Summary
Domestic animals and animal products are the source of pathogenic Campylobacter jejuni and C. coli in industrialized countries, yet little is known about the transmission of these bacteria in developing countries. Guinea pigs (Cavia porcellus) are commonly raised for food in the Andean region of South America, however, limited research has characterized this rodent as a reservoir of zoonotic enteric pathogens. In this study, we examined the prevalence of Campylobacter spp. in 203 fecal samples from domestic animals of 59 households in a semi-rural parish of Quito, Ecuador. Of the twelve animal species studied, guinea pigs showed the highest prevalence of C. jejuni (n = 39/40; 97.5%). Multilocus sequence typing (MLST) was used to characterize the genetic relationship of C. jejuni from domestic animals and 21 sequence types (STs) were identified. The majority of STs from guinea pigs appeared to form new clonal complexes that were not related to STs of C. jejuni isolated from other animal species and shared only a few alleles with other C. jejuni previously characterized. The study identifies guinea pigs as a major reservoir of C. jejuni and suggests that some C. jejuni strains are adapted to this animal species.
Introduction
Campylobacter jejuni and C. coli are transmitted to humans from domestic animals in developed countries, yet little is known about transmission of these bacteria in developing countries where unusual animal species (such as guinea pigs) are raised for food (Dasti et al., 2010). The domestication of guinea pigs occurred more than 2,000 years ago in the Andean region (Wing, 1986); currently there is an estimated population of greater than 35 million guinea pigs (de Zaldivar, 1995). Guinea pigs are generally used for companionship or for research in Europe and North America (Pigiere et al., 2012), while in South America, the majority of guinea pigs are raised for their meat, as well as minor uses in traditional Andean rituals and medicine (Spotorno et al., 2007). In 2009, the Instituto Nacional Autónomo de Investigaciones Agropecuarias (INIAP) estimated that 710,000 Ecuadorian households raised guinea pigs, producing nearly 21 million guinea pigs that year. Demand for guinea pigs, for consumption inside and outside the country, is increasing and annual increases in production have been observed.
Research has documented a variety of zoonotic disease risks associated with guinea pigs raised under different contexts (Lutz-Wohlgroth et al., 2006; Amman et al., 2007; Fredriksson-Ahomaa, 2007; Walther et al., 2012; Gruszynski et al., 2015). Studies among laboratory research animals have found guinea pigs to carry Campylobacter spp. (Weber et al., 1982; Muto et al., 1983; Fakir, 1986; Bartholomew et al., 2014; Komba et al., 2014). In the Andean Region, guinea pigs have been found to carry important zoonotic pathogens such as Fasciola hepatica (Carolina Gonzalez et al., 2011) and Yersinia pestis, (Gabastou et al., 2000), as well as Trypanosoma cruzi (Levy et al., 2006). Given that guinea pigs in South America are often raised in the home, a better understanding of their role as potential reservoirs of other zoonotic diseases is critical.
The zoonotic enteropathogen Campylobacter spp. is the most common cause of bacterial gastrointestinal illness globally – estimated to result in 7.5 million disability-adjusted life years in 2010 (Wagenaar et al., 2015). Campylobacter jejuni and C. coli are the species most often implicated in human disease (Wagenaar et al., 2015). In low- and middle-income countries (LMICs) the burden of C. jejuni and C. coli infections among humans is not well documented and is likely underestimated due to the lack of diagnosis. A recent case-control study of paediatric diarrhoea in a poor urban neighbourhood and a poor rural community of Ecuador found a small number of infections with Campylobacter spp. among symptomatic and asymptomatic participants (Vasco et al., 2014).
Campylobacter have been identified in a range of animal hosts including domestic and wild animal species (Mughini Gras et al., 2013; Wagenaar et al., 2015). Chickens are considered an important source of C. jejuni and C. coli, and several studies have analyzed household poultry production as a risk for human infection (Grados et al., 1988; Georgescourbot et al., 1990; Marquis et al., 1990; Oberhelman et al., 2003). No study to the authors’ knowledge has assessed the prevalence and diversity of Campylobacter spp. in guinea pigs raised for food, a prevalent practice in the Andean Region. The aims of the study were to: (i) test guinea pigs and other domestic animals for the presence of Campylobacter spp.; (ii) compare the prevalence of C. jejuni and C. coli carriage between guinea pigs and different animal species raised for food in the study area and (iii) to study the transmission and/or host association of C. jejuni and C. coli among animal species.
Results and discussion
Twelve different species of animals, including both livestock and domestic pets, were present among the fifty-nine households studied. The range of species present among any one household ranged from one to eight. Chickens were the primary animal raised, present in 42 households (71.2%). Forty of the households (67.8%) raised guinea pigs for food, primarily for household consumption. There were 32 (54.2%) households that had both chickens and guinea pigs and eight households (13.6%) that had guinea pigs and no chickens. Dogs, pigs, and rabbits were also commonly owned by households at 66.1%, 61.0% and 33.9%, respectively. The average number of guinea pigs raised by each household was 12 (range: 2–40). All of the households housed the guinea pigs outside the home and reported to reuse the guinea pig faecal waste on their land (Supporting Information Table S1).
Using culture based methods, the prevalence of C. jejuni was highest in samples taken from guinea pig cages (72.5%), followed by chicken cages (59.5%) (Table 1). At much lower prevalence levels, C. jejuni was identified in dogs (25.0%), rabbit cages (10.0%), cows (14.0%), cats (33.3%), ducks (20.0%) and quail (50.0%). Three pigs were positive for C. jejuni (8.3%); pigs, however, were more often positive for C. coli (38.9%) and C. hyointestinalis (27.8%). In comparing the prevalence of C. jejuni among the different animal species (Supporting Information Table S2), samples from both guinea pigs and chickens had significantly higher levels of C. jejuni than other domestic animals. Other animal pairs were not compared due to small sample sizes. The two sheep and one cow sampled were negative for C. jejuni. Campylobacter coli was found in pigs (38.9%), chickens (16.7%), guinea pigs (5.0%), dogs (2.6%), cows (14.3%), cats (16.7%), sheep (50.0%) and ducks (20.0%). We decided to analyze further C. jejuni isolates because they were present in larger numbers and in most faecal samples from the animal species analyzed, whereas most C. coli isolates were obtained from pigs (Table 1).
Table 1.
Prevalence of Campylobacter jejuni and Campylobacter coli identified in domestic animals from a semi-rural parish of Quito, Ecuador.
| Source | No. Samples | C. jejuni (%) | C. coli (%) | ||
|---|---|---|---|---|---|
| Guinea pigs | 40 | 29 | (72.5) | 2 | (5.0) |
| Chickens | 42 | 25 | (59.5) | 7 | (16.7) |
| Dogs | 40 | 10 | (25.0) | 1 | (2.5) |
| Pigs | 36 | 3 | (8.3) | 14 | (38.9) |
| Rabbits | 20 | 2 | (10.0) | 0 | (0) |
| Cattle | 7 | 1 | (14.3) | 1 | (14.3) |
| Cats | 6 | 2 | (33.3) | 1 | (16.7) |
| Ducks | 5 | 1 | (20.0) | 1 | (20.0) |
| Quail | 3 | 2 | (66.7) | 0 | (0) |
| Sheep | 2 | 0 | (0) | 1 | (50.0) |
| Total | 265 | 75 | (28.3) | 28 | (10.5) |
The gene pgm was amplified and sequenced from all recovered C. jejuni isolates (this gene is the most variable of the seven in the MLST set); all isolates which shared an identical DNA sequence with at least another isolate, were further analyzed using the following two loci, glyA and tkt (44 out of 65). Finally, the samples with the same glyA-pgm-tkt profile were sequenced for the remaining four MLST loci (44 out of 48). Among 44 C. jejuni isolates analyzed using MLST, we identified 21 sequence types (STs) (Table 2 and Fig. 1). Ten STs were identified in isolates from guinea pigs; 9 of which were novel (i.e. not described previously). Guinea pig STs formed three clusters (potential clonal complexes, i.e. STs that matched the central genotype at four or more loci). Cluster 1 comprised ST 7775 (n = 4), ST7777 (n = 1), ST7778 (n = 1), ST7781 (n = 3) and ST7789 (n = 1); cluster 2 was formed by ST7779 (n = 2) and ST7780 (n = 2); and cluster 3 comprised ST 7759 (n = 6), ST 7775 (n = 1) (Table 2 and Fig. 1). Remarkably, all STs from cluster 1 and most STs from clusters 2 and 3 seemed to have exchanged alleles only with strains isolated from guinea pigs (Fig. 1), which may indicate genetic isolation; allele similarity is indication of horizontal gene transfer rather than the emergence of independent mutations (Sheppard and Maiden, 2015). Campylobacter jejuni STs can be either adapted to one animal species or generalists (i.e. equally able to infect different animal species) (Sheppard et al., 2013). Our data (Fig. 1) seem to show that C. jejuni isolates from guinea pigs belong to STs adapted to this animal species.
Table 2.
Campylobacter jejuni sequence types (STs) and clonal complexes (CCs) identified in guinea pigs (Cavia porcellus) and other domestic animals in a semi-urban community close to Quito.
| CC | STa | Animal | No. of isolates | Alleleb
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | ||||
| Unassigned | 7759 | Guinea pigs | 6 | 4 | 7 | 455 | 62 | 731 | 25 | 104 |
| 7760 | Guinea pigs | 1 | 4 | 7 | 455 | 62 | 733 | 25 | 104 | |
| 7775 | Guinea pigs | 4 | 394 | 538 | 454 | 601 | 729 | 582 | 462 | |
| 7777 | Guinea pigs | 1 | 394 | 537 | 454 | 601 | 729 | 582 | 463 | |
| 7778 | Guinea pigs | 1 | 394 | 538 | 454 | 601 | 729 | 582 | 464 | |
| 7779 | Guinea pigs | 2 | 393 | 536 | 454 | 62 | 734 | 583 | 104 | |
| 7780 | Guinea pigs | 2 | 393 | 536 | 454 | 62 | 13 | 25 | 104 | |
| 7781 | Guinea pigs | 3 | 394 | 538 | 454 | 601 | 730 | 582 | 462 | |
| 7789 | Guinea pigs | 1 | 394 | 538 | 454 | 601 | 729 | 582 | 463 | |
| 7671 | Dogs | 1 | 8 | 113 | 5 | 121 | 11 | 25 | 6 | |
| 7672 | Cats | 1 | 2 | 114 | 5 | 298 | 13 | 61 | 460 | |
| 7672 | Dogs | 1 | 2 | 114 | 5 | 298 | 13 | 61 | 460 | |
| 464 | 464 | Chickens | 1 | 24 | 2 | 2 | 2 | 10 | 3 | 1 |
| 464 | Rabbits | 1 | 24 | 2 | 2 | 2 | 10 | 3 | 1 | |
| 45 | 137 | Rabbit | 1 | 4 | 7 | 10 | 4 | 42 | 7 | 1 |
| 353 | 1233 | Chickens | 2 | 7 | 17 | 5 | 10 | 10 | 177 | 6 |
| 1233 | Guinea pigs | 1 | 7 | 17 | 5 | 10 | 10 | 177 | 6 | |
| 3515 | Chickens | 1 | 7 | 17 | 2 | 2 | 10 | 3 | 6 | |
| 7643 | Quail | 1 | 7 | 17 | 5 | 2 | 10 | 3 | 54 | |
| 7643 | Dogs | 1 | 7 | 17 | 5 | 2 | 10 | 3 | 54 | |
| 354 | 354 | Chickens | 1 | 8 | 10 | 2 | 2 | 11 | 12 | 6 |
| 7662 | Quail | 1 | 390 | 2 | 2 | 2 | 11 | 5 | 6 | |
| 7662 | Chickens | 1 | 390 | 2 | 2 | 2 | 11 | 5 | 6 | |
| 7669 | Chickens | 1 | 8 | 10 | 95 | 2 | 11 | 12 | 6 | |
| 607 | 607 | Chickens | 2 | 8 | 2 | 5 | 53 | 11 | 3 | 1 |
| 607 | Dogs | 2 | 8 | 2 | 5 | 53 | 11 | 3 | 1 | |
| 1212 | Chickens | 1 | 8 | 2 | 5 | 53 | 11 | 3 | 105 | |
| 1212 | Cattle | 1 | 8 | 2 | 5 | 53 | 11 | 3 | 105 | |
| 1212 | Pigs | 1 | 8 | 2 | 5 | 53 | 11 | 3 | 105 | |
Sequence types and alleles in bold are new.
Fig. 1.
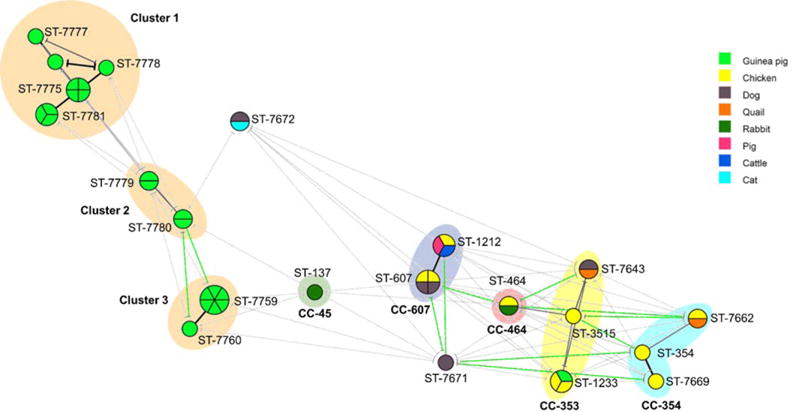
Minimum spanning tree analysis of 44 C. jejuni isolates (from a semi-rural parish of Quito, Ecuador) based on MLST profile and according to animal source. Each circle represents the sequence type (ST), the size of the circle and circle divisions indicate the number of isolates within any given ST. Line colours indicate the following information: black=5–6 shared alleles; grey 3–4 shared alleles; and gree n=1–2 shared alleles. The colour of the circle indicates the animal species.
Seven STs were present in multiple animal species: quails and chickens (ST 7662); dogs and chickens (ST 607); cattle, chickens and pigs (ST 1212); guinea pigs and chickens (ST 1233); quails and dogs (ST 7643); rabbits and chickens (ST 464); and cats and dogs (ST 7672). Twenty-three strains (46.9%) from animals other than guinea pigs belonged to four major clonal complexes (CC) previously described: CC-607, CC-353, CC-354, CC-464 and CC-45 (Fig. 1). Seven isolates (16%) belonged to ST-607, the founder sequence type of CC-607; 6 isolates (13.6%) to CC-353, 4 isolates (9%) to CC-354, 2 isolates (4.5%) to CC-464 and 1 isolate (2.2%) to CC-45 (Fig. 1). Unlike guinea pigs, isolates from other animal species seemed to share alleles with STs and CCs previously described. The complete description of alleles from all isolates is shown in (Table 2; Supporting Information Fig. S1), all nucleotide sequences are available at Genbank, accession numbers are: KU728723 to KU728740.
This study identified high levels of Campylobacter jejuni in guinea pigs raised for food in Ecuador, however C. jejuni infection (worldwide) has been attributed to chickens (50–80%), cattle (20–30%) and to a lesser extent sheep, pigs, wild animals, water and unpasteurized milk (Michaud et al., 2004; Wagenaar et al., 2015). Detecting C. jejuni from 97.5% of guinea pig fecal samples tested, using PCR, contrasts with the results of studies looking at other domestic and wild animals: 19.4% – 72.5% in poultry; 16–90% in cattle (Kaakoush et al., 2015), 7.9–90.0% in dogs (Marks et al., 2011; Gras et al., 2013; Ramonaite et al., 2014), 35.4% in free-living birds (Ramonaite et al., 2014). The difference in the prevalence of C. jejuni was not statistically significant between guinea pigs and chickens. Both of these animal species, however, had significantly higher levels than those of dogs, pigs and rabbits. There were no differences in the prevalence of C. jejuni in guinea pigs raised with chickens versus guinea pigs raised without chickens and these two animal species did not share STs except in the case of ST-1233 in guinea pigs and chickens from the same household.
Surveillance programs that track C. jejuni are limited (Scallan et al., 2011) and control measures have focused on poultry because they are generally considered the primary reservoir of C. jejuni and cause outbreaks in humans in industrialized countries (Kaakoush et al., 2015; Wagenaar et al., 2015). Little is known about the epidemiology of C. jejuni infections in humans living in developing countries where unusual animal species are used as food and where other domestic animals are raised under different conditions. In this case, the risk of C. jejuni infection may include not only households raising guinea pigs but the whole community who consumes crops raised in the area. The use of domestic animal faecal waste to fertilize crops could potentially be affecting the microbiological safety of strawberries, the primary edible crop raised in the area. More research is needed to understand how the use of guinea pig feces as a fertilizer could result in transmission of C. jejuni through contaminated crops.
It is unclear why guinea pigs in this region have such a high prevalence of C. jejuni. In this study, many of the households applied fresh guinea pig faecal waste directly to the crops (including alfalfa used for feeding guinea pigs) (Supporting Information Table S1); this practice may favour the cycling of C. jejuni in guinea pigs if the fertilized alfalfa is then fed back to local guinea pigs. Although Campylobacter jejuni is considered fragile in contrast to many other bacterial pathogens, research has found Campylobacter to survive in the environment between three to ten months, especially under wet conditions, including faecal slurries, contaminated waters and stored manure (Inglis et al., 2010). It will be important to determine the prevalence of C. jejuni in guinea pigs in other parts of Ecuador and in guinea pigs kept as pets in other parts of the world.
In this study, isolates from chickens belonged to 8 STs which were present in other animal species or have been described previously in other animal species (STs: 607, 1212, 464, 3515, 1233, 354, 7662 and 7669) (Fig. 1). This finding underscores the importance of avian species as reservoirs of genetically diverse and generalist C. jejuni.
Given the limited scope of the study – 59 households, 40 of which raised guinea pigs – it is important to treat the uncertainties in a transparent manner. First, the generalizability of the findings is limited and it will be important for future research to determine how C. jejuni varies in guinea pigs across ecologically separate niches in Ecuador. Second, only a subset of the C. jejuni isolates – 44 of the 65 (67.7%) – were analyzed using MLST due to resource limitations. Furthermore, this study was not able to assess seasonal variation or the variation stemming from animal management practices, such as housing, diet and crowding. We did not investigate the relationship between five large-scale poultry operations in the area and the high rate of C. jejuni colonization in guinea pigs.
This study highlights the complexity of C. jejuni transmission in the environment where not only local agricultural practices but also C. jejuni genetics are involved. It will be important to study the temporal patterns of ST distribution (in different animal species) which may indicate how stable different STs are overtime in one region. These type of studies may show the potential transient emergence of C. jejuni STs with different aptitude to cross-infect different animal species (Boes et al., 2005).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the Fogarty International Center of the National Institutes of Health under Award Number K01 TW 009484. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank Valeria Garzon and the rest of the members of the field team in Yaruqui.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
References
- Amman BR, Pavlin BI, Albarino CG, Comer JA, Erickson BR, Oliver JB, et al. Pet rodents and fatal lymphocytic choriomeningitis in transplant patients. Emerg Infect Dis. 2007;13:719–725. doi: 10.3201/eid1305.061269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew ML, Heffernan RT, Wright JG, Klos RF, Monson T, Khan S, et al. Multistate Outbreak of Salmonella enterica serotype enteritidis infection associated with pet guinea pigs. Vector Borne Zoonotic Dis. 2014;14:414–421. doi: 10.1089/vbz.2013.1506. [DOI] [PubMed] [Google Scholar]
- Boes J, Nersting L, Nielsen E, Kranker S, Enøe C, Wachmann H, Baggesen DL. Prevalence and diversity of Campylobacter jejuni in pig herds on farms with and without cattle or poultry. J Food Prot. 2005;68:722–727. doi: 10.4315/0362-028x-68.4.722. [DOI] [PubMed] [Google Scholar]
- Dasti JI, Tareen AM, Lugert R, Zautner AE, Groß U. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol. 2010;300:205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- González LC, Esteban JG, Bargues MD, Valero MA, Ortiz P, Náquira C, Mas-Coma S. Hyperendemic human fascioliasis in Andean valleys: An altitudinal transect analysis in children of Cajamarca province, Peru. Acta Trop. 2011;120:119–129. doi: 10.1016/j.actatropica.2011.07.002. [DOI] [PubMed] [Google Scholar]
- de Zaldívar LC. Producción de cuyes (Cavia porcellus) Food & Agriculture Org. 1997;138 https://books.google.com/books?hl=en&lr=&id=VxLVzsZ5HWcC&oi=fnd&pg=PP9&dq=de+Zald%C3%ADvar,+Lilia+Chauca.+Producci%C3%B3n+de+cuyes+(Cavia+porcellus).+Vol.+138.+Food+%26+Agriculture+Org.,+1997&ots=XNacZsMdGm&sig=rDiL0t_WoDRrTsY8L2diMn8Rz0M#v=onepage&q&f=false. [Google Scholar]
- Fakir JD. A study of thermophilic Campylobacter in cattle, sheep and laboratory animals: a thesis presented in partial (70%) fulfilment of the requirements for the degree of Master of Philosophy in Veterinary Microbiology at Massey University (Doctoral Dissertation) 1986. [Google Scholar]
- Fredriksson-Ahomaa M. Yersinia enterocolitica and Yersinia pseudotuberculosis. In: Simjee S, editor. Foodborne Diseases. New York City: Humana Press; 2007. pp. 79–113. [Google Scholar]
- Gabastou JM, Proaño J, Vimos A, Jaramillo G, Hayes E, Gage K, et al. An outbreak of plague including cases with probable pneumonic infection, Ecuador, 1998. Trans R Soc Trop Med Hyg. 2000;94:387–391. doi: 10.1016/s0035-9203(00)90114-7. [DOI] [PubMed] [Google Scholar]
- Georgescourbot MC, Casselberaud AM, Gouandjika I, Monges J, Georges AJ. A cohort study of enteric Campylobacter infection in children from birth to 2 years in Bangui (Central African Republic) Trans R Soc Trop Med Hyg. 1990;84:122–125. doi: 10.1016/0035-9203(90)90402-z. [DOI] [PubMed] [Google Scholar]
- Grados O, Bravo N, Black RE, Butzler JP. Pediatric Campylobacter diarrhea from household exposure to live chickens in Lima, Peru. Bull World Health Organ. 1988;66:369–374. [PMC free article] [PubMed] [Google Scholar]
- Gras LM, Smid JH, Wagenaar JA, Koene MGJ, Havelaar AH, Friesema IHM, et al. Increased risk for Campylobacter jejuni and C. coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol Infect. 2013;141:2526–2535. doi: 10.1017/S0950268813000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszynski K, Young A, Levine SJ, Garvin JP, Brown S, Turner L, et al. Streptococcus equi subsp zooepidemicus infections associated with guinea pigs. Emerg Infect Dis. 2015;21:156–158. doi: 10.3201/eid2101.140640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis GD, McAllister TA, Larney FJ, Topp E. Prolonged survival of Campylobacter species in bovine manure compost. Appl Environ Microbiol. 2010;76:1110–1119. doi: 10.1128/AEM.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komba E, Mdegela R, Msoffe P, Matowo D, Maro M. Occurrence, species distribution and antimicrobial resistance of thermophilic Campylobacter isolates from farm and laboratory animals in Morogoro, Tanzania. Vet World. 2014;7:559–565. [Google Scholar]
- Levy MZ, Bowman NM, Kawai V, Waller LA, Del Carpio JGC, Benzaquen EC, et al. Periurban Trypanosoma cruzi-infected Triatoma infestans, Arequipa, Peru. Emerg Infect Dis. 2006;12:1345–1352. doi: 10.3201/eid1209.051662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz-Wohlgroth L, Becker A, Brugnera E, Huat ZL, Zimmermann D, Grimm F, et al. Chlamydiales in guinea-pigs and their zoonotic potential. J Vet Med A Physiol Pathol Clin Med. 2006;53:185–193. doi: 10.1111/j.1439-0442.2006.00819.x. [DOI] [PubMed] [Google Scholar]
- Marks SL, Rankin SC, Byrne BA, Weese JS. Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J Vet Intern Med. 2011;25:1195–1208. doi: 10.1111/j.1939-1676.2011.00821.x. [DOI] [PubMed] [Google Scholar]
- Marquis GS, Ventura G, Gilman RH, Porras E, Miranda E, Carbajal L, Pentafiel M. Fecal contamination of shantytown toddlers in households with noncorralled poultry, Lima, Peru. Am J Public Health. 1990;80:146–149. doi: 10.2105/ajph.80.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Ménard S, Arbeit RD. Campylobacteriosis, Eastern Townships, Québec. Emerg Infect Dis. 2004;10:1844–1847. doi: 10.3201/eid1010.040228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughini Gras L, Smid JH, Wagenaar JA, Koene MGJ, Havelaar AH, Friesema IHM, et al. Increased risk for Campylobacter jejuni and C. coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol Infect. 2013;141:2526–2535. doi: 10.1017/S0950268813000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Noguchi Y, Suzuki K, Zaw KM. Adenomatous intestinal hyperplasia in guinea pigs associated with Campylobacter-like bacteria. Jpn J Med Sci Biol. 1983;36:337–342. doi: 10.7883/yoken1952.36.337. [DOI] [PubMed] [Google Scholar]
- Oberhelman RA, Gilman RH, Cabrera L, Meza R, Sheen P, Cordova J, et al. Corraling of free-ranging chickens for control of Campylobacter diarrhea in a Peruvian shantytown community. Am J Trop Med Hyg. 2003;69:417–418. [PubMed] [Google Scholar]
- Pigiere F, Van Neer W, Ansieau C, Denis M. New archaeozoological evidence for the introduction of the guinea pig to Europe. J Archaeol Sci. 2012;39:1020–1024. [Google Scholar]
- Ramonaite S, Kudirkiene E, Tamuleviciene E, Leviniene G, Malakauskas A, Goelz G, et al. Prevalence and genotypes of Campylobacter jejuni from urban environmental sources in comparison with clinical isolates from children. J Med Microbiol. 2014;63:1205–1213. doi: 10.1099/jmm.0.072892-0. [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Maiden MC. The Evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb Perspect Biol. 2015;7:a018119. doi: 10.1101/cshperspect.a018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, et al. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol. 2013;22:1051–1064. doi: 10.1111/mec.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotorno AE, Manriguez G, FernandezL A, Marin JC, Gonzalez F, Wheeler J. Domestication of guinea pigs from a southern Peru-northern Chile wild species and their middle pre-Columbian mummies. Univ Calif Publ Zool. 2007;134:367–388. [Google Scholar]
- Vasco G, Trueba G, Atherton R, Calvopiña M, Cevallos W, Andrade T, et al. Identifying etiological agents causing diarrhea in low income Ecuadorian communities. Am J Trop Med Hyg. 2014;91:563–569. doi: 10.4269/ajtmh.13-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar JA, Newell DG, Kalupahana RS, Mughini-Gras L. Campylobacter: Animal reservoirs, human infections, and options for control. In: Sing A, editor. Zoonoses-Infections Affecting Humans and Animals. Dordrecht: Springer Netherlands; 2015. pp. 159–177. [Google Scholar]
- Walther B, Wieler LH, Vincze S, Antao EM, Brandenburg A, Stamm I, et al. MRSA variant in companion animals. Emerg Infect Dis. 2012;18:2017–2020. doi: 10.3201/eid1812.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Lembke C, Schäfer R. The occurrence of Campylobacter jejuni in rabbits, guinea pigs, rats and mice in the laboratory animal unit. Berl Munch Tierarztl Wochenschr. 1982;95:488. [PubMed] [Google Scholar]
- Wing ES. Domestication of Andean mammals. In: Vuilleunier F, Monasterio M, editors. High Altitude Tropical Biogeography. New York: Oxford University Press; 1986. pp. 246–264. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


