Abstract
Myosin light chain kinase (MLCK) phosphorylates S19 of the myosin regulatory light chain (RLC), which is required to activate myosin's ATPase activity and contraction. Smooth muscles are known to display plasticity in response to factors such as inflammation, developmental stage, or stress, which lead to differential expression of nonmuscle and smooth muscle isoforms. Here, we compare steady-state kinetics parameters for phosphorylation of different MLCK substrates: (1) nonmuscle RLC, (2) smooth muscle RLC, and heavy meromyosin subfragments of (3) nonmuscle myosin IIB, and (4) smooth muscle myosin II. We show that MLCK has a ~2-fold higher kcat for both smooth muscle myosin II substrates compared with nonmuscle myosin IIB substrates, whereas Km values were very similar. Myosin light chain kinase has a 1.6-fold and 1.5-fold higher specificity (kcat/Km) for smooth versus nonmuscle-free RLC and heavy meromyosin, respectively, suggesting that differences in specificity are dictated by RLC sequences. Of the 10 non-identical RLC residues, we ruled out 7 as possible underlying causes of different MLCK kinetics. The remaining 3 residues were found to be surface exposed in the N-terminal half of the RLC, consistent with their importance in substrate recognition. These data are consistent with prior deletion/chimera studies and significantly add to understanding of MLCK myosin interactions.
Keywords: myosin light chain kinase, myosin regulatory light chain, nonmuscle myosin, smooth muscle, smooth muscle myosin, steady-state kinetics, phosphorylation
1 INTRODUCTION
Myosin light chain kinase (MLCK; EC 2.7.11.18) is a ubiquitous Ca2+/calmodulin (CaM)-activated kinase found in smooth, cardiac, and skeletal muscle as well as in mammalian nonmuscle cells. In humans, the MYLK1 gene encodes both the so-called nonmuscle (long) and the smooth muscle isoforms (short) of MLCK in addition to the nonkinase telokin by alternative initiation sites.1 The nonmuscle and smooth muscle isoforms share identical sequences across the 2 major domains that are known to be important to MLCK-myosin interactions, the kinase and telokin (C-terminal IgG) domains. Therefore, it is expected that the nonmuscle and smooth muscle MLCK will have very similar myosin phosphorylation kinetics.
Nonmuscle myosin (NMM) and smooth muscle myosin (SMM) are the only known substrates of MLCK in vivo.2,3 Phosphorylation of myosin on S19 of the RLC (also called LC20) activates myosin ATPase activity in the presence of actin, which is necessary and sufficient for muscle contraction. In knockout mice, the lack of MLCK-mediated phosphorylation of myosin in tracheal, bronchial,4 and gastrointestinal5 smooth muscle cannot be rescued by other kinases, demonstrating its pivotal position in signaling pathways that regulate force generation.
All smooth muscles express two forms of myosin II, SMM (most abundant in adult tissues; includes 4 alternatively spliced variants derived from a single SMM heavy chain gene: 1A, 1B, 2A, and 2B) and NMM (IIA, IIB, and IIC minor). While more studies are required in a wider variety of muscles, to date, several lines of evidence suggest that NMM IIB may participate in a physiologically relevant pathway for force maintenance in smooth muscle.3,6–11 Interestingly, the expression levels of NMM relative to SMM varies depending upon cell type and conditions and are thought to be altered in response to disease and are also developmentally regulated.11–13 Specifically, the expression of NMM IIB in certain smooth muscles could be one of the molecular mechanisms leading to the tonic contractile phenotype.8
The substrate specificity of smooth muscle MLCK has been relatively well-characterized by steady-state kinetics using the smooth muscle RLC14,15 and peptides derived from the smooth muscle RLC2,14,16 as substrates. However, to our knowledge, there are no published corresponding studies for any nonmuscle RLC. The relative activity of MLCK toward these two types of myosin is an important question because the phosphorylated forms of these myosins have very different ATPase kinetics, ADP release kinetics, processivity, and ability to generate force.9,10,17–21 Therefore, the relative amounts of the phosphorylated forms of the 2 myosins could tune muscle contraction parameters in important ways.
In this work, we have compared steady-state kinetic parameters of MLCK toward 4 different substrates: the 2-headed heavy meromyosin (HMM) soluble subfragments of both SMM and NMM IIB, each containing their respective smooth or nonmuscle RLCs, and the 2 types of free RLCs. We found that the kcat for the smRLC was ~2-fold higher than for the nmRLC. The relative differences were maintained in the respective HMM subfragments. This suggests that the sequences of the RLCs alone are sufficient to dictate the different kinetic properties of MLCK. By comparing the sequences of the RLCs, we propose that 3 closely spaced and surface-exposed residues probably interact with MLCK and these interactions underlie the different kinetic properties.
2 MATERIALS AND METHODS
2.1 Proteins
Smooth muscle MLCK was isolated from frozen chicken gizzards22 and stored at −80°C in the presence of 10mM dithiothreitol. This MLCK is the so-called short isoform (P11799-1, often called MLCK-108). The mass of the protein is 110 600 g mol−1 and the E0.1% = 1.14 at 280 nm. It begins with the sequence DFRDILGKKVSTK.
Smooth muscle myosin was isolated from frozen chicken gizzards23 except that the last polymerization-depolymerization step was omitted. It was used without freezing within 2 weeks of purification and stored on ice. Smooth muscle HMM was prepared from purified SMM by proteolytic digestion24 and was never frozen. This digestion procedure was used because the RLC are not degraded and the HMM retains normal regulation by phosphorylation.25 The concentration of HMM was determined at 280 nm using E0.1% = 0.65 and was multiplied by 2 to give the [head], which equals the [RLC]. Native smooth RLC was purified from frozen purified SMM.26 The smooth RLC sequence corresponds exactly to the gi|45384118| “chicken smooth major sequence” in the database (Figure 2) and has an E0.1% = 0.337 at 277 nm.26
FIGURE 2.
Aligned regulatory light chain (RLC) sequences. Top sequence: smooth muscle chicken NCBI reference NP_990609.1, also known as “smooth muscle major.” Bottom sequence: nonmuscle chicken NCBI reference NP_990609.1, also known as “smooth muscle minor.” The phosphorylated serine residue (S19) is underlined and in bold. Residues in red bold are not identical. Residues in blue are the basic amino acids known to confer a low Km toward myosin light chain kinase (MLCK). For arrow designation, see text. Amino acid numbers are shown above
Nonmuscle myosin IIBHMMwas expressed in insect cells as previously described17 and was purified by affinity chromatography on an antiflag column (Sigma). Each heavy chain contained a C-terminal green fluorescent protein and flag tag. The [HMM] was determined at 488 nm using a molar extinction coefficient of 112 000M−1 cm−1 (for 2 green fluorescent proteins/HMM). The [HMM heads] = 2[HMM]. Like smooth HMM, nonmuscle HMM contains 3 subunits, each in 2 copies and is therefore a hexamer with 2 heads and a portion of the coiled-coil tail. The heavy chain is from chicken, GenBank™ accession number M93676, no splice insert; (residues 1–1228). The nmRLC is Accession: NP_990672.1 or gi|45384410| “chicken smooth minor,” which is now known to represent the nonmuscle-type RLC from chicken.27. Note that the RLC is not the same as that erroneously reported in Zavodny et al,28 which is P02612|MLRM_CHICK myosin regulatory light chain 2, smooth muscle major isoform. The nonmuscle essential light chain (ELC) subunit is the nonmuscle ELC (myosin light polypeptide 6 [Bos taurus] NCBI Reference Sequence: NP_786974.1) originally cloned from bovine aortic smooth muscle and referred to as ELC 17b.29
For the free nonmuscle RLC (not bound to a heavy chain), the chicken nonmuscle RLC (Genbank P24032) construct used here was a gift from Frank Brozovich. The construct in the pAED4 vector was expressed in Escherichia coli and purified as described.3 It has an identical sequence to the nonmuscle RLC incorporated into the NMM IIB (Figure 2) and an E0.1% = 0.3 at 280 nm.
2.2 Steady-state MLCK activity
For activity measurements, 150-µL aliquots of MLCK were frozen at 1 µg mL−1 in 10 mg mL−1 body surface area in assay buffer. Aliquots were thawed immediately prior to each assay set, and samples were not refrozen. In this way, activity was reproducible from sample to sample. Myosin light chain kinase activity was measured in assay buffer (10mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid) (HEPES), pH 7.6, 125mM NaCl, 5mM MgCl2, 1mM CaCl2, 0.1mM Ethylene glycol-bis (2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA), 5mM dithiothreitol, and 30nM NaN3) with 4µM CaM, 0.01µM MLCK, and with indicated concentrations of MLCK substrates. The reaction (25°C) was initiated by adding [γ-32P]ATP (PerkinElmer, EasyTides, BLU502A001MC) to 0.2mM. The specific activity of the [γ-32P]ATP was determined by counting 20 µL of the reaction mix in 4 mL scintillation fluid (typically 200 cpm pmol−1). At 1, 2, 3, 4, and 5 minutes, aliquots (20 or 10 µL) were pipetted onto paper discs (GE Health Sciences, Whatman P81 3698-325 or 3698-915) to quench the reaction. After all time points were quenched, the discs were washed 3 times with 0.5% (v/v) H3PO4 and once with acetone, each for 5 minutes. Wash volumes were 500 mL. To wash, the discs were placed into a 250-mL plastic beaker in which holes were punched, and that beaker was placed into an 800-mL beaker containing a stir bar and the washing solution. Upon drying in a fume hood on a Styrofoam surface for 0.5 hours; the discs were placed in scintillation vials along with 4 mL of scintillation fluid (ICN Biomedical, Echolume, 882470) and counted in a TriCarb 2900TR Liquid Scintillation Analyzer. The specific activity of the MLCK was calculated from the slope of a linear fit to a plot of counts per minute versus time (cpm min−1) multiplied by the specific activity of the [γ-32P]ATP divided by the milligram of MLCK present in the quenched aliquots. Data were converted from µmol min−1 mg−1 to s−1 using a molecular mass of 110 600 g mol−1 for MLCK. The MLCK copurifies with SMM,30 but it is removed during the preparation of HMM by gel filtration. Control experiments without added kinase showed no detectable activity for all preparations using 40µM smooth RLC as the substrate (data not shown).
3 RESULTS AND DISCUSSION
Two MLCK substrates, SMM and NMM IIB, were prepared as the 2-headed HMM subfragments. The HMM subfragments are soluble at physiological ionic strength because they lack the C-terminal 2/3 of the tail domain and therefore cannot form filaments. However, because HMM contains 2 heads, it retains normal up-regulation of its activity upon MLCK phosphorylation.20,31 Each HMM head contains 1 RLC and 1 ELC, both of which correspond to their respective myosin heavy chain origin, ie, smooth or nonmuscle. We also prepared the respective RLCs as free proteins not incorporated into the HMM subfragment. All substrates could be phosphorylated to completion (1 mol phosphate/mol RLC) by MLCK as measured using urea gels as described.32
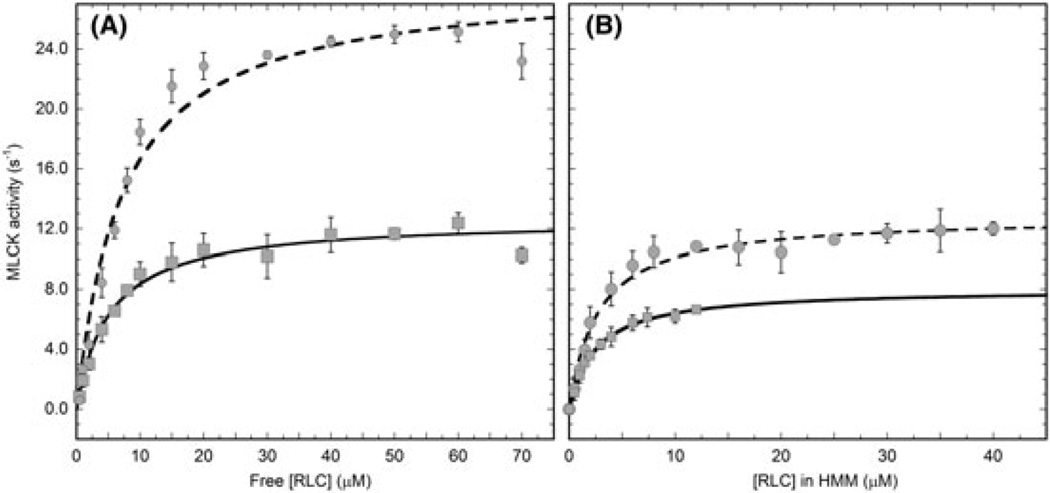
We chose to study the kinetics of the smooth muscle MLCK or the short isoform because it is the predominant MLCK in smooth muscle.33 Under our assay conditions, only S19 is phosphorylated by MLCK and MLCK was maximally active because the [CaM] and [Ca2+] were both much greater than KCaM and KCa, respectively. Figure 1A shows a comparison of the MLCK activity (s−1) versus the substrate concentration of both free smooth RLC (circles) and free nonmuscle RLC (squares). The corresponding data for the respective RLCs as subunits of either smooth HMM (circles) or nonmuscle HMM IIB (squares) are shown in Figure 1B. For both the free and HMM-bound forms, the MLCK phosphorylated the smooth RLC faster than the nonmuscle RLC. Data were fit to the Michaelis-Menten equation giving kcat and Km representing the maximal turnover rate and the substrate concentration at one-half kcat (Table 1). The values for the free smooth RLC are close to those previously reported (kcat = 51 ± 5.7 s−1, Km = 8.3 ± 1.3) under much lower ionic strength conditions.15 Our lower value for kcat is consistent with prior data showing that MLCK is inhibited as the ionic strength increases.34 For both the smooth and nonmuscle proteins, the kcat was lower when the RLC was bound to the heavy chain, as was the Km. The larger kcat observed for smooth RLC relative to nonmuscle RLC was preserved when the 2 proteins were incorporated as subunits of HMM and the smooth kcat/Km/nonmuscle kcat/Km was similar for free and bound substrates. Importantly, the kinetic differences between smooth RLC and nonmuscle RLC appeared to be inherent to the respective RLCs sequences.
FIGURE 1.
Determination of kcat and Km for myosin substrates of myosin light chain kinase (MLCK). A, Free regulatory light chains (RLCs) and B, RLCs as subunits of their respective heavy meromyosin (HMM) molecules. Circles are smooth muscle RLC or smooth muscle HMM, and squares are nonmuscle RLC or nonmuscle HMM IIB. The solid lines are fits to the Michaelis-Menten equation. See Table 1 for results of the fits. Error bars represent the standard deviation of between 3 and 5 determinations at each substrate concentration. Note that HMM concentration in Figure 1B is expressed as the concentration of RLC (two RLC per HMM)
TABLE 1.
Summary of kinetic constants resulting from the fits to the data in Figure 1
| MLCK Substrate | kcat (s−1) | Km (µM) |
kcat/Km (µM−1 s−1) |
|---|---|---|---|
| Smooth muscle RLC | 28.6 ± 1.2 | 7.2 ± 1.1 | 4.0 |
| Nonmuscle RLC | 12.6 ± 0.4 | 5.1 ± 0.7 | 2.5 |
| Smooth muscle HMM | 12.8 ± 0.3 | 2.6 ± 0.3 | 4.9 |
| Nonmuscle HMM IIB | 8.0 ± 0.2 | 2.5 ± 0.1 | 3.2 |
Abbreviations: HMM, heavy meromyosin; MLCK, myosin light chain kinase; RLC, regulatory light chain.
kcat is the maximal MLCK activity, Km is the substrate concentration at one-half maximal MLCK activity, and kcat/Km is a measure of the catalytic efficiency or preference for substrate. Errors are the errors in the fits to the Michaelis-Menten equation.
To understand the source of these kinetic differences, we compared the smooth and nonmuscle RLC sequences (Figure 2). The phosphorylated S19 (in bold and underlined) is immediately C-terminal of the several basic amino acids known to confer a low Km toward MLCK (blue residues),14,16,35,36 which are identical in both sequences. The consensus sequence for substrate recognition of the smooth RLC is K-K-R-X-X-R-X-T-S-X.37 The 10 nonidentical amino acids are shown in bold red. Using information from prior chimera and deletion studies, we can exclude several of these amino acids as residues that affect MLCK activity. If the N-terminal first 102 residues of the smooth RLC are present, the remaining amino acids can be substituted with the divergent skeletal RLC sequence without significantly affecting either kcat or Km.15 From this, we predict that none of the 5 amino acid differences in the second row of the sequence (Figure 2) are likely to underlie the differences in MLCK activity. Also, previous work has shown that peptides containing the smooth RLC sequence 1–23, 6–23, and 8–23 have very similar kcat and Km,14 and deletion of the first 7 residues of the full-length RLC did not change either kcat or Km.37 These data suggest that the A7 versus T7 residue (Figure 2) is unlikely to underlie the differences in kcat observed here (Table 1).
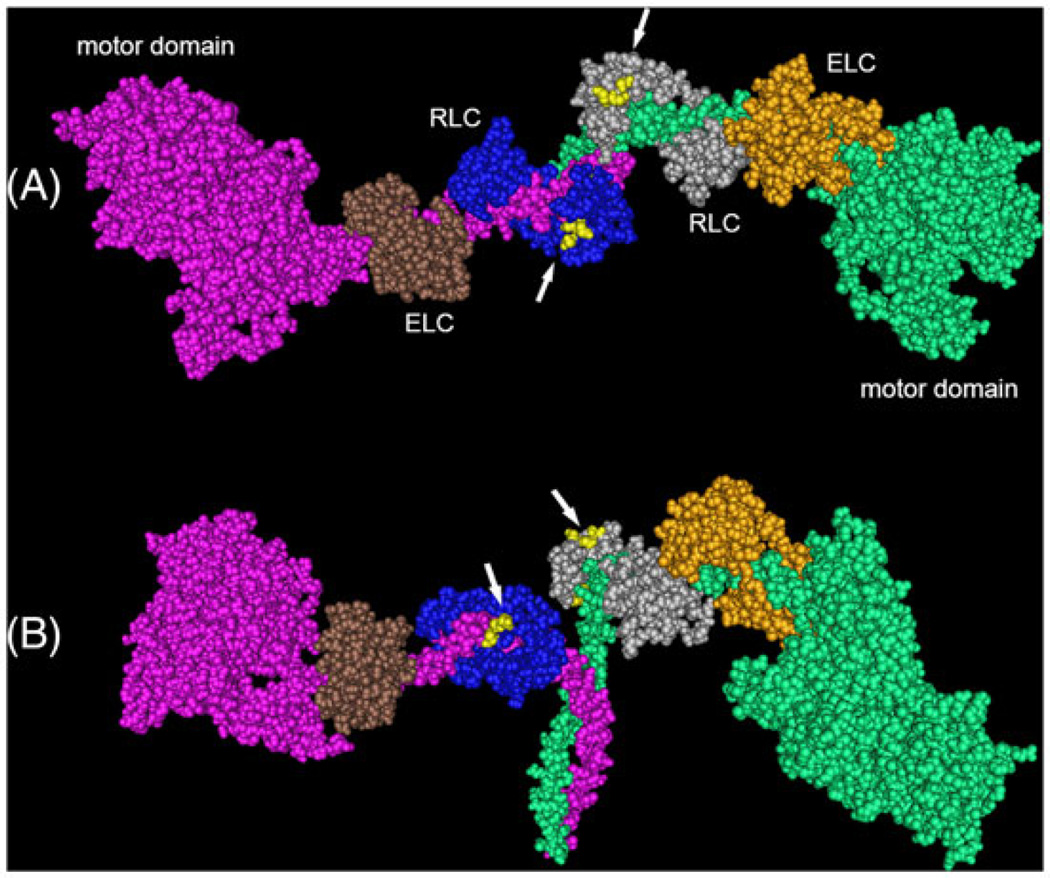
To predict which of the remaining possible amino acid differences are important for MLCK activity, we examined the location of M60, D70, G71, and S74 on the phosphorylated smooth HMM structure (Figure 3; PDB 3J04).38 All 4 residues are within the N-terminal lobe of the RLC, which is the one closest to the head-tail junction of myosin. M60 (not visible) is buried at the interface with the heavy chain and is therefore probably inaccessible to MLCK during catalysis. In contrast, D70, G71, and S74 (yellow) all localize to a single patch on the surface of the RLC and therefore may represent an important interface with MLCK during catalysis. Unfortunately, their spatial relationship to the phosphorylatable S19 cannot be visualized in this structure (structure begins at F25), because of the known dynamic nature of this region of the RLC.39
FIGURE 3.
Space filling model of phosphorylated chicken smooth muscle heavy meromyosin (PDB 3J04).38 Subunits by color are as labeled. RLC residues E70, G71, and S74 (yellow residues indicated by white arrows) are surface exposed. View A is positioned with the coiled-coil heavy chains facing away from the viewer. View B is positioned with the coiledcoil facing down. RLC, regulatory light chain; ELC, essential light chain
It is known that the kinetics of phosphorylation of peptides containing the N-terminal RLC residues (amino acids 1–23) are very different from those for the full-length RLCs. The kcat values obtained with peptides are much lower, being 10% or less compared with the native smooth RLC.14 This suggests that there are other regions of the RLC structure that allow MLCK to properly recognize its substrate. The general location of these other region(s) has been defined by prior studies with chimeras of the smooth and skeletal RLCs.15 The skeletal RLC is a poor substrate for smooth MLCK, and replacing the first 29 residues of the skeletal RLC with the smooth RLC sequence was not sufficient to achieve the high kcat characteristic of the native smooth RLC. As mentioned earlier, a chimera containing the entire N-terminal one-half (amino acids 1–102) of the smooth RLC sequence connected to the C-terminal one-half of the skeletal RLC showed essentially the same kinetics as the native smooth RLC.15 These data strongly suggest that some region within the N-terminal half of the RLC, but excluding the first 29 residues, is critically important for MLCK substrate recognition. Our data are consistent with this and further suggest that the surface patch formed by D70, G71, and S74 represent at least a portion of this previously uncharacterized region.
We compared all the confirmed nonmuscle sequences (corresponding to the human 12A or 12B gene) with the smooth muscle sequences (corresponding to the human ML9 gene) in the database. The residues that sort selectively into their respective classes are indicated by double arrows in Figure 2. Importantly, all smooth RLC contain a G71 and S74 residue, whereas all nonmuscle RLC contain an A71 and N74 residue. The M60/L60 and the E70/D70 (single arrows) are highly variable within either RLC class, that is, many of the smooth RLC contain an L rather than and M, and vice versa. Although, all other residues in Figure 2 indicated by a double arrow also sort selectively into classes, they are unlikely to be important to MLCK kinetics, but rather may relate to some other structural difference between the 2 myosin classes, such as binding to the heavy chain.
The kinase activity toward the smooth and nonmuscle substrates differs by only about a factor of 2. However, this small difference is probably highly significant physiologically. Myosin light chain kinase is a low-abundance kinase,30,40 and it is known that changes of similar magnitude (2 fold) in MLCK activity in cells are correlated to many disease states. Selective over expression of MLCK in asthmatic airway smooth muscle contributes to airway hyper-responsiveness observed in asthma,41,42 and variants in the enzyme are associated with severe asthma43 and increased susceptibility to sepsis.44 Increased MLCK activity has been shown to be necessary and sufficient for several barrier dysfunctions (lung and intestinal epithelium45–47 and microvasculature48). These dysfunctions are the cause of or lead to important human diseases and conditions such as colitis,49 Crohn's disease, inflammatory bowel disease,50,51 diarrhea,52 and increased vascular permeability.
The implications of our findings with regard to the kinetic preference of MLCK for smooth over NMM IIB remain to be elucidated. However, they support the idea that SMM may be phosphorylated faster than NMM in the muscle following agonist stimulation or depolarization. Delaying phosphorylation of NMM IIB until after SMM phosphorylation may present an advantage. Smooth muscle myosin is a faster myosin and would be able to support the rapid early phase of contraction unimpeded by the slower NMM IIB. But during the later force maintenance phase in tonic muscle, the more slowly phosphorylated NMM may have had time to attain a sufficient level of phosphorylation to maintain force.
Significance of the study.
Phosphorylation of nonmuscle and smooth muscle myosin by myosin light chain kinase (MLCK) is required for activation of myosin's ATPase activity. In smooth muscles, nonmuscle myosin coexists with smooth muscle myosin, but the two myosins have very different chemo-mechanical properties relating to their ability to maintain force. Differences in specificity of MLCK for different myosin isoforms had not been previously investigated. We show that the MLCK prefers smooth muscle myosin by a significant factor. These data suggest that nonmuscle myosin is phosphorylated more slowly than smooth muscle myosin during a contraction cycle.
Acknowledgments
We thank Frank Brozovich for the nonmuscle RLC construct and Michael Walsh for MLCK.
ABBREVIATIONS
- HMM
heavy meromyosin subfragment of full-length myosin
- SMM
smooth muscle myosin
- NMM
nonmuscle myosin
- RLC
regulatory or phosphorylatable light chain of myosin
- ELC
essential light chain of myosin
Footnotes
CONFLICT OF INTEREST
The authors verify that they have no conflicts of interest.
REFERENCES
- 1.Lazar V, Garcia JG. A single human myosin light chain kinase gene (MLCK; MYLK) Genomics. 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 2.Gallagher PJ, Herring BP, Stull JT. Myosin light chain kinases. J Muscle Res Cell Motil. 1997;18:1–16. doi: 10.1023/a:1018616814417. [DOI] [PubMed] [Google Scholar]
- 3.Yuen SL, Ogut O, Brozovich FV. Nonmuscle myosin is regulated during smooth muscle contraction. Am J Physiol Heart Circ Physiol. 2009;297:H191–H199. doi: 10.1152/ajpheart.00132.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang WC, Peng YJ, Zhang GS, et al. Myosin light chain kinase is necessary for tonic airway smooth muscle contraction. J Biol Chem. 2010;285:5522–5531. doi: 10.1074/jbc.M109.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He WQ, Peng YJ, Zhang WC, et al. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology. 2008;135:610–620. doi: 10.1053/j.gastro.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lofgren M, Ekblad E, Morano I, Arner A. Nonmuscle myosin motor of smooth muscle. J Gen Physiol. 2003;121:301–310. doi: 10.1085/jgp.200208720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morano I, Chai GX, Baltas LG, et al. Smooth-muscle contraction without smooth-muscle myosin. Nat Cell Biol. 2000;2:371–375. doi: 10.1038/35014065. [DOI] [PubMed] [Google Scholar]
- 8.Rhee AY, Ogut O, Brozovich FV. Nonmuscle myosin, force maintenance, and the tonic contractile phenotype in smooth muscle. Pflugers Arch. 2006;452:766–774. doi: 10.1007/s00424-006-0091-4. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci U S A. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 11.Eddinger TJ, Meer DP. Myosin II isoforms in smooth muscle: heterogeneity and function. Am J Physiol Cell Physiol. 2007;293:C493–C508. doi: 10.1152/ajpcell.00131.2007. [DOI] [PubMed] [Google Scholar]
- 12.Gaylinn BD, Eddinger TJ, Martino PA, Monical PL, Hunt DF, Murphy RA. Expression of nonmuscle myosin heavy and light chains in smooth muscle. Am J Physiol. 1989;257:C997–C1004. doi: 10.1152/ajpcell.1989.257.5.C997. [DOI] [PubMed] [Google Scholar]
- 13.Eddinger TJ, Murphy RA. Two smooth muscle myosin heavy chains differ in their light meromyosin fragment. Biochemistry. 1988;27:3807–3811. doi: 10.1021/bi00410a043. [DOI] [PubMed] [Google Scholar]
- 14.Kemp BE, Pearson RB, House C. Role of basic residues in the phosphorylation of synthetic peptides by myosin light chain kinase. Proc Natl Acad Sci U S A. 1983;80:7471–7475. doi: 10.1073/pnas.80.24.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhi G, Herring BP, Stull JT. Structural requirements for phosphorylation of myosin regulatory light chain from smooth muscle. J Biol Chem. 1994;269:24723–24727. [PubMed] [Google Scholar]
- 16.Kemp BE, Pearson RB. Spatial requirements for location of basic residues in peptide substrates for smooth muscle myosin light chain kinase. J Biol Chem. 1985;260:3355–3359. [PubMed] [Google Scholar]
- 17.Norstrom MF, Smithback PA, Rock RS. Unconventional processive mechanics of non-muscle myosin IIB. J Biol Chem. 2010;285:26326–26334. doi: 10.1074/jbc.M110.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy A, Takagi Y, Billington N, et al. Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J Biol Chem. 2013;288:709–722. doi: 10.1074/jbc.M112.424671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cremo CR, Sellers JR, Facemyer KC. Two heads are required for phosphorylation- dependent regulation of smooth muscle myosin. J Biol Chem. 1995;270:2171–2175. doi: 10.1074/jbc.270.5.2171. [DOI] [PubMed] [Google Scholar]
- 21.Cremo CR, Geeves MA. Interaction of actin and ADP with the head domain of smooth muscle myosin: implications for strain-dependent ADP release in smooth muscle. Biochemistry. 1998;37:1969–1978. doi: 10.1021/bi9722406. [DOI] [PubMed] [Google Scholar]
- 22.Ngai PK, Carruthers CA, Walsh MP. Isolation of the native form of chicken gizzard myosin light-chain kinase. Biochem J. 1984;218:863–870. doi: 10.1042/bj2180863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikebe M, Hartshorne DJ. Effects of Ca2+ on the conformation and enzymatic activity of smooth muscle myosin. J Biol Chem. 1985;260:13146–13153. [PubMed] [Google Scholar]
- 24.Ikebe M, Hartshorne DJ. Proteolysis of smooth muscle myosin by Staphylococcus aureus protease: preparation of heavy meromyosin and subfragment 1 with intact 20,000-dalton light chains. Biochemistry. 1985;24:2380–2387. doi: 10.1021/bi00330a038. [DOI] [PubMed] [Google Scholar]
- 25.Ellison PA, Sellers JR, Cremo CR. Kinetics of smooth muscle heavy meromyosin with one thiophosphorylated head. J Biol Chem. 2000;275:15142–15151. doi: 10.1074/jbc.275.20.15142. [DOI] [PubMed] [Google Scholar]
- 26.Facemyer KC, Cremo CR. A new method to specifically label thiophosphorylatable proteins with extrinsic probes. labeling of serine-19 of the regulatory light chain of smooth muscle myosin. Bioconj Chem. 1992;3:408–413. doi: 10.1021/bc00017a009. [DOI] [PubMed] [Google Scholar]
- 27.Zavodny PJ, Petro ME, Lonial HK, et al. Cloning and characterization of a vertebrate cellular myosin regulatory light chain complementary DNA. Circ Res. 1990;67:933–940. doi: 10.1161/01.res.67.4.933. [DOI] [PubMed] [Google Scholar]
- 28.Zavodny PJ, Petro ME, Kumar CC, et al. The nucleotide sequence of chicken smooth muscle myosin light chain two. Nucleic Acids Res. 1988;16:1214. doi: 10.1093/nar/16.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lash JA, Helper DJ, Klug M, Nicolozakes AW, Hathaway DR. Nucleotide and deduced amino acid sequence of cDNAs encoding two isoforms for the 17,000 dalton myosin light chain in bovine aortic smooth muscle. Nucleic Acids Res. 1990;18:7176. doi: 10.1093/nar/18.23.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong F, Haldeman BD, John OA, et al. Characterization of tightly associated smooth muscle myosin-myosin light-chain kinase-calmodulin complexes. J Mol Biol. 2009;390:879–892. doi: 10.1016/j.jmb.2009.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cremo CR, Wang F, Facemyer K, Sellers JR. Phosphorylation-dependent regulation is absent in a nonmuscle heavy meromyosin construct with one complete head and one head lacking the motor domain. J Biol Chem. 2001;276:41465–41472. doi: 10.1074/jbc.M107103200. [DOI] [PubMed] [Google Scholar]
- 32.Haldeman BD, Brizendine RK, Facemyer KC, Baker JE, Cremo CR. The kinetics underlying the velocity of smooth muscle myosin filament sliding on actin filaments in vitro. J Biol Chem. 2014;289:21055–21070. doi: 10.1074/jbc.M114.564740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong F, Haldeman BD, Jackson D, Carter M, Baker JE, Cremo CR. Biochemistry of smooth muscle myosin light chain kinase. Arch Biochem Biophys. 2011;510:135–146. doi: 10.1016/j.abb.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikebe M, Inagaki M, Naka M, Hidaka H. Correlation of conformation and phosphorylation and dephosphorylation of smooth muscle myosin. J Biol Chem. 1988;263:10698–10704. [PubMed] [Google Scholar]
- 35.Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem. Sci. 1990;15:345–349. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 36.Michnoff CH, Kemp BE, Stull JT. Phosphorylation of synthetic peptides by skeletal muscle myosin light chain kinases. J Biol Chem. 1986;261:8320–8326. [PubMed] [Google Scholar]
- 37.Ikebe M, Reardon S, Schwonek JP, Sanders CR, 2nd, Ikebe R. Structural requirement of the regulatory light chain of smooth muscle myosin as a substrate for myosin light chain kinase. J Biol Chem. 1994;269:28165–28172. [PubMed] [Google Scholar]
- 38.Baumann BA, Taylor DW, Huang Z, et al. Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation. J Mol Biol. 2012;415:274–287. doi: 10.1016/j.jmb.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vileno B, Chamoun J, Liang H, et al. Broad disorder and the allosteric mechanism of myosin II regulation by phosphorylation. Proc Natl Acad Sci U S A. 2011;108:8218–8223. doi: 10.1073/pnas.1014137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong F, Brizendine RK, Carter MS, et al. Diffusion of myosin light chain kinase on actin: a mechanism to enhance myosin phosphorylation rates in smooth muscle. J Gen Physiol. 2015;146:267–280. doi: 10.1085/jgp.201511483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leguillette R, Laviolette M, Bergeron C, et al. Myosin, transgelin, and myosin light chain kinase: expression and function in asthma. Am J Respir Crit Care Med. 2009;179:194–204. doi: 10.1164/rccm.200609-1367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang H, Rao K, Halayko AJ, Liu X, Stephens NL. Ragweed sensitization-induced increase of myosin light chain kinase content in canine airway smooth muscle. Am J Respir Cell Mol Biol. 1992;7:567–573. doi: 10.1165/ajrcmb/7.6.567. [DOI] [PubMed] [Google Scholar]
- 43.Flores C, Ma SF, Maresso K, Ober C, Garcia JG. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol. 2007;31:296–305. doi: 10.1002/gepi.20210. [DOI] [PubMed] [Google Scholar]
- 44.Gao L, Grant AV, Rafaels N, et al. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol. 2007;119:1111–1118. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen L, Black ED, Witkowski ED, et al. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J Cell Sci. 2006;119:2095–2106. doi: 10.1242/jcs.02915. [DOI] [PubMed] [Google Scholar]
- 47.Shen L, Su L, Turner JR. Mechanisms and functional implications of intestinal barrier defects. Dig. Dis. 2009;27:443–449. doi: 10.1159/000233282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia JG, Verin AD, Schaphorst K, et al. Regulation of endothelial cell myosin light chain kinase by Rho, cortactin, and p60(src) Am J Physiol. 1999;276:L989–L998. doi: 10.1152/ajplung.1999.276.6.L989. [DOI] [PubMed] [Google Scholar]
- 49.Su L, Shen L, Clayburgh DR, et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber CR, Turner JR. Inflammatory bowel disease: is it really just another break in the wall? Gut. 2007;56:6–8. doi: 10.1136/gut.2006.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blair SA, Kane SV, Clayburgh DR, Turner JR. Epithelial myosin light chain kinase expression and activity are upregulated in inflammatory bowel disease. Lab Invest. 2006;86:191–201. doi: 10.1038/labinvest.3700373. [DOI] [PubMed] [Google Scholar]
- 52.Clayburgh DR, Barrett TA, Tang Y, et al. Epithelial myosin light chain kinase-dependent barrier dysfunction mediates T cell activation-induced diarrhea in vivo. J Clin Invest. 2005;115:2702–2715. doi: 10.1172/JCI24970. [DOI] [PMC free article] [PubMed] [Google Scholar]