue.
DNA extraction
DNA was extracted from the sectioned blocks using two different extraction methods: 1) A method developed at CDC using the Qiagen blood and tissue kit with an extra heating step (5) and 2) the xylene-based gold standard method (6). The extra heating step has been reported to result in an 8.2 fold increase in PCR-amplifiable cellular DNA and a decrease in inadequate results (5.3%, down from 19.3%) (5).
One tube from each FFPE sample was extracted with xylene. The paraffin was removed by incubation with 1 mL xylene in 50°C for 30 min followed by vortexing and centrifugation 3000 × g, 10 min, the supernatant was removed with a sterile transfer pipette. This step was repeated once. After removing the xylene the samples was washed twice with pure ethanol and air dried. The air dried pellet was incubated with 100 μL Digestion buffer (50 mM Tris HCl,1 mM EDTA, pH 8,5) with Proteinase K (50 mg/mL) in 37°C for about 24 h. After incubation the samples were boiled in 100°C for 10 min to inactivate Proteinase K.
The other tube from each FFPE sample was extracted with the Qiagen/heating method. 180 μL ATL buffer was added to the tube and high-heat treated in 120°C for 20 min to melt the paraffin. Within the 5 first minutes the tubes were mixed by tapping the tube to make sure that all of the paraffin was under the surface. After 20 minutes the samples were incubated at room temperature for 3 minutes, followed by a quick centrifugation. 20 μL proteinase K was added, briefly vortex and incubated in 65°C for 16 hours (the manufacturer’s protocol describes 56°C, but a previous study found 65°C to be better (5)). The tubes where quickly centrifuged. A solution of 200 μL buffer AL and 200 μL ethanol, per sample, was prepared. After adding 400 μL of the prepared solution immediate vortex was needed. The mixture was added to a DNeasy Mini spin column and centrifuged 1 min at 8000 rpm. The following steps were performed according to the manufacturer’s protocol except for the volume in the elution step that was changed to 100 μL AE buffer. In total 200 extractions were performed.
Quantitative real-time PCR for beta-globin and HPV16
All samples were tested for both beta-globin and HPV16 in real-time PCR, as previously described (7, 8). Samples from the two extraction methods were tested in parallel in both beta-globin and HPV16, undiluted and diluted 1/10 in water.
Briefly, one μL extracted sample was used in the PCR of a total volume of 25 μL. A standard program was used, 50°C, 2.0 min, 95°C, 10.0 min; 95°C 15.0 sec, 60°C 1.0 min for 40 cycles. The following primers and probe were used for HPV16, HPV16 E7 M forward 5′AGCTCAGAGGAGGAGGATGAA 3′, HPV16 E7 M reverse 5′ GGTTACAATATTGTAATGGGCTC 3′ and HPV16 E7 M probe 5′-FAM-CCAGCTGGACAAGCAGAACCGG-TAMRA-3′.
All samples were analyzed in parallel for quantitative HPV16 real-time PCR, quantitative beta-globin real-time PCR and HPV-genotyping. Samples from each individual case-block, extracted with the two different extraction methods, were analyzed in parallel in the same run in all assays with the same standard curve and positive controls. One μL extracted sample was used in all the assays to avoid PCR inhibitors.
For a valid result, the blank-block had to be negative in all three tests and the case-block positive for beta-globin, where a measurement of one copy was considered positive. FFPE samples are known to contain PCR inhibitors and to avoid this, all 50 samples were tested first undiluted and then diluted 1 to 10.
HPV genotyping
HPV detection and genotyping of HPV was done using modified general primers, MGP-PCR and Luminex as previously described (9). Briefly, one μL was used as input in the MGP-PCR in a total volume of 25 μL. The probes used in the multiplex Luminex were, HPV6, 11, 16, 18, 18.6650 G variant, 26, 30, 31, 33.2, 35, 35.6624:A variant, 39, 40, 42, 43, 45, 51, 52, 53, 54, 56, 58.1, 59, 61, 66, 67, 68 prototype (discovered by Gerard Orth, also called HPV68a), 68ME.1 (also called HPV68b), 69, 70, 73, 74:911664, 81, 82, 83, 86, 87, 89, 90, 91, universal1 and universal2 (for probes, see Table 1). Samples positive for probe universal1 and/or universal2 were planned to be sequenced to obtain the correct HPV-type.
Table 1.
Probes used for the different HPV genotypes in Luminex assay.
| Probes | Sequence |
|---|---|
| HPV 18.6650 G | 5′amino-modified C12- TGC GTC TAC ACA GTC TCC T 3′ |
| HPV 26 | 5′amino-modified C12- GTA CAT TAT CTG CAG CAT C 3′ |
| HPV 30 | 5′amino-modified C12- TCT GCA ACC ACA CAA ACG TT 3′ |
| HPV 33.2 | 5′amino-modified C12- ACA AGT AAC TAG TGA CAG TAC 3′ |
| HPV 35.6624A | 5′amino-modified C12- CTG CTG TGT CTA CTA GTG A 3′ |
| HPV 40 | 5′amino-modified C12- AGT CCC CCA CAC CAA CC 3′ |
| HPV 53 | 5′amino-modified C12- TGT CTA CAT ATA ATT CAA AGC 3′ |
| HPV 54 | 5′amino-modified C12- CAC GCA GGA TAG CTT TAA T 3′ |
| HPV 58.1 | 5′amino-modified C12- CTA ATA TGA CAT TAT GCA CTG AA 3′ |
| HPV 61 | 5′amino-modified C12- CCC TGT ATC TGA ATA TAA AGC 3′ |
| HPV 67 | 5′amino-modified C12- CTA CAT ACA AAA ATG AAA AC 3′ |
| HPV 68 (Orth) | 5′amino-modified C12- GCT GTG TAT GAT TCT AAT AAA T 3′ |
| HPV 68 ME.1 | 5′amino-modified C12- CCA ATT TTA CTT TGT CTA CTA CTA C |
| HPV 69 | 5′amino-modified C12- CAT CTG CCA CTT TTA AAC C 3′ |
| HPV 74 | 5′amino-modified C12- CAG ACT ACA AAC AAT ACA TG 3′ |
| HPV 81 | 5′amino-modified C12- GCT ACA TCT GCT GCT GC 3′ |
| HPV 83 | 5′amino-modified C12- TGC TGC TAC ACA GGC TAA 3′ |
| HPV 86 | 5′amino-modified C12- ATT AGT GCC GCT ACC CAG AA 3′ |
| HPV 87 | 5′amino-modified C12- CCA CTG AAT ATG ACC CCA 3′ |
| HPV 89 | 5′amino-modified C12- AAT ACA GTT CTA CAC GCT 3′ |
| HPV 90 | 5′amino-modified C12- CAC ATA CAA GGC TTC CAA TT 3′ |
| HPV 91 | 5′amino-modified C12- TAC CTA CTA CAT ATG ACA AC 3′ |
| Universal 2 | 5′amino-modified C12-GMC AYR CAG ARG AAT ATG A 3′ |
Statistics
Descriptive data on number and proportion of betaglobin and HPV-positive samples for each extraction method were obtained. McNemar’s test was used for paired categorical data, and paired t-tests were used for paired measurements of continuous data. Mann-Whitney-Wilcoxon test was used for the comparison of tumor size and amplifiability..
All tests were two-tailed and a P-value of <0.05 was considered statistically significant.
Results
One case-block out of 50 extracted with the Qiagen/heating was negative for undiluted beta-globin compared to 9/50 case-blocks extracted with the xylene method (p=0.008). All the 50 case-blocks were valid with both extraction methods after 1/10 dilution. Requirement for dilution did correlate with size of tumor, with positivity only after dilution being found more often in large tissue specimens (p=0.015).
In the HPV16 real-time PCR, one Qiagen/heating-extracted case-block became positive after dilution, compared to 4 case-blocks extracted with xylene. Two of the cases extracted with the Qiagen/heating method and that were found HPV16-positive had xylene-extracted sections that remained negative in HPV16 real-time PCR even after dilution.
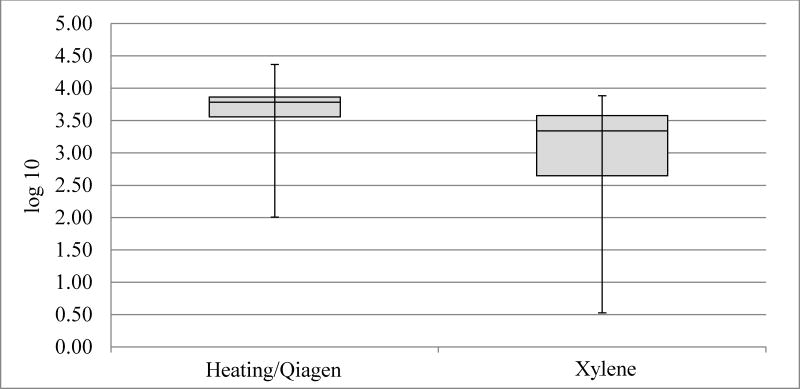
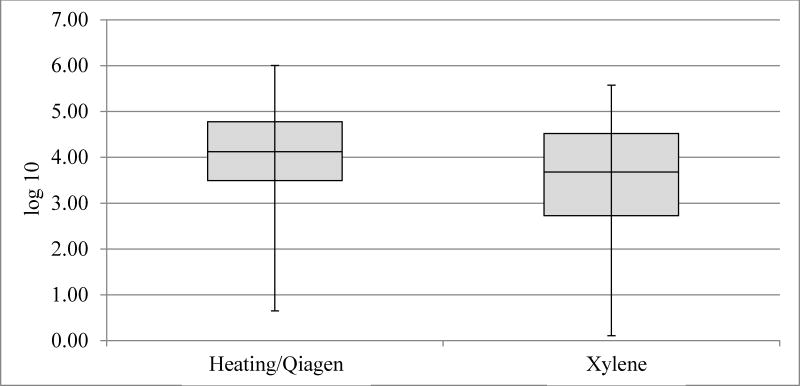
HPV16 real-time PCR and genotyping for HPV16 with Luminex had a 96–98% concordance for the two extraction methods (data not shown). Mean undiluted copy numbers for both beta-globin and HPV16 were about twice as high for Qiagen/heating-extracted case-blocks compared to xylene extracted case-blocks; HPV16 Qiagen/heating extraction method resulted in 104273 copies compared to 50133 copies with xylene extraction. Beta-globin Qiagen/heating extraction gave 5525 copies vs 2554 copies with the xylene extraction, (p<0.0001 and p=0.036, respectively; Figure 1 and Figure 2). All HPV-negative specimens were positive for Beta-globin. HPV negativity tended to associate with large tumors, albeit not significantly so (p=0.1).
Figure 1.
Box plot for betaglobin count in log10 (copies/microliter). Inadequate samples/pairs removed (n=9).
Figure 2.
Box plot for HPV16 copy number in log10(copies/microliter). Inadequate samples/pairs removed (n=5).
The most common HPV-type found in the samples extracted by the Qiagen/heating method was HPV16 (48%). The second most common type was HPV18 (16%), followed by HPV59 (6%), (Table 2). 10/50 samples were HPV positive with the Qiagen/heating method but negative with xylene extraction; 2/50 xylene-extracted samples were HPV positive but negative with Qiagen/heating extraction. 5/50 (10%) of the xylene-extracted case-blocks had to be diluted to get a positive HPV result compared to 0/50 of the Qiagen/heating-extracted (p=0.06). Overall, detection was improved with the Qiagen/heating method and the amount of HPV-positive samples increased from 70% to 86% (35/50 samples extracted with xylene versus 43/50 samples extracted with the Qiagen/heating method, p=0.039). Requirement for dilution did correlate with size of tumor (p=0.015). HPV negativity tended to associate with large tumors, albeit not significantly so (p=0.1).
Table 2.
Number (n) and proportion of positivity for HPV genotypes (as well as HPV negative samples) in the 50 FFPE samples extracted.
| Qiagen/heating method (Positives) | Xylene method (Positives) | |||
|---|---|---|---|---|
|
| ||||
| HPV type | Number | % | Number | % |
| 6 | 1 | 2% | 1 | 2% |
| 16 | 23 | 46% | 22 | 44% |
| 18 | 8 | 16% | 5 | 10% |
| 33 | 2 | 4% | 2 | 4% |
| 35 | 1 | 2% | 0 | 0% |
| 39 | 1 | 2% | 1 | 2% |
| 45 | 2 | 4% | 2 | 4% |
| 52 | 1 | 2% | 1 | 2% |
| 56 | 1 | 2% | 1 | 2% |
| 59 | 2 | 4% | 0 | 0% |
| Multiple types | 1 | 2% | 0 | 0% |
| HPV Negative | 7 | 14% | 15 | 30% |
|
| ||||
| Total | 50 | 100% | 50 | 100% |
Discussion
In a systematic comparison of two extraction protocols designed for FFPE tissue; standard xylene-based and a method using Qiagen columns augmented with a heat treatment, we find that the Qiagen/heating extraction method was superior in all comparisons made, along with the added benefit of a less labor-intensive method and fewer health hazards due to the fact that no use of xylene is required. The Qiagen kit is faster to work with and requires less handling after extraction, making it more time efficient. The xylene method requires more work (including dilution). Hazardous waste is produced, which increases the waste handling costs. Overall, the cost of the xylene method when considering consumables only was 0,32 USD and for the new method 3,34 USD. However, when also considering the technician time required (valued at 24 USD/hour), the xylene method cost 1,4 USD/sample and the new method 0,9 USD/sample.
It has been reported that the new method can be used in 96 well plates, amenable to automation (5). However, we did not use 96 well plates in this paper as minimizing the risk for contamination was a prime concern.
The prime focus of this paper was to investigate robustness and transferability using the key type of tissue (cervical cancer) of interest for HPV detection. The original method description (5) included tissues other than cervical. Automated use of the new method for non-cervical tissues has also been reported (10).
Although the method per se has been published before (5), this report provides an independent confirmation of the method improvement and shows that the method is robust and can readily be transferred to other laboratories in other countries. This robust and improved extraction protocol should be of use in several aspects of monitoring of HPV-based prevention of cervical cancer, for example impact of HPV vaccination.
With the xylene method, we found only 70% HPV-positive specimens, which increased to 86% with the new method. While it is generally accepted that cervical cancer “should” be HPV positive, studies using formalin-fixed paraffin embedded tissues typically report that a significant minority of samples are negative (11). Proposed explanations include cross-linked and degraded DNA, poor preservation, presence of inhibitors, as well misclassification of primary site (confusion between endometrial and endocervical origin), presence of HPV types not included in the assay, loss or variant changes in the region amplified and rare true negative tumors (6, 11). The detection of HPV is increased when very short amplicons are used, but even with this additional testing, in large systematic studies, not all cervical cancers have detectable HPV (11). FFPE material is becoming widely used in molecular studies and xylene-based extraction has since long been the gold standard despite its disadvantages. Other extraction methods have been compared to xylene but not with as satisfying results as the Qiagen/heating method (4, 12, 13). Ours was a stringently designed validation study where, in order to maximize constancy of other study variables and generalizability of results, all 50 case-blocks in the extraction method comparison were randomly collected from one biobank in the national study audit. All blocks were sectioned at the same accredited laboratory. Furthermore, the same experienced pathologist re-reviewed all slides to ensure that representative tumor material was present, before initializing DNA extraction protocols. Then, all tests were run in parallel in exactly the same fashion, using the same standard curves and dilution protocols such that the only differing factor was the extraction method used.
The fact that the Qiagen/heating method does not include xylene is an improvement for both the personnel and the environment. The purification step in the Qiagen/heating method also leads to fewer inhibitors in the extracted DNA, resulting in fewer steps in the HPV genotyping analysis. A xylene-free and robust extraction method for HPV-DNA typing in FFPE material currently is in great demand. Our proposed standard method appears to be generally useful. Of note, the CDC laboratory has transitioned the Qiagen/heating method to an automated extraction platform. The high temperature dewaxing, along with overnight lysis, appear to make the biggest difference in extraction efficiency. High temperatures have been used to disrupt formalin cross-links, and this may contribute to the improved yield. (14)
Acknowledgments
Funding
This study was supported by the Swedish foundation for strategic research.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.”
Competing interest
None.
Ethical approval
Ethical approval for this study was granted by the Regional ethical review board of Stockholm, Sweden, Dnr: 2011/1026-31/4.
References
- 1.International Agency for Research on Cancer. Monographs. Lyon, France: International Agency for Research on Cancer; 2012. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [PMC free article] [PubMed] [Google Scholar]
- 2.Bzhalava D, Eklund C, Dillner J. International standardization and classification of human papillomavirus types. Virology. 2015;476:341–4. doi: 10.1016/j.virol.2014.12.028. Epub 2015/01/13. [DOI] [PubMed] [Google Scholar]
- 3.Soderlund-Strand A, Uhnoo I, Dillner J. Change in population prevalences of human papillomavirus after initiation of vaccination: the high-throughput HPV monitoring study. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23(12):2757–64. doi: 10.1158/1055-9965.EPI-14-0687. Epub 2014/11/09. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez-Aldana A, Martinez JW, Sepulveda-Arias JC. Comparison of five protocols to extract DNA from paraffin-embedded tissues for the detection of human papillomavirus. Pathology, research and practice. 2015;211(2):150–5. doi: 10.1016/j.prp.2014.10.011. Epub 2014/12/03. [DOI] [PubMed] [Google Scholar]
- 5.Steinau M, Patel SS, Unger ER. Efficient DNA extraction for HPV genotyping in formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics: JMD. 2011;13(4):377–81. doi: 10.1016/j.jmoldx.2011.03.007. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillner J, Unger ER. WHO Human Papillomavirus Laboratory Manual. 2009. [Google Scholar]
- 7.Sundstrom K, Ploner A, Dahlstrom LA, Palmgren J, Dillner J, Adami HO, et al. Prospective study of HPV16 viral load and risk of in situ and invasive squamous cervical cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013;22(1):150–8. doi: 10.1158/1055-9965.EPI-12-0953-T. Epub 2012/11/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science (New York, NY) 1985;230(4732):1350–4. doi: 10.1126/science.2999980. Epub 1985/12/20. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. Bead-based multiplex genotyping of human papillomaviruses. Journal of clinical microbiology. 2006;44(2):504–12. doi: 10.1128/JCM.44.2.504-512.2006. Epub 2006/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saraiya M, Unger ER, Thompson TD, Lynch CF, Hernandez BY, Lyu CW, et al. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. Journal of the National Cancer Institute. 2015;107(6):djv086. doi: 10.1093/jnci/djv086. Epub 2015/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopenhayn C, Christian A, Christian WJ, Watson M, Unger ER, Lynch CF, et al. Prevalence of human papillomavirus types in invasive cervical cancers from 7 US cancer registries before vaccine introduction. Journal of lower genital tract disease. 2014;18(2):182–9. doi: 10.1097/LGT.0b013e3182a577c7. Epub 2014/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janecka A, Adamczyk A, Gasinska A. Comparison of eight commercially available kits for DNA extraction from formalin-fixed paraffin-embedded tissues. Analytical biochemistry. 2015;476:8–10. doi: 10.1016/j.ab.2015.01.019. Epub 2015/02/03. [DOI] [PubMed] [Google Scholar]
- 13.Rabelo-Goncalves E, Roesler B, Guardia AC, Milan A, Hara N, Escanhoela C, et al. Evaluation of five DNA extraction methods for detection of H. pylori in formalin-fixed paraffin-embedded (FFPE) liver tissue from patients with hepatocellular carcinoma. Pathology, research and practice. 2014;210(3):142–6. doi: 10.1016/j.prp.2013.11.003. Epub 2013/12/21. [DOI] [PubMed] [Google Scholar]
- 14.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y, et al. DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2002;50(8):1005–11. doi: 10.1177/002215540205000802. Epub 2002/07/23. [DOI] [PubMed] [Google Scholar]