Abstract
Heterotypic interactions in cancer microenvironments play important roles in disease initiation, progression, and spread. Co-culture is the predominant approach used in dissecting paracrine interactions between tumor and stromal cells, but functional results from simple co-cultures frequently fail to correlate to in vivo conditions. Though complex heterotypic in vitro models have improved functional relevance, there is little systematic knowledge of how multi-culture parameters influence this recapitulation. We therefore have employed a more iterative approach to investigate the influence of increasing model complexity; increased heterotypic complexity specifically. Here we describe how the compartmentalized and microscale elements of our multi-culture device allowed us to obtain gene expression data from one cell type at a time in a heterotypic culture where cells communicated through paracrine interactions. With our device we generated a large dataset comprised of cell type specific gene-expression patterns for cultures of increasing complexity (three cell types in mono-, co-, or tri-culture) not readily accessible in other systems. Principal component analysis indicated that gene expression was changed in co-culture but was often more strongly altered in tri-culture as compared to mono-culture. Our analysis revealed that cell type identity and the complexity around it (mono-, co-, or tri-culture) influence gene regulation. We also observed evidence of complementary regulation between cell types in the same heterotypic culture. Here we demonstrate the utility of our platform in providing insight into how tumor and stromal cells respond to microenvironments of varying complexities highlighting the expanding importance of heterotypic cultures that go beyond conventional co-culture.
Keywords: Heterotypic interactions, Principal component analysis, Multi-culture, Compartmentalization, Microfluidic
1 Introduction
Cancer is a complex disease which results from both cancer cell autonomous deregulation of growth and from non-cell autonomous interactions of cancer cells with their environments. While advances in cancer cell biology have led to diagnostic and therapeutic benefit (ie. targeted therapies such as tamoxifen, traztuzimab), an understanding of the crosstalk between cancer and non-cancer cells has proved to be less forthcoming in part because of limited models that allow dissection of the individual contributions of a complex cellular milieu. Simple co-culture models have been successfully implemented to identify factors mediating interactions between cancer and stromal compartments and point toward phenotypic changes that occur as a result (Wang et al 2015; Furukawa et al 2015; Wan et al 2014; Hollmén et al 2015; Rozenchan et al 2009; Straussman et al 2012). However, more complex in vitro models incorporating aspects of the microenvironment such as dimensionality (Weigelt et al 2014; Thoma et al 2014; Sung et al 2013; Krishnan et al 2011; Bin Kim et al 2004) and structure (Bischel et al 2015; Pisano et al 2015; Zervantonakis et al 2012; Choi et al 2015) have more successfully recreated functional responses of breast cancer seen in vivo. Heterotypic culture including three or more cell-types has also proven to be an aspect of in vitro model design that has significantly impacted model relevance when recapitulating in vivo microenvironments (Choi et al 2014; Stadler et al 2015; Balkwill and Hagemann 2012). Advances in modeling breast cancer in vitro using multi-culture techniques has recently been reviewed (Regier et al 2016). Though less common than mono- and co-culture models, heterotypic models comprised of breast cancer cells with two or more other cell types have successfully generated in vitro functional recapitulation of in vivo processes including migration (Torisawa et al 2010), intravasation (Zervantonakis et al 2012), and extravasation (Jeon et al 2015) as well as other critical functions such as angiogenesis induction (Hielscher et al 2012; Hielscher et al 2013), and micrometastasis formation (Bersini et al 2014). However, the role of the increase in heterotypic complexity in the success of these models is difficult to define for two primary reasons. First, most standard and custom platforms for heterotypic culture include a single compartment or two connected compartments limiting the manner in which multiple cell type interactions can be studied. To date, in vitro models that include three or more cell types have been used to generate almost exclusively functional and morphological measures as readouts (Torisawa et al 2010; Zervantonakis et al 2012; Jeon et al 2015; Cavnar et al 2014). Second, most multi-culture models include other varied aspects of microenvironmental complexity that make direct assessment of the effect of increasing heterotypic interactions difficult to parse (Bersini et al 2014; Choi et al 2015; Kim et al 2013a, 2013b; Chandrasekaran et al 2012). As a result, cell-type specific characterization of transcriptional changes in response to multi-culture has not been studied previously. To address the need for a more complete view of the effects of heterotypic complexity, we describe a compartmentalized multi-culture technique to measure gene expression changes across a range of breast cancer model configurations.
2 Results and discussion
2.1 Design of the Compartmentalized Micro Multi-Culture Device
We have used a compartmentalized approach to develop a platform with the advantages of straightforward operation (it is operated using a standard pipette and eliminates the need for cell sorting upstream of cell-type specific gene expression readouts) and sufficient throughput to generate twenty-four gene expression profiles where each experimental condition represented triplicate experiments. These design considerations were made to allow us to generate models with diverse configurations including various cell types in combinations of increasing complexity and to identify the effects of these changes in culture setup on the individual cell type components. The primary aim was to develop and query a device that allowed for the investigation of the effect of increasing heterotypic complexity rather than to dissect a specific in vitro or in vivo microenvironment. We therefore designed our study using cell types that were likely to influence each other’s gene expression when in co- and tri-culture. Because interactions between cancer, mesenchymal (Downey et al 2014; Amornsupak et al 2014; Luo et al 2015), and immune compartments (Nagalla et al 2013; Maley et al 2015; Stewart et al 2012; Tabariès et al 2015; Goswami et al 2005) play critical roles in a number of aspects of cellular phenotypic and functional modification in cancer progression, our models included combinations of one of two distinct breast cancers cell lines, SKBR3 and BT474, with the bone marrow stromal cell line, HS5, and ThP1 monocytes differentiated to M2, TAM-like macrophages. SKBR3 and BT474 are representative of the HER2-expressing subtype of breast cancer which comprises approximately 20 percentage of breast cancers and in which a stromal-derived gene signature was shown to have strongest prognostic value in clinical samples (Finak et al 2008).
We conducted the study described here with the goal of developing an easy to use platform that allows cell type-by-cell type analysis of cells in mono-, co-, tri-, and potentially higher level heterotypic culture. To the authors’ knowledge, no previous study has demonstrated the cell-type specific gene expression profiles for more than two cell types in a single culture without sorting upstream of cell lysis. It is important to note that the added steps of dissociation and isolation complicate procedures and involve more manipulation of and potential gene expression modification to cells. For example, different cell isolation processes have been shown to cause variable changes in gene expression profiles (Beliakova-Bethell et al 2014). Compartmentalized microscale systems have been used to maintain separate populations that interact through paracrine signals (Berry et al 2014; Domenech et al 2012; Domenech et al 2009; Torisawa et al 2010; Young et al 2012; Lang et al 2013). Diffusion ports between compartments allow soluble signals to spread by diffusion while maintaining separation of the individual cell populations. In addition, the scale prevents flow between compartments and maintains media content separation long enough for one cell population to be seeded or lysed without disturbing the other population(s) (Berry et al 2014; Lang et al 2013; Domenech et al 2009). Unique to this device is that it allows for cell type configurations with increasing heterotypic complexity, such that we could compare mono-, co-, and tri-culture gene associated regulation patterns in all three individual cell types.
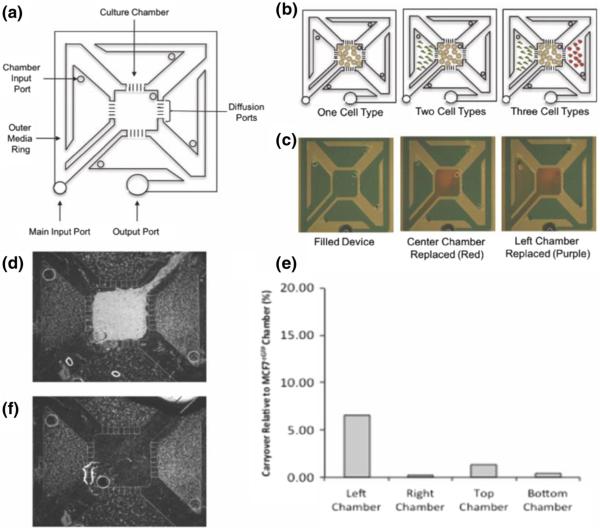
Finally, we required sufficient throughput so that all combinations of cancer, immune, and mesenchymal cell types in mono-, co-, and tri-culture could be tested. To this end the compartmentalized microscale multi-culture device was modified from a previously developed multi-chamber co-culture device (Domenech et al 2009). Both devices consisted of a center chamber and four individual culture chambers that were separated by diffusion ports ~15 μm in height, allowing for soluble factor exchange between the culture chambers while maintaining isolated cell populations. We modified the compartmentalized micro multi-culture device from a circular to a quadrilateral design to connect the main input port and output port, creating a one-push-to-fill device and eliminating the potential for air to trap within the device during filling (Fig. 1a). This modification permitted ease of use through simple pipetting and “passive pumping”(Walker and Beebe 2002). Cells were seeded into the individual chambers through the culture chamber input ports to ensure that there was no mixing of the various cell types within the culture chambers (Fig. 1b). Media changes were accomplished via the main input port, which replaces the fluid in the outer ring and by replacing media in each chamber input port. In Fig. 1c we have loaded dyes into the individual culture chambers for visualization of the addressability of individual compartments. The fluorescence micrograph in Fig. 1d demonstrates the maintenance of compartmentalization during culture between MCF7eGFP cells in the center chamber and HS5 cells in all four side chambers, whereas the micrograph in Fig. 1f shows the same device after the removal of only the MCF7eGFP cells, illustrating our capability to access cells from the different chambers selectively.
Fig. 1.
Schematic drawing of the compartmentalized micro multi-culture platform and validation of limited cross-contamination. a The micro multiculture device consists of five culture chambers separated by diffusion ports. The device holds a total volume of 50 μL with each culture chamber containing 4 μL. b Cells are seeded into the individual compartments through the chamber input port. Media was changed by replacing the media in the outer ring or in each chamber input port. c Green food grade dye was used to fill the device (50 μL) then 4 μL red dye was pipetted into the center chamber, and 4 μL purple dye was pipetted into the left culture chambers for visualization of addressability of individual compartments. d Fluorescence micrograph of MCF7eGFP cells in the center chamber with HS5 cells in all four side chambers demonstrating maintenance of cell compartmentalization. e Percentage of carryover of cellular lysate in the culture chambers. eGFP mRNA expression levels were measured in HS5 samples after co-culture with MCF7eGFP cells, percentage of carryover over was determined by comparing eGFP expression detected in HS5 cells relative to MCF7eGFP cells at the intermediate time point of 48 h. Between <0.5 % - 6 % cross-contamination was detected, indicating the compartmentalized micro multi-culture device can effectively isolate cellular lysate from the individual culture chambers without the need for downstream separation techniques (n = 2). f Fluorescence micrograph of HS5 cells in the side chambers after MCF7eGFP cells have been selectively removed
2.2 Individual cell types can Be assayed without downstream separation techniques
The objective for the implementation of this device was to measure individual cell type gene expression such that we could obtain cell type-specific molecular profile data for multi-culture models. To ensure the efficacy of the compartmentalized microscale multi-culture, it is important to have little to no cross-contamination of cellular lysate between culture chambers within the device when collecting RNA from individual cell types. Green fluorescent protein mRNA (eGFP) was measured using RT-qPCR to assess cross-contamination of RNA lysate into the individual culture chambers of the compartmentalized micro multi-culture device. MCF7eGFP cells, stably expressing eGFP, were seeded in the center chamber and co-cultured with HS5 cells in all of the possible compartmentalized co-culture patterns. Any eGFP gene transcripts detected in the HS5 samples would be derived from the MCF7eGFP labeled cells. These cells express high levels of GFP to bias detection of GFP of any contamination between compartments. Cross-contamination of individual chambers in the compartmentalized micro multi-culture device under these conditions ranged between <0.5 % - 6 % (Fig. 1e). The co-culture between the center and left chambers resulted in the highest percentage of contamination due to the close proximity of the center and left chamber channel arms. However, the right, top and bottom culture chambers demonstrated <1.3 % of carryover from the center. The low rates of cross-contamination detected in this ‘worst case scenario’ (where one cell-type very strongly expresses a transcript while the other exhibits no expression) allow us to conclude that compartmentalized micro multi-culture platform can effectively isolate cellular lysate from individual cell types without the need for downstream separation techniques.
2.3 Cell type-, heterotypic complexity-, and time-dependent regulation was apparent
Twenty gene transcript targets known to be dysregulated in breast cancer were selected for measurement by RT-qPCR. We chose to measure the expression of a subset of genes that were likely to be modulated in response to increasing heterotypic complexity with these cell types. We adopted a targeted approach rather than a microarray screen as the goal of our study was to characterize expression modulation of important genes as a result of heterotypic interactions, not to discover new interactions. We chose targets known to be involved in a number of cancer related functions and are listed in Table 1. These genes include a broad range of functions and encode ligands, enzymes, and receptors. Measuring expression levels for the same genes in all three cell types as well as ligand/receptor pairs allowed us to form hypotheses regarding complementary regulation and autocrine signaling. Assaying gene expression across each cell type in monoculture, co-culture combinations, and tri-culture resulted in twelve sets of gene expression profiles for the seven culture conditions (Table 2). The greater number of gene expression profiles than conditions is due to the ability for each cell type’s gene expression profile to be measured in the co- and tri-culture conditions, e.g. for the tri-culture condition there were the SKBR3 dataset (SK/HS/M2), the HS5 dataset (HS/SK/M2), and the ThP1-M2 dataset (M2/SK/HS). Because lysate from each individual cell type was extracted from the device and profiled, each co-culture setup gave two cell type-specific profiles and the tri-culture experiments yielded three.
Table1.
Target genes assessed in the compartmentalized micro multi-culture device. Target genes, known to be deregulated, in breast cancer were selected based on review of the literature. This panel includes gene targets involved in proliferation, angiogenesis, and invasion and migration in tumor development and progression
| List of Target Genes | |||
|---|---|---|---|
| Function | Abbrev. | Encoded Protein | Ref. |
| Proliferation | TGFB1 | Transforming Growth Factor Beta 1 | (Tobin et al. 2002; Muraoka et al. 2002) |
| Ligand | FGF2 | Basic Fibroblast Growth Factor | (Penault-Llorca et al. 1995) |
| FGF1 | Acidic Fibroblast Growth Factor | (Penault-Llorca et al. 1995; Yoshimura et al. 1998) | |
| HGF | Hepatocyte Growth Factor | (Tyan et al. 2011; Edakuni et al. 2001) | |
| Angiogenesis | VEGFa | Vascular Endothelial Growth Factor A | (Weigand et al. 2005) |
| Ligand | PDGFα | Platelet-derived Growth Factor alpha | (Shao et al. 2000; Yu et al. 2015) |
| PDGFβ | Platelet-derived Growth Factor beta | (Shao et al. 2000; Frings et al. 2013) | |
| Invasion & | MMP9 | Matrix Metalloproteinase 9 | (Fleming et al. 2012; Nanda et al. 2013) |
| Migration | MMP3 | Matrix Metalloproteinase 3 | (Nozaki et al. 2000) |
| MMP7 | Matrix Metalloproteinase 7 | (Fernandez-Garcia et al. 2014; DeLassus et al. 2011) | |
| MMP1 | Matrix Metalloproteinase 1 | (DeLassus et al. 2011; Boström et al. 2011) | |
| Invasion | TIMP1 | Tissue Inhibitor of Metalloproteinases 1 | (Fernandez-Garcia et al. 2014; Peng et al. 2011) |
| Inhibitor | TIMP2 | Tissue Inhibitor of Metalloproteinases 2 | (Fernandez-Garcia et al. 2014; Hsu et al. 2012) |
| Proliferation | FGFR1 | Fibroblast Growth Factor Receptor 1 | (Penault-Llorca et al. 1995; Jacquemier et al. 1994) |
| Receptor | FGFR2 | Fibroblast Growth Factor Receptor 2 | (Sungeun Kim et al. 2013a, 2013b) |
| TGFBR1 | Transforming Growth Factor Beta Receptor 1 | (Guido et al. 2012) | |
| TGFBR2 | Transforming Growth Factor Beta Receptor 2 | (Busch et al. 2015; Tobin et al. 2002) | |
| MET | Met Proto-oncogene | (Edakuni et al. 2001) | |
| Angiogenesis | FLT1 | FMS-related Tyrosine Kinase 1 | (Weigand et al. 2005) |
| Receptor | KDR | Kinase Insert Domain Receptor | (Guo et al. 2010) |
Table 2.
Experimental culturing conditions representing a, ER−, HER2+ breast cancer tumor microenvironment using the compartmentalized micro multi-culture device. The cell lines SKBR3, HS5 and ThP1-M2 were used to represent the cancer, mesenchymal, and immune components of a tumor microenvironment, respectively. Cell lines were cultured in tri-culture as well as mono- and co-culture combinations for controls. The same experimental conditions were used with 24-h and 72-h endpoints and substituting BT474 (ER+ HER2+) cancer cells with both endpoints
| Experimental Culturing Conditions | |||
|---|---|---|---|
|
| |||
| Cancer Cell Line | Stromal Cell Line | Immune Cell Line | |
| Mono-culture | |||
| 1 | SKBR3 | – | – |
| 2 | – | HS5 | – |
| 3 | – | – | ThP1-M2 |
| Co-culture | |||
| 1 | SKBR3 | HS5 | – |
| 2 | SKBR3 | – | ThP1-M2 |
| 3 | – | HS5 | ThP1-M2 |
| Multi-culture | |||
| 1 | SKBR3 | HS5 | ThP1-M2 |
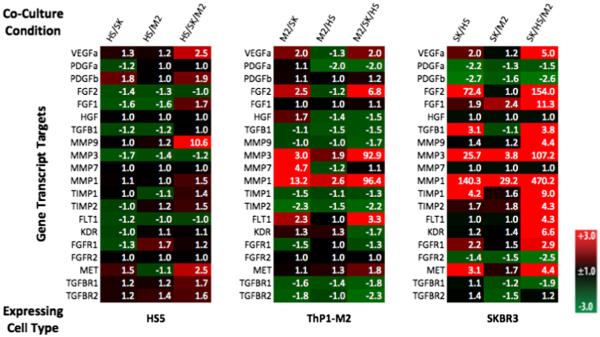
Patterns were recognizable by simple inspection of the mono-culture normalized heat maps (Fig. 2, Supp Fig. 1A, 2A, 3A). There was strong induction in heterotypic cultures specifically in breast cancer cells. For the breast cancer cell lines, strongest inductions of gene expression occurred in tri-culture, with the same trend frequently holding in ThP1-M2 and HS5 cells but typically to a less pronounced extent (i.e. more genes showed strong changes in expression in cancer cells in tri-cultures than in cancer cells in co-cultures and than in stromal cells in tri-culture). For all datasets (Fig. 2, Supp. Figure 1A, 2A, & 3A), HS5 cells demonstrated much less gene expression modulation in response to co- and tri-cultures that the other two cell types. This is consistent with previous co-culture studies in which breast cancer and normal epithelial cells paired with normal and cancer-associated fibroblasts were shown to exhibit more widespread differences in gene expression than the stromal compartment (Rozenchan et al 2009). This pattern of transcriptional effects was recreated in our cancer/stromal co-cultures conditions, but tri-culture appears to amplify the effect (Fig. 2, Supp. Figure 1A, 2A, & 3A). Increased sensitivity to paracrine interactions has been demonstrated in microscale culture where signaling factors are more concentrated relative to macroscale culture (Domenech et al 2009). The amplification of gene modulation in tri-culture may similarly make key regulations in the TME that result from heterotypic interactions more detectable.
Fig. 2.
Relative gene expression of target genes of interest across experimental conditions for experiments utilizing SKBR3, HS5, and ThP1-M2 cells with a 72-h endpoint. Gene expression levels of selected target genes known to be dysregulated in cancer were measured using RT-qPCR. The heat-map was generated by normalizing the expression of each gene to a reference gene and to monoculture for the expressing cell type and calculating the fold-change using the 2^-ΔΔCt method. Fold changes are listed in the heat-map for each condition and target gene with a false reference. Red indicates up-regulation, green down-regulation, and black no change in expression detected. Note: Throughout the figures the cell types in co- or tri-culture are listed and separated by slashes to indicate the configuration of the experiment. To indicate which cell type a gene regulation profile corresponds to, the expressing cell-type is listed first (ie SK/HS is a dataset derived from SKBR3 lysate in a configuration where the SKBR3 cells were co-cultured with HS5 cells) with all other cell types in the culture condition listed after the assayed cell type and separated by slashes.
Time-dependent patterns were exemplified in the 72-h expression patterns for ThP1-M2 and cancer cell types, which demonstrate a higher degree of gene expression modulation than was seen at 24 h (Fig. 2, Supp. Figure 1A, 2A, & 3A). Dynamic regulation of number of genes transcript and protein levels including TNF-α and MMP7 and VEGF has been demonstrated elsewhere for macrophage co-culture with ovarian and breast cancer cells (Hagemann et al 2006; Hagemann et al 2004). Our data suggests that modulation of gene expression over time is also influenced by the transition to tri-culture. For example, we found heterotypic complexity-related changes (co-culture with cancer cells compared to tri-culture) in both when and to what degree genes such as MMP1, MMP3, and MMP7 were differentially expressed in ThP1-M2 s relative to mono-culture (Fig. 2, Supp. Figure 1A, 2A, & 3A). The temporal nature of these interactions and gene expression modulation patterns may have particular impact in in vitro models where cell introductions to the model occur in sequence, such as in models of metastasis.
Also recognizable were the variety of expression modulation patterns with the change in complexity. Several genes demonstrate a gradient effect in one or more cell types where gene expression was lowest in monoculture, higher in co-cultures, and highest in tri-cultures, e.g. genes such as FGF1, TIMP2, MET in SKBR3 cells at 72 h as well as MMP1 and MMP3 in M2 and SKBR3 cells from the 72-h dataset (Fig. 2). Another potential profile represented in some genes is a relative additive effect. FGFR1 in HS5 cells, which was down regulated in co-culture with ThP1-M2 cells, up regulated in co-culture with SKBR3 cells, and showed an intermediate up regulation in tri-culture. For genes such as FGF2 in ThP1-M2 cells, however, expression showed a mixed pattern rather than being additively regulated. FGF2 expression was down regulated in HS5 co-culture, up regulated in SKBR3 co-culture, and strongly up regulated in tri-culture for ThP1-M2 s. This variety of expression modulation patterns highlights the complexity of heterotypic interactions in vitro and would be difficult to parse in a system without access to each individual cell type’s lysate.
The easily observable changes in gene expression regulation corresponding to changes in heterotypic configurations demonstrate the capability of our device to capture effects of microenvironmental complexity. It should be noted that our device limits heterotypic interactions to paracrine signal exchange, whereas in vivo interactions include juxtacrine signals, which have been shown in other cancer/stroma co-culture models to increase expression transcripts involved in proliferation and invasion beyond expression in indirect co-cultures (Che et al 2006; Fujita et al 2009). Accordingly, direct tri-cultures, while lacking the ease of use and increased throughput inherent to our platform, also merit investigation in terms of gene expression regulation. However, our platform enables increased throughput (no downstream sorting) while still raising and addressing questions of heterotypic paracrine interactions in the cancer microenvironments. Interesting, but difficult to interpret, are the diverse regulation patterns of specific genes (e.g. additive or mixed) with increasing complexity. Nevertheless, these different gene-, cell type-, time-, and complexity-dependent patterns illustrate the incomplete nature of many conventional co-culture experiments in capturing the contribution of the diverse cell types in the tumor and metastatic microenvironments to gene regulation in the stromal and cancer compartments.
2.4 Principle component analysis (PCA) effectively captures variation while reducing dimensionality
To address the complexity of the dataset, we employed a multivariate data analysis technique for dimension reduction, PCA, to identify important aspects of the data and to make more systematic and detailed observations. PCA is often capable of reducing high dimensional data to three or fewer dimensions (i.e. principal components) while capturing the majority of the variation in the original dataset, simplifying data visualization and interpretation.
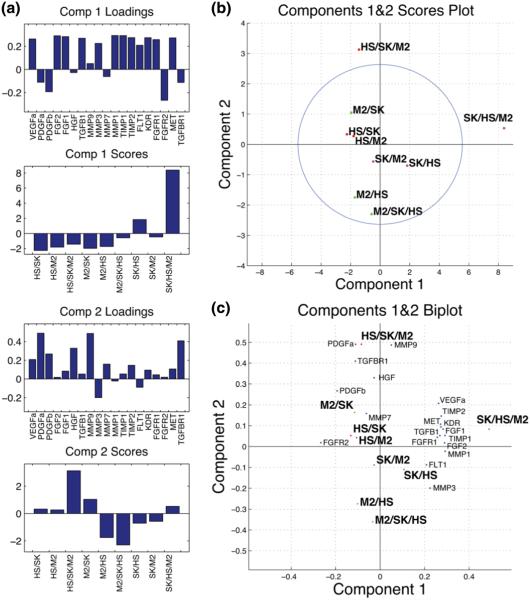
Using the princomp function in Matlab, we were able to generate scores and loading matrices that captured relationships within the culture conditions (observations) and within the gene transcript targets (variables), as well as between culture conditions and transcript levels (observations and variables). Because the analysis was able to capture 55 % of the variation in the data in the first principal component and an additional 21 % of the variation in the second principal component (Fig. 3), these two composite variables were used to map the original variables and data points to a 2D space allowing the transformed dataset to be visualized and interpreted while maintaining the majority of the information from the original dataset. For datasets from 24 h in BT474 and SKBR3 experiments and from 72 h in BT474 experiments, the first two components captured at least 67 % of the variation in the corresponding dataset (Supp Fig. 4A, 5A, & 6A). While elimination of variables from the model may have eliminated noise and improved model fit and explained variation (Krzanowski 2000; Jolliffe 2002), we chose to analyze the entire dataset to demonstrate the feasibility of deriving information and hypotheses from the datasets with minimal intervention.
Fig. 3.
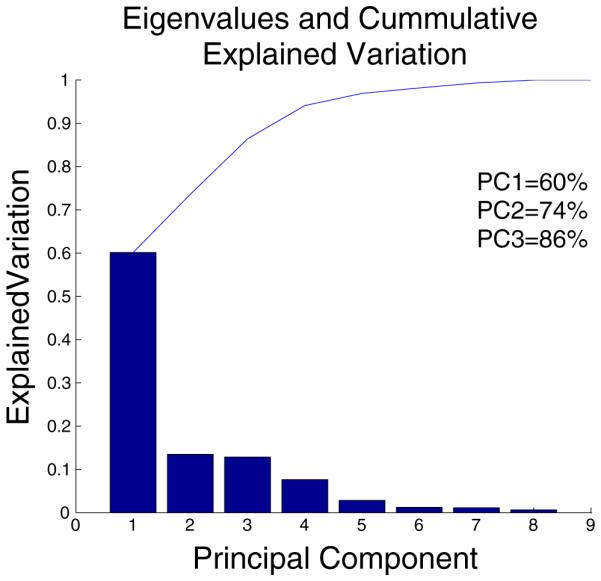
PCA explained variation plot depicting the individual (bar) and cumulative (line) explained variation for each principal component when considering all culture conditions and transcripts
2.5 Component one identifies gene regulation in cancer cells as the primary source of variation
Interestingly, all PCA score graphs for component one revealed a consistent pattern (Fig. 4A, Supp Fig. 4D, 5D, and 6D). These graphs depict a strong contribution for cancer cells in tri-culture, a lower contribution with similar directionality for cancer cells co-cultured with HS5s, and a minimal or opposing contribution for all other conditions. This pattern is unsurprising as it was evident from the heat map that expression was generally most extreme for cancer cells in tri-cultures. This observation of the heat maps corroborates the component one identification of the cancer cell-lines as the major source of variation (Fig. 4b & Fig. 2).
Fig. 4.
PCA summary figures depicting, a bar graphs containing the scores for each culture condition and loadings for each transcript in the first two principle components (Comp 1&2) b the scores for all nine heterotypic culture gene expression profiles normalized to monoculture plotted with the 95 % confidence Hotelling’s T2 ellipse, and c a biplot depicting the relationships between variable loadings and the scaled observation scores from b
Inspection of the variable (gene transcript) projections in the first component for experiments with different cancer cell types and endpoints reveals that FGF1, FGF2, TGFB1, MMP3, MMP1, and TIMP1 consistently project with or are up-regulated in cancer cells in tri-culture and to a lesser degree in cancer cells co-cultured with HS5 cells. PDGFa, PDGFb, FGFR1, and TGFBR1 are down-regulated across cancer cell regulation patterns in tri-culture. Conversely, comparison of component one projections of the BT474 and SKBR3 experiments reveal that VEGFa, MET, and KDR are differentially regulated in the two cancer cell-lines in response to tri-culture (Fig. 4a, Supp Figs 4D, 5D, & 6D). These patterns suggest that this type of model may help define conserved and divergent gene regulation by paracrine signaling in different cancer cell-lines and patient samples.
2.6 PCA identifies cancer gene regulation in tri-culture as unique
Gene expression analysis of heterotypic cultures among complex pairings of cells, like tumors, are challenged in that cells commonly have both overlapping as well as distinct gene expression profiles which are not easily teased apart or interpreted. Exploiting the compartmentalization, we were able to describe gene changes in individual compartments, and additionally. This was of particular enabling given the gene set of interest as it did not contain cell type specific targets. We took advantage of principle component analyses PCA to inform the interpretation of the gene changes across the three cell types. The PCA derived score plots, including a Hotellings T2 ellipse representing the 95 % confidence region, consistently indicate that the tri-culture conditions for breast cancer cells are outliers or fundamentally different from the other conditions (Hotelling 1933). An outlier falling outside of the Hotelling’s region is capable of skewing the model toward itself and thus is often removed from PCA analysis (Krzanowski 2000; Jolliffe 2002). We have chosen to include these observations here as their scores show a trend toward strong variation occurring in the transition from co-culture to tri-culture. This analysis reiterates the potential of tri-culture to elicit divergent or enhanced gene expression changes in breast cancer cells and in fibroblasts and macrophages in tri-culture, in certain conditions (Fig. 4b, Supp Fig. 4b).
2.7 Components are dominated by different cell types
Taking the data from one expressing cell type at a time, trends were observable in which the co-culture and tri-culture conditions for that cell type fall roughly on a line along a principal component axis or in a cluster. Because the different cancer expression scores fall along the x-axis, one could describe principal component one as the cancer component. It separated expression in the three culture conditions for the cancer cell lines and isolated cancer cells in tri-culture from the other conditions (Abdi and Williams 2010; Jolliffe 2002; Krzanowski 2000). This pattern of projections in component one was consistent across datasets for the two cancer cell lines and for the two endpoints. The second principal component could be termed the stromal component. The mesenchymal and immune cell types’ responses to co-culture and tri-culture were delineated along that axis. Unlike the scores for cancer cells in tri-culture in component one, the tri-culture scores for ThP1-M2 s and HS5s were not always the strongest projectors in component two. Instead, the pattern of scores for stromal cells in this component was highly variable, being time dependent and dependent on which cancer cell line was in co-culture (Fig. 4a, Supp Figs 4D, 5D, & 6D). These observations raise the possibility that in our model, the cancer cells were strongly and stably induced toward a specific program of gene expression to a certain degree in co-culture with HS5 cells but to a greater extent in tri-culture. Conversely the mesenchymal and immune components were more variably influenced by their context. It has been demonstrated in co-culture that gene expression in the epithelial compartments was more uniformly regulated in response to co-culture than in the corresponding fibroblast compartments for co-cultures of MDA-MB-231 and MCF10A cell lines with normal and cancer-associated fibroblasts (Rozenchan et al 2009), but the effect of tri-culture in this regard has not been demonstrated previously.
2.8 Co-culture is limitedly predictive of tri-culture gene expression patterns
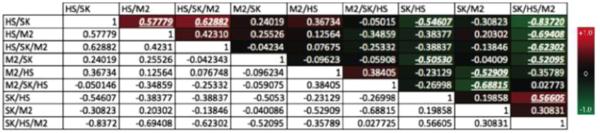
Importantly, we found that co-cultures showed different degrees of correlation with tri-culture. As an example, the cancer cell-lines in co-culture with HS5s consistently project with the same direction but with a lesser magnitude in component one as compared to the cancer cells in tri-culture. The Pearson’s correlation coefficient consistently identified the profiles for these conditions as being significantly correlated (Fig. 5, Supp Figs 1B&C, 2B&C, 3B&C, & 7) indicating that co-culture with HS5s could be at least partially predictive of the gene expression regulation in tri-culture where ThP1-M2 s are added. In component two the score for ThP1-M2 s in co-culture with cancer cells consistently projects with ThP1-M2’s tri-culture score (Fig. 4b, Supp Figs 4B, 5B, & 6B). This relationship between their contributions to components one and two was not reflected in Pearson’s correlation coefficients, which did not always indicate a significant correlation (Fig. 5, Supp Figs 1B&C, 2B&C, 3B&C, & 7). Therefore, it cannot be concluded that the overall expression pattern from co-culture conditions was uniformly indicative of expression patterns in tri-culture. This observation supports a unique role for models with increased heterotypic complexity in elucidating interactions and cellular responses not evident in more traditional co-culture.
Fig. 5.
Matrix containing Pearson’s correlation rho values for the mean centered, unit variance scaled gene regulation (fold change) profiles of each culture condition compared with each other culture condition in the SKBR3 72-h dataset for comparison of gene expression changes in response to co- or tri-culture conditions. Significant (p < 0.05) rho values are bolded, italicized, and underlined
2.9 Autocrine and complimentary regulation occurs in tri-culture
As previously mentioned the genes for the ligand and receptors involved in VEGF signaling were differentially regulated in the two cancer cell lines. Interestingly, both the ligand, VEGFa, and receptor, FLT1 and KDR, genes were upregulated in tri-culture for SKBR3s but not BT474s ((Fig. 2 and Suppl. Figs 1A, 2A, 3A)., which suggests enhanced autocrine signaling for SKBR3s in response to tri-culture. Autocrine signaling through the VEGF axis has been demonstrated to enhance invasion and survival both in vivo and in vitro (Luo et al 2016; Bachelder et al 2001). Based on our results may also be magnified in response to certain heterotypic culture conditions. There was also evidence of an increase in TGF-β paracrine signaling between SKBR3s (ligand) and HS5s (receptors) in tri-culture (Fig. 2 and Suppl. Figure 3A). More generally, there was widespread and frequently significant anti-correlation between the cancer cell regulation profiles and those of the stromal cells across cancer types and co- and tri-culture combinations (Fig. 5, Suppl Figs 1B, 2B, 3B). This type of complimentary regulation has been noted elsewhere for co-ordinated metabolic programming in co-cultures of breast cancer cells with fibroblasts (Ueno et al 2015; Fiaschi et al 2012; Rattigan et al 2012). Our dataset expands this observed phenomenon of cooperative regulation to potential involvement in ligand, MMP, and receptor gene expression. Additionally, in our dataset many of the strongest, significant anti-correlations involve cancer cells in tri-culture suggesting that this effect may be further magnified in tri-culture.
3 Conclusions
This study presents a new approach to studying heterotypic interactions in in vitro models of breast cancer. Exploiting the maintenance of compartmentalization of cells and their lysates in micro-scale, we have measured gene expression across three cell types in monocultures, co-culture combinations, and tri-culture. By simple observation, we noted cell type-, time-, and complexity-dependent classes of gene expression changes. Using PCA supplemented with Pearson’s correlations, we were able to observe the strong effect of tri-culture on gene expression and to form hypotheses concerning the transition from mono-culture to co- and tri-culture. For example, we identified the cancer cells as the potential primary regulators of the global gene expression patterns as they achieve the most consistent and increasing expression pattern with changes in complexity, while the stromal compartments may be differentially regulated under different configurations to “tune” or “stabilize” changing cancer cell regulation. We also found evidence in our data that supports an enhanced effect in autocrine and paracrine signaling and a complementary regulation effect across compartments in tri-culture. These examples of general hypotheses could be investigated in more specific model settings, and more directed hypotheses could be formed by catering the experimental setup to a more specific set of interactions and gene targets.
This study used cell lines as proof of principle, but extrapolating to primary samples, this might also explain the heterogeneity seen in analysis of gene arrays of the tumor microenvironments as unique cancer components may direct the tumor microenvironment. Experimental tools similar to our compartmentalized multi-culture device and PCA (or other data analysis techniques) may generate hypotheses concerning the fundamental and higher level influences of heterotypic complexity. Incorporation of additional aspects of microenvironmental complexity such as dimensionality and ECM components, mechanics, and architecture may provide even more insight into the integration and response of cells to diverse signals in their microenvironment. Data obtained through the straightforward operation of a compartmentalized microfluidic devices, such as that presented here, has the potential give both depth and breadth of information concerning the effects of elements of complexity in cancer microenvironments, which may in turn drive more informed model design, drug development, and treatment selection.
4 Methods
4.1 Compartmentalized Micro Multi-Culture Device Fabrication
Compartmentalized micro multi-culture devices were made using previously described techniques (Xia and Whitesides 1998; Duffy et al 1998). Briefly, poly-(dimethylsiloxane) (PDMS) (Dow Corning Corporation, Midland, MI, USA) devices were cross-linked on a SU-8 wafer. To make the SU-8 wafer, an initial layer of SU-8 10 (MicroChem™) was spin-coated onto a silicon wafer (University Wafers) to create the 15 μm height diffusion port layer. A second layer of SU-8100 (MicroChem™) was then used to create the 250 μm height culture chambers. Finally, a third layer of SU-8100 was spin-coated to create the 500 μm height port holes. The SU-8 wafer was exposed to UV light between each spin-coated layer to cross-link the SU-8. After spin-coating and exposing all the layers, the SU-8 wafer was then developed in propylene glycol monomethyl ether acetate (Sigma-Aldrich®) for 40 min followed by washing twice with Acetone (VWR®) and 2-Propanol (Thermo Fisher Scientific, Inc.). Once the PDMS devices were cross-linked onto the SU-8 wafer, any remaining non-cross-linked PDMS monomers were next extracted from the devices using a 500 mL Soxhlet extractor containing 100 % ethyl alcohol heated to boiling for a minimum of 4 h. Compartmentalized microfluidic multi-culture devices were then cut into 3 × 3 arrays (9 devices/array), plasma treated (Diener Electronic) in the presence of O2, bonded to a 75×50×1mm pre-cleaned micro glass slide (Corning®), and heated for 5 min at 135 °C to secure the bond.
4.2 Cell culture
The breast cancer epithelial cell lines SKBR3 and BT474 (ATCC, Rockville, MD, USA), and human ThP1 monocytes (ThP1) (ATCC, Rockville, MD, USA) were maintained in RPMI 1640 with L-glutamine (CellGro, Corning®) containing 10 % fetal bovine serum (Gibco®, LifeTechnologies™) supplemented with 1 % penicillin (10,000 units/mL) /streptomycin (10,000 μg/mL) (Gibco®, LifeTechnologies™) and 1 % sodium pyruvate (100 mM) (Gibco®, LifeTechnologies™). ThP1 cells were also supplemented with 0.1 % beta-2-mercaptoethanol (50 mM) to avoid clumping (Sigma-Aldrich®). HS5 bone marrow cells (HS5) (ATCC, Rockville, MD, USA) and the breast epithelial cell line MCF7 (ATCC, Rockville, MD, USA) previously transfected with the green fluorescent protein eGFP (MCF7eGFP) were maintained in high glucose DMEM (CellGro, Corning®) containing 10 % fetal bovine serum (Gibco®, LifeTechnologies™) supplemented with 1 % penicillin (10,000 units/mL) /streptomycin (10,000 μg/mL) (Gibco®, LifeTechnologies™) and 1 % sodium pyruvate (100 mM) (Gibco®, LifeTechnologies™). All cell lines were cultured at 37 °C in 5 % CO2. SKBR3, MCF7, and HS5 cell lines were passaged with 0.05 % trypsin-EDTA solution (Invitrogen™, LifeTechnologies™). ThP1 cells were passaged in 6-well dishes. When cells reached a confluence of 1×106 cells/mL, 1 mL of cell suspension was plated into a separate 6-well dish containing 2mLs of complete media. Differentiation and polarization of the ThP1 cells into M2 TAM-like macrophages (ThP1-M2) were conducted using previously described techniques (Tjiu et al 2009). After differentiation and polarization, ThP1-M2 cells rested for 3 days prior to starting experiments.
4.3 Compartmentalized Micro Multi-Culture Experimental Design Set-Up
Bonded device slides (3 × 3 array (9 devices total)) for each experimental condition were placed into sterile omni-trays (NUNC™) and 50 μL of 0.1 % gelatin in 1× PBS was pipetted into each of the devices through passive pumping (Walker and Beebe 2002) using a standard manual pipet to coat the surface for cell attachment. The gelatin solution was then immediately removed from the devices and allowed to dry under a biological safety cabinet for 20 min. Following gelatin coating, 50 μL of complete media containing 1 % fetal bovine serum was pipetted into individual devices. Prior to seeding, cells were washed twice in 1 % fetal bovine serum containing media. Individual cell suspensions were pipetted into device chambers by placing a 4 μL droplet in the input port of each respective culture chamber. A 10 μL droplet of complete media was placed at the output port to reduce evaporation. SKBR3 or BT474 cells (center chamber) were seeded at a density of 5000 cells/chamber, HS5 cells (left-side chamber) were seeded at a density of 2500 cells/chamber and ThP1-M2 cells (right-side chamber) were seeded at a density of 5000 cells/chamber due to their loss of proliferation after the polarization process. These seeding densities prevented overgrowth and nutrient depletion over the culture period while providing sufficient mRNA for analysis from nine pooled devices for each of three replicates. Cells were monitored for viablility, and normal morphologies were noted throughout the course of all experiments. The remaining two compartments were filled with media only to focus our investigation on the effects of tri-culture in the individual cell types. The extra compartments will enable future experiments to include up to five different cell types for investigation of the tumor microenvironment. End-points for the individual cell types were collected at 24 or 72 h.
4.4 RNA extraction and isolation, and cDNA synthesis
Total RNA was extracted using the RNeasy Micro Plus Kit (Qiagen, Inc. USA). Briefly, 4 μL of buffer RLT-Plus containing 1 % beta-2-mercaptoethanol (Sigma-Aldrich®) was pipetted into individual culture chambers using a similar method to cell seeding. Lysate was disrupted within the culture chamber by pipetting up and down several times and transferred into a microcentrifuge tube. Nine biological replicates for each experimental condition of each individual cell population were pooled together to obtain a sufficient concentration of RNA, additional lysis buffer was then added to the microcentrifuge tube for a total manufacturer recommended volume of 350 μL. Samples were then vortexed to homogenize the lysate and mixed with an equal volume of 70 % ethanol. To eliminate any genomic DNA, the lysate mixture was pipetted into a supplied gDNA spin-column and centrifuged for 30 s at 10,000 g. The samples were then pipetted into a miniElute spin-column and total RNA was then isolated according to manufacturer ’s instructions. cDNA synthesis was conducted using the High Capacity RNA to cDNA kit (LifeTechnologies™) where 25 ng of total RNA was transcribed in 20 μL reactions per sample. Concentration of total RNA was measured using the NanoDrop™ spectrophotometer (ThermoScienfic).
4.5 Real-time reverse transcription-PCR (RT-qPCR)
RT-qPCR was conducted using Locked Nucleic Acid (LNA™) probes selected from the Universal Probe Library (Roche Applied Sciences). Gene primers were designed using ProbeFinder software (Roche Applied Sciences). Primer sequences will be made available upon request. Each reaction was carried out in 384-well plates in triplicate at a 10 μL volume containing 5 μL of 2× gene expression master mix (Roche Applied Sciences), 0.15 μL of forward and reverse primers (20 μM) (Invitrogen™, LifeTechnologies™), 0.1 μL of LNA™ probe (Roche Applied Sciences), 2.6 μL of nuclease-free water and 2 μL (25 ng RNA) of template. For amplification and detection of target genes the Roche LightCycler 480 was used under the following cycling conditions, 95 °C for 10 min to initiate the Taq-polymerase enzyme, then 40-cycles of 95 °C for 10 s for denaturation, 60 °C for 30 s for annealing and 72 °C for 1 s for extension, followed by 40 °C for 30 s to cool the plate. The average of two selected reference genes for each cell type was used as endogenous control. Nuclease-free water served as a no template control.
4.6 Cross-Contamination Experiments
MCF7eGFP and HS5 cells were seeded into the compartmentalized micro multi-culture devices as described above at a density of 5000 cells/chamber (MCF7eGFP) and 2500 cells/chamber (HS5). MCF7eGFP cells were cultured in the center chamber with HS5 cells cultured in co-culture patterns around the four outer chambers. Total RNA was extracted from each individual chamber after 48 h and converted to cDNA using previously described methods. eGFP gene expression was measured using RT-qPCR in both the MCF7 and HS5 samples. Percentage of eGFP expression was then calculated by normalizing HS5 samples to MCF7 samples using the ΔΔCt method.
4.7 Principal component analysis
The 20 variables were variably scaled and consolidated into two composite components by PCA, permitting the mapping of the dataset to two-dimensional space, and making visualization possible. PCA was conducted using the princomp function in MATLAB v8.1. Prior to analysis the data for each analyte was mean centered and autoscaled. Our principal component space, the transformed two dimensional dataset, was represented in a score plots and biplot where the calculated loadings in the first two principal components were graphed with the scores of the observations, which were scaled to unity with the maximum value of the scores and then scaled to the length of the loadings. Scaling in this manner maintained the relative magnitude of the scores between one another and their directionality with respect to each other and the loadings (Krzanowski 2000; Jolliffe 2002). To calculate the cumulative explained variation, the latent vector, a vector containing the eigenvalues of the covariance matrix of the gene expression dataset, was summed up to a given principal component and divided by the total sum for all principal components. Hence, the cumulative variation explained increased proportionally to the amount of variation captured in each principal component, until 100 % of the variation was explained at some component ≤ the ninth principal component (Krzanowski 2000; Jolliffe 2002).
4.8 Pearson’s correlations
Pearson’s correlations were calculated for the each dataset after mean centering and unit variance scaling each variable. The corr() function in MATLAB v8.1 was used to output the rho and p-value matrices. This data was imported to Excel and a color scale was applied using conditional formatting. Correlation rho’s with p-values ≤0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was funded by University of Wisconsin Carbone Cancer Center Cancer Center Support Grant NIH P30 CA014520, NIH R33 CA160344, NIH R01 CA159578, NIH T32 GM008349, NIH T32 CA157322.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10544-016-0083-x) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Disclosure of potential conflicts of interest David J. Beebe holds equity in Bellbrook Labs, LLC, Tasso, Inc., Stacks to the Future, LLC, and Salus Discovery, LLC. Scott M. Berry holds equity in Salus Discovery, LLC.
References
- Abdi H, Williams LJ. Principal component analysis. Wiley Interdisciplinary Reviews: Computational Statistics. 2010;2(4):433–459. [Google Scholar]
- Amornsupak K, et al. Cancer-associated fibroblasts induce high mobility group box 1 and contribute to resistance to doxorubicin in breast cancer cells. BMC Cancer. 2014;14:955. doi: 10.1186/1471-2407-14-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelder RE, et al. Vascular endothelial growth factor is an autocrine survival factor for neuropilin-expressing breast carcinoma cells. Cancer Res. 2001;61(15):5736–5740. [PubMed] [Google Scholar]
- Balkwill FR, Hagemann T. The tumor microenvironment at a glance. J. Cell Sci. 2012;125(23):5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- Beliakova-Bethell N, et al. The effect of cell subset isolation method on gene expression in leukocytes. Cytometry Part A. 2014;85(1):94–104. doi: 10.1002/cyto.a.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SM, et al. Streamlining gene expression analysis: integration of co-culture and mRNA purification. Integrative Biology. 2014;6(2):224–231. doi: 10.1039/c3ib40136g. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24413730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersini S, et al. A microfluidic 3D invitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35(8):2454–2461. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Kim J, Stein R, O’Hare M. Three-dimensional in vitro tissue culture models of breast cancer - a review. Breast Cancer Res. Treat. 2004;85:281–291. doi: 10.1023/B:BREA.0000025418.88785.2b. Available at: http://discovery.ucl.ac.uk/36475/ [DOI] [PubMed] [Google Scholar]
- Bischel LL, Beebe DJ, Sung KE. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer. 2015;15(1):1–10. doi: 10.1186/s12885-015-1007-5. Available at: http://www.biomedcentral.com/1471-2407/15/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström P, et al. MMP-1 expression has an independent prognostic value in breast cancer. BMC Cancer. 2011;11(1):1. doi: 10.1186/1471-2407-11-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S, et al. TGF-beta receptor type-2 expression in cancer-associated fibroblasts regulates breast cancer cell growth and survival and is a prognostic marker in pre-menopausal breast cancer. Oncogene. 2015;34(1):27–38. doi: 10.1038/onc.2013.527. [DOI] [PubMed] [Google Scholar]
- Cavnar SP, et al. Microfluidic source-sink model reveals effects of biophysically distinct CXCL12 isoforms in breast cancer chemotaxis. Integr. Biol. 2014;6(5):564–576. doi: 10.1039/c4ib00015c. doi:10.1039/C4IB00015C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, et al. Effect of homotypic and heterotypic interaction in 3D on the E-selectin mediated adhesive properties of breast cancer cell lines. Biomaterials. 2012;33(35):9037–9048. doi: 10.1016/j.biomaterials.2012.08.052. doi:10.1016/j.biomaterials.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che ZM, et al. Collagen-based co-culture for invasive study on cancer cells-fibroblasts interaction. Biochem. Biophys. Res. Commun. 2006;346(1):268–275. doi: 10.1016/j.bbrc.2006.05.111. [DOI] [PubMed] [Google Scholar]
- Choi SYC, et al. Lessons from patient-derived xenografts for better in vitro modeling of human cancer. Adv. Drug Deliv. Rev. 2014;79-80:222–237. doi: 10.1016/j.addr.2014.09.009. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0169409X14002075. [DOI] [PubMed] [Google Scholar]
- Choi Y, et al. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015;15(16):3350–3357. doi: 10.1039/c5lc00514k. doi:10.1039/C5LC00514K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLassus GS, Cho H, Eliceiri GL. New signaling pathways from cancer progression modulators to mRNA expression of matrix metalloproteinases in breast cancer cells. J. Cell. Physiol. 2011;226(12):3378–3384. doi: 10.1002/jcp.22694. [DOI] [PubMed] [Google Scholar]
- Domenech M, et al. Cellular observations enabled by microculture: paracrine signaling and population demographics. Integrative biology: quantitative biosciences from nano to macro. 2009;1(3):267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech M, et al. Hedgehog signaling in myofibroblasts directly promotes prostate tumor cell growth. Integrative Biology. 2012;4(2):142. doi: 10.1039/c1ib00104c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey CL, et al. The prognostic significance of tumour – stroma ratio in oestrogen receptor-positive breast cancer. Br. J. Cancer. 2014;110(7):1744–1747. doi: 10.1038/bjc.2014.69. doi:10.1038/bjc.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DC, et al. Rapid prototyping of microfluidic systems in poly (dimethylsiloxane) Analytical …. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. doi:10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Edakuni G, et al. Expression of the hepatocyte growth factor/c-met pathway is increased at the cancer front in breast carcinoma. Pathol. Int. 2001;51(3):172–178. doi: 10.1046/j.1440-1827.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia B, et al. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology. 2014;64(4):512–522. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- Fiaschi T, et al. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. 2012;72(19):5130–5140. doi: 10.1158/0008-5472.CAN-12-1949. [DOI] [PubMed] [Google Scholar]
- Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Fleming JM, et al. Paracrine interactions between primary human macrophages and human fibroblasts enhance murine mammary gland humanization in vivo. Breast Cancer Res. 2012;14(3):R97. doi: 10.1186/bcr3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frings O, et al. Prognostic significance in breast cancer of a Gene signature capturing stromal PDGF signaling. Am. J. Pathol. 2013;182(6):2037–2047. doi: 10.1016/j.ajpath.2013.02.018. Available at: http://www.sciencedirect.com/science/article/pii/S000294401300196X. [DOI] [PubMed] [Google Scholar]
- Fujita H, et al. Tumor-stromal interactions with direct cell contacts enhance proliferation of human pancreatic carcinoma cells. Cancer Sci. 2009;100(12):2309–2317. doi: 10.1111/j.1349-7006.2009.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, et al. Lung epithelial cells induce both phenotype alteration and senescence in breast cancer cells. PloS one. 2015;10(1):e0118060. doi: 10.1371/journal.pone.0118060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S, et al. Macrophages promote the invasion of breast carcinoma cells via a Colony-stimulating factor-1 / epidermal growth factor paracrine loop. Cancer Res. 2005;65(12):5278–5284. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- Guido C, et al. Metabolic reprogramming of cancer-associated fibroblasts by TGF-β drives tumor growth: connecting TGF-β signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle. 2012;11(16):3019–3035. doi: 10.4161/cc.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, et al. Vascular endothelial growth factor receptor-2 in breast cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2010;1806(1):108–121. doi: 10.1016/j.bbcan.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann T, et al. Enhanced invasiveness of breast cancer cell lines upon co-cultivation with macrophages is due to TNF-α dependent up-regulation of matrix metalloproteases. Carcinogenesis. 2004;25(8):1543–1549. doi: 10.1093/carcin/bgh146. [DOI] [PubMed] [Google Scholar]
- Hagemann T, et al. Ovarian cancer cells polarize macrophages toward a tumor-associated phenotype. J. Immunol. 2006;176(8):5023–5032. doi: 10.4049/jimmunol.176.8.5023. [DOI] [PubMed] [Google Scholar]
- Hielscher AC, Qiu C, Gerecht S. Breast cancer cell-derived matrix supports vascular morphogenesis. American journal of physiology. Cell physiology. 2012;302(8):C1243–C1256. doi: 10.1152/ajpcell.00011.2012. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3774551{&}tool=pmcentrez{&}rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hielscher A, et al. Hypoxia affects the structure of breast cancer cell-derived matrix to support Angiogenic responses of endothelial cells. J Carcinog Mutagen. 2013;18(9):1199–1216. doi: 10.4172/2157-2518.S13-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmén M, et al. Characterization of macrophage-cancer cell crosstalk in estrogen receptor positive and triple-negative breast cancer. Scientific reports. 2015;5 doi: 10.1038/srep09188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933;24(6):417–441. [Google Scholar]
- Hsu C-H, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2(3):568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- Jacquemier J, et al. Expression of the FGFR1 gene in human breast-carcinoma cells. Int. J. Cancer. 1994;59(3):373–378. doi: 10.1002/ijc.2910590314. [DOI] [PubMed] [Google Scholar]
- Jeon JS, et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. 2015;112(1):214–219. doi: 10.1073/pnas.1417115112. doi:10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT. Principal component analysis. 2nd. Springer-Verlag; New York: Available at: https://search.library.wisc.edu/catalog/9999358336021212002. [Google Scholar]
- Kim S, et al. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013a;13(8):1489–1500. doi: 10.1039/c3lc41320a. Available at: http://pubs.rsc.org.globalproxy.cvt.dk/en/content/articlehtml/2013/lc/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- Kim S, et al. FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells. PloS one. 2013b;8(1):e51671. doi: 10.1371/journal.pone.0051671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, et al. Dynamic interaction between breast cancer cells and osteoblastic tissue: comparison of two-and three-dimensional cultures. J. Cell. Physiol. 2011;226(8):2150–2158. doi: 10.1002/jcp.22550. [DOI] [PubMed] [Google Scholar]
- Krzanowski WJ. Principles of multivariate analysis: a user’s perspective. Oxford University Press; New York: 2000. 2000. rev. ed. Oxford [Oxfordshire] Available at: https://search.library.wisc.edu/catalog/999932692202121. [Google Scholar]
- Lang JD, et al. Hormonally responsive breast cancer cells in a microfluidic co-culture model as a sensor of microenvironmental activity. Integrative Biology. 2013;5(5):807–816. doi: 10.1039/c3ib20265h. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23559098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, et al. Cancer-associated fibroblasts: a multifaceted driver of breast cancer progression. Cancer Lett. 2015;361(2):155–163. doi: 10.1016/j.canlet.2015.02.018. doi:10.1016/j.canlet.2015.02.018. [DOI] [PubMed] [Google Scholar]
- Luo M, et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-κB and β-catenin. Cancer letters. 2016;373(1):1–11. doi: 10.1016/j.canlet.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Maley CC, et al. An ecological measure of immune-cancer colocalization as a prognostic factor for breast cancer. Breast Cancer Research. 2015;17(1):1. doi: 10.1186/s13058-015-0638-4. doi:10.1186/s13058-015-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka RS, et al. Blockade of TGF-β inhibits mammary tumor cell viability, migration, and metastases. J. Clin. Invest. 2002;109(12):1551–1559. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalla S, et al. Interactions between immunity , proliferation and molecular subtype in breast cancer prognosis. Genome Biology. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda DP, et al. Matrix metalloproteinase-9 as a potential tumor marker in breast cancer. J. Environ. Pathol. Toxicol. Oncol. 2013;32(2):115–129. doi: 10.1615/jenvironpatholtoxicoloncol.2013008166. [DOI] [PubMed] [Google Scholar]
- Nozaki S, Sledge GW, Nakshatri H. Cancer cell-derived interleukin 1α contributes to autocrine and paracrine induction of prometastatic genes in breast cancer. Biochem. Biophys. Res. Commun. 2000;275(1):60–62. doi: 10.1006/bbrc.2000.3241. [DOI] [PubMed] [Google Scholar]
- Penault-Llorca F, et al. Expression of FGF and FGF receptor genes in human breast cancer. Int. J. Cancer. 1995;61(2):170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- Peng L, et al. A fine balance between CCNL1 and TIMP1 contributes to the development of breast cancer cells. Biochem. Biophys. Res. Commun. 2011;409(2):344–349. doi: 10.1016/j.bbrc.2011.05.021. [DOI] [PubMed] [Google Scholar]
- Pisano M, et al. An in vitro model of the tumor-lymphatic microenvironment with simultaneous transendothelial and luminal flows reveals mechanisms of flow enhanced invasion. Integrative biology. 2015;26(1):B5. doi: 10.1039/c5ib00085h. doi:10.1039/c5ib00085h. [DOI] [PubMed] [Google Scholar]
- Rattigan YI, et al. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp. Cell Res. 2012;318(4):326–335. doi: 10.1016/j.yexcr.2011.11.014. Available at: http://www.sciencedirect.com/science/article/pii/S001448271100471X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier MC, Alarid ET, Beebe D. Progress towards understanding heterotypic interactions in multi-culture models of breast cancer. Integr. Biol. 2016;8:684–692. doi: 10.1039/c6ib00001k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenchan PB, et al. Reciprocal changes in gene expression profiles of cocultured breast epithelial cells and primary fibroblasts. Int. J. Cancer. 2009;125(12):2767–2777. doi: 10.1002/ijc.24646. [DOI] [PubMed] [Google Scholar]
- Shao Z-M, Nguyen M, Barsky SH. Human breast carcinoma desmoplasia is PDGF initiated. Oncogene. 2000;19(38) doi: 10.1038/sj.onc.1203785. [DOI] [PubMed] [Google Scholar]
- Stadler M, et al. Increased complexity in carcinomas : analyzing and modeling the interaction of human cancer cells with their microenvironment. Semin. Cancer Biol. 2015;35:107–124. doi: 10.1016/j.semcancer.2015.08.007. doi:10.1016/j.semcancer.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Stewart DA, et al. Basal-like Breast Cancer Cells Induce Phenotypic and Genomic Changes in Macrophages. Mol. Cancer Res. 2012;10(6):727–739. doi: 10.1158/1541-7786.MCR-11-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung KE, et al. Understanding the impact of 2D and 3D fibroblast cultures on in vitro breast cancer models. PLoS One. 2013;8(10):1–13. doi: 10.1371/journal.pone.0076373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabariès S, et al. Granulocytic immune infiltrates are essential for the efficient formation of breast cancer liver metastases. Breast Cancer Res. 2015;17(1):1. doi: 10.1186/s13058-015-0558-3. doi:10.1186/s13058-015-0558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma CR, et al. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014;69-70:29–41. doi: 10.1016/j.addr.2014.03.001. doi:10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Tjiu J-W, et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. The Journal of investigative dermatology. 2009;129(4):1016–1025. doi: 10.1038/jid.2008.310. [DOI] [PubMed] [Google Scholar]
- Tobin SW, et al. Consequences of altered TGF-beta expression and responsiveness in breast cancer: evidence for autocrine and paracrine effects. Oncogene. 2002;21(1):108–118. doi: 10.1038/sj.onc.1205026. [DOI] [PubMed] [Google Scholar]
- Torisawa Y-S, et al. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Integrative biology: quantitative biosciences from nano to macro. 2010;2(11–12):680–686. doi: 10.1039/c0ib00041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyan S-W, et al. Breast cancer cells induce cancer-associated fibroblasts to secrete hepatocyte growth factor to enhance breast tumorigenesis. PLoS One. 2011;6(1):1–9. doi: 10.1371/journal.pone.0015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, et al. Characteristic Gene expression profiles of human fibroblasts and breast cancer cells in a newly developed bilateral Coculture system. BioMed Research International. 2015;2015:960840. doi: 10.1155/2015/960840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- Wan S, et al. BMP9 regulates cross-talk between breast cancer cells and bone marrow-derived mesenchymal stem cells. Cell. Oncol. 2014;37(5):363–375. doi: 10.1007/s13402-014-0197-1. doi:10.1007/s13402-014-0197-1. [DOI] [PubMed] [Google Scholar]
- Wang C, et al. Human Adipocytes Stimulate Invasion of Breast Cancer MCF-7 Cells by Secreting IGFBP-2. PloS one. 2015;10(3):e0119348. doi: 10.1371/journal.pone.0119348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand M, et al. Autocrine vascular endothelial growth factor signalling in breast cancer. Evidence from cell lines and primary breast cancer cultures in vitro. Angiogenesis. 2005;8(3):197–204. doi: 10.1007/s10456-005-9010-0. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Ghajar CM, Bissell MJ. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv. Drug Deliv. Rev. 2014;69-70:42–51. doi: 10.1016/j.addr.2014.01.001. doi:10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia YN, Whitesides GM. Soft lithography. Annu. Rev. Mater. Sci. 1998;37(5):551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. Available at: http://apps.isiknowledge.com/InboundService.do?product=WOS&action=retrieve&SrcApp=Papers&UT=000075395600009&SID=4 AeDcEnmon6P1nPEIJo&Init=Yes&SrcAuth=mekentosj&mode=FullRecord&customersID=mekentosj&DestFail=http://access.isiproducts.com/custom_images/wok_f. [DOI] [PubMed] [Google Scholar]
- Yoshimura N, et al. The expression and localization of fibroblast growth factor-1 (FGF-1) and FGF receptor-1 (FGFR-1) in human breast cancer. Clin. Immunol. Immunopathol. 1998;89(1):28–34. doi: 10.1006/clin.1998.4551. [DOI] [PubMed] [Google Scholar]
- Young EWK, et al. Microscale functional cytomics for studying hematologic cancers. Blood. 2012;119(10):76–86. doi: 10.1182/blood-2011-10-384347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, et al. FoxM1 promotes breast tumorigenesis by activating PDGF-A and forming a positive feedback loop with the PDGF/AKT signaling pathway. Oncotarget. 2015;6(13):11281. doi: 10.18632/oncotarget.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervantonakis IK, et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. 2012;109(34):13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.