We measured mRNA expression by RT-PCR of 91 genes reflecting tight junction proteins, chemokines, innate immunity, ion channels, and transmitters in small intestinal mucosa from 15 IBS-D and 7 controls (biopsies negative for celiac disease). The following genes were significantly upregulated (q < 0.05) in irritable bowel syndrome with diarrhea (IBS-D): INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15. Protein expression of PPP2R5C in nuclear lysates was greater in patients with IBS-D compared with controls. Intestinal mucosal function deserves further study in IBS-D.
Keywords: cytokines, barrier, immune, secretion
Abstract
Prior studies in with irritable bowel syndrome with diarrhea (IBS-D) patients showed immune activation, secretion, and barrier dysfunction in jejunal or colorectal mucosa. We measured mRNA expression by RT-PCR of 91 genes reflecting tight junction proteins, chemokines, innate immunity, ion channels, transmitters, housekeeping genes, and controls for DNA contamination and PCR efficiency in small intestinal mucosa from 15 IBS-D and 7 controls (biopsies negative for celiac disease). Fold change was calculated using 2(−ΔΔCT) formula. Nominal P values (P < 0.05) were interpreted with false detection rate (FDR) correction (q value). Cluster analysis with Lens for Enrichment and Network Studies (LENS) explored connectivity of mechanisms. Upregulated genes (uncorrected P < 0.05) were related to ion transport (INADL, MAGI1, and SONS1), barrier (TJP1, 2, and 3 and CLDN) or immune functions (TLR3, IL15, and MAPKAPK5), or histamine metabolism (HNMT); downregulated genes were related to immune function (IL-1β, TGF-β1, and CCL20) or antigen detection (TLR1 and 8). The following genes were significantly upregulated (q < 0.05) in IBS-D: INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15. Among the 14 nominally upregulated genes, there was clustering of barrier and PDZ domains (TJP1, TJP2, TJP3, CLDN4, INADL, and MAGI1) and clustering of downregulated genes (CCL20, TLR1, IL1B, and TLR8). Protein expression of PPP2R5C in nuclear lysates was greater in patients with IBS-D and controls. There was increase in INADL protein (median 9.4 ng/ml) in patients with IBS-D relative to controls (median 5.8 ng/ml, P > 0.05). In conclusion, altered transcriptome (and to lesser extent protein) expression of ion transport, barrier, immune, and mast cell mechanisms in small bowel may reflect different alterations in function and deserves further study in IBS-D.
NEW & NOTEWORTHY
We measured mRNA expression by RT-PCR of 91 genes reflecting tight junction proteins, chemokines, innate immunity, ion channels, and transmitters in small intestinal mucosa from 15 IBS-D and 7 controls (biopsies negative for celiac disease). The following genes were significantly upregulated (q < 0.05) in irritable bowel syndrome with diarrhea (IBS-D): INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15. Protein expression of PPP2R5C in nuclear lysates was greater in patients with IBS-D compared with controls. Intestinal mucosal function deserves further study in IBS-D.
the causes of loose bowel movements in patients with irritable bowel syndrome with diarrhea (IBS-D) include organ level alterations of function such as acceleration of colonic transit, documented in ∼45% of patients with IBS-D (13), and intestinal secretory mechanisms (reviewed in Ref. 9). The latter include increased duodenal and rectosigmoid expression of secretory transmitters (e.g., 5-HT) measured by morphological studies, reduced expression of the serotonin reuptake protein, and fecal excretion of secretogranins or chromogranins (18, 19, 38). There was also evidence of reduced expression of proabsorption mechanisms (e.g., mucosal PYY, somatostatin, and neuropeptide Y).
IBS has also been associated with changes in rectosigmoid mucosal mRNA expression of immune factors, barrier function, and mucus secretion (2, 5, 6, 10, 12, 14, 26, 43, 46, 48, 50, 51, 53). The upregulated mechanisms measured initially by RNA sequencing in 9 patients with IBS-D were associated with changes in ion transport and included PDZD3 and GUCA2B (12); these results were largely confirmed in a replication study using RT-PCR in 47 patients with IBS-D (11). PDZ adapter proteins are involved in multiple ion transport functions in the intestine, and GUCA2B is the gene controlling the endogenous uroguanylin that induces chloride secretion. We also demonstrated [with false detection rate correction (FDR)] abnormal expression of mRNA in rectosigmoid mucosal biopsies of receptors or neurotransmitters (P2RY4 and VIP), cytokines and complement components (C4BPA and CCL20), immune function (e.g., TNFSF15), and mucosal repair and cell adhesion (TFF1 and FN1).
Differences in jejunal mucosal expression (at gene and protein levels) and in the distribution of apical junction complex proteins (34, 35), as well as reduced ZO-1 expression in HLA DQ2/8-positive patients with non-celiac IBS-D (46), support the observed alterations in barrier mechanisms seen in colonic mucosa in patients with IBS-D.
In this study, our aim was to compare, in small intestinal mucosa of patients with IBS-D and controls with normal small intestinal mucosa, the expression of a larger complement of genes potentially associated with the pathobiology of IBS-D, including tight junction proteins, chemokines, innate immunity, ion channels, and transmitters.
METHODS
Ethical approval.
The study was approved by Mayo Clinic Institutional Review Board on June 3, 2014 (IRB No. 14-002151), and an amendment was approved on October 28, 2015. Written informed consent was received from participants before inclusion in the study.
Study design.
We appraised bowel functions and descending duodenal mucosal mRNA expression in 15 patients with IBS-D (by Rome III criteria) and 7 controls who had undergone clinically indicated duodenal biopsies (predominantly to exclude celiac disease in patients with iron deficiency), which were morphologically normal.
Patient selection.
Patients were recruited by public advertisement or by invitation to participate from a database of ∼1,200 patients with IBS living within ∼120 miles of the Mayo Clinic in Rochester, MN. Inclusion criteria were based on symptoms using validated diary questionnaire that characterized IBS symptoms and particularly bowel functions (44). Participants also completed the Hospital Anxiety and Depression Inventory (52). These patients had been evaluated at Mayo Clinic, and alternative diagnoses such as inflammatory bowel disease, cancer, and celiac disease were excluded. The main exclusion criterium was intake of medications that could cause bleeding (owing to the need for duodenal biopsies). During a 14-day period (±4 days), the patients with known IBS-D completed a daily bowel pattern diary [that included the 7-point Bristol Stool Form Scale (30), ranging from 1 or hard lumpy stool to 7 or watery diarrhea] to record their bowel habits; they also recorded daily their average abdominal pain severity and worst pain severity using a 100-mm visual analog scale. Clinical details of the seven controls who underwent clinically indicated duodenal biopsies are shown in results (see Table 2).
Table 2.
Clinical features and diagnosis
| A. Clinical Features of Patients with IBS-D (at baseline) | |
|---|---|
| Daily Score | Mean (SE) |
| #Stools/day | 2.35 (0.203) |
| Stool form (Bristol Stool Form Scale, 1–7: 1 = hard lumps; 7 = watery) | 5.13 (0.146) |
| Stool ease of passage (based on scale 1–7: 1 = manual disimpaction; 7 = incontinence) | 4.70 (0.095) |
| Proportion incomplete evacuation (0 = no; 1 = yes) | 0.37 (0.070) |
| Average pain severity (pain scales 0–100 mm, visual analog scale: 1 = none; 100 = worst ever) | 18.88 (4.425) |
| Worst pain severity (pain scales 0–100 mm, visual analog scale: 1 = none; 100 = worst ever) | 24.38 (4.561) |
| Fasting serum FGF-19, pg/ml | 115 (17) |
| Fasting serum 7αC4, ng/ml | 33.7 (6.9) |
| B. Controls Undergoing Duodenal Biopsy for Clinical Diagnosis (All Negative for Celiac Disease) | ||||
|---|---|---|---|---|
| Control Patients | Age yr | Gender | BMI, kg/m2 | Symptoms |
| 1 | 24 | M | 21.8 | Diarrhea (loose stools) |
| 2 | 18 | F | 23.9 | Dizziness, postive anti-gliadin IgG and IgA antibodies; no gastrointestinal symptoms |
| 3 | 67 | F | 19.3 | Diarrhea, weight loss, epigastric pain/discomfort |
| 4 | 26 | F | 27.5 | Diarrhea (48-h stool weight 800 g), abdominal pain; Past medical illness: rheumatoid arthritis |
| 5 | 49 | F | 15.9 | Diarrhea, weight loss, iron deficiency anemia |
| 6 | 54 | M | 20.8 | Vomiting, diarrhea, colon polyps, weight loss, chronic nausea |
| 7 | 47 | F | 15.2 | Diarrhea, bloating, burping, nausea, weight loss |
Normal fasting serum 7α C4 >47.1 ng/ml and serum FGF-19 <131pg/ml, based on 90th percentile of healthy volunteers data. IBS-D, irritable bowl syndrome with diarrhea; BMI, body mass index; M, male; F, female.
Stored biospecimens.
The biopsies from patients with IBS-D were obtained at baseline (before treatment) in a study of the effects of therapy with serum bovine immunoglobulin (results to be published elsewhere).
We used stored samples from seven patients who had consented to the use of biospecimens (for future research) in prior studies (Principal Investigator: Joseph A. Murray) conducted at Mayo Clinic in Rochester, MN. These samples were from patients who underwent biopsies for clinical indications such as iron deficiency, with no evidence of small intestinal abnormality on biopsy. We chose to study “disease controls,” most of whom had diarrhea that was not caused by celiac disease and did not fulfill criteria for IBS-D, to control for the presence of diarrhea. In this pilot study, we considered it was important to identify differences in expression that were not caused by the diarrhea per se.
Biopsies were preserved in a solution of RNAlater and stored at −80°C until the time of assay of gene expression.
Selection of genes of interest.
We developed a custom profile including 89 genes (reflecting tight junction proteins, chemokines, innate immunity, ion channels, and transmitters; Table 1), extending from the previous 19 gene profile (12) as well as housekeeping (HKG) genes for normalization (B2M, ACTB, and GAPDH). Subsequently, we included SCN9A (NHE3) and CFTR expression in view of their known roles in the absorption of Na+ and secretion of Cl− ions, respectively. In addition, controls monitored DNA contamination, and first strand synthesis and PCR efficiency were included to check for sample quality and reaction quality.
Table 1.
Genes of interest included in the RT-PCR analysis of duodenal mucosa
| Gene Symbol | Refseq No. | Official Full Name |
|---|---|---|
| ACTB | NM_001101 | Actin, beta (house-keeping gene [HKG]) |
| C4BPA | NM_000715 | Complement component 4 binding protein, alpha |
| CCL20 | NM_004591 | Chemokine (C-C motif) ligand 20 |
| CLDN1 | NM_021101 | Claudin 1 |
| FGFR4 | NM_002011 | Fibroblast growth factor receptor 4 |
| FN1 | NM_002026 | Fibronectin 1 |
| GAPDH | NM_002046 | Glyceraldehyde-3-phosphate dehydrogenase (HKG) |
| GPBAR1 | NM_170699 | G protein-coupled bile acid receptor 1 |
| GUCA2B | NM_007102 | Guanylate cyclase activator 2B (uroguanylin) |
| HGDC | SA_00105 | Human genomic DNA contamination |
| IFIT3 | NM_001549 | Interferon-induced protein with tetratricopeptide repeats 3 |
| NR1H4 | NM_005123 | Nuclear receptor subfamily 1, group H, member 4 |
| OCLN | NM_002538 | Occludin |
| P2RY4 | NM_002565 | Pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| PDZD3 | NM_024791 | PDZ domain containing 3 |
| PPC | SA_00103 | Positive PCR control |
| RBP2 | NM_004164 | Retinol binding protein 2, cellular |
| RTC | SA_00104 | Reverse transcription control |
| SLC6A4 | NM_001045 | 5-HT transporter |
| SLC10A2 | NM_000452 | Solute carrier family 10 member 2 (sodium/bile acid cotransporter) |
| TFF1 | NM_003225 | Trefoil factor 1 |
| TJP1 | NM_175610 | Tight junction protein 1 (zona occludens 1) |
| TNFSF15 | NM_005118 | Tumor necrosis factor (ligand) superfamily, member 15 |
| VIP | NM_003381 | Vasoactive intestinal peptide |
| IFNG | NM_000619 | Interferon-gamma |
| MYLK | NM_053025 | Myosin light chain kinase |
| SLC9A1 | NM_003047 | Na+-H+-exchange protein |
| B2M | NM_004048 | Beta 2 microglobulin (HKG) |
| CALR | NM_004343 | CALReticulin |
| CD3E | NM_000733 | CD3-epsilon chain |
| CD74 | NM_004355 | HLA-DR antigen-associated invariant chain |
| CLDN2 | NM_020384 | Claudin 2 |
| CLDN3 | NM_001306 | Claudin 3 |
| CLDN4 | NM_001305 | Claudin 4 |
| CLDN7 | NM_001307 | Claudin 7 |
| CLDN12 | NM_012129 | Claudin 12 |
| CLDN15 | NM_014343 | Claudin 15 |
| CLDN16 | NM_006580 | Claudin 16 |
| CPSF1 | NM_013291 | Cleavage and polyadenylation specific factor 1 |
| CTNNA1 | NM_001903 | Catenin (cadherin-associated protein), alpha 1 |
| CTNNB1 | NM_001904 | Catenin (cadherin-asociated protein), beta1 |
| DLG1 | NM_004087 | Discs, large homolog 1 (Drosophila) |
| FOXP3 | NM_014009 | Forkhead box p3 |
| HAAO | NM_012205 | Kydroxyanthranilic acid oxygenase (3-hydroxyanthranilate 3,4-dioxygenase) |
| HNMT | NM_006895 | Histamine N-methyltransferase |
| HRH1 | NM_000861 | Histamine receptor 1 |
| HRH2 | NM_022304 | Histamine receptor 2 |
| IDO1 | NM_002164 | Indoleamine 2,3-dioxygenase |
| IDO2 | NM_194294 | Indoleamine 2,3-dioxygenase 2 |
| IL1B | NM_000576 | Interleukin-1beta |
| IL2RA | NM_000417 | (CD25) Interleukin-2 receptor subunit alpha |
| IL4 | NM_000589 | Interleukin 4 |
| IL6 | NM_000600 | Interleukin-6 |
| IL8 | NM_000584 | Interleukin-8 |
| IL10 | NM_000572 | Interleukin-10 |
| IL13 | NM_002188 | Interleukin-13 |
| IL15 | NM_000585 | Interleukin-15 |
| IL17A | NM_002190 | Interleukin-17A |
| INADL | NM_176877 | InaD-like (Drosophila) |
| AADAT | NM_182662 | Kynurenine aminotransferase 2 (alias KAT2) |
| CCBL2 | NM_001008661 | Kynurenine aminotransferase 3 (alias KAT3) |
| GOT2 | NM_002080 | Kynurenine aminotransferase 4 (alias KAT4) |
| KITLG | NM_003994 | Kit-ligand, stem cell factor |
| KMO | NM_003679 | Kynurenine 3-monooxygenase |
| KYNU | NM_003937 | Kyureninase |
| MAGI1 | NM_004742 | Membrane-associated guanylate kinase, WW and PDZ domain containing 1 |
| MPP5 | NM_022474 | Membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| MPP7 | NM_173496 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| PPP1CB | NM_002709 | Protein phosphatase 1, catalytic subunit, beta isozyme |
| PPP2R5C | NM_001161725 | Protein phosphatase 2, regulatory subunit beta |
| PRG2 | NM_002728 | Major basic protein |
| PVRL3 | NM_015480 | Poliovirus signaling-related 3 |
| QPRT | NM_014298 | Quinolinic acid phosphoribosyltransferase |
| TDO2 | NM_005651 | Tryptophan 2,3-dioxygenase |
| TGFB1 | NM_000660 | Transforming growth factor beta |
| TJP2 | NM_004817 | Zona occludens 2 |
| TJP3 | NM_014428 | Zona occludens 3 |
| TLR1 | NM_003263 | Toll-like receptor 1 |
| TLR2 | NM_003264 | Toll-like receptor 2 |
| TLR3 | NM_003265 | Toll-like receptor 3 |
| TLR4 | NM_138554 | Toll-like receptor 4 |
| TLR5 | NM_003268 | Toll-like receptor 5 |
| TLR6 | NM_006068 | Toll-like receptor 6 |
| TLR7 | NM_016562 | Toll-like receptor 7 |
| TLR8 | NM_138636 | Toll-like receptor 8 |
| TLR9 | NM_017442 | Toll-like receptor 9 |
| TNF | NM_000594 | Tumor necrosis factor-alpha (TNF-a) |
| TNFSF14 | NM_003807 | LIGHT/tumor necrosis factor superfamily 14 |
| TPH1 | NM_004179 | Tryptophan hydroxylase 1 |
| TPSAB1 | NM_003294 | Tryptase |
| TPSB2 | NM_024164 | Tryptase beta 2 (gene/pseudogene) |
| AHR | NM_001621 | Aryl hydrocarbon Rrceptor |
| SOS1 | NM_005633 | Son of Sevenless homolog 1 (Drosophila) |
| MAPKAPK5 | NM_003668 | Mitogen-activated protein kinase-activated protein kinase 5 |
| MKNK2 | NM_017572 | MAP kinase interacting serine/threonine kinase 2 |
| SLC9A3 | NM_004174 | Solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| CFTR | NM_000492 | Cystic fibrosis transmembrane conductance regulator (ATP-binding cassette subfamily c, member 7) |
Thus, the RT2-Profiler-PCR-Array data examined 96 genes on 22 samples (n = 15 IBS-D patients and 7 healthy controls). Out of the 96 genes, two were identical (CCBL2, used for quality control), three were the housekeeping genes (B2M, ACTB, and GAPDH), and another three (HGDC, RTC, and PPC) were assay controls. Therefore, the custom array analysis focused on 89 genes.
Gene expression method by RT2 PCR array.
Mature RNA was isolated using the RNAeasy Extraction Kit (Qiagen) according to the manufacturer's instructions. All RNA integrity number values (measured using Agilent QC) were 9.5–10, confirming excellent RNA quality. RNA (500 ng) was reverse transcribed using a cDNA conversion kit, and cDNA in combination with RT2 SYBR Green qPCR Mastermix (Qiagen) was used on a Custom RT2 Profiler PCR Array. PCR was performed on a ViiA7 thermocycler (Applied Biosystems, Life Technologies, Grand Island, NY) and, finally, relative expression was determined using data from the real-time cycler and the ΔΔCT method.
Individual RT2 qPCR Primer Assays (Qiagen) for SCN9A (NHE3) and CFTR expression were used under the same conditions as the custom array plate (i.e., starting amount of RNA, cDNA synthesis, thermocycling conditions, and analysis software). The following assays were applied for the individual assays: SCN9A (PPH11615E), CFTR (PPH01387F), B2M (PPH01094E), ACTB (PPH00073G), and GAPDH (PPH00150F).
Thus a total of 91 genes of interest were evaluated in the mucosal biopsies.
To control for DNA contamination introduced during reaction setup, a no template control (NTC) reaction replacing template with water as well as a no reverse transcription (NRT) reaction were run with the assays.
The mRNA expressions were assayed in triplicate, and the mean value was used for the statistical analysis.
Data and statistical analyses.
Calculation of the threshold cycle (CT) was determined for each well. Briefly, using the ViiA7 Software on the real-time machine, baseline was defined by choosing the automated baseline option, and threshold was defined manually using the log view of the amplification plots. The threshold value was chosen above the background signal, but within the lower one-third to lower one-half of the linear phase of the amplification plot. In our study, the threshold was chosen at 0.16 for all plates. CT values for all wells were exported to Excel and analyzed through Qiagen's Data Analysis Center (http://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page).
Delta CT (ΔCT) was calculated between gene of interest and an average of housekeeping genes (B2M, ACTB, and GAPDH), followed by delta delta CT (ΔΔCT) calculations [ΔCT(IBS-D) − ΔCT(health)]. Fold change was calculated using 2−ΔΔCT formula. The P values (P < 0.05) were reported based on t-test, and the q values were computed to reflect statistical significance with FDR correction (42).
Protein expression in duodenal mucosa.
Nuclear and cytosolic lysates were isolated from human colonic biopsies using a subcellular fractionation protocol supplied by Abcam. Briefly, tissue biopsies were homogenized in RIPA Lysis buffer with added inhibitors (Santa Cruz Biotechnology, Dallas, TX) and centrifuged at 720 G. The pelleted nuclear proteins were then sonicated in additional RIPA lysis buffer to resuspend. The nuclear pellet supernatant was recentrifuged at 10,000 g and the resulting supernatant contained the cytosolic and membrane fraction. Concentrations were determined using the Bradford reagent according to the instructions of the supplier (Sigma, St. Louis, MO).
PPP2R5C protein measurements by Western blots.
Nuclear lysates were separated using 4–15% Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA) and blotted onto nitrocellulose membranes. The membranes were blocked with 5% milk in phosphate buffered saline (PBS)/0.2% Tween, after which PPP2R5C primary antibody (ab94633_1:1,000; Abcam, Cambridge, MA) was applied overnight at 4°C. α-Actin (A2066_1:1,000; Sigma) was used for normalization of protein loading. Membranes were washed and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (donkey or goat anti-rabbit; Santa Cruz Biotechnology) and visualized with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Pittsburgh, PA) and autoradiography. Band densities were quantified with the ImageJ (40).
Enzyme-linked immunosorbent assay.
All duodenal biopsy extracts were used at a common concentration (1 μg/μl). Fifty microliters were used to assay INADL, MAPKAPK5, and MAGI1, (MyBioSource, San Diego, CA) whereas 100 μl was used for the housekeeping gene phosphoglycerate kinase 1 (PGK1; LifeSpan Biosciences, Seattle, WA), which has been recommended as the preferred gene [over β-actin (ACTB) and β2-microglobulin (β2M)] in the context of inflammatory bowel diseases (29). Enzyme-linked immunosorbent assay (ELISA) kits were used for all these protein assays following the manufacturer's instructions. Samples were run in duplicate and spike recoveries were prepared in kit assay buffer at two concentration levels to verify performance.
Pathway and cluster analyses.
We used a publicly available informatics tool to appraise the potential pathways or clustered mechanisms that are altered in the duodenal mucosa of patients with IBS-D, relative to controls. The Lens for Enrichment and Network Studies (LENS) of proteins provides interactome analysis of the list of human genes examined in this study to explore the network of protein-protein interactions that connects them (23). LENS computes associations of the genes in the interactome to pathways and assesses statistics of network connectivity of these genes compared against connectivity of randomly selected genes. Sources of data for the network generation are direct (biophysical) interactions from HPRD and BioGRID, and pathways are obtained from Reactome (http://severus.dbmi.pitt.edu/LENS/index.php).
Significance analysis of microarray for gene sets analysis of gene expression and pathways.
Gene-set analysis evaluates the expression of biological pathways, or a priori defined gene sets rather than that of single genes, in association with a binary phenotype, and it is of great biologic interest in many DNA microarray studies. Gene Set Enrichment Analysis (GSEA) has been applied widely as a tool for gene set analyses, but it was not applicable for a small gene set in our study. We used an alternative method, more appropriate for targeted gene studies, by extending the single gene analysis method, Significance Analysis of Microarray (SAM), to Gene-Set Analyses (SAM-GS). Briefly, CT data were converted to expression data as follows: expression = [2^(−Avg.(ΔCT)], where Δ(CT) = CT (Gene Of Interest) − Ave.[CT(Housekeeping Genes)].
For pathway investigation, we also included the gene set subcatalogs C1 and C2 from MolecularSignatures Database (MSigDB) v5.1, a collection of annotated gene sets for use with GSEA software (http://www.broad.mit.edu/gsea). The C1 catalog includes gene sets corresponding to human chromosomes and cytogenetic bands, while the C2 catalog includes gene sets that are involved in specific metabolic and signaling pathways collected from various sources such as online pathway databases, publications in PubMed, and knowledge of domain experts. SAM-GS divides the C1 and C2 catalogs into 308 pathways, which were included in our analyses, so that, in total, we had 150 pathways/gene sets based on the 89 genes under consideration (16, 17, 32).
RESULTS
Patients.
We studied 15 patients with IBS-D [Rome III positive: 2 male (M), 13 female (F); mean age 40.3 ± 2.3 yr; mean body mass index (BMI) 34.3 ± 3.0 kg/m2] and 7 controls (2M,5F; 40.7 ± 6.9 yr; P = ns) who were negative for celiac disease. Bowel function and abdominal pain scores of the patients with IBS-D are shown in Table 2A. Clinical details of the seven controls who underwent clinically-indicated duodenal biopsies are shown in Table 2B.
mRNA fold change in duodenal mucosa from IBS relative to healthy controls.
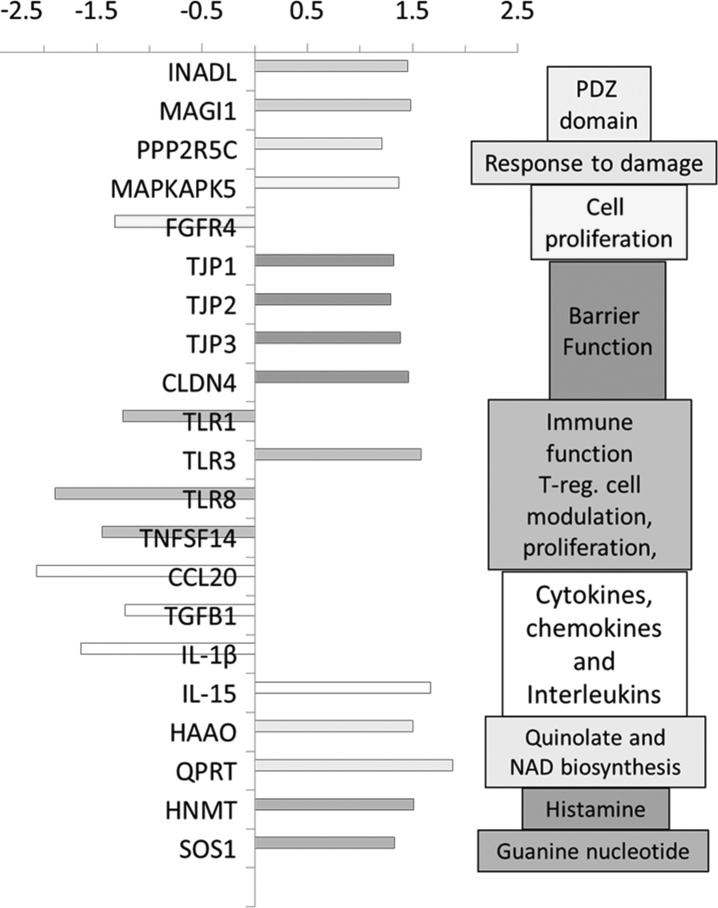
The mean (and 95% CI) fold changes (based on 2−ΔΔ CT) in IBS-D and subgroups of IBS-D patients relative to healthy controls are illustrated in Table 3 and Fig. 1. Table 3 shows gene fold expressions (P < 0.05): upregulated [INADL and MAGI1 (possibly related to ion transport)] and barrier (TJP2 and 3) or immune functions (TLR3, IL15, and MAPKAPK5); or downregulated including factors involved in bile acid pathway (FGFR4), immune function (TGFB1 and CCL20), or antigen detection (TLR7).
Table 3.
Duodenal mucosal mRNA fold expression in patients with IBS-D and controls
| mRNA Regulation IBS-D/Controls |
||||||
|---|---|---|---|---|---|---|
| Gene Symbol | Official Name | Summary of Functions | Fold | 95% CI | P* | q* |
| INADL | InaD-like (Drosophila) | Gene encoding multiple PDZ domain protein | 1.45 | 1.13, 1.78 | 0.00291 | 0.0419 |
| MAGI1 | Membrane-associated guanylate kinase | Cell junction, interaction with ASIC and PDZ domain | 1.48 | 1.05, 1.90 | 0.00607 | 0.0435 |
| PPP2R5C | Protein phosphatase 2, regulatory subunit beta | Regulation of damage response, cell proliferation, apoptosis | 1.24 | 1.06, 1.41 | 0.00529 | 0.0435 |
| TJP1 | Tight junction protein ZO-1 | Barrier function | 1.32 | 1.00, 1.64 | 0.0250 | 0.0673 |
| TJP2 | Tight junction protein ZO-2 | 1.29 | 1.02, 1.56 | 0.0325 | 0.0781 | |
| TJP3 | Tight junction protein ZO-3 | 1.38 | 1.04, 1.72 | 0.0170 | 0.0632 | |
| CLDN4 | Claudin 4 | 1.46 | 1.01, 1.92 | 0.0167 | 0.0632 | |
| MAPK-APK5 | Mitogen-activated protein kinase-activated protein kinase 5 | Kinase involved in growth factor stimulated cell proliferation and muscle cell differentiation | 1.37 | 1.09, 1.65 | 0.0028 | 0.0419 |
| TLR1 | Toll-like receptor 1 | Modulation of T-reg cells | −1.25 | 0.63, 0.98 | 0.0235 | 0.0673 |
| TLR3 | Toll-like receptor 3 | 1.58 | 1.13, 2.03 | 0.0029 | 0.0419 | |
| TLR8 | Toll-like receptor 8 | −1.90 | 0.18, 0.87 | 0.0357 | 0.0781 | |
| IL-1β | Interleukin-1β | Immune function | −1.65 | 0.18, 1.03 | 0.0240 | 0.0673 |
| IL-15 | Interleukin-15 | 1.67 | 1.18, 2.17 | 0.0049 | 0.0435 | |
| FGFR4 | Fibroblast growth factor receptor 4 | Tyrosine-protein kinase activity in cell proliferation, differentiation and migration | −1.33 | 0.53, 0.97 | 0.03477 | 0.0781 |
| CCL20 | Chemokine (C-C motif) ligand 20 | Immunoregulatory and inflammatory processes | −2.07 | 0.00001, 1.14 | 0.01259 | 0.0632 |
| TGFB1 | Transforming growth factor-β1 | Cytokine regulation of proliferation, differentiation, adhesion, migration, other functions in many cell types | −1.23 | 0.63, 1.00 | 0.0363 | 0.0781 |
| TNFSF 14 | Tumor necrosis factor ligand superfamily member 14 | Apoptosis and T-cell proliferation/function | −1.45 | 0.45, 0.93 | 0.0156 | 0.0632 |
| HAAO | 3-Hydroxyanthranilate 3,4-dioxygenase | Synthesis of quinolinic acid, which participates in pathogenesis of neurologic and inflammatory disorders | 1.50 | 0.96, 2.04 | 0.0224 | 0.0673 |
| QPRT | Quinolinate phosphoribosyltransferase | Biosynthesis of NAD | 1.88 | 0.93, 2.83 | 0.0176 | 0.0632 |
| HNMT | Histamine N-methyl transferase | Metabolism of histamine | 1.51 | 1.02, 2.00 | 0.0383 | 0.0784 |
| SOS1 | Sons of Sevenless (Drosophila) homolog 1 | Guanine nucleotide exchange factor | 1.33 | 1.01, 1.66 | 0.0102 | 0.0624 |
| SLC9A3 | Sodium hydrogen exchange type 3 channel | Sodium absorption | 1.54 | 0.81, 2.28 | 0.0963 | 0.1525 |
| CFTR | Cystic fibrosis transmembrane regulator | Chloride secretion | 1.46 | 0.55, 2.37 | 0.3834 | 0.2917 |
ASIC, acid-sensing ion channel; PDZ, common structural domain of 80–90 amino-acids found in signaling proteins that anchor receptor proteins in the membrane to cytoskeletal components; + Numbers, upregulated; − numbers, downregulated.
P values are Student's t-test performed on IBS-D patients compared with controls before false detection rate (FDR) correction;
q values are FDR adjusted P values.
Fig. 1.
Fold changes in duodenal mucosa of patients with irritable bowel syndrome with diarrhea (IBS-D) compared with controls. mRNA fold expression of genes associated with different functions is shown (P < 0.05 without FDR correction). With false detection rate correction (FDR) correction, the following genes were significantly upregulated (q < 0.05): INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15 in IBS-D relative to controls.
With FDR correction, the following genes were significant (q < 0.05): INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15. All are upregulated in patients with IBS-D relative to controls.
There were other genes that showed numerical (nonsignificant) changes in expression (P > 0.05, < 0.1). Thus there was numerically increased expression in duodenal mucosa from IBS-D patients compared with controls for the following:
1) SLC10A2 (the gene for the bile acid transporter, 2.01-fold, P = 0.093);
2) CLDN1 (claudin, a tight junction protein,1.43-fold, P = 0.066);
3) KITLG (kit ligand, a determinant of development of interstitial cells of Cajal, 1.38-fold, P = 0.074);
4) MPP5 (membrane protein, palmitoylated 5, involved in polarization of differentiating cells, 1.35 fold, P = 0.070);
5) PVRL3 (poliovirus receptor-related 3, 1.22-fold, P = 0.075); and
6) AHR (aryl hydrocarbon receptor, which regulates xenobiotic-metabolizing enzymessuch as cytochrome P450, 1.22-fold, P = 0.053).
In contrast, the following were associated with numerically (not significantly) reduced expression in IBS-D duodenal mucosa compared with controls: CD3E (CD3-epsilon chain involved in T cell surface glycoprotein, −1.38-fold, 0.0.088); and TPSAB1 (tryptase alpha/beta 1, −1.61-fold, P = 0.097).
Clustering analysis of pathways and mechanisms.
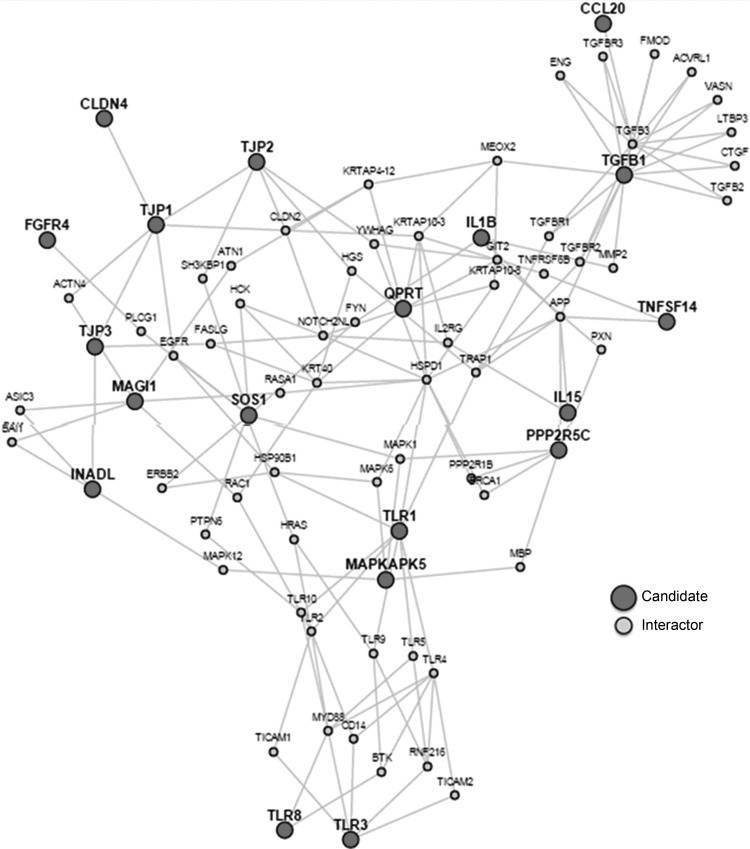
Having identified the nominally, univariately significant upregulation or downregulation of genes in the mucosa, we applied LENS analysis, and plotted connectivity among the genes associated with barrier, T lymphocytes, and cytokines (Fig. 2), focusing only on the genes that were nominally (P < 0.05) upregulated or downregulated. Among the 14 nominally upregulated genes, clustering of the barrier-associated mechanisms and related PDZ domains (TJP1, TJP2, TJP3, CLDN4, INADL, and MAGI1) was noted; among the seven downregulated genes, a cluster of immune function (chemokine, Toll-like receptor, and interleukin), specifically CCL20, TLR1, IL1B, and TLR8, was observed. It is relevant to note that FDR-corrected increased expression was identified in INADL and MAGI1.
Fig. 2.
Lens for Enrichment and Network Studies (LENS) interactome of candidate genes evaluated in this study that showed increased or decreased expression. Note the connections between elements involved in ion transport (INADL and MAGI1), barrier function or epithelial integrity (e.g., MAPKAPK5, TJPs), and immune function (e.g., TLR-3 and IL15).
SAM-GS analysis.
The analysis results (Table 4) show the P value and q value of each gene set, based on the permutation test for no association between the gene expression of the gene set and the binary phenotype (IBS-D and control). The results are shown in order of the nominal P value.
Table 4.
SAM-GS results (only top 10 rows are shown)
| Gene Set Name/Pathway | GS Size | GS P Value (0 ↔ <1/nb Permutations) | GS q Value | oGS Size | Genes Evaluated in Our Study Belonging to the Stated Pathway |
|---|---|---|---|---|---|
| SIG_CD40 PATHWAY MAP | 1 | 0.02 | 0.386 | 34 | MAPKAPK5 |
| ST_p38_MAPK_Pathway | 1 | 0.02 | 0.386 | 34 | MAPKAPK5 |
| FRASOR_ER_ DOWN | 1 | 0.045 | 0.386 | 68 | KYNU |
| HTERT_DOWN | 1 | 0.045 | 0.386 | 64 | KYNU |
| NFKB_ INDUCED | 7 | 0.08 | 0.386 | 106 | CCL20/FN1/IL15/IL1B/IL6/IL8/TNF |
| X41bbPathway | 2 | 0.08 | 0.386 | 18 | IFNG/IL4 |
| MAP00380_ Tryptophan_ metabolism | 3 | 0.085 | 0.386 | 42 | HAAO/KYNU/TDO2 |
| StemPathway | 3 | 0.09 | 0.386 | 15 | IL4/IL6/IL8 |
| CR_IMMUNE_ FUNCTION | 6 | 0.105 | 0.386 | 50 | IFNG/IL10/IL13/IL15/IL1B/IL4 |
| MAP00340_ Histidine_ metabolism | 1 | 0.12 | 0.386 | 20 | HNMT |
| 89 genes in current study | 89 | 0.185 | 0.386 | 89 | AADAT/AHR/C4BPA/CALR/CCBL2/CCL20/ CD3E/CD74/CLDN1/CLDN12/CLDN15/ CLDN16/CLDN2/CLDN3/CLDN4/CLDN7/ CPSF1/CTNNA1/CTNNB1/DLG1/FGFR4/ FN1/FOXP3/GOT2/GPBAR1/GUCA2B/ HAAO/HNMT/HRH1/HRH2/IDO1/IDO2/ IFIT3/IFNG/IL10/IL13/IL15/IL17A/IL1B/ IL2RA/IL4/IL6/IL8/INADL/KITLG/KMO/ KYNU/MAGI1/MAPKAPK5/MKNK2/MPP5/ MPP7/MYLK/NR1H4/OCLN/P2RY4/ PDZD3/PPP1CB/PPP2R5C/PRG2/PVRL3/ QPRT/RBP2/SLC10A2/SLC6A4/SLC9A1/ SOS1/TDO2/TFF1/TGFB1/TJP1/TJP2/TJP3/ TLR1/TLR2/TLR3/TLR4/TLR5/TLR6/TLR7/ TLR8/TLR9/TNF/TNFSF14/TNFSF15/ TPH1/TPSAB1/TPSB2/VIP |
SAM-GS, Significance Analysis of Microarray to Gene-Set Analyses; GS size, number of genes in the gene set/pathway; GS P value, permutation test for no association between the gene expression of the gene set and the binary phenotype (IBS-D compared with controls); GS q value, to account for multiple comparisons when multiple gene sets are tested, one might consider FDR instead of type I error probability; oGS, original number of genes in the pathway/gene set (for example, there were 106 genes in the NFKB_INDUCED pathway, where 7 genes from our study were found); Genes in Pathway, the genes in our study found in the pathway/gene set (for example, there are 7 genes from our study overlapping with NFKB_INDUCED pathway; the genes are CCL20, FN1, IL15, IL1B, IL6, IL8, and TNF).
The P value (P = 0.185) of the test evaluating the entire 89 gene set shows no association between the gene expression of the gene set and phenotype.
However, there were several genes with univariate significance (see q values in Table 3) that were represented in pathways identified in the SAM-GS analysis, specifically in immune function (IL-15) and MAP kinase-activated protein kinase (MAPAPK5). These genes, found in gene-sets by SAM-GS, showed a trend (P values between 0.02 and 0.105) but not a statistically significant association to the phenotype.
Mucosal protein expression.
Western blot analysis of duodenal mucosa from six healthy controls and seven patients with IBS-D who randomly selected from the same patient group was used to assess protein expression of PPP2R5C. This protein was selected as it showed increased mRNA fold expression (see Table 3). Figure 3 shows the increased protein expression including the appearance of the expression of PPP2R5C relative to the housekeeping protein, actin. The difference in expression between IBS-D group and biopsy-negative controls was statistically significant (P = 0.044).
Fig. 3.
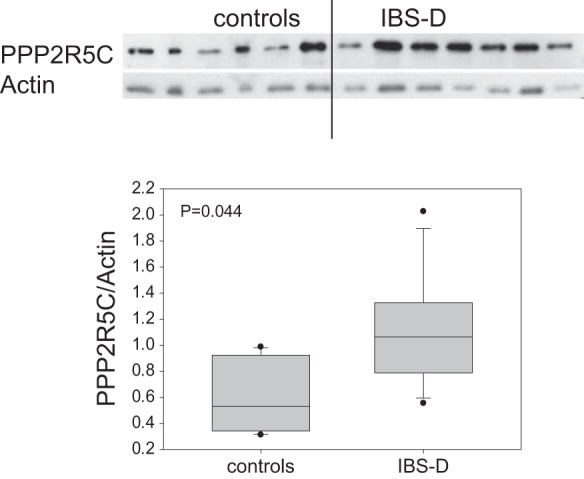
Western blot analysis of protein expression of PPP2R5C in nuclear lysates from duodenal mucosal biopsies of patients with IBS-D and controls. Note the increase in protein (corrected for the housekeeping protein actin) in the patients with IBS-D.
Expression of other mucosal proteins (INADL, MAGI1, and MAPKAPK5) using ELISA measurement was applied to duodenal biopsy extracts from 15 patients with IBS-D and 6 controls (Table 5). There were very similar concentrations of the housekeeping gene PGK1 in the patients and controls. Numerical differences in protein expression of one of those tested, INADL, did not reach statistical significance: median 9.4 ng/ml in patients with IBS-D relative to controls, median 5.8 ng/ml (P > 0.05).
Table 5.
Measurements of mucosal proteins by ELISA
| Protein | IBS-D (n = 15) | Control Patients (n = 6) | Estimated Sample Size per Group to Observe 2.5-Fold Difference in Protein Concentration, Based on Pooled SD of Each Protein |
|---|---|---|---|
| INADL | 9.39 (2.68, 25.74) | 5.83 (2.9, 23.2) | 30 |
| MAGI1 | 0.83 (0.00, 2.75) | 0.79 (0.00, 2.26) | 13 |
| MAPKAPK5 | 0.54 (0.04, 1.96) | 0.73 (0.00, 1.60) | 15 |
| PGK1 | 4.17 (3.71, 4.53) | 4.38 (3.69, 4.83) | Housekeeping gene |
Data are median and interquartile range (IQR); all comparisons are not statistically significant; PGK1 is the housekeeping gene. There was insufficient mucosa for protein measurements in 1 control patient. SD, standard deviation.
DISCUSSION
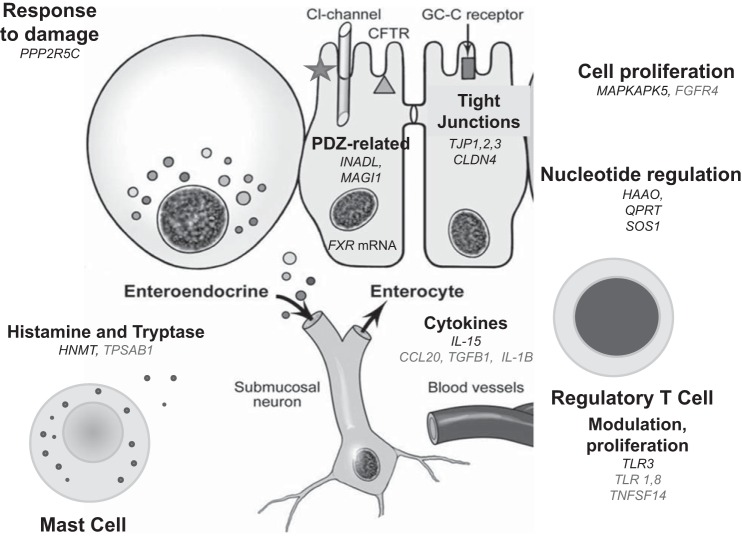
Our study evaluated expression in small intestinal mucosa of genes related to tight junctions, chemokines, innate immunity, ion channels, and transmitters in 15 patients with IBS-D and 7 controls with proven normal biopsies based on histopathology. With FDR correction, the following genes were significant (q < 0.05): INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15, all of which were upregulated in IBS-D relative to controls. The fold expression alterations in mechanisms of ion transport (INADL and MAGI1), epithelial response to damage (PPP2R5C), and immune mechanisms (TLR3 and IL15) in small bowel mucosa suggest that differential expression of small intestinal mucosa in IBS-D may be important in the biological mechanisms in IBS-D and certainly deserve further study. The findings on LENS cluster analysis of barrier and ion transport (both upregulated) and immune function (downregulated) are summarized in Fig. 4. Although the q values in the pathway analysis using SAM-GS do not show significant associations of pathways with phenotype, it is relevant to note that the MAPKAPK5 pathways and CR-immune function pathways show interesting trends (not statistically significant) and include upregulated genes in the comparisons between IBS-D and controls (specifically MAPKAPK5 and IL15).
Fig. 4.
Cartoon summarizing altered mRNA expression in cellular elements of the descending duodenal mucosa in patients with IBS-D, based on nominal significance (P < 0.05) or statistical significance (q < 0.05: INADL, MAGI1, PPP2R5C, MAPKAPK5, TLR3, and IL-15) and including TPSBA1 (P < 0.10). TJ, tight junction; GC-C, guanylate cyclase; CFTR, cystic fibrosis transmembrane regulator. Fold upregulated mRNA is in black; downregulated is in gray.
The upregulated or downregulated gene expressions suggest, in some cases, that the changes may represent reciprocal or compensatory changes within the same biological functions. One example of reciprocal changes in expression is downregulation of TLR1, TLR8, and CD3E in contrast to the upregulation of TLR3; another example where the changes may simply reflect reciprocal changes in expression is downregulated IL-1B and upregulated IL-15. On the other hand, there is consistent upregulation of barrier function gene expression, as shown by the results for ZO-1, 2, and 3, claudin-1 and -4, and MAPKAPK5. ZO and claudin genes are directly related to intercellular junction proteins, whereas MAPKAPK5 controls a kinase involved in growth factor-stimulated cell proliferation and muscle cell differentiation. IL-15 plays a key role in intraepithelial lymphocyte function and in some forms of intestinal inflammation (1, 33).
The biological significance of the upregulation of barrier function is unclear. There are other changes in expression that may conceivably be associated with increased fluid or electrolyte secretion, such as the upregulation of the following:
1) InaD-like (Drosophila) gene, for which there is a human analog INADL gene producing a protein (39), which encodes multiple PDZ domain proteins that may enhance ion transport, and a related tight junction-associated multi-PDZ protein PATJ (PALS1-associated TJ protein) that connects and stabilizes apical and lateral components of tight junctions in human intestinal cells (36);
2) Membrane-associated guanylate kinase MAGI1, which influences cell junction, interaction with acid-sensing ion channel (sensation) and PDZ domain (ion transport);
3) SONS1 [Sons of Sevenless (Drosophila) homolog] 1, which is a guanine nucleotide exchange factor; it is worth noting that guanine nucleotide exchange factor, Epac (Exchange protein directly activated by cAMP), appears to be involved in intestinal secretion (22, 25, 41). However, there is no evidence in the literature that this gene has functional effects in humans.
4) SLC9A3 [Solute Carrier Family 9, Subfamily A, Member 3), which encodes for the NHE3 channel is possibly upregulated (fold change 1.54, q = 0.153). If the increased expression results in increased protein, it would be expected that there would be greater Na+ absorption which may conceivably reduce fluid secretion in contrast to some of the other secretory mechanisms that may be upregulated.
Overall, our current studies support upregulation of intestinal ion transport mechanisms. This is consistent with increased mRNA expression of PDZ-related mechanisms previously demonstrated in colonic mucosa (11, 12). These data suggest increased ion transport, possibly leading to water and electrolyte secretion, that is partly counteracted by the increased expression of barrier function mechanisms. These observations emphasize the importance of thorough characterization of the phenotype in patients with IBS-D, given the different peripheral mechanisms that result in the phenotype of IBS-D (8, 9). Future studies will be enhanced by further characterization of the baseline pathophysiological phenotype in patients with the symptom phenotype of IBS-D, such as bile acid diarrhea, rapid small bowel or colonic transit, and upregulated mucosal immune mechanisms.
In addition to the changes in mRNA expression, we also observed significantly increased expression of PPP2R5C as demonstrated by Western blot, and a numerical increase in the protein level of INADL by ELISA, although this did not reach statistical significance. The concentrations of the housekeeping gene in the duodenal biopsy lysates used from the two groups were similar in the ELISA assays, suggesting that the absence of statistically significant differences in the protein concentrations is not the result of technical error in the measurements. On the other hand, the pooled standard deviation of INADL in the two groups was large (19.7 pg/ml), and the estimated sample size to prove statistical difference for 2.5- or 1.5-fold differences in concentration of INADL relative to control (median 5.83 pg/ml) would be respectively 30 and 81 per patient group, respectively. Therefore, our study was underpowered to appraise differences in the proteins measured by ELISA.
The intestinal mucosa contains a pool of histamine that rapidly turns over and that appears to be derived largely from a non-mast cell source. Histamine inhibits chloride absorption, inducing bicarbonate secretion in ileal mucosa and influencing chloride/bicarbonate exchange, largely by actions mediated by histamine H1 receptors (31). Chromogranin A (ChgA), an acidic protein found in large dense-core secretory vesicles, functions as a monoamine-binding protein that facilitates the regulated endocrine secretion of large amounts of monoamines from enteroendocrine cells (20). The increased gene expression of the histamine metabolizing enzyme, HNMT, may also serve as a compensatory mechanism to reduce the effects of histamine released from enteroendocrine cells or from mast cells whose density is increased in the jejunal mucosa of non-atopic IBS-D patients (47).
The latter investigation by Vicario et al. (47) identified a number of immune-modulating mechanisms, suggesting increased humoral immune function in the jejunal mucosa of patients with IBS-D. This is consistent with several alterations in gene expression for immune regulation and T-cell function in our current study. In addition, the increased expression for the enzyme metabolizing histamine is consistent with increased release or supernantant content of histamine from colorectal mucosal biopsies from patients with IBS (3, 4, 7, 15, 28). This overexpression of HNMT may conceivably provide a natural metabolism of histamine, which may be produced and secreted in larger quantities by the increased numbers of mast cells. This natural reduction in histamine may serve as an alternative to achieve the beneficial effects on symptoms of the mast cell stabilizer and antihistaminic medication ketotifen (28) and the selective H1 receptor antagonist ebastine (45). The numerically increased gene expression for Kit ligand is of significant interest in view of the role of the gene in the function of this tyrosine kinase receptor gene in responding to growth factors such as insulin-like growth factor-1 (IGF-1) in the development of interstitial cells of Cajal, the pacemakers of the gastrointestinal tract.
The numerical downregulation of TPSAB1 (P = 0.097 and q = 0.144, which controls the synthesis of alpha and beta tryptases) and KYNU (P = 0.131 and q = 0.166 kynureninase, which metabolizes tryptophan) is similarly intriguing in the context of IBS. Beta tryptases appear to be the main isoenzymes expressed in mast cells; whereas, in basophils, alpha tryptases predominate. Tryptases are one of the main products of mast cells. Our data suggest the hypothesis that there is downregulation in the synthesis of tryptase, in contrast to several prior publications that have documented increased expression of tryptase in mucosa from patients with IBS, specifically in colonic mucosa (3, 4, 7, 15, 49) and in mRNA (21), as well as protein expression (35) in jejunal mucosal biopsies of IBS patients. The mechanism resulting in this hypothetical reduction in tryptases is the observed numerical downregulation of TPSAB1; however, it is important to confirm these observations given the other data showing increased tryptase and other proteases in supernatants from (particularly) colonic biopsies from patients with IBS-D.
Kynureninase metabolizes tryptophan, and the synthesis of NAD (or NADP) and quinolic acid from tryptophan involves a series of enzymes and the formation of a number of intermediates which are collectively called “kynurenines.” Kynurenic acid is a neuroprotective N-methyl d-aspartate (NMDA) antagonist (e.g., on enteric glutamatergic neurons) and an α7-nicotinic cholinergic agonist, potentially dysregulating intestinal motility and altering mechanosensitivity (27). In mononuclear cells, including tissue macrophages, quinolinic acid is the main end product of the kynurenine pathway and plays a role in immunoregulatory processes (37). Recent evidence shows kyruneninase plays a role in inflammation (24). Reduced expression of KYNU may conceivably result in greater motor dysregulation and inflammation in IBS-D due to enhanced tryptophan metabolism through the kynurenine pathway. It is also interesting to note that there were other upregulated genes in the quinolinate pathway (HAAO and QPRT).
In summary, our results extend the prior observations recorded in jejunal mucosa (35), confirm the potential upregulation of ion transport processes observed in colonic mucosa (11, 12), and identify potential compensatory changes in immune and barrier functions in small intestinal mucosa. However, our current studies should be regarded as pilot data that require replication in larger numbers, as well as control for BMI, enriching the current study sample with patients with a group of IBS -D patients with normal BMI, since there is evidence that obesity is associated with gastrointestinal symptoms, an inflammatory diathesis in general, as well as possible changes in intestinal permeability or bile acid malabsorption. Neverthelss, this pilot study identifies mechanisms that deserve further study and identifies the fold expression and coefficient of variation that will allow planning with adequate sample sizes in future studies. In fact, our planned hypothesis-testing study will include 60 patients with IBS-D, 30 with IBS-C, and 30 healthy controls. In addition, given the wide variations in protein expression observed, it appears that biological significance of these different mechanisms expressed in intestinal mucosa from patients with IBS-D may be interrogated in future studies using enteroids. These have the potential to provide new insights on the pathobiological mechanisms, allowing different mechanisms and pathways can be interrogated in greater depth. The observations in the current study provide a step to further understand potential etiopathogenetic mediators as well as biological pathways involved in IBS-D.
GRANTS
M. Camilleri is supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK92179; this study (and permission to use data from duodenal biopsies in patients) was supported in part by research funds from EnteraHealth for an ancillary study.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.C. conception and design of research; M.C., J.N., and E.W.K. analyzed data; M.C. interpreted results of experiments; M.C. drafted manuscript; M.C., P.C., N.V., A.A., J.O., R.D., J.N., E.W.K., and J.A.M. edited and revised manuscript; M.C. approved final version of manuscript; P.C., N.V., A.A., J.O., D.J.E., R.D., and J.A.M. performed experiments.
ACKNOWLEDGMENTS
We thank Cindy Stanislav for excellent secretarial assistance. This publication was supported in part by Grant UL1TR000135 from the National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Heatlh.
REFERENCES
- 1.Abadie V, Jabri B. IL-15: a central regulator of celiac disease immunopathology. Immunol Rev 260: 221–234, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aerssens J, Camilleri M, Talloen W, Thielemans L, Göhlmann HW, Van Den Wyngaert I, Thielemans T, De Hoogt R, Andrews CN, Bharucha AE, Carlson PJ, Busciglio I, Burton DD, Smyrk T, Urrutia R, Coulie B. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 194–205, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, Collins SM, Corinaldesi R. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology 126: 693–702, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Barbara G, Wang B, Stanghellini V, de Giorgio R, Cremon C, Di Nardo G, Trevisani M, Campi B, Geppetti P, Tonini M, Bunnett NW, Grundy D, Corinaldesi R. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology 132: 26–37, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Belmonte L, Beutheu Youmba S, Bertiaux-Vandaële N, Antonietti M, Lecleire S, Zalar A, Gourcerol G, Leroi AM, Déchelotte P, Coëffier M, Ducrotté P. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One 7: e42777–e42786, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertiaux-Vandaële N, Youmba SB, Belmonte L, Lecleire S, Antonietti M, Gourcerol G, Leroi AM, Déchelotte P, Ménard JF, Ducrotté P, Coëffier M. The expression and the cellular distribution of the tight junction proteins are altered in irritable bowel syndrome patients with differences according to the disease subtype. Am J Gastroenterol 106: 2165–2173, 2011. [DOI] [PubMed] [Google Scholar]
- 7.Buhner S, Li Q, Vignali S, Barbara G, De Giorgio R, Stanghellini V, Cremon C, Zeller F, Langer R, Daniel H, Michel K, Schemann M. Activation of human enteric neurons by supernatants of colonic biopsy specimens from patients with irritable bowel syndrome. Gastroenterology 137: 1425–1434, 2009. [DOI] [PubMed] [Google Scholar]
- 8.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 367: 1626–1635, 2012. [DOI] [PubMed] [Google Scholar]
- 9.Camilleri M. Physiological underpinnings of irritable bowel syndrome: neurohormonal mechanisms. J Physiol 592: 2967–2980, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilleri M, Andrews CN, Bharucha AE, Carlson PJ, Ferber I, Stephens D, Smyrk TC, Urrutia R, Aerssens J, Thielemans L, Göhlmann H, van den Wyngaert I, Coulie B. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology 132: 17–25, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camilleri M, Carlson P, Acosta A, Busciglio I. Colonic mucosal gene expression and genotype in irritable bowel syndrome patients with normal or elevated fecal bile acid excretion. Am J Physiol Gastrointest Liver Physiol 309: G10–G20, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camilleri M, Carlson P, Acosta A, Busciglio I, Nair AA, Gibbons SJ, Farrugia G, Klee EW. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea: a pilot case-control study. Am J Physiol Gastrointest Liver Physiol 306: G1089–G1098, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camilleri M, McKinzie S, Busciglio I, Low PA, Sweetser S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Prospective study of motor, sensory, psychologic, and autonomic functions in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 772–781, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, Moses PL. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology 126: 1657–1664, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Cremon C, Carini G, Wang B, Vasina V, Cogliandro RF, De Giorgio R, Stanghellini V, Grundy D, Tonini M, De Ponti F, Corinaldesi R, Barbara G. Intestinal serotonin release, sensory neuron activation, and abdominal pain in irritable bowel syndrome. Am J Gastroenterol 106: 1290–1298, 2011. [DOI] [PubMed] [Google Scholar]
- 16.Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Gene-set analysis and reduction. Brief Bioinform 10: 24–34, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Improving gene set analysis of microarray data by SAM-GS. BMC Bioinform 8: 242–254, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Salhy M, Gundersen D, Hatlebakk JG, Gilja OH, Hausken T. Abnormal rectal endocrine cells in patients with irritable bowel syndrome. Regul Pept 188: 60–65, 2014. [DOI] [PubMed] [Google Scholar]
- 19.El-Salhy M, Wendelbo I, Gundersen D. Serotonin and serotonin transporter in the rectum of patients with irritable bowel disease. Mol Med Rep 8: 451–455, 2013. [DOI] [PubMed] [Google Scholar]
- 20.Engelstoft MS, Lund ML, Grunddal KV, Egerod KL, Osborne-Lawrence S, Poulsen SS, Zigman JM, Schwartz TW. Research Resource: a chromogranin A reporter for serotonin and histamine secreting enteroendocrine cells. Mol Endocrinol 29: 1658–1671, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guilarte M, Santos J, de Torres I, Alonso C, Vicario M, Ramos L, Martinez C, Casellas F, Saperas E, Malagelada JR. Diarrhoea-predominant IBS patients show mast cell activation and hyperplasia in the jejunum. Gut 56: 203–209, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halm ST, Zhang J, Halm DR. beta-Adrenergic activation of electrogenic K+ and Cl− secretion in guinea pig distal colonic epithelium proceeds via separate cAMP signaling pathways. Am J Physiol Gastrointest Liver Physiol 299: G81–G95, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handen A, Ganapathiraju MK. LENS: web-based lens for enrichment and network studies of human proteins. BMC Med Genomics 8, Suppl 4: S2, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harden JL, Lewis SM, Lish SR, Suárez-Fariñas M, Gareau D, Lentini T, Johnson-Huang LM, Krueger JG, Lowes MA. The tryptophan metabolism enzyme l-kynureninase is a novel inflammatory factor in psoriasis and other inflammatory diseases. J Allergy Clin Immunol 137: 1830–1840, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoque KM, Woodward OM, van Rossum DB, Zachos NC, Chen L, Leung GP, Guggino WB, Guggino SE, Tse CM. Epac1 mediates protein kinase A-independent mechanism of forskolin-activated intestinal chloride secretion. J Gen Physiol 135: 43–58, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerckhoffs AP, ter Linde JJ, Akkermans LM, Samsom M. SERT and TPH-1 mRNA expression are reduced in irritable bowel syndrome patients regardless of visceral sensitivity state in large intestine. Am J Physiol Gastrointest Liver Physiol 302: G1053–G1060, 2012. [DOI] [PubMed] [Google Scholar]
- 27.Keszthelyi D, Troost FJ, Masclee AA. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil 21: 1239–1249, 2009. [DOI] [PubMed] [Google Scholar]
- 28.Klooker TK, Braak B, Koopman KE, Welting O, Wouters MM, van der Heide S, Schemann M, Bischoff SC, van den Wijngaard RM, Boeckxstaens GE. The mast cell stabiliser ketotifen decreases visceral hypersensitivity and improves intestinal symptoms in patients with irritable bowel syndrome. Gut 59: 1213–1221, 2010. [DOI] [PubMed] [Google Scholar]
- 29.Krzystek-Korpacka M, Diakowska D, Bania J, Gamian A. Expression stability of common housekeeping genes is differently affected by bowel inflammation and cancer: implications for finding suitable normalizers for inflammatory bowel disease studies. Inflamm Bowel Dis 20: 1147–1156, 2014. [DOI] [PubMed] [Google Scholar]
- 30.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 32: 920–924, 1997. [DOI] [PubMed] [Google Scholar]
- 31.Linaker DB, McKay TJ, Higgs NB, Turnberg LA. Mechanisms of histamine stimulated secretion in rabbit ileal mucosa. Gut 22: 964–970, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Q, Dinu I, Adewale AJ, Potter JD, Yasui Y. Comparative evaluation of gene-set analysis methods. BMC Bioinform 8: 431–450, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marietta EV, Nadeau AM, Cartee AK, Singh I, Rishi A, Choung RS, Wu TT, Rubio-Tapia A, Murray JA. Immunopathogenesis of olmesartan-associated enteropathy. Aliment Pharmacol Ther 42: 1303–1314, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez C, Lobo B, Pigrau M, Ramos L, González-Castro AM, Alonso C, Guilarte M, Guilá M, de Torres I, Azpiroz F, Santos J, Vicario M. Diarrhoea-predominant irritable bowel syndrome: an organic disorder with structural abnormalities in the jejunal epithelial barrier. Gut 62: 1160–1168, 2013. [DOI] [PubMed] [Google Scholar]
- 35.Martínez C, Vicario M, Ramos L, Lobo B, Mosquera JL, Alonso C, Sánchez A, Guilarte M, Antolín M, de Torres I, González-Castro AM, Pigrau M, Saperas E, Azpiroz F, Santos J. The jejunum of diarrhea-predominant irritable bowel syndrome shows molecular alterations in the tight junction signaling pathway that are associated with mucosal pathobiology and clinical manifestations. Am J Gastroenterol 107: 736–746, 2012. [DOI] [PubMed] [Google Scholar]
- 36.Michel D, Arsanto JP, Massey-Harroche D, Béclin C, Wijnholds J, Le Bivic A. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J Cell Sci 118: 4049–4057, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Moffett JR, Namboodiri MA. Tryptophan and the immuneresponse. Immunol Cell Biol 81: 247–265, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Ohman L, Stridsberg M, Isaksson S, Jerlstad P, Simrén M. Altered levels of fecal chromogranins and secretogranins in IBS: relevance for pathophysiology and symptoms? Am J Gastroenterol 107: 440–447, 2012. [DOI] [PubMed] [Google Scholar]
- 39.Philipp S, Flockerzi V. Molecular characterization of a novel human PDZ domain protein with homology to INAD from Drosophila melanogaster". FEBS Lett 413: 243–248, 1997. [DOI] [PubMed] [Google Scholar]
- 40.Rasband WS. ImageJ (Online) Bethesda, MD: National Institutes of Health; http://imagej.nih.gov/ij/ [1997–2016]. [Google Scholar]
- 41.Sheikh IA, Koley H, Chakrabarti MK, Hoque KM. The Epac1 signaling pathway regulates Cl− secretion via modulation of apical KCNN4c channels in diarrhea. J Biol Chem 288: 20404–20415, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, Neal KR, Whorwell PJ, Hall IP, Spiller RC. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut 62: 985–994, 2013. [DOI] [PubMed] [Google Scholar]
- 44.Talley NJ, Phillips SF, Wiltgen CM, Zinsmeister AR, Melton LJ 3rd. Assessment of functional gastrointestinal disease: the bowel disease questionnaire. Mayo Clin Proc 65: 1456–1479, 1990. [DOI] [PubMed] [Google Scholar]
- 45.van Wanrooij S, Wouters MM, Van Oudenhove L, Vermeire S, Rutgeerts PJ, Boeckxstaens GE. Effect of the H1-receptor antagonist ebastin on visceral perception and clinical symptoms in IBS. Gastroenterology 144: S160, 2013. [Google Scholar]
- 46.Vazquez-Roque MI, Camilleri M, Smyrk T, Murray JA, O'Neill J, Carlson P, Lamsam J, Eckert D, Janzow D, Burton D, Ryks M, Rhoten D, Zinsmeister AR. Association of HLA-DQ gene with bowel transit, barrier function and inflammation in irritable bowel syndrome with diarrhea. Am J Physiol Gastrointest Liver Physiol 303: G1262–G1269, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicario M, González-Castro AM, Martínez C, Lobo B, Pigrau M, Guilarte M, de Torres I, Mosquera JL, Fortea M, Sevillano-Aguilera C, Salvo-Romero E, Alonso C, Rodiño-Janeiro BK, Söderholm JD, Azpiroz F, Santos J. Increased humoral immunity in the jejunum of diarrhoea-predominant irritable bowel syndrome associated with clinical manifestations. Gut 64: 1379–1388, 2015. [DOI] [PubMed] [Google Scholar]
- 48.Villani AC, Lemire M, Thabane M, Belisle A, Geneau G, Garg AX, Clark WF, Moayyedi P, Collins SM, Franchimont D, Marshall JK. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology 138: 1502–1513, 2010. [DOI] [PubMed] [Google Scholar]
- 49.Vivinus-Nébot M, Dainese R, Anty R, Saint-Paul MC, Nano JL, Gonthier N, Marjoux S, Frin-Mathy G, Bernard G, Hebuterne X, Tran A, Theodorou V, Piche T. Combination of allergic factors can worsen diarrheic irritable bowel syndrome: role of barrier defects and mast cells. Am J Gastroenterol 107: 75–81, 2012. [DOI] [PubMed] [Google Scholar]
- 50.Wang YM, Chang Y, Chang YY, Cheng J, Li J, Wang T, Zhang QY, Liang DC, Sun B, Wang BM. Serotonin transporter gene promoter region polymorphisms and serotonin transporter expression in the colonic mucosa of irritable bowel syndrome patients. Neurogastroenterol Motil 24: 560–565, e254–e255, 2012. [DOI] [PubMed] [Google Scholar]
- 51.Wouters MM, Lambrechts D, Knapp M, Cleynen I, Whorwell P, Agréus L, Dlugosz A, Schmidt PT, Halfvarson J, Simrén M, Ohlsson B, Karling P, Van Wanrooy S, Mondelaers S, Vermeire S, Lindberg G, Spiller R, Dukes G, D'Amato M, Boeckxstaens G. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut 63: 1103–1111, 2014. [DOI] [PubMed] [Google Scholar]
- 52.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand 67: 361–370, 1983. [DOI] [PubMed] [Google Scholar]
- 53.Zucchelli M, Camilleri M, Nixon Andreasson A, Bresso F, Dlugosz A, Halfvarson J, Törkvist L, Schmidt PT, Karling P, Ohlsson B, Duerr RH, Simren M, Lindberg G, Agreus L, Carlson P, Zinsmeister AR, D'Amato M. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 60: 1671–1677, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]