Long noncoding RNA (lncRNA) biology represents an exciting new field with implications in gene regulation. This field has only recently been investigated in the pathogenesis of ulcerative colitis. By utilizing high-throughput technologies on clinical samples, a lncRNA signature was generated and the lncRNA IFNG-AS1 was shown to regulate the important inflammatory cytokine interferon-γ. This work represents one of the first mechanistic studies of lncRNA biology in inflammatory bowel disease.
Keywords: inflammatory bowel disease, ulcerative colitis, lncRNA, inflammation, interferon γ
Abstract
High-throughput technologies revealed new categories of genes, including the long noncoding RNAs (lncRNAs), involved in the pathogenesis of human disease; however, the role of lncRNAs in the ulcerative colitis (UC) has not been evaluated. Gene expression profiling was used to develop lncRNA signatures in UC samples. Jurkat T cells were activated by PMA/ionomycin subsequently interferon-γ (IFNG) and tumor necrosis factor (TNF)-α protein levels were assessed by ELISA. Anti-sense molecules were designed to block IFNG-AS1 expression. A unique set of lncRNAs was differentially expressed between UC and control samples. Of these, IFNG-AS1 was among the highest statistically significant lncRNAs (fold change: 5.27, P value: 7.07E−06). Bioinformatic analysis showed that IFNG-AS1 was associated with the IBD susceptibility loci SNP rs7134599 and its genomic location is adjacent to the inflammatory cytokine IFNG. In mouse models of colitis, active colitis samples had increased colonic expression of this lncRNA. Utilizing the Jurkat T cell model, we found IFNG-AS1 to positively regulate IFNG expression. Novel lncRNA signatures differentiate UC patients with active disease, patients in remission, and control subjects. A subset of these lncRNAs was found to be associated with the clinically validated IBD susceptibility loci. IFNG-AS1 was one of these differentially expressed lncRNAs in UC patients and found to regulate the key inflammatory cytokine, IFNG, in CD4 T cells. Taking these findings together, our study revealed novel lncRNA signatures deregulated in UC and identified IFNG-AS1 as a novel regulator of IFNG inflammatory responses, suggesting the potential importance of noncoding RNA mechanisms on regulation of inflammatory bowel disease-related inflammatory responses.
NEW & NOTEWORTHY
Long noncoding RNA (lncRNA) biology represents an exciting new field with implications in gene regulation. This field has only recently been investigated in the pathogenesis of ulcerative colitis. By utilizing high-throughput technologies on clinical samples, a lncRNA signature was generated and the lncRNA IFNG-AS1 was shown to regulate the important inflammatory cytokine interferon-γ. This work represents one of the first mechanistic studies of lncRNA biology in inflammatory bowel disease.
inflammatory bowel diseases (IBDs) are common gastrointestinal disorders that are clinically comprised of ulcerative colitis (UC) and Crohn's disease (CD) (14, 24). These diseases have a devastating toll on a patient's morbidity and quality of life (27). The etiology of IBD, however, still remains elusive. Studies have shown that IBD may result from an inappropriate inflammatory response to intestinal microbes and foreign antigens in genetically susceptible hosts (8, 40). Key mediators of this inflammatory process such as TNF-α, IL-23, IL-12, and interferon-γ (IFNG) have been implicated in the pathogenesis of UC (1, 6, 10, 20, 25, 26). Therapies directed against these cytokines have been relatively successful in treating UC symptoms; however, a subpopulation of patients may fail to respond to these treatments (32, 33, 35). In addition to these cytokines, researchers have also focused on the genetic underpinnings of the disease. Familial clustering and twin studies have shown a role for genetic factors for UC (3, 14). Indeed, meta-analyses of multiple genome-wide association studies have implicated 163 genetic loci in both CD and UC pathogenesis (14). However, the mechanisms of action for many of the genes in close association with these loci have yet to be determined.
The IBD susceptibility loci point to key genomic positions and highlight areas of the genome for further mechanistic studies. In addition to the associated protein-coding transcripts previously reported, noncoding transcripts may be associated with these loci. Recently, investigators have begun to address the noncoding transcripts and their role in disease pathogenesis. One such category of noncoding transcript are the long noncoding RNAs (lncRNAs) (21). The central dogma, wherein “DNA makes RNA makes protein,” has long relegated RNA as an intermediary between gene and protein. Recent research has challenged the notion that the noncoding RNAs are innocent bystanders and may indeed be critical to gene regulation (30, 31). Long noncoding RNAs are defined as sequences of greater than 200 nucleotides in length. They can express introns and exons, which can be alternatively spliced and generally lack open reading frames for protein translation (29). lncRNAs appear to have a diverse set of functions in chromatin remodeling, telomere activity, and subcellular structural organization (4). They can mediate epigenetic changes by recruiting chromatin-remodeling complexes to specific loci in the genome. As an example, the lncRNA HOTAIR has been implicated in inducing a repressive chromatin state by recruiting the Polycomb chromatin remodeling complex PRC2 (12). In addition, lncRNAs can regulate transcription by altering RNA polymerase II activity and thereby influence promoter activation (30). Finally, lncRNAs have also been implicated in various steps in the posttranscriptional processing of messenger RNAs, including splicing, editing, transport, translation, and degradation (21).
Based on these above findings, it has been suggested that noncoding RNAs play a role in the pathogenesis of UC (15, 16, 28). In addition, lncRNAs have been shown to participate in multiple inflammatory processes and contribute to the pathogenesis of autoimmune and immune-related disorders; however, their role in UC has yet to be fully determined (2, 13, 18, 23). To this end, we have profiled the expression of both coding and noncoding transcripts in UC patients. By comparing their expression to those of control patients, we have identified a set of differentially expressed lncRNAs in adult UC patients. To evaluate the relationship of this set of lncRNAs with known IBD genomic alterations, we compared the chromosomal location of the lncRNAs with those of the IBD susceptibility loci. The lncRNA IFNG-AS1 was associated with the IBD susceptibility loci SNP rs7134599 and is in close proximity to the inflammatory cytokine IFNG. It was found to be among the most statistically significant differentially expressed lncRNAs. Given the close proximity to IFNG within the human genome, we investigated its cellular mechanism of action and found a positive regulatory mechanism between IFNG-AS1 and IFNG expression.
MATERIALS AND METHODS
Human tissue collection and RNA extraction.
Colonic tissues were obtained from two sources. For the microarray analysis, RNA extracted from colonic surgical samples were obtained from Origene. A total of 50 anonymized samples, 19 for the initial profiling experiment and 31 for the validation cohort, were included in this study. For the profiling experiment, 7 control patients, 8 UC-active (UCA), and 4 UC-inactive (UCI) were collected. Diagnoses were confirmed by an independent set of pathologists associated with Origene. The samples were obtained through institutional review board (IRB) protocols and with documented patient consent, all from accredited U.S.-based medical institutions (www.origene.com). A second set of colonic samples was obtained from colonic surgeries performed at Cedars-Sinai Medical Center, Los Angeles, CA. These samples were collected under an IRB-approved protocol. Patients that participated in the IRB tissue recruitment were undergoing colonic resections as part of their regular medical care. A total of 31 anonymized patient samples were included in this part of the study, 16 control patients and 15 UC patients. A segment of the colon resection was flash frozen in liquid nitrogen and stored at −80°C. The diagnoses were confirmed on histopathology. Available patient information is summarized in Table 1.
Table 1.
Patient information
| Sample | Age | Sex | Location |
|---|---|---|---|
| Con1 | 31 | Male | Colon |
| Con2 | 45 | Female | colon |
| Con3 | 26 | Male | Colon: sigmoid |
| Con4 | 46 | Male | Colon |
| Con5 | 50 | Female | Colon |
| Con6 | 60 | Female | Colon |
| Con7 | 56 | Male | Colon |
| UCA1 | 30 | Female | Colon |
| UCA2 | 56 | Female | Colon: left |
| UCA3 | 26 | Female | Colon |
| UCA4 | 46 | Female | Colon |
| UCA5 | 29 | Male | Colon |
| UCA6 | 24 | Male | Colon |
| UCA7 | 45 | Female | Colon |
| UCA8 | 26 | Female | Colon |
| UCI1 | 31 | Male | Colon |
| UCI2 | 71 | Female | Colon |
| UCI3 | 51 | Male | Colon |
| UCI4 | 33 | Female | Colon |
RNA extraction.
Total RNA was extracted from frozen colonic tissues and cells in culture using TRIzol (Qiagen, Valencia, CA; cat. no. 79306) and miRNeasy Mini Kit (Qiagen; cat. no. 217004) following the product instructions and utilizing DNase I digestion (cat. no. 79254). RNA was stored at −80°C and measured with a NanoDrop 1000 Spectrophotometer (Thermo Scientific). The integrity of the RNA submitted for microarray analysis was assessed with either the Agilent Bioanalyzer RNA 6000 LabChip kit (Agilent Technologies) or gel analysis.
Microarray analysis for coding and noncoding transcripts.
For microarray analysis, the RNA samples were submitted to Arraystar for coding and noncoding RNA expression profiling. An Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method (Arraystar Flash RNA Labeling Kit, Arraystar). The labeled cRNAs were hybridized onto the Human LncRNA Array v3.0 (8 × 60 K, Arraystar). After slides were washed, the arrays were scanned by the Agilent Scanner G2505C.
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies). After quantile normalization of the raw data, lncRNAs and mRNAs that at least 19 samples have flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis. Differentially expressed lncRNAs and mRNAs were identified using Student's t-test between two groups with a fold change cut-off >2 and P < 0.05. Pathway analysis and GO analysis were applied to determine the roles of these differentially expressed mRNAs played in these biological pathways or GO terms. Finally, Hierarchical Clustering was performed to show the distinguishable lncRNAs and mRNAs expression patterns among samples.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (9) and are accessible through GEO Series accession number GSE77013.
Quantitative-real-time PCR for lncRNA and mRNA.
To analyze human gene expression, 500 ng of RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad; cat. no. 1708891). Real-time PCR was performed in triplicate using the SYBR Green mastermix (Bio-Rad; cat. no. 203450). qPCR primers were designed for the reference genes ACTIN and GAPDH, as well as IFNG-AS1, and IFNG. The SYBR-Green primers were ordered from Integrated DNA Technologies. The sequences are ACTIN-F: 5′-cccagcacaatgaagatcaa-3′; ACTIN-R: 5′-acatctgctggaaggtggac-3′; GAPDH-F: 5′-atgttcgtcatgggtgtgaa-3′; GAPDH-R: 5′-ggtgctaagcagttggtggt-3′; IFNG-AS1-F: 5′-tgcctaggtgctctctgatg-3′; IFNG-AS1-R: 5′-ggcacaacccatggaagac-3′; IFNG-F: 5′-gagtgtggagaccatcaagga-3′; IFNG-R: 5′-gtattgctttgcgttggaca-3′. The qPCRs were performed in a CFX384 Real Time PCR detection system (Bio-Rad). For gene expression of mouse genes 18S and Tmevpg1, TaqMan master mix (Applied Biosystems; cat. no. 4352042) was used and primers were ordered from Applied Biosystems (18S:Mm03928990_g1; Tmevpg1: Mm01161206_m1).
Cell culture.
Jurkat CD4 T cells were generously provided by Dr. Otto Yang's laboratory at UCLA. The cells were grown in RPMI+10%FBS+1% penicillin+1% streptomycin. Experiments were performed in 24-well plates with 1 million cells plated in 1 ml of media. Activation of T cells was achieved by 6 h incubation at 37°C with phorbol-12-myristate-13-acetate (PMA) at a final concentration of 50 ng/ml and ionomycin at 1 μM concentration (PMA: Sigma-Aldrich; cat. no. P8139. Ionomycin: Sigma-Aldrich; cat. no. I3909).
Peripheral mononuclear blood cells were obtained from blood draws at Cedars-Sinai Medical Center, Los Angeles, CA. The samples were obtained from control patients under the IRB no. 23705. CD4 cells were isolated using the EasySep Human CD4+ T cell enrichment kit (Stemcell; cat. no. 19052).
Knockdown and overexpression of lncRNA IFNG-AS1.
Reductions in IFNG-AS1 lncRNA expression were achieved using two custom-made siRNA constructs: siIFNG-AS1#1 sense, caaucuuaaggauacagaatt; siIFNG-AS1#2 sense, caaugcauuuauuaauucutt (Ambion). siRNA control was transfected for comparison (Ambion; cat. no. AM4613). Transfections were performed using Lipofectamine RNAiMax (Invitrogen; cat. no. 13778-075) following the product instructions. Briefly, 5 μl of a 20 μM concentration of siRNA was mixed with 7.5 μl of Lipofectamine RNAiMax in 200 μl of Optimem media (Thermo Scientific; cat. no. 31985-062). The sample was incubated for 15 min and then added to 1 million Jurkat cells for 24 h. Overexpression of the lncRNA was achieved by transfecting 4 μg of pCMV6-AC-IRES expression vector with IFNG-AS1 (Blue Heron). Control cells were transfected with the expression vector alone without insert.
Protein analysis.
IFNG protein production was quantified using the Quantikine ELISA (R&D Systems; cat. no. DIF50). TNF-α protein production was assayed using the Milliplex Human High Sensitivity T cell Magnetic Bed Panel (Millipore; cat. no. HSTCMAG28SPMX13). Jurkat cells were grown in 24-well plates in 1 ml of volume. The cells were activated with PMA/ionomycin for 6 h and medium was collected 24 h after activation. The medium was centrifuged to remove excess cells. Manufacturer's instructions were followed for quantification of protein using both kits. The quantity of protein expression was normalized to cell numbers at the time of collection.
Mouse models of colitis.
Colitis was induced in mice via trinitrobenzenesulfonic acid (TNBS) treatment with modifications of previously described protocols (17). All animal studies were approved by the IACUC. Total RNA from the colonic tissues of the TNBS model was purified by use of the miReasy Mini Kit (Qiagen). Tissue sections were scored for histopathology analyses in a double-blinded manner, as previously reported (17). Animals were maintained at University of California-Los Angeles animal research facility and received standard pelleted chow and water ad libitum. The 6- to 8-wk-old male C57BL/6J mice (n = 8 per group) received a 50-μl intracolonic injection of 40 mg/kg TNBS (Fluka) in 30% ethanol, using a 1-ml syringe (Becton Dickinson) fitted with a polyethylene cannula (Intramedic PE-20 tubing). The control groups were injected with 50 μl of 30% ethanol alone into the colon. The mice were held head down for 1 min after the enema administration to ensure the containment of the TNBS solution into the colon. The colons were fixed in 10% buffered formalin for histological analysis. Paraffin sections were stained with hematoxylin/eosin.
IL10−/− C57BL/6 male (n = 8) and female (n = 8) mice were purchased from Jackson Laboratories (Bar Harbor, ME). Animals were kept under specific pathogen-free conditions and fed a standard diet and water ad libitum, under the approval by the institutional animal care and use committee (IACUC) at the Department of Veteran Affairs. Mice were weighed weekly and monitored for appearance of diarrhea and blood in the stools. Mice that developed severe rectal prolapse that was evident at all times were euthanized. Control mice were of the same background and euthanized at the same age. After euthanasia, the entire colon was excised and a portion of the proximal and distal colon was fixed in 10% buffered formalin for histological analysis. Paraffin sections were stained with hematoxylin/eosin.
Experimental statistical analysis.
Student's t-test for two-group comparisons was performed to determine any statistically significant differences between the experimental groups. P < 0.05 was considered statistically significant. All results are expressed as means ± standard error of the mean.
RESULTS
lncRNA signatures in UC colonic tissues.
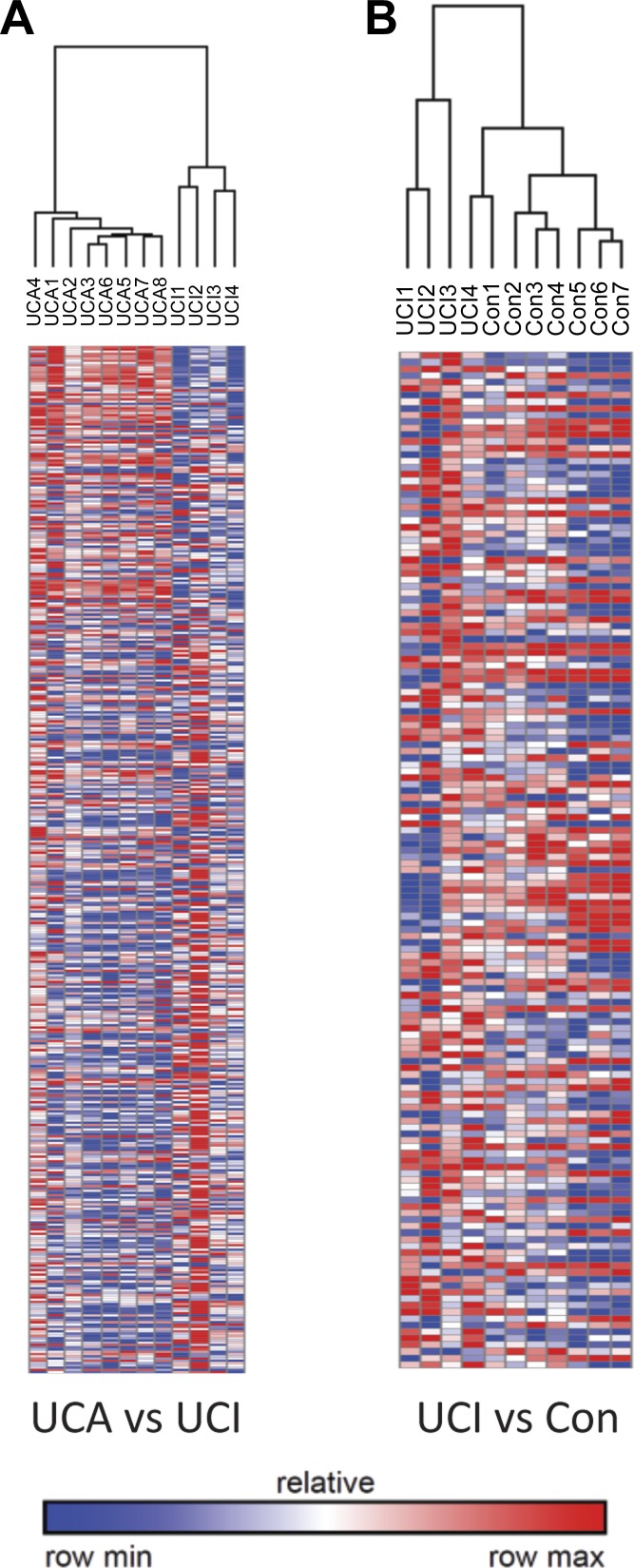
Clinical samples from a total of 19 patients consisting of colon resections from 8 UCA, 4 UCI, and 7 control patients (Table 1), were used to perform lncRNA expression profiling analysis. The lncRNA expression profiling was performed using the Arraystar lncRNA microarray V3.0, which consists of ∼30,600 lncRNAs. This analysis (>2-fold and P < 0.05) revealed that there were 1,931 differentially expressed lncRNAs between UCA and control, while 1,361 lcnRNAs were differentially expressed between UCA and UCI colonic tissues (Figs. 1A and 2). Furthermore, 287 lncRNAs were found to be differentially expressed in UCI relative to control tissues (Fig. 2).
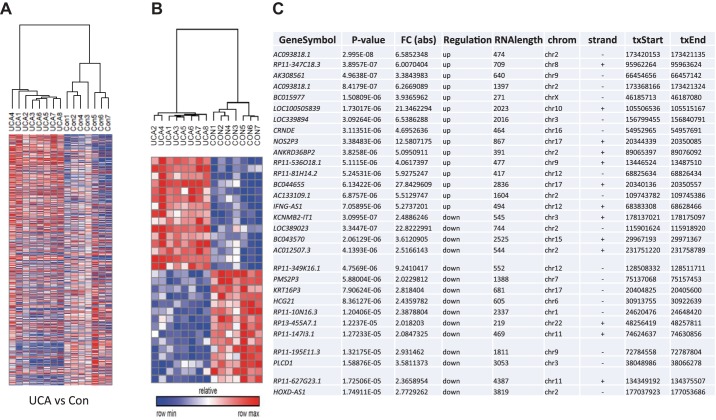
Fig. 1.
A: heat map representation of expression changes between UCA and control (Con) groups with a fold change greater than twofold and P value less than 0.05. B: heat map representation of the top 15 upregulated and 15 downregulated lncRNAs in the UCA vs. Con comparison. C: table shows the identity of the lncRNAs along with their associated P value, fold change, direction of regulation, RNA length, chromosomal location (chrom), strand location (strand), and transcriptional start (txStart) and end (txEnd) positions. Row min and max refer to the minimum and maximum expression levels.
Fig. 2.
Heat map representations of expression changes in lncRNA transcripts between UCA vs. UCI, UCI vs. Con. The number of differentially expression lncRNAs with a fold change cutoff of 2 and a P value cutoff of 0.05.
A focus was placed on the comparison between UCA vs. control samples due to the robust statistical differences seen between these groups as well as access to a separate cohort of UC and control samples for validation studies. As a representative sample of these lncRNAs, Fig. 1, B and C demonstrates the top 15 most statistically significant upregulated and 15 downregulated lncRNAs differentially expressed between the UCA and control samples. As shown, the expression of these 30 lncRNAs enables the clustering of the UCA samples away from the control samples, indicating that these IBD samples express a unique lncRNA expression signature that is distinct from control samples. Figure 1C identifies the 30 differentially expressed lncRNAs and shows the range of expression changes from over 20-fold upregulated to over 20-fold downregulated between the UCA and control patients. These results demonstrate that lncRNAs are significantly differentially expressed between UCA samples and controls and that the identity of the majority of these differentially expressed lncRNAs is unique between these comparisons. Taken together, these analyses show novel lncRNA signatures that were deregulated between UCA, UCI, and control colonic tissues.
Differentially expressed lncRNAs are associated with known IBD loci.
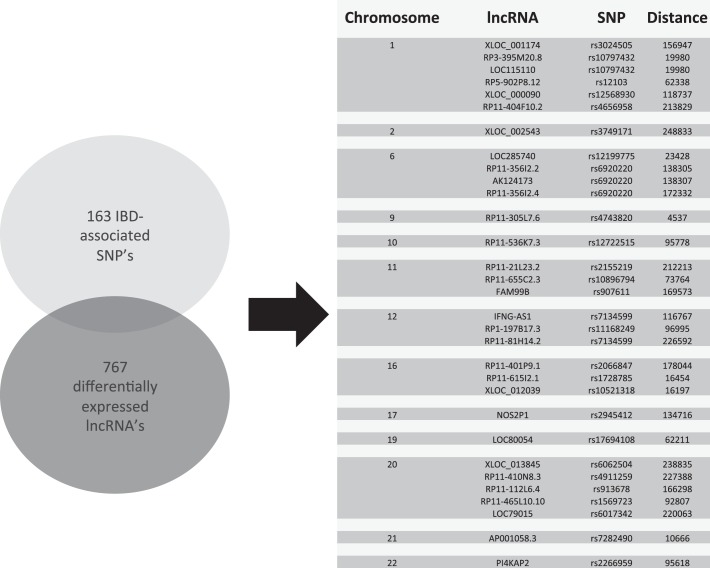
The list of lncRNAs differentially expressed in UCA samples compared with control samples spans the human genome. Previous research has identified 163 distinct single nucleotide polymorphism (SNP) loci as IBD susceptibility loci (14). A comparison of the chromosomal locations was performed between the known IBD susceptibility loci and a list of lncRNAs differentially expressed in IBD samples. A list of lncRNAs was generated to compare the common differentially expressed lncRNAs between the UCA samples and the controls as well as the UCA and the inactive UC samples. This comparison was performed to assess for lncRNAs that were associated with active inflammation in UC samples. The analysis revealed that, of the 767 lncRNAs evaluated, only 31 lncRNAs were found to be within 250 kilobases from the IBD loci (Fig. 3). The 250-kb distance was selected based on the analysis performed by Jostins et al. (14) where a locus was defined as at 500-kb region with the SNP at its center. This comparison demonstrates that a portion of the lncRNAs identified as differentially expressed transcripts between IBD samples and control samples are closely associated with clinically validated IBD susceptibility loci.
Fig. 3.
Graphical representation of the overlap between the 163 IBD associated SNPs and the lncRNAs commonly differentially expressed between UCA vs. Con and UCA vs. UCI. The chromosomal location, lncRNA identity, associated SNP, and genomic distance from the SNPs are presented.
Validating lncRNA expression in a second cohort of UC patient samples.
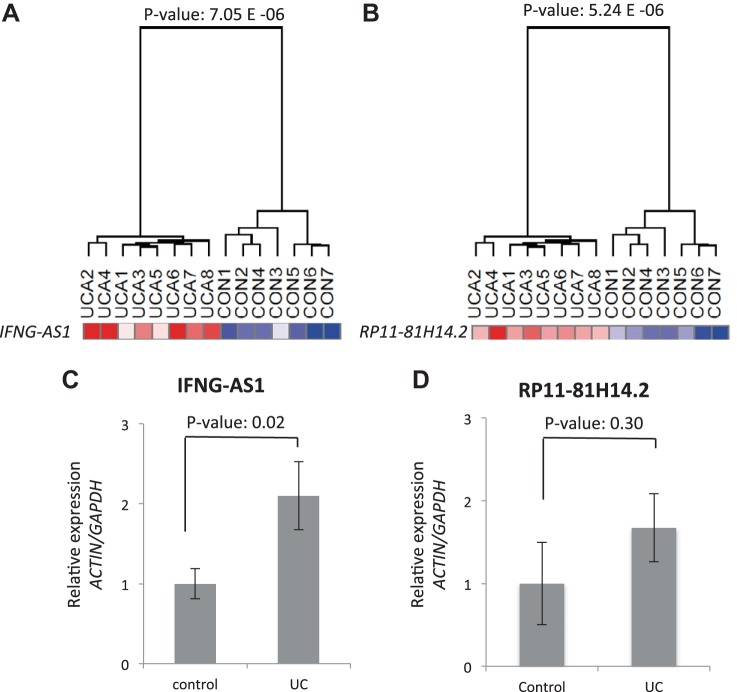
The lncRNA expression differences seen on the microarray analysis showed robust expression changes between the comparison groups. To validate these findings, a separate second cohort of UC samples was assayed for lncRNA expression. The samples consisted of 15 UCA samples and 16 control colonic biopsy samples (Table 2). Total RNA from these samples was extracted and the level of expression for a select number of lncRNAs was performed using quantitative real-time polymerase chain reaction (qRT-PCR). We chose to validate the lncRNAs that were associated with the IBD susceptibility loci and were among the most statistically significant differentially expressed lncRNAs (Fig. 3). The lncRNAs RP11-81H14.2 and IFNG-AS1 were selected for validation (Fig. 4, A and B). RP11-81H14.2 expression analysis did not reveal a statistically significant difference between UC and control samples (Fig. 4D). IFNG-AS1, however, was found to be statistically different between the two sample cohorts and was chosen as a lncRNA of interest for further analysis (Fig. 4C).
Table 2.
Ulcerative colitis and control samples
| Disease | Age | Sex | Pathology | |
|---|---|---|---|---|
| Control | ||||
| 1 | diverticulosis | n/a | n/a | n/a |
| 2 | rectal cancer | 50 | F | benign colonic tissue |
| 3 | rectal villous adenoma | 68 | F | nondysplastic mucosa |
| 4 | metastatic colon cancer | 53 | M | colonic tissue with no histopathological abnormality |
| 5 | colon cancer, cecum | 76 | F | invasive adenocarcinoma of cecum with adajcent normal colon |
| 6 | colon cancer, rectum | 70 | M | colonic adenocarcinoma with adjacent normal colon |
| 7 | colon-normal | n/a | n/a | n/a |
| 8 | colon-normal | n/a | n/a | n/a |
| 9 | colon-normal | n/a | n/a | n/a |
| 10 | colon-normal | n/a | n/a | n/a |
| 11 | colon-normal | n/a | n/a | n/a |
| 12 | colon-normal | 42 | F | foci of endometriosis, large intestine colon without significant change |
| 13 | colon-normal | n/a | n/a | n/a |
| 14 | colon cancer | n/a | n/a | n/a |
| 15 | perforation | n/a | n/a | n/a |
| 16 | vaginal prolapse | n/a | n/a | n/a |
| UC | ||||
| 1 | ulcerative colitis | 61 | F | moderate to severely active ulcerative colitis, negative for dysplasia |
| 2 | ulcerative colitis | 49 | M | chronic pancolitis with severe activity, extensive ulcerations |
| 3 | ulcerative colitis | 52 | F | moderate to severe diffuse active pancolitis negative for dysplasia |
| 4 | ulcerative colitis | 35 | M | moderate to severe activity, chronic pancolitis, negative dysplasia |
| 5 | ulcerative colitis | 68 | F | severely active ulcerative colitis |
| 6 | ulcerative colitis | 31 | M | chronic active colitis with foci of ulceration |
| 7 | ulcerative colitis | 43 | M | n/a |
| 8 | ulcerative colitis | 57 | M | chronic colitis with moderate to severe activity and extensive ulcerations |
| 9 | ulcerative colitis | 39 | F | moderately active ulcerative colitis, negative dysplasia |
| 10 | ulcerative colitis | 30 | F | colon with moderate chronic colitis, no dysplasia |
| 11 | ulcerative colitis | 36 | M | chronic pancolitis with moderate to severe activity, ujlcration and inflammatory polyps |
| 12 | ulcerative colitis | 49 | F | n/a |
| 13 | ulcerative colitis | 64 | M | severe inflammatory bowel disease type indeterminate |
| 14 | ulcerative colitis | 34 | F | rectosigmoid, involved |
| 15 | ulcerative colitis | n/a | n/a | n/a |
n/a, Not available.
Fig. 4.
A and B: microarray expression data for the lncRNAs IFNG-AS1 and RP11-81H14.2 in the analysis comparing UCA vs. Con. C and D: qPCR expression for IFNG-AS1 and RP11-81H14.2 in the validation cohort (control N = 16, UC N = 15). ACTIN and GAPDH were used as control housekeeping genes. Relative expression was to the average expression in the control samples.
lncRNA-associated gene pathway analysis.
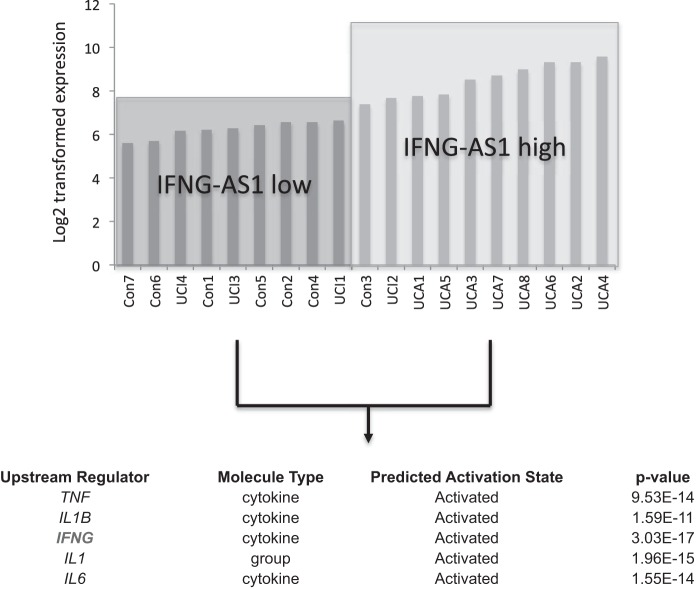
To understand how changes in lncRNA expression may affect phenotypic changes in UC, an analysis of the changes in coding transcript expression was performed. It was hypothesized that changes in long noncoding RNA expression may affect coding transcript expression and as a result alter distinct gene pathways. To address this, the coding transcripts of each of the UC samples and control samples were assayed using the microarray platform from Arraystar. Nineteen patient samples were then divided into two groups: those expressing relatively high quantities of IFNG-AS1 and those expressing relatively low amounts IFNG-AS1 as determined by their expression level on the microarray analysis (Fig. 5). In dividing the clinical samples into these two groups, the coding transcripts could then be assessed for differences in expression between the two groups. A threshold of a fold change of 1.5 upregulated and 2 downregulated was used to generate the coding transcript list. A total of 426 coding transcripts were differentially expressed using these cutoffs. By averaging the expression of the differentially expressed coding transcripts, a gene signature associated with relative IFNG-AS1 expression could be generated. Taking this list of differentially expressed coding transcripts, a bioinformatics gene pathway analysis was performed using Ingenuity Pathway Analysis software. Based on this analysis, several upstream regulators were shown to be activated in the IFNG-AS1 high samples including IFNG, IL1, IL6, and TNF-α (Fig. 5). Of particular interest, was the enhancement of IFNG signaling in the group that expresses higher levels of IFNG-AS1 as this had the lowest associated P value (Fig. 5).
Fig. 5.
Expression of IFNG-AS1 based on microarray expression across all UC and control samples. Two groups were created showing relative high and low levels of IFNG-AS1. Bioinformatic analysis was performed on the associated coding transcript expression changes between the two groups. A list of upstream regulators found to be activated in the IFNG-AS1 high group are listed along with the predicated activation state and the associated P value.
IFNG-AS1 expression in CD4 T cells.
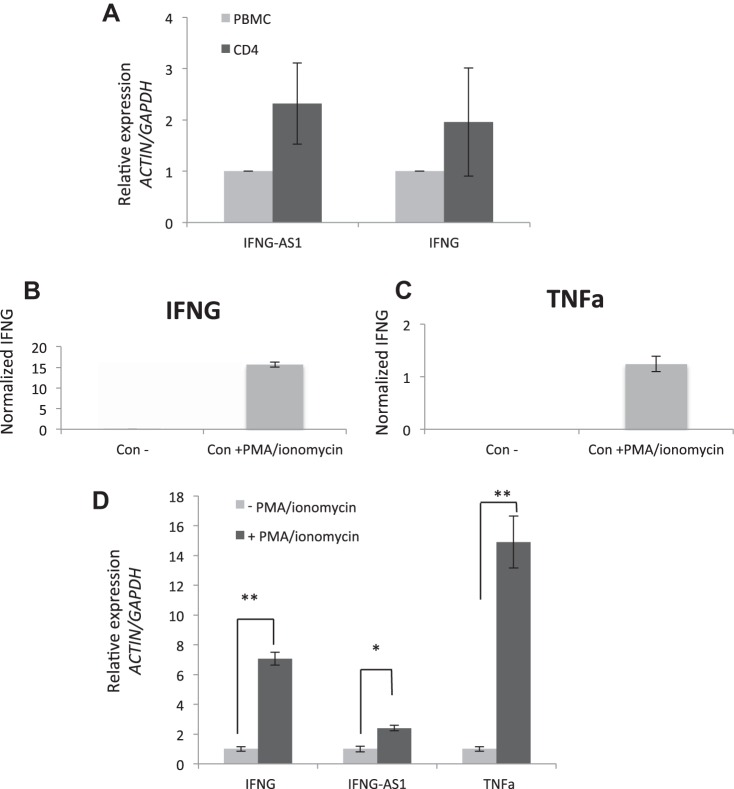
The lncRNA IFNG-AS1 has previously been implicated in immune cell biology in both mouse and human studies (11, 39). In an attempt to identify a target cell for our selected lncRNA, we first examined peripheral CD4 T cells isolated from control patients. The expression of IFNG-AS1 was compared in the total peripheral mononuclear cells with the selected CD4 T subpopulation. Figure 6A shows qRT-PCR data showing that CD4 T subpopulations have enriched expression of IFNG-AS1 consistent with recently published data that IFNG-AS1 is expressed in the Th1 immune cells (39).
Fig. 6.
A: qPCR expression of IFNG-AS1 and IFGN among CD4 cells and total PBMC (N = 4). B and C: Jurkat T cells activated with PMA/ionomycin for 6 h and cultured media collected after 24 h show inductions of IFNG and TNF-α protein, respectively. D: Jurkat T cells activated with PMA/ionomycin for 6 h; qPCR analysis of IFNG, IFNG-AS1, and TNF-α. *P value < 0.05, **P value < 0.01; n = 3.
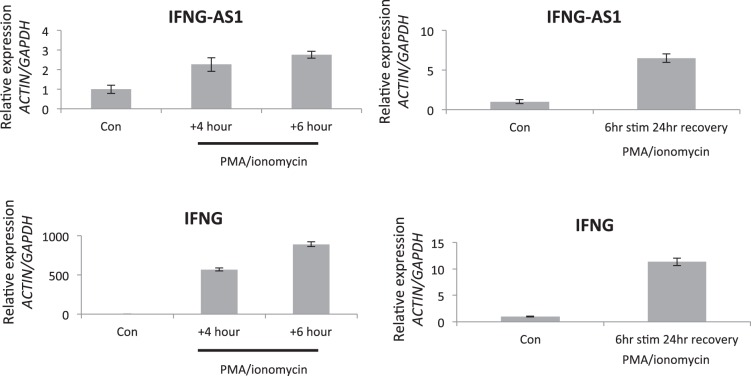
Given these results, we chose to study the function of IFNG-AS1 using the Jurkat immune T cell model. Jurkat CD4 T cells can be activated via PMA and ionomycin stimulation and can be induced to secrete a variety of immune mediators such as IFNG and TNF-α (Fig. 6, B and C). Analyzing the RNA expression, PMA/ionomycin was found to induce IFNG-AS1 as well as IFNG and TNF-α (Fig. 6D). Figure 7 highlights the time course of IFNG-AS1 and IFNG expression with IFNG-AS1 reaching maximal induction 24 h after stimulation with PMA/ionomycin.
Fig. 7.
qPCR expression for a time course evaluating the expression IFNG-AS1 and IFNG in Jurkat T cells.
IFNG-AS1 regulates IFNG expression at the RNA and protein level.
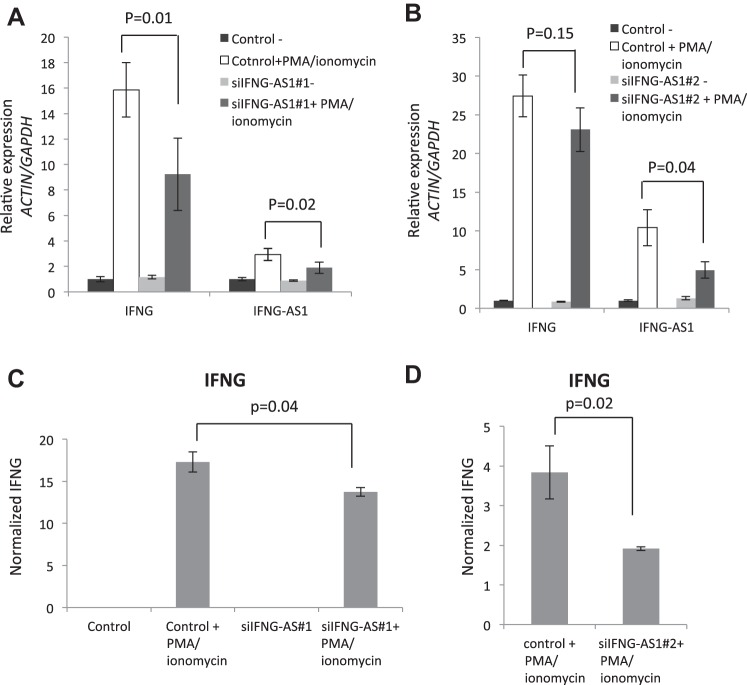
Previous studies in mouse immune cells have suggested that Ifng-as1 can directly regulate the expression of its genomic neighbor, Ifng (11, 36). These studies reveal a pattern of histone methyltransferase recruitment to the Ifng promoter resulting in positive regulation of Ifng expression. We sought to show that the human ortholog, IFNG-AS1, could regulate IFNG production in a similar positive relationship. Using two siRNA constructs, we transiently transfected Jurkat T cells and assessed the expression of IFNG-AS1 and IFNG in the context of T cell activation via PMA/ionomycin. The induction of IFNG-AS1 was abrogated in both siRNAs by ∼30–40% and resulted in reduced expression of IFNG RNA (Fig. 8, A and B).
Fig. 8.
A and B: qPCR analysis of Jurkat T cells treated with PMA/ionomycin for 6 h followed by a 24-h recovery period. Cells were treated with 2 independent antisense siRNAs against IFNG-AS1 (n = 3). C and D: protein analysis for IFNG was assessed by measuring the culture medium (n = 3). The siRNAs refer to small interfering RNAs with siIFNG-AS1#1 and siIFNG-AS1#2 referring to two independent small interfering RNAs directed against IFNG-AS1.
Protein expression was then analyzed with an IFNG-specific ELISA kit. IFNG protein levels were similarly abrogated when IFNG-AS1 expression was reduced (Fig. 8, C and D).
In addition to the knockdown studies, we generated an IFNG-AS1 overexpression construct and transiently transfected it to Jurkat T cells. Figure 9A demonstrates the level of overexpression of IFNG-AS1 achieved with this construct. When transfected into cells and induced with PMA/ionomycin, there was an increase in IFNG RNA levels as early as 4 h of PMA/ionomycin induction (Fig. 9B). Under these conditions, protein expression of IFNG was increased by an average of 45% (Fig. 9C). These data suggest that IFNG-AS1 has a direct relationship with the expression of IFNG within immune cells.
Fig. 9.
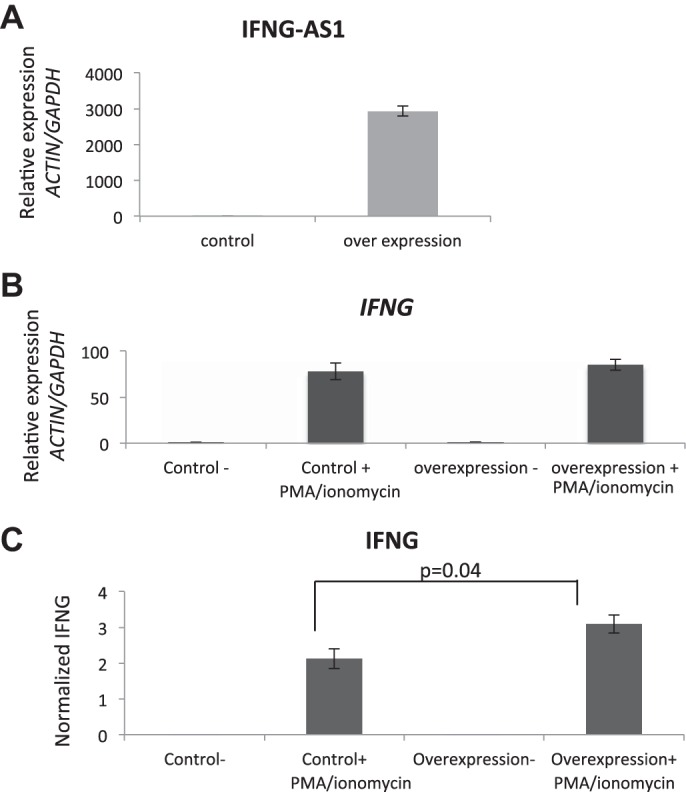
A: qPCR expression of Jurkat T cells transiently overexpressing IFNG-AS1. B: qPCR analysis of Jurkat T cells with and without transient transfection of an IFNG-AS1-overexpressing construct. Cells were activated with PMA/ionomycin for 4 h (n = 3). C: IFNG protein was measured from cultured media (n = 3).
Mouse models of colitis have increased Ifng-as1 levels.
We then assessed whether colitis models would show a similar increase in the expression of the mouse ortholog Ifng-as1. We utilized two mouse models of colitis, the acute TNBS colitis model at 48 h post-TNBS administration as well as the IL10 knockout mouse model of spontaneous colitis at age 12 wk. Colonic tissues were obtained from each mouse model and compared with controls. Expression of the lncRNA was then assessed by qRT-PCR. In both models, the mouse ortholog Ifng-as1 was found to be significantly elevated in the colitis setting compared with control (Fig. 10, A and B). These results are consistent with our human clinical data showing increased IFNG-AS1 expression in colitis tissues.
Fig. 10.
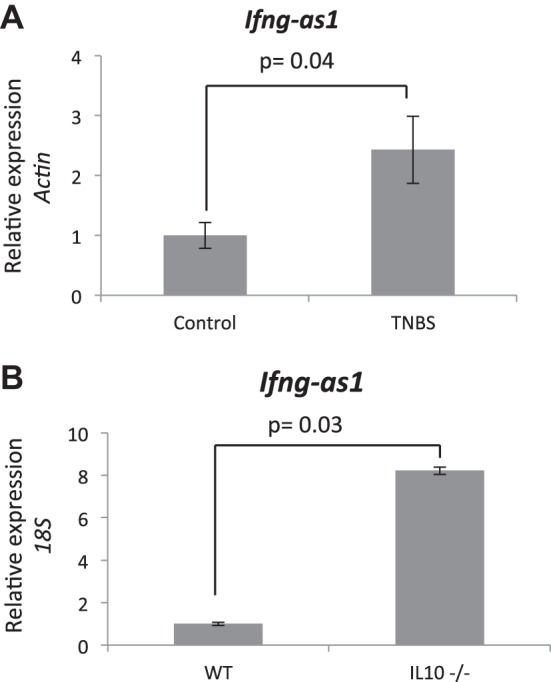
A: Ifgn-as1 qPCR expression analysis. Whole colon samples from TNBS-treated animals vs. controls (N = 5) were assays for Ifng-as1 expression with actin as the control gene. B: Ifng-as1 qPCR analysis from colon tissue derived from IL10 knockout animals compared with controls (N = 4).
DISCUSSION
The diagnosis and treatment of IBD faces many challenges that can only be addressed by gaining a deeper understanding of the molecular mechanisms involved in the pathogenesis of the disease. Noncoding RNA biology has emerged as a new and potent form of gene regulation and has only recently began to be evaluated in IBD (7, 42). In the past year, Mirza et al. (22) performed the first transcriptomic profiling of lncRNAs in IBD patients and revealed hundreds of differentially expressed lncRNAs between UC, CD, and control patients. Additionally, Wu et al. (41) also reported a profiling analysis of lncRNAs differentially expressed in UC patients vs. controls. They evaluated over 60 patients with and without IBD and identified 455 lncRNAs that were differentially expressed. This group focused on one of the differentially expressed lncRNAs, BC012900, and showed that it can regulate epithelial cell apoptosis. Our IBD studies investigated differentially expressed lncRNAs among UC patients and in addition uncovered new potential mechanisms of action. Through a noncoding RNA expression analysis, we identified a set of lncRNAs that are differentially expressed between UC samples and controls. By focusing on those lncRNAs differentially expressed in UC patients, we then filtered our list to identify lncRNAs that were associated with the clinically validated IBD susceptibility loci. We were able to validate the differential expression of one of these lncRNAs, IFNG-AS1, with the use of a separate clinical cohort of colonic samples. Of note, Mirza et al. also found this lncRNA to be differentially expressed in their IBD patient samples, thereby providing further validation (22). Given its discovery on two independent analyses, IFNG-AS1 presents itself as an ideal candidate for further study. As one of the first mechanistic studies for lncRNAs in the IBD field, this work focused on IFNG-AS1. Future studies will be needed to investigate additional lncRNAs involved in IBD pathophysiology.
IFNG-AS1 is a noncoding transcript located on chromosome 12 and adjacent to the IFNG gene. Prior studies have suggested that IFNG-AS1 can positively regulate IFNG expression; however, much of the data implies function from the mouse ortholog ifng-as1/NeST/tmevpg1. These previous investigations have shown that ifng-as1 is expressed in immune cells such as CD8 and CD4 T cells (5, 11, 37). The prior studies highlighted a mechanism wherein ifng-as1 recruits a subunit of MLL/SET1 H3K4 methyltransferase complex to the ifng promoter and lays down an activating histone methylation signal that promotes expression of ifng in mouse immune cells (11). This mechanism, however, has yet to be validated in the human context. Indeed, the human studies of IFNG-AS1 by Collier et al. (5) in the context of general immune cell biology and Wang et al. (39) in the context of Sjogren syndrome have shown expression of this noncoding transcript within CD4+ T cells. Our own studies show that a site of expression of IFNG-AS1 is within these CD4+ cells. Using knockdown and overexpression constructs in human T cells, we directly show that IFNG-AS1 expression can positively regulate IFNG production. We suspect the relatively low abundance of the IFNG-AS1 transcript results in modest knockdown efficiencies seen in our model system. However, our data suggest that within immune cells there is a mechanistic link between the overexpression of this noncoding RNA in colitis samples and the production of a key inflammatory cytokine. In addition, lncRNAs have also been implicated in the stability of mRNA transcripts, thereby providing additional modes by which IFNG-AS1 could potentially regulate its targets (38).
Through this work, we have identified a novel set of lncRNAs implicated in UC pathophysiology. Additional studies that are focused not only on IFNG-AS1 but also on the other noted lncRNAs presented in this work will be essential in expanding our knowledge of UC pathogenesis. lncRNAs have served as novel targets in preclinical studies from other fields of medicine such as cancer biology and may serve as important therapeutic targets in UC (19). Additionally, lncRNAs could also serve as important biomarkers with a range of uses (34). lncRNA biology is a nascent field in IBD and much work is needed to expand its potential applications. Future studies evaluating the effects of the IBD susceptibility SNPs on expression of lncRNAs will help to link the genotypic changes seen in patients with specific SNPs with expression changes in SNP-associated genes. Additionally, mouse models of colitis will need to be employed to better define the mechanisms for lncRNA biology. To this end, this study has identified that the murine ortholog ifgn-as1 is also overexpressed in the context of mouse models of colitis. This opens up additional avenues of research to carefully dissect other mechanisms of action for this noncoding RNA. The generation of ifng-as1 knockout and transgenic animals provides additional tools to investigate its function (11). All told, the lncRNA biology serves as an additional layer of gene regulation adding to the complexity of IBD pathophysiology but also providing new tissue-specific targets.
GRANTS
This research was supported by NIH/NIDDK T32 DK007180-41 grant (D. Padua), and R01 DK060729 (C. Pothoulakis). Support was also provided by NIH/NIDDK P30 DK 41301 Center Grant (D. Iliopoulos and C. Pothoulakis).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
D.M.P., C. Polytarchou, and D.I. conception and design of research; D.M.P., I.K.M.L., J.P.V., and D.Q.S. performed experiments; D.M.P., S.M.-J., and I.K.M.L. analyzed data; D.M.P. interpreted results of experiments; D.M.P. prepared figures; D.M.P. drafted manuscript; D.M.P., J.R.P., D.Q.S., D.I., and C. Pothoulakis edited and revised manuscript; D.I. and C. Pothoulakis approved final version of manuscript.
ACKNOWLEDGMENTS
We thank I. Karagiannides, K. Bakirtzi, M. Hatziapostolou, C. Vorvis, and M. Koutsioumpa for helpful discussions and technical suggestions. We also acknowledge E. Faure-Kumar in the Center for Systems for Biomedicine for expert technical assistance.
REFERENCES
- 1.Bank S, Andersen PS, Burisch J, Pedersen N, Roug S, Galsgaard J, Ydegaard Turino S, Brodersen JB, Rashid S, Kaiser Rasmussen B, Avlund S, Bastholm Olesen T, Hoffmann HJ, Andersen Nexø B, Sode J, Vogel U, Andersen V. Polymorphisms in the Toll-like receptor and the IL-23/IL-17 pathways were associated with susceptibility to inflammatory bowel disease in a Danish cohort. PLoS One 10: e0145302, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O'Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341: 789–792, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho JH, Weaver CT. The genetics of inflammatory bowel disease. Gastroenterology 133: 1327–1339, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 189: 2084–2088, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui D, Huang G, Yang D, Huang B, An B. Efficacy and safety of interferon-γ-targeted therapy in Crohn's disease: a systematic review and meta-analysis of randomized controlled trials. Clin Res Hepatol Gastroenterol 37: 507–513, 2013. [DOI] [PubMed] [Google Scholar]
- 7.Dalal SR, Kwon JH. The role of microRNA in inflammatory bowel disease. Gastroenterol Hepatol (N Y) 6: 714–722, 2010. [PMC free article] [PubMed] [Google Scholar]
- 8.Eckburg PB, Relman DA. The role of microbes in Crohn's disease. Clin Infect Dis 44: 256–262, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S, Chaudhary R, Carpani M, Playford R. Interfering with interferons in inflammatory bowel disease. Gut 55: 1071–1073, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152: 743–754, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, Lund RJ, Li Y, Ullah U, Modderman R, Abdulahad W, Lahdesmaki H, Franke L, Lahesmaa R, Wijmenga C, Withoff S. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med 6: 88, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H; International IBD Genetics Consortium (IIBDGC), Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koukos G, Polytarchou C, Kaplan JL, Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M, Pothoulakis C, Winter HS, Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology 145: 842–852.e2, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koukos G, Polytarchou C, Kaplan JL, Oikonomopoulos A, Ziring D, Hommes DW, Wahed R, Kokkotou E, Pothoulakis C, Winter HS, Iliopoulos D. A microRNA signature in pediatric ulcerative colitis: deregulation of the miR-4284/CXCL5 pathway in the intestinal epithelium. Inflamm Bowel Dis 21: 996–1005, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law IK, Bakirtzi K, Polytarchou C, Oikonomopoulos A, Hommes D, Iliopoulos D, Pothoulakis C. Neurotensin-regulated miR-133α is involved in proinflammatory signalling in human colonic epithelial cells and in experimental colitis. Gut 64: 1095–1104, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci USA 111: 1002–1007, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, Subramaniyan S, Carley A, Niknafs YS, Sahu A, Han S, Ma T, Liu M, Asangani IA, Jing X, Cao X, Dhanasekaran SM, Robinson DR, Feng FY, Chinnaiyan AM. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res 12: 1081–1087, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marafini I, Angelucci E, Pallone F, Monteleone G. The IL-12/23/STAT axis as a therapeutic target in inflammatory bowel disease: mechanisms and evidence in man. Dig Dis 33, Suppl 1: 113–119, 2015. [DOI] [PubMed] [Google Scholar]
- 21.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159, 2009. [DOI] [PubMed] [Google Scholar]
- 22.Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, Gorodkin J, Pociot F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med 7: 39, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, Gorodkin J, Pociot F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med 7: 39, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142: 46–54.e42; quiz e30, 2012. [DOI] [PubMed] [Google Scholar]
- 25.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol 14: 329–342, 2014. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen OH, Ainsworth MA. Tumor necrosis factor inhibitors for inflammatory bowel disease. N Engl J Med 369: 754–762, 2013. [DOI] [PubMed] [Google Scholar]
- 27.Peyrin-Biroulet L, Loftus EV Jr, Colombel JF, Sandborn WJ. Long-term complications, extraintestinal manifestations, and mortality in adult Crohn's disease in population-based cohorts. Inflamm Bowel Dis 17: 471–478, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, van der Meulen-de Jong AE, Oikonomopoulos A, van Deen WK, Vorvis C, Serebrennikova OB, Birli E, Choi J, Chang L, Anton PA, Tsichlis PN, Pothoulakis C, Verspaget HW, Iliopoulos D. MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology 149: 981–992.e11, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 1: 391–407, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29: 742–749, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 45: 1392–1398, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR, de Villiers WJ, Present D, Sands BE, Colombel JF. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med 353: 2462–2476, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Sandborn WJ, van Assche G, Reinisch W, Colombel JF, D'Haens G, Wolf DC, Kron M, Tighe MB, Lazar A, Thakkar RB. Adalimumab induces and maintains clinical remission in patients with moderate-to-severe ulcerative colitis. Gastroenterology 142: 257–265.e1, e3, 2012. [DOI] [PubMed] [Google Scholar]
- 34.Shang C, Guo Y, Zhang H, Xue YX. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol 77: 507–513, 2016. [DOI] [PubMed] [Google Scholar]
- 35.Tillack C, Ehmann LM, Friedrich M, Laubender RP, Papay P, Vogelsang H, Stallhofer J, Beigel F, Bedynek A, Wetzke M, Maier H, Koburger M, Wagner J, Glas J, Diegelmann J, Koglin S, Dombrowski Y, Schauber J, Wollenberg A, Brand S. Anti-TNF antibody-induced psoriasiform skin lesions in patients with inflammatory bowel disease are characterised by interferon-γ-expressing Th1 cells and IL-17A/IL-22-expressing Th17 cells and respond to anti-IL-12/IL-23 antibody treatment. Gut 63: 567–577, 2014. [DOI] [PubMed] [Google Scholar]
- 36.Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of γ interferon. J Virol 7: 5632–5638, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of γ interferon. J Virol 77: 5632–5638, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang GQ, Wang Y, Xiong Y, Chen XC, Ma ML, Cai R, Gao Y, Sun YM, Yang GS, Pang WJ. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep 6: 21865, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Peng H, Tian J, Ma J, Tang X, Rui K, Tian X, Wang Y, Chen J, Lu L, Xu H, Wang S. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjogren syndrome. Immunol Res 64: 489–496, 2016. [DOI] [PubMed] [Google Scholar]
- 40.Weinstock JV. Helminths and mucosal immune modulation. Ann NY Acad Sci 1072: 356–364, 2006. [DOI] [PubMed] [Google Scholar]
- 41.Wu F, Huang Y, Dong F, Kwon JH. Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis. Inflamm Bowel Dis 22: 782–795, 2016. [DOI] [PubMed] [Google Scholar]
- 42.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 α. Gastroenterology 135: 1624–1635.e24, 2008. [DOI] [PubMed] [Google Scholar]