Changes in the microbiome early in life may affect host physiology across the life span. Early life interaction between host and luminal microbes was investigated using a mouse model in which germ-free mice were transfaunated with fecal lysates from human preterm infants. Our data demonstrate that microbial communities affect differentiation of intestinal epithelial cell lineages, which may lead to significant effects on developmental, defensive, and physiological processes of the gastrointestinal epithelium.
Keywords: development, gnotobiotic, gut, microbiota, preterm
Abstract
Development of the infant small intestine is influenced by bacterial colonization. To promote establishment of optimal microbial communities in preterm infants, knowledge of the beneficial functions of the early gut microbiota on intestinal development is needed. The purpose of this study was to investigate the impact of early preterm infant microbiota on host gut development using a gnotobiotic mouse model. Histological assessment of intestinal development was performed. The differentiation of four epithelial cell lineages (enterocytes, goblet cells, Paneth cells, enteroendocrine cells) and tight junction (TJ) formation was examined. Using weight gain as a surrogate marker for health, we found that early microbiota from a preterm infant with normal weight gain (MPI-H) induced increased villus height and crypt depth, increased cell proliferation, increased numbers of goblet cells and Paneth cells, and enhanced TJs compared with the changes induced by early microbiota from a poor weight gain preterm infant (MPI-L). Laser capture microdissection (LCM) plus qRT-PCR further revealed, in MPI-H mice, a higher expression of stem cell marker Lgr5 and Paneth cell markers Lyz1 and Cryptdin5 in crypt populations, along with higher expression of the goblet cell and mature enterocyte marker Muc3 in villus populations. In contrast, MPI-L microbiota failed to induce the aforementioned changes and presented intestinal characteristics comparable to a germ-free host. Our data demonstrate that microbial communities have differential effects on intestinal development. Future studies to identify pioneer settlers in neonatal microbial communities necessary to induce maturation may provide new insights for preterm infant microbial ecosystem therapeutics.
NEW & NOTEWORTHY
Changes in the microbiome early in life may affect host physiology across the life span. Early life interaction between host and luminal microbes was investigated using a mouse model in which germ-free mice were transfaunated with fecal lysates from human preterm infants. Our data demonstrate that microbial communities affect differentiation of intestinal epithelial cell lineages, which may lead to significant effects on developmental, defensive, and physiological processes of the gastrointestinal epithelium.
the intestinal tract harbors the most abundant and complex microbial community in the human body and has a profound impact on human physiology, immune responses, and metabolism (5). The traditional view of the gut microbiota as only a pathogenic threat has been fundamentally replaced by an appreciation of its many beneficial influences on human health. Recent studies have found that this human host-gut microbiota symbiotic relationship starts early in life. Infants may be colonized even before birth, possibly through prenatal maternal microbial transmission (10, 21, 34).
In normal full-term infants, establishment of the gut microbiota proceeds in a stepwise manner with facultative anaerobes as the pioneer settlers (13). Within weeks of life, strict anaerobes become abundant with a dominant presence of Bifidobacterium sp. (14). The diversity of the infant gut microbiota increases over time with a major shift at weaning, which coincides with rapid morphological and functional gut maturation (12, 28). In contrast, colonization of the preterm infant gut is perturbed by many factors including antibiotic treatment to mother/infant, delayed enteral feeding, and prolonged hospitalization (31). Exposures to those prenatal and postnatal insults are associated with reduced diversity and microbial communities that may contribute to disease pathogenesis in preterm infants (29, 50).
Increasing evidence suggests that intestinal development not only depends on genetic factors but is also determined by the gut microbiota (19). Several studies have revealed a regulatory role of the microbiota on gut development and morphogenesis by promoting epithelial cell regeneration (2), modulating intestinal epithelial permeability (4), remodeling the intestinal vascular system, and promoting angiogenesis (38, 42). Impaired colonization in preterm infants has been proposed as contributing to the dysmotility of the intestinal tract, immature epithelial barrier function, and uncontrolled immune responses that lead to disease states including neonatal necrotizing enterocolitis (NEC) (20). Intestinal development in the preterm infant may depend on optimization of gut microbial colonization in preterm infants. Characterization of an “ideal” early colonization pattern is needed based on an understanding of the “beneficial” functions of the early gut microbial residents on intestinal development.
Previously, we utilized a humanized microbiome gnotobiotic mouse model and cDNA microarray profiling to dissect the function of early microbiota colonization on global gene expression in the host. Using weight gain as a surrogate marker for health, we found that early microbiota from a preterm infant with poor weight gain (MPI-L) induced a baseline increased inflammation phenotype in the transfaunated germ free mice, whereas early microbiota from a preterm infant with normal weight gain (MPI-H) induced a baseline decreased inflammation phenotype in the host. These studies suggested the existence of an “optimal” early microbiota community in preterm infants that may alter inflammation phenotypes and improve health outcomes (26). The purpose of this present study was to further investigate the functions of early preterm infant microbiota on host gut development and maturation.
We hypothesized that early colonization of the developing preterm gut influences small intestine development and maturation. We utilized our model of germ-free mice transfaunated with microbiota from fecal samples of human preterm infants with different growth rates. SPF (specific pathogen free with endogenous murine microbiota) and GF (germ free without microbiota) mice served as controls. Since nutrient absorption primarily occurs in the small intestine, and the microbiota gradient increases dramatically from duodenum to ileum, our studies focused on the ileum to investigate the trophic effect of microbiota on gut development. Gut development was evaluated by the parameters of villus height, crypt depth, epithelium cell proliferation, and apoptosis. Additionally, the microbial impact on differentiation of the four epithelial cell lineages (enterocytes, goblet cells, Paneth cells, enteroendocrine cells) as well as tight junction (TJ) formation was examined. Laser capture microdissection (LCM) to recover cell populations from either villus or crypt cell populations was used to examine effects on specific intestinal cell subsets by detecting marker gene expression of the four cell lineages with real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Our data demonstrate that different microbial communities specifically affect villus height, crypt depth, enterocyte proliferation, and the number of goblet cells and Paneth cells. Comparisons of functional marker gene expression in villus and crypt populations further revealed specific upregulations of mucin 3 (Muc3) in MPI-H villi; and Lysozyme 1(Lyz1) and Cryptdin 5 (Crp5) in MPI-H crypts compared with MPI-L. These data demonstrate that different microbial communities have differential effects on intestinal development. Effects of MPI-L were notably similar to those from GF mice, suggesting that certain microbial communities may fail to induce maturation. Differences in the initial microbiota colonization and the resulting impact on intestinal maturation may contribute to preterm infant health, further supporting the existence of an optimal early preterm infant microbial colonization community.
MATERIALS AND METHODS
Subjects
This study was designed to investigate the influence of the early microbiota on the developing gut using gnotobiotic mice transplanted with the early microbiota from preterm infants. Subjects were recruited from the neonatal intensive care unit (NICU) at The Comer Children's Hospital of the University of Chicago. As growth can be used as a marker of health in infants, we arbitrarily selected two preterm human infants with normal (>10 g·kg−1·day−1) weight gain or decreased (<10 g·kg−1·day−1) weight gain as the donors for microbiota transplantation. The two preterm infants were comparable in gestational age (27-wk GA), mode of delivery (Caesarean section), feeding pattern (breast fed), and exposure to antibiotics (48 h administration of ampicillin and gentamicin immediately after birth). Based on our previously published work demonstrating a temporal progression of preterm infant microbiota with distinct clustering at <2 wk of life (6), the early microbiota (<2 wk of life) from preterm infants with normal growth (MPI-H) and low growth (MPI-L) were collected for further analysis and transfaunation studies. At the time the samples were obtained, both were receiving total parenteral nutrition (TPN) intravenously, as well as their own mother's nonfortified breast milk from frozen stores via nasogastric tube (26).
Subject Compliance/Protection and Informed Consent
Specimen collection procedures involved minimal physical risk to subjects. The procedures were approved by the Institutional Review Board (IRB), approval 14991B, and written informed consent was obtained from patient parents.
Mice
All procedures were carried out in accordance with Institute guidelines at the University of Chicago. Germ-free (GF) C57BL/6J mice were maintained in the gnotobiotic facility of the Digestive Disease Research Core Center (DDRCC) at the University of Chicago. GF colonies are routinely tested for microbes and parasites by the facility's staff to ensure germ-free conditions. Conventional C57BL6J mice (from the Jackson Laboratory) were bred and housed in the animal care facilities of the University of Chicago under specific pathogen-free environment (SPF) conditions. All groups of mice were allowed ad libitum access to Harlan Teklad 7012 (SPF) or its autoclaved equivalent NIH 31 (GF, MPI) chow. Small intestine tissues were obtained from mice at 3 wk of age. All animal studies were approved by the Institutional Animal Care and Use Committee of the University of Chicago (protocol number 71703).
Colonization Experiments
To initiate microbial colonization, pregnant GF female 8- to 9-wk-old mice (estimated between E15 and E17) were gavaged with 0.25 ml of freshly prepared fecal homogenate from frozen fecal samples of the preterm human infant donors (MPI-L, MPI-H), n = 3 dams for each gavage group. Each dam had several liters. Each of the litters was small (ranging from 3–7 pups), which in our experience is typical for germ-free or gnotobiotic litters. In each experiment 4–6 pups were randomly picked from different litters. Pups delivered spontaneously and naturally acquired the microbiota of interest. Litters remained with the mother to allow natural passage of intestinal microbes. These pups were studied in parallel with age-matched SPF and GF controls.
Histochemistry, Immunohistochemistry, and Immunofluorescence
Morphology (small intestine length, villi height, and crypt depth).
The small intestine was dissected from the pylorus to the cecum. The distance between pylorus and cecum was measured as the small intestine length. Ileum was fixed with buffered formalin and embedded in paraffin. Serial histological sections of 4 μm thickness were cut, deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E) for morphometric analysis under a light microscope. Villus height and crypt depth were measured in the ileum of GF, SPF, and MPI mice using ImageJ software. At least 100 well-oriented villi and crypts were measured in at least three individual mice from each group for this study.
Proliferation and apoptosis.
Ileum sections were blocked in 10% normal goat serum (Sigma, St. Louis, MO) for 1 h at room temperature and then incubated with rabbit monoclonal Ki67 antibody (Abcam, Cambridge, MA) at 1:25 dilution, followed by an Alexa Fluor 594-conjugated secondary antibody (Invitrogen, Camarillo, CA). Nuclei were labeled with 4′, 6-diamidino-2-phenylindole (DAPI) (100 ng/ml) (Sigma, St. Louis, MO). Coverslips were mounted on slides using SlowFade Gold anti-fade reagent (Invitrogen, Grand Island, NY). Images were acquired by Olympus TIRF microscope (Olympus of the Americas, Center Valley, PA) and analyzed using Slidebook 5.0 software (Intelligent Imaging Innovations, Denver, CO). Cell proliferation was quantified by assessing Ki67-positive nuclei as a percentage of total nuclei in each high-power field (minimum 12 HPFs were counted per mouse). At least three mice were used for each group.
Formalin-fixed, paraffin-embedded ileum sections were assessed for apoptotic cells by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay using the In Situ Cell Death Detection Kit, TMR Red (Roche, Indianapolis, IN). The nuclei were counterstained with DAPI. Slides were examined with a Leica TCS SP2 Confocal Microscope. ImageJ software was used to illustrate the localization of TUNEL positive nuclei.
Goblet cell staining.
For goblet cells, ileum sections were stained with H&E to assess cellular morphology and with periodic acid-Schiff (PAS) (Newcomer Supply, Middleton, WI) to visualize mucin-containing goblet cells. The total number of PAS-positive cells per villus crypt unit was determined.
Paneth cell staining.
The population of Paneth cells present in intestinal crypts was detected using the phloxine-tartrazine technique (23). Nuclei in ileal sections were stained with hematoxylin for 45 s. After a brief wash in water, sections were stained in phloxine solution (0.5 g of phloxine B and 0.5 g of calcium chloride in 100 ml of distilled water) for 30 min, successively rinsed in water, and differentiated with a saturated tartrazine solution (2.5 g of tartrazine in 100 ml of Cellosolve) for 5 min. Sections were rinsed, mounted, and evaluated using light microscopy wherein Paneth cell granules appear in red, nuclei in blue, and other tissue in yellow. The number of positive Paneth cells was assessed by counting phloxine-tartrazine positive cells per crypt in 10 representative microscopic fields (200×) for each ileum sample.
Chromogranin A staining of enteroendocrine cells.
Chromogranin A (ChrgA) is found in the secretory granules of a wide variety of endocrine cells (35). Each ileal section was stained with polyclonal Rabbit anti-ChrgA antibody (1:500 dilution) (Abcam, Cambridge, MA); an anti-rabbit HRP (Dako, Carpinteria, CA) was applied as the secondary antibody. Positive staining was visualized with DAB chromogen and nuclei counterstain was performed with hematoxylin. Enteroendocrine cells appear brown on the background of hematoxylin and eosin staining. ChrgA-positive cells were counted in the ten most stained microscopic fields per slide using a ×40 objective. The numbers of villus/crypt units (VCU) were also counted manually in each field. Numbers of ChrgA positive cells were expressed per VCU for each field. Measurements were made in a blinded fashion, and conducted on a minimum of 3 animals per group.
TJ proteins staining.
Formalin-fixed, paraffin-embedded ileal sections were blocked with 10% goat serum. Tissue sections were incubated with Alexa Fluor 594 conjugated occludin mouse monoclonal antibody (Invitrogen, Camarillo, CA) and Alexa Fluor 488 conjugated ZO-1 mouse monoclonal antibody (Invitrogen, Camarillo, CA). After overnight incubation at 4°C, the sections were washed three times with PBS. The nuclei were stained with DAPI (100 ng/ml) (Sigma, St. Louis, MO). Coverslips were mounted on slides using SlowFade Gold anti-fade reagent (Invitrogen, Grand Island, NY). Images were acquired by Olympus TIRF microscope (Olympus of the Americas, Center Valley, PA) and analyzed using Slidebook 5.0 software (Intelligent Imaging Innovations, Denver, CO).
Laser Capture Microdissection (LCM) Plus qRT-PCR
Two-centimeter-long segments from the ileum were prepared for LCM using protocols described previously (43). All sectioning and subsequent specimen handling procedures were conducted under RNase free conditions. Well-oriented crypts or villi from nuclear Fast Red-stained, 7-μm-thick cryostat sections were harvested using a Leica LMD 6500 System (Leica Microsystems, Buffalo Grove, IL) (n ≥ 50 crypt or villi per mouse; 3 mice per group were evaluated). Total cellular RNA was extracted using the Arcturus PicoPure RNA isolation kit (Arcturus, Mountain View, CA). Quantity and quality of RNA were analyzed using an RNA LabChip and BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA). cDNA corresponding to 1 ng of purified RNA was added to each 10-μl quantitative RT-PCR (qRT-PCR) mixture, which also contained 5 μl of 2× SYBR green master mix (Applied Biosystems, Carlsbad, CA), and 500 nM gene-specific primers (see Table 1). A melting curve was used to identify a temperature where the amplicon (and not primer-dimers) were the source of SYBR green-bound fluorescence. Each assay per compartment per mouse was performed in triplicate and the data were normalized to 18S RNA level by delta-delta Ct (ΔΔCT) calculation (ABI 7900HT Sequence Detection System) (Applied Biosystems, Foster City, CA).
Table 1.
Oligonucleotide primer pairs used for PCR measurements
| Product | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| 18S | GCAATTATTCCCCATGAACG | GGCCTCACTAAACCATCCAA |
| Villin | AAGTCTTCGGTGGACAGGTG | CGTTTTCACTGCCAATACCA |
| Lgr5 | CCTACTCGAAGACTTACCCAGT | GCATTGGGGTGAATGATAGCA |
| Crp5 | AGGCTGATCCTATCCACAAAACAG | TGAAGAGCAGACCCTTCTTGGC |
| Lyz1 | GAGACCGAAGCACCGACTATG | CGGTTTTGACATTGTGTTCGC |
| Abst | TGTCTGTCCCCCAAATGCA | TGCATTGAAGTTGCTCTCAGGTA |
| Muc2 | ACCTCCAGGTTCAACACCAG | ATGGCAGTCCAGAGAGCAGT |
| Muc3 | GAGACATGCAAGAAGGAGGC | CCAAGTCCATACACCAGGCT |
Statistics
ANOVA analysis and a post hoc Tukey honest significance (HSD) test were used to compare the differences among groups. Findings are presented as means ± SD, and P < 0.05 was considered statistically significant.
RESULTS
Preterm Infant Microbiota Transfaunation in GF Mice Influences Small Intestine Growth and Development
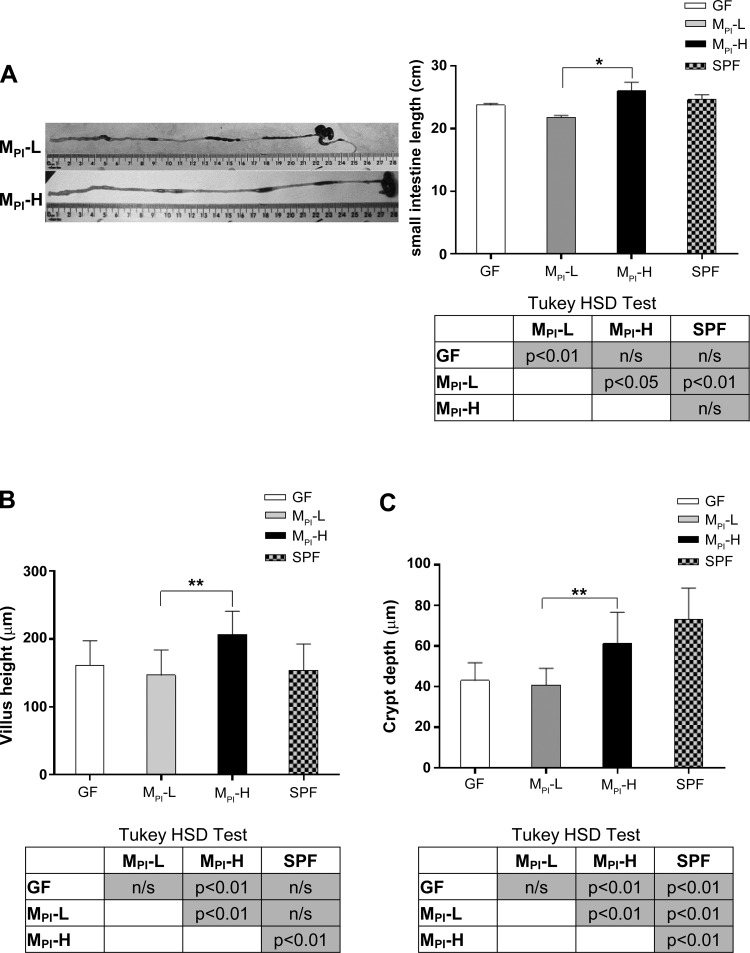
In our previous study, we found that human microbiota transfaunated mouse pups could recapitulate a human infant growth phenotype. The mouse pups born to a mother transfaunated with microbiota from an infant with poor weight gain (MPI-L) had a lower weight at weaning than pups receiving microbiota from an infant with normal weight gain (MPI-H) (26). The small intestine is the major site of nutrient absorption and its length represents the surface area available for nutrient uptake. To characterize the effects of preterm infant microbiota on the morphology of developing small intestine, 3-wk-old mouse pups transfaunated with preterm infant microbiota (MPI-L and MPI-H) were examined. We measured small intestine length to examine whether the weight gain pattern correlated with small intestinal growth. Age-matched GF and SPF mice were used as reference controls. We found that the small intestine length was significantly greater in MPI-H compared with MPI-L pups (P < 0.05) (Fig. 1A). We next measured ileal villus height and crypt depth. Both villus height and crypt depth were significantly greater in MPI-H mice compared with MPI-L mice (P < 0.01) (Fig. 1, B and C).
Fig. 1.
Effect of MPI transfaunation on intestine length, villus height, and crypt depth in ileum. A: total small intestine length was measured in GF, MPI-L, MPI-H, and SPF groups (n = 3–6 mice). B: mean ileal villus height was measured in GF, MPI-L, MPI-H, and SPF mice (n = 3–6 mice). C: mean crypt depth was measured in GF, MPI-L, MPI-H, and SPF mice (n = 3–6 mice). Results are presented as means ± SD. One-way ANOVA with post hoc Tukey's HSD test was used to compare the groups. *P < 0.05, **P < 0.01.
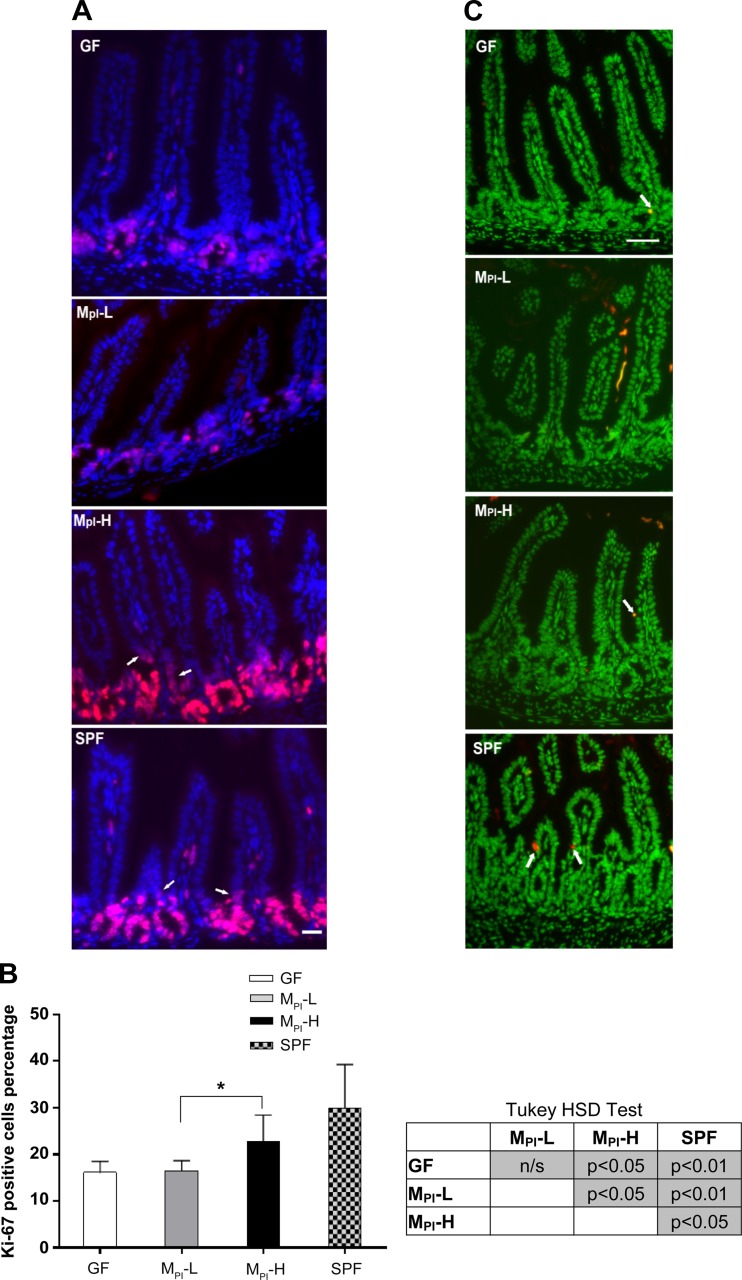
To determine the cellular basis for the increase in villus height and crypt depth, we measured the number of Ki-67 positive (proliferating cells) in the ileum by immunofluorescence staining. As shown in Fig. 2, A and B, the numbers of proliferating cells were significantly higher in MPI-H (P < 0.05) and SPF (P < 0.01) ileum compared with MPI-L and GF mice. Ki-67 staining showed similar proliferation rates for GF and MPI-L mice (16.10 ± 2.37% and 16.48 ± 2.17%), whereas proliferation rate increased to 22.79 ± 5.65% in MPI-H mice and to 29.93 ± 9.34% in SPF mice (Fig. 2B).
Fig. 2.
Effect of MPI transfaunation on cell proliferation and apoptosis in ileum. A: immunofluorescence detection of Ki-67 positive cells in ileum of GF, MPI-L, MPI-H, and SPF mice (n = 3–6 mice); arrows refer to the proliferative cells in the lower villus area. B: one-way ANOVA with post hoc Tukey's HSD test was used to compare the groups. *P < 0.05. C: representative TUNEL stained ileum sections in GF, MPI-L, MPI-H, and SPF mice (n = 3–6 mice); arrows refer to the TUNEL positive cells. Bar = 50 μm.
To determine if increased growth was associated with altered cell destruction in addition to altered proliferation, we investigated the differences in cell death by TUNEL staining for apoptotic cells. There were very few TUNEL positive signals present in any of the ileal tissues examined. There was no significant difference among groups (Fig. 2C).
Early Human Microbiota Affects Small Intestinal Cell Lineage Differentiation in Gnotobiotic Mice
The small intestinal epithelium consists of four principal cell types deriving from one multipotent stem cell: enterocytes, goblet cells, enteroendocrine cells, and Paneth cells. To investigate the effect of different early human microbiota on the differentiation of intestinal epithelial cell lineages, we next compared the numbers of goblet cells, Paneth cells, and enteroendocrine cells among GF, MPI-L, MPI-H, and SPF mouse ileum.
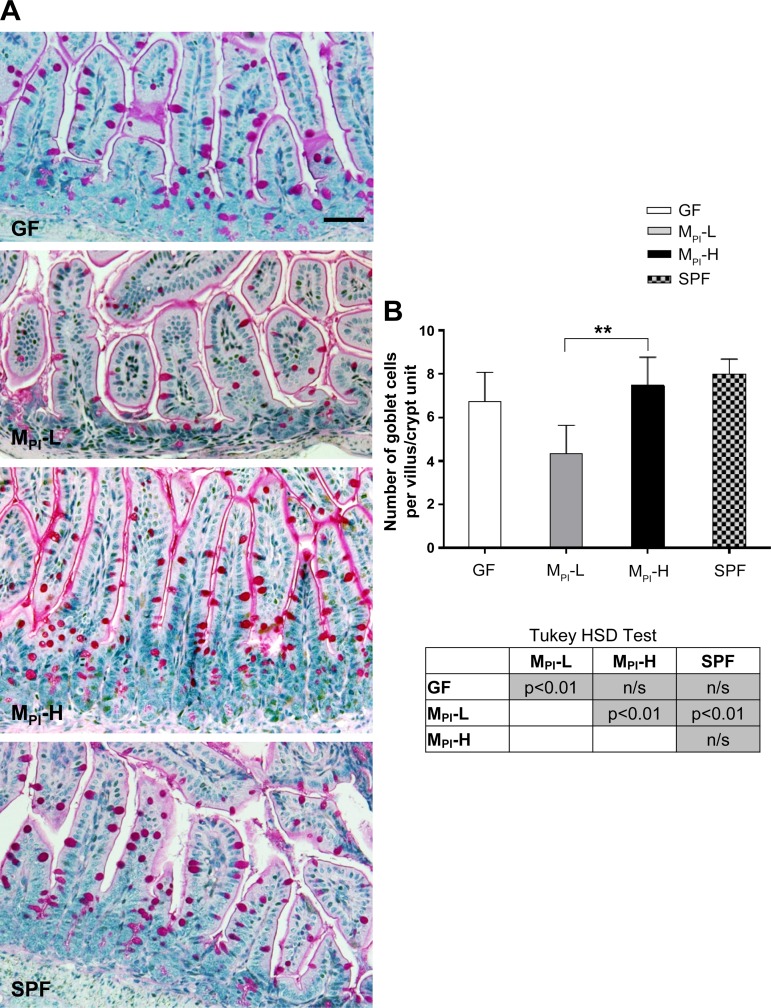
PAS staining showed comparable numbers of goblet cells in GF, MPI-H, and SPF ileum. However, MPI-L colonized ileum showed significantly fewer goblet cells (Fig. 3, A and B).
Fig. 3.
Effects of MPI transfaunation on goblet cell number in ileum. A: higher magnification views of PAS positive goblet cells in ileum villi and crypts in GF, MPI-L, MPI-H, and SPF mice. B: quantification of goblet cells in ileum in more than 10 fields per group (n = 4–6 mice). Results are presented as means ± SD. One-way ANOVA with post hoc Tukey's HSD test was used to compare the groups. **P < 0.01. Bar = 50 μm.
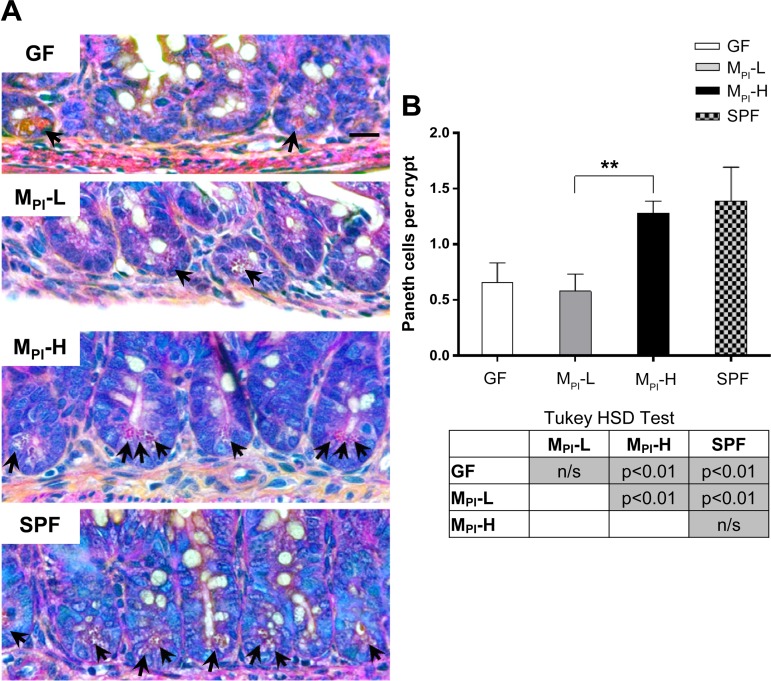
Phloxine-tartrazine staining was performed to investigate Paneth cell development. Paneth cells were identified by the distinct secretory granules (red stained) at the base region of the crypts (Fig. 4A, arrows). Phloxine-tartrazine positive cells in ileal sections of GF, MPI, and SPF mice were quantified and demonstrated a marked increase in MPI-H mouse ileum compared with that of MPI-L ileum (1.28 ± 0.11 vs. 0.58 ± 0.15, P < 0.01, Fig. 4B). No difference was observed between MPI-L and GF mice (Fig. 4B).
Fig. 4.
Effects of MPI transfaunation on Paneth cell number in ileum crypt. A: Paneth cells granules were revealed by phloxine-tartrazine staining in GF, MPI-L, MPI-H, and SPF mice (arrows) (n = 4–6 mice). B: quantification of the number of Paneth cells revealed a significant induction of Paneth cells at MPI-H crypts compared with MPI-L. Results are presented as means ± SD. One-way ANOVA with post hoc Tukey's HSD test was used to compare the groups. **P < 0.01. Bar = 50 μm.
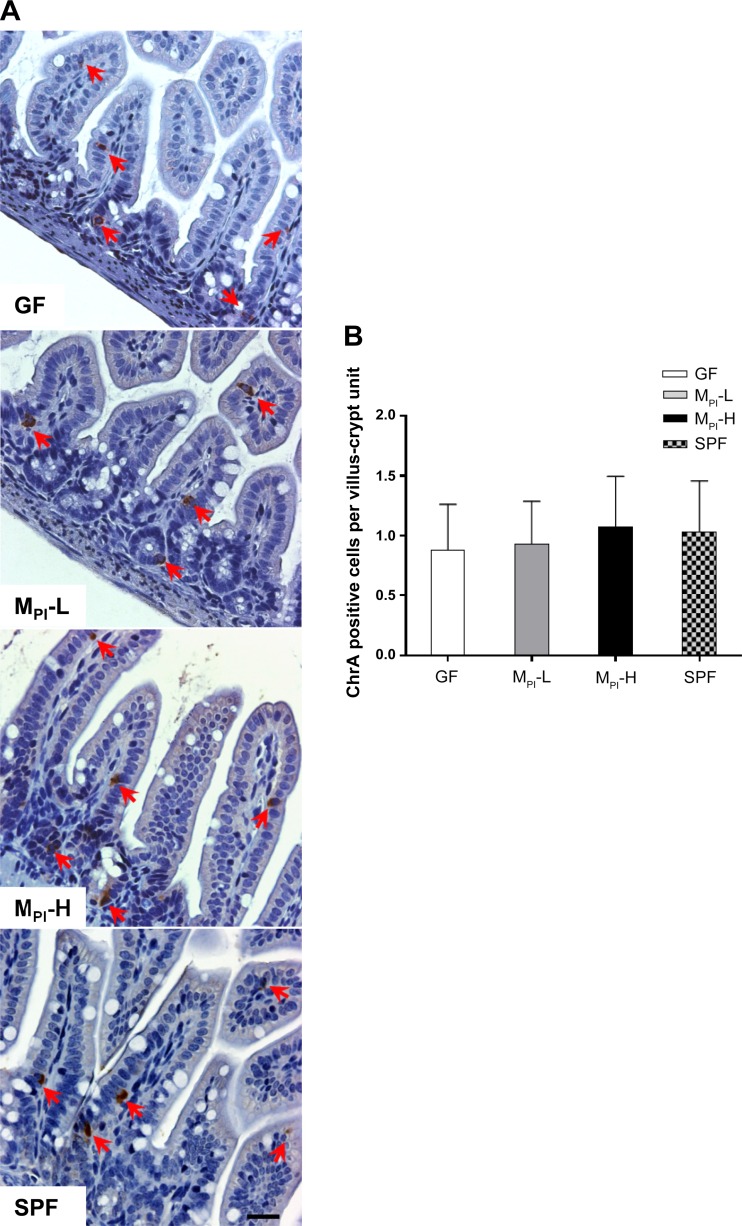
Enteroendocrine cells are distributed in the epithelium as single, scattered cells surrounded by enterocytes and comprise less than 1% of the total epithelial cells (44). Immunostaining for the pan-endocrine marker chromogranin A (ChrgA) revealed no significant difference in enteroendocrine cell numbers among groups (representative images shown in Fig. 5).
Fig. 5.
Effects of MPI transfaunation on enteroendocrine cell number in ileum. A: chromogranin A (ChrA) staining for enteroendocrine cells in GF, MPI-L, MPI-H, and SPF ileum (red arrows) (n = 4–6 mice). B: quantification of the number of enteroendocrine cells in ileum revealed no difference among groups. Bar = 50 μm.
Altered Epithelial Barrier Integrity in Preterm Infant Microbiota Transfaunated GF Mice
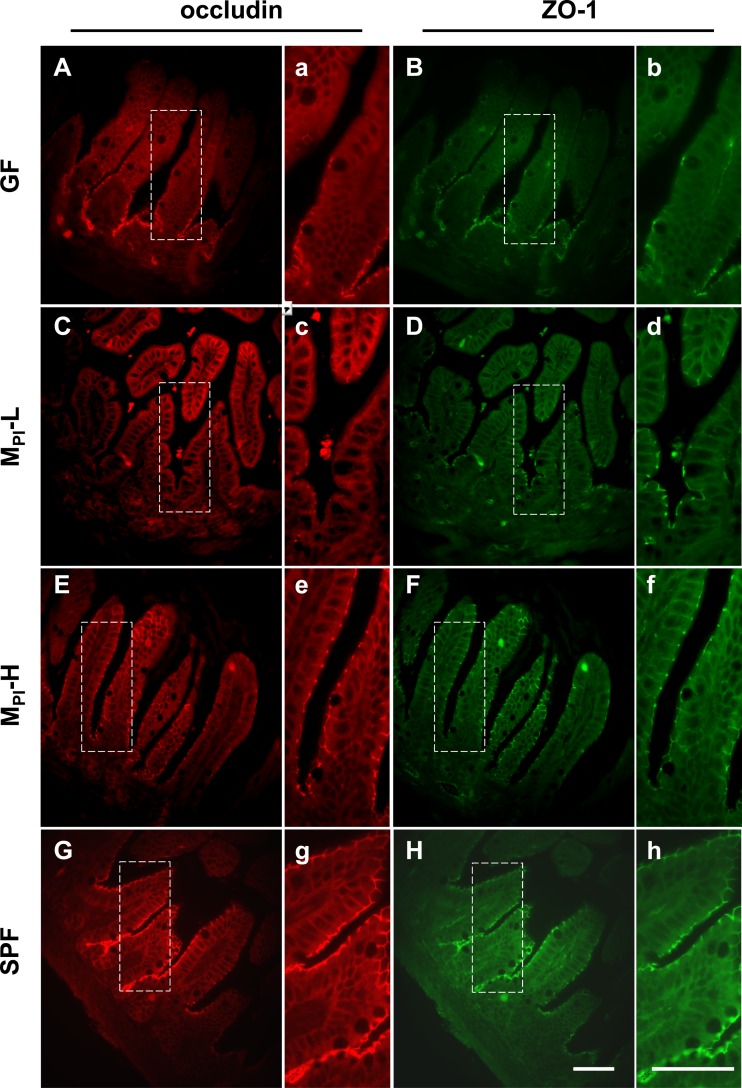
The single layer of intestinal epithelial cells forms an effective barrier between the inner and outer environment via the structure of tight junctions (TJs), desmosomes, adhesion junctions, and gap junctions (45). TJs are responsible for controlling the paracellular flux between the epithelial cells, hence the permeability of the epithelial barrier (45). Immunofluorescence staining for the tight junction proteins occludin and ZO-1 was performed. We compared the expression and localization of these proteins among groups in 3-wk-old mouse ileum. Both occludin and ZO-1 are located at the apical region and form a continuous staining pattern around the whole enterocyte layer in MPI-H and SPF mice (Fig. 6, E–H). In contrast, in MPI-L and GF mice, there is lower expression of occludin and ZO-1 in the tissue, mostly in lower villus and crypt areas (Fig. 6, A–D). The discontinuous staining pattern at the villus surface in MPI-L and GF mice suggests a less well-formed TJ assembly.
Fig. 6.
Effects of MPI transfaunation on TJ protein expression. In ileum from GF and MPI-L mice, occludin and ZO-1 staining was decreased with only residual staining remaining in the crypts region. In MPI-H and SPF mice ileum, staining of both proteins was continuous along the villus epithelium layer. Panels with small letters (a–h) represent higher magnification of the selected area (outlined with dashed line) in panels with capital letters (A–H). Sections from at least 3 mice were examined for each group. Bar = 50 μm.
Microbial Colonization Altered Gene Expression in LCM Isolated Intestinal Cells from Villi/Crypts of GF, MPI-L, MPI-H, and SPF Ileum
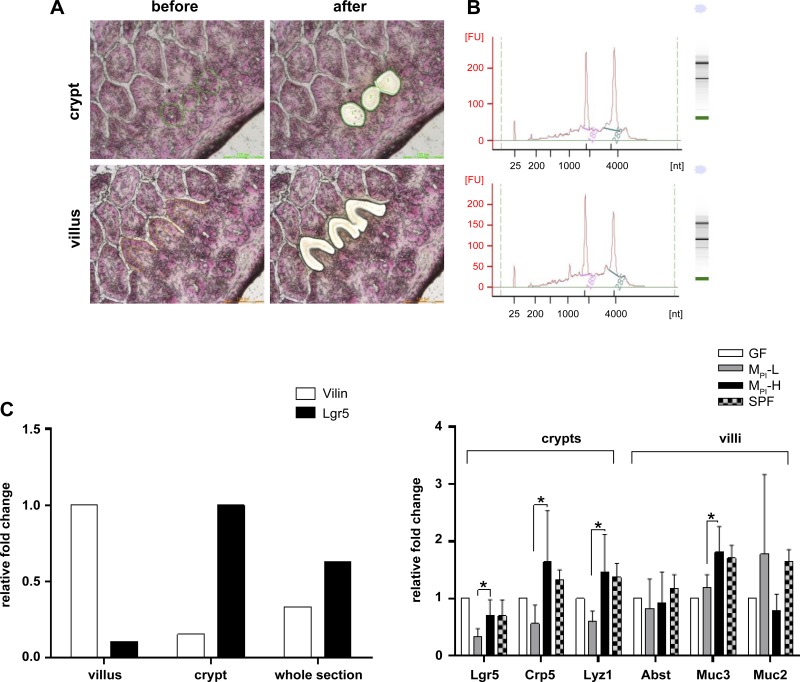
LCM was used to investigate specific local gene expression responses to microbiota colonization in the small intestine. LCM permitted selective isolation of intestinal villus or crypt cells (Fig. 7A). The expected 18S and 28S ribosomal peaks indicated good integrity of the RNAs (Fig. 7B). The purity of LCM villi cells/crypt cells was established by measuring the distribution of Villin and Lgr5 mRNAs, respectively. Samples from villi had a strong villus specific signal (Villin) and no significant signal for intestinal Lgr5 (a crypt-specific marker) by RT-PCR, indicating minimal contamination by mRNA of crypt cells and vice versa for all the samples from crypts (Fig. 7C).
Fig. 7.
Altered gene expression in isolated ileal villi or crypts by LCM. A: the morphology of the extracted villi and crypts during LCM. B: electropherograms of total RNAs from LCM isolated villus or crypt population. C: RT-PCR of cell compartment-specific genes (Villin for villus population and Lgr5 for crypt population) for cDNA made from total RNA isolated from LCM-extracted villus or crypt cells; total RNA from whole section was added as control. D: marker genes expression in villus or crypt in GF, MPI-L, MPI-H, and SPF groups were analyzed by normalizing to 18S RNA (n = 3–6 mice). Data are means ± SD. One-way ANOVA with post hoc Tukey's HSD test was used to compare the groups. *P < 0.05.
The expression of specific marker genes of the four epithelial cell lineages was determined to assess the responses of different epithelial cell types to microbiota colonization. The expression of markers for enterocytes, goblet cells, and enteroendocrine cells were maximally present in the villus population, while markers for Paneth cells and stem cells were expressed maximally in the crypt population. The gene expression for enterocytes markers, mainly for absorptive and nutritional functions [e.g., apical sodium-dependent bile acid transporter (Abst)], was similar among all groups (Fig. 7D); markers for enteroendocrine cells were not identified (e.g., Ngn3, Gastrin), possibly due to the low percentage of this cell type (data not shown). The increase in Paneth cell markers lysozyme 1(Lyz1), Cryptdin 5 in the crypt population was detected in MPI-H group compared with MPI-L group, correlating with the increase in Paneth cells in MPI-H. A major cell surface bound mucin, Mucin 3 (Muc3) mRNA abundance in villi was increased significantly in MPI-H compared with that of MPI-L. Muc2, a major secreted mucin gene in the intestines was highly expressed, but no significant difference was observed among the groups examined (Fig. 7D).
DISCUSSION
We previously reported that transfaunation of human gut microbiota in gnotobiotic mice is a useful model for dissecting the role of preterm infant microbiota in modulating inflammatory responses in the developing gut. Using this approach, we now investigate the effect of early human preterm infant microbiota on the growth and differentiation of immature intestinal epithelium. The impact of gut bacteria on intestinal development has been indicated by the phenotypic differences between GF and SPF animals (1). The decreased intestinal surface (16), reduced crypt depth (15), and decreased epithelial cell turnover in GF animals all suggest that gut microbiota is essential for gut development (22). However, our study is the first to investigate the effects of early preterm microbiota on intestinal morphology, cell lineage differentiation, and gene expression.
The villus height and crypt depth in MPI-H pups were significantly greater than that of MPI-L pups (Fig. 1, B and C). Furthermore, we found that the ileum of MPI-H mice had higher expression of the intestinal stem cell marker Lgr5 and increased numbers of proliferating cells in the crypt and transit-amplifying region as indicated by significantly higher number of positive Ki-67 stained cells in MPI-H and SPF groups compared with MPI-L and GF pups (Fig. 2, A and B). These data suggest that MPI-H microbiota impact intestinal development by modulating intestinal stem cells and proliferating transit amplifying cells that differentiate into mature cell lineages (absorptive, goblet, and enteroendocrine cells). We correspondingly found higher numbers of goblet cell and Paneth cells in MPI-H ileum. Goblet cells and Paneth cells are key components of the mucosal barrier and play important roles in intestinal stem cell homeostasis (7, 42, 41), host defense, and regulation of the intestinal microbiota (40, 48, 39, 47). Goblet cells synthesize and secrete secretory and membrane bound mucins forming a barrier to prevent direct contact between intestinal contents and the epithelial cell layer (46). In addition, studies also show the involvement of mucins in fetal development, epithelial renewal, differentiation, and integrity (35) (9, 27, 32). Muc2, a major secretory mucin in the intestine, was highly expressed but exhibited no significant expression difference among the four groups. In contrast, we found that Muc3, the most abundantly expressed membrane mucin in the small intestine, was differentially expressed with significantly higher expression in MPI-H pups compared with the GF and MPI-L pups (Fig. 7D). This indicates that Muc3 can be differentially regulated by the gut microbiota in the ileum. Moreover, the mouse mucin Muc3 has been shown to inhibit apoptosis and stimulate cell migration, implying a bioactive role in maintaining the integrity of the surface epithelial layer (18).
Paneth cells produce antimicrobial compounds such as lysozymes, lactoferrin, and α- and β-defensins, which limit bacterial growth and regulate microbiota composition (52). As shown in Fig. 4 and Fig. 7, there were significantly more Paneth cells and an upregulated expression of Cryptdin 5 and Lyz1 in MPI-H crypts compared with MPI-L. In humans, the number of Paneth cells is lower in preterm infants compared with term infants (30, 40), and reduced Paneth cell number is associated with NEC infants (53), suggesting that Paneth cell number in general can be used as a developmental parameter of small intestine maturity. Furthermore, Human DEF5, and its mouse counterpart Cryptidin 5, has been shown to protect the host against pathogens and shape microbiota composition (39, 49). Furthermore, increased DEF5 expression in Paneth cells has also been reported in term infants compared with preterm infants (30) and increased lysozyme staining has been found in normal control infants compared with preterm NEC infants (8). Our findings indicate that there is a better-developed mucosal defense in MPI-H pups. The evidence that an increase in Paneth cell number and function is associated with specific microbial communities may have significant health implications for the preterm infant.
Using LCM analysis, we demonstrate a higher expression of stem cell marker Lgr5 in MPI-H crypts (Fig. 7). This is consistent with the finding that there was a higher proliferative rate in MPI-H mouse ileum. Furthermore, in these MPI-H mice, there was higher expression of the goblet cell and mature enterocyte marker, Muc3, as well as the Paneth cell markers Lyz1 and Cryptdin 5 compared with MPI-L mice (Fig. 7D). These data suggest that the interaction between the early microbial community and the preterm gut in its most naïve and immature state may significantly influence intestinal development including stem cell self-renewal and differentiation of the intestinal epithelium in the gnotobiotic mouse.
Over the past decade, a number of diseases, such as NEC (36), IBD (25), obesity, and metabolic disorders (33), have been found to be associated with decreased intestinal barrier function and increased intestinal permeability. Alterations in barrier integrity are associated with changes in TJ protein expression and distribution. In our study, we found that both occludin and ZO-1 formed a continuous apical staining pattern in MPI-H and SPF mice, contrasted by the discontinuous expression patterns seen in MPI-L and GF mice (Fig. 6). We speculate that the upregulation of occludin and ZO-1 and more stabilized tight junction formation on the ileum epithelial surface may limit bacteria translocation and strengthen the mucosal barrier in MPI-H and SPF mice.
Our findings are limited by the inclusion of only two microbial communities. Furthermore, we have intentionally chosen to use samples from individual patients rather than pooled samples as combining organisms from different patients to create an artificial community of organisms not originally found together would have unpredictable functional results. Our findings suggest that certain microbial communities functionally promote maturation in epithelial cell lineages, while others may fail to promote growth and impact host innate immune development, hence leaving preterm infants vulnerable to disease. Thus this study highlights specific roles for beneficial preterm infant communities that warrant further investigation. Although the identification of the optimal preterm microbiota community is largely unknown, our previous study showed that at the phylum level, there is a greater contribution of Bacteroidetes (8.30 vs. 3.42) in MPI-H colonized mice compared with MPI-L mice (26). One bacterium species, Bacteroides thetaiotaomicron, has been demonstrated to stimulate intestinal developmental changes during weaning (42). Future research must focus on identification of pioneer settlers that contribute to the establishment of the optimal community and their metabolites such as SCFA (butyrate), folate, bile acids, or vitamins, which may have trophic effects on gut development. Tandem analysis of preterm intestinal microbial transcriptomic, proteomic, metabolomic profiles should be performed to further identify bacterial signals stimulating intestinal maturation, intestinal lymphoid structure development, immune cell differentiation, and immune mediator production. Understanding the key functions of microbial communities is an important foundation for identifying clinical interventions to optimize and protect developing preterm infant microbial communities.
GRANTS
This work was supported by National Institute Health of Child Health and Human Development Grants R01-HD-059123 (E. C. Claud) and R01-HD-083481 (E. C. Claud) and Digestive Disease Research Core Center of the University of Chicago Grant P30-DK-42086.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Y.Y., L.L., and E.C.C. conception and design of research; Y.Y. performed experiments; Y.Y. analyzed data; Y.Y., L.L., and E.C.C. interpreted results of experiments; Y.Y. and E.C.C. prepared figures; Y.Y. and E.C.C. drafted manuscript; Y.Y., L.L., J.S., E.O.P., and E.C.C. edited and revised manuscript; Y.Y., L.L., J.S., E.O.P., and E.C.C. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Christine Labno and Shirley Bond from the Integrated Microscopy Core Facility for microscopy assistance, and the Digestive Disease Research Core Center of the University of Chicago for providing core facilities and services used for this study.
REFERENCES
- 1.Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest 12: 355–364, 1963. [PubMed] [Google Scholar]
- 2.Banasaz M, Norin E, Holma R, Midtvedt T. Increased enterocyte production in gnotobiotic rats mono-associated with Lactobacillus rhamnosus GG. Appl Environ Microbiol 68: 3031–3034, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, Muccioli GG, Delzenne NM. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58: 1091–1103, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 132: 1359–1374, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet 13: 260–270, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claud EC, Keegan KP, Brulc JM, Lu L, Bartels D, Glass E, Chang EB, Meyer F, Antonopoulos DA. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 1: 20, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol 75: 289–311, 2013. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho HB, da Mota HC, Coutinho VB, Robalinho TI, Furtado AF, Walker E, King G, Mahida YR, Sewell HF, Wakelin D. Absence of lysozyme (muramidase) in the intestinal Paneth cells of newborn infants with necrotising enterocolitis. J Clin Pathol 51: 512–514, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr 73: 1131S–1141S, 2001. [DOI] [PubMed] [Google Scholar]
- 10.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PloS one 3: e3056, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison RT 3rd, Giehl TJ. Killing of gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest 88: 1080–1091, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev 62: 1157–1170, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr 91: 48–55, 2003. [DOI] [PubMed] [Google Scholar]
- 14.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9: 577–589, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Glaister JR. Factors affecting the lymphoid cells in the small intestinal epithelium of the mouse. Int Arch Allergy Appl Immunol 45: 719–730, 1973. [DOI] [PubMed] [Google Scholar]
- 16.Gordon HA, Bruckner-Kardoss E. Effect of normal microbial flora on intestinal surface area. Am J Physiol 201: 175–178, 1961.13707165 [Google Scholar]
- 17.Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, Gilbert JA. Development of the preterm infant gut microbiome: a research priority. Microbiome 2: 38, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho SB, Dvorak LA, Moor RE, Jacobson AC, Frey MR, Corredor J, Polk DB, Shekels LL. Cysteine-rich domains of muc3 intestinal mucin promote cell migration, inhibit apoptosis, and accelerate wound healing. Gastroenterology 131: 1501–1517, 2006. [DOI] [PubMed] [Google Scholar]
- 19.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol 12: 129–134, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Hunter CJ, Upperman JS, Ford HR, Camerini V. Understanding the susceptibility of the premature infant to necrotizing enterocolitis (NEC). Pediatr Res 63: 117–123, 2008. [DOI] [PubMed] [Google Scholar]
- 21.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, Fernandez L, Rodriguez JM. Is meconium from healthy newborns actually sterile? Res Microbiol 159: 187–193, 2008. [DOI] [PubMed] [Google Scholar]
- 22.Khoury KA, Floch MH, Hersh T. Small intestinal mucosal cell proliferation and bacterial flora in the conventionalization of the germfree mouse. J Exp Med 130: 659–670, 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lendrum AC. The phloxin-tartrazine method as a general histological stain and for the demonstration of inclusion bodies. J Pathol Bacteriol 59: 399–404, 1947. [Google Scholar]
- 24.Leser TD, Molbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol 11: 2194–2206, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Li N, Neu J. Tight junctions, leaky intestines, and pediatric diseases. Acta Paediatr 94: 386–393, 2005. [DOI] [PubMed] [Google Scholar]
- 26.Lu L, Yu Y, Guo Y, Wang Y, Chang EB, Claud EC. Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model. PLos One 10: e0124504, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol Gastrointest Liver Physiol 276: G941–G950, 1999. [DOI] [PubMed] [Google Scholar]
- 28.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 69: 1035S–1045S, 1999. [DOI] [PubMed] [Google Scholar]
- 29.Magne F, Suau A, Pochart P, Desjeux JF. Fecal microbial community in preterm infants. J Pediatr Gastroenterol Nutr 41: 386–392, 2005. [DOI] [PubMed] [Google Scholar]
- 30.Mallow EB, Harris A, Salzman N, Russell JP, DeBerardinis RJ, Ruchelli E, Bevins CL. Human enteric defensins. Gene structure and developmental expression. J Biol Chem 271: 4038–4045, 1996. [DOI] [PubMed] [Google Scholar]
- 31.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol 21: 167–173, 2013. [DOI] [PubMed] [Google Scholar]
- 32.Mattar AF, Teitelbaum DH, Drongowski RA, Yongyi F, Harmon CM, Coran AG. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr Surg Int 18: 586–590, 2002. [DOI] [PubMed] [Google Scholar]
- 33.Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, Masciana R, Forgione A, Gabrieli ML, Perotti G, Vecchio FM, Rapaccini G, Gasbarrini G, Day CP, Grieco A. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 49: 1877–1887, 2009. [DOI] [PubMed] [Google Scholar]
- 34.Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, Fernandez L, Rodriguez JM, Jimenez E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLos One 8: e66986, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connor DT, Burton D, Deftos LJ. Chromogranin A: immunohistology reveals its universal occurrence in normal polypeptide hormone producing endocrine glands. Life Sci 33: 1657–1663, 1983. [DOI] [PubMed] [Google Scholar]
- 36.Piena-Spoel M, Albers MJ, ten Kate J, Tibboel D. Intestinal permeability in newborns with necrotizing enterocolitis and controls: Does the sugar absorption test provide guidelines for the time to (re-)introduce enteral nutrition? J Pediatr Surg 36: 587–592, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Potten CS, Merritt A, Hickman J, Hall P, Faranda A. Characterization of radiation-induced apoptosis in the small intestine and its biological implications. Int J Radiat Biol 65: 71–78, 1994. [DOI] [PubMed] [Google Scholar]
- 38.Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Ostergren-Lunden G, Petersen LC, Ruf W, Backhed F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 483: 627–631, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11: 76–83, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol 19: 70–83, 2007. [DOI] [PubMed] [Google Scholar]
- 41.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA 99: 15451–15455, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stappenbeck TS, Hooper LV, Manchester JK, Wong MH, Gordon JI. Laser capture microdissection of mouse intestine: characterizing mRNA and protein expression, and profiling intermediary metabolism in specified cell populations. Methods Enzymol 356: 167–196, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Sternini C, Anselmi L, Rozengurt E. Enteroendocrine cells: a site of ‘taste’ in gastrointestinal chemosensing. Curr Opin Endocrinol Diabet Obes 15: 73–78, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki T. Regulation of intestinal epithelial permeability by tight junctions. Cell Mol Life Sci 70: 631–659, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9: 799–809, 2009. [DOI] [PubMed] [Google Scholar]
- 47.Verburg M, Renes IB, Meijer HP, Taminiau JA, Buller HA, Einerhand AW, Dekker J. Selective sparing of goblet cells and Paneth cells in the intestine of methotrexate-treated rats. Am J Physiol Gastrointest Liver Physiol 279: G1037–G1047, 2000. [DOI] [PubMed] [Google Scholar]
- 48.Watson AJ, Pritchard DM. Lessons from genetically engineered animal models. VII. Apoptosis in intestinal epithelium: lessons from transgenic and knockout mice. Am J Physiol Gastrointest Liver Physiol 278: G1–G5, 2000. [DOI] [PubMed] [Google Scholar]
- 49.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H Jr, Fellermann K, Ganz T, Stange EF, Bevins CL. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA 102: 18129–18134, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr 25: 361–368, 2006. [DOI] [PubMed] [Google Scholar]
- 51.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Prot Sci 3: 1362–1373, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature 415: 389–395, 2002. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, Sherman MP, Prince LS, Bader D, Weitkamp JH, Slaughter JC, McElroy SJ. Paneth cell ablation in the presence of Klebsiella pneumoniae induces necrotizing enterocolitis (NEC)-like injury in the small intestine of immature mice. Dis Models Mech 5: 522–532, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]