Highlights
-
•
A balance of phosphorylation and K63-linked ubiquitination regulates RIG-I.
-
•
RIG-I and MAVS functions are modulated by alternative splicing and translation.
-
•
Subcellular localization and autophagy regulate RLR signaling.
-
•
Viral pathogens manipulate the regulatory steps of RLR–MAVS activation.
Abstract
Mammalian cells have the intrinsic capacity to detect viral pathogens and to initiate an antiviral response that is characterized by the induction of interferons (IFNs) and proinflammatory cytokines. A delicate regulation of the signaling pathways that lead to cytokine production is needed to ensure effective clearance of the virus, while preventing tissue damage caused by excessive cytokine release. Here, we focus on the mechanisms that modulate the signal transduction triggered by RIG-I-like receptors (RLRs) and their adaptor protein MAVS, key components of the host machinery for sensing foreign RNA. Specifically, we summarize recent advances in understanding how RLR signaling is regulated by posttranslational and posttranscriptional mechanisms, microRNAs (miRNAs) and autophagy. We further discuss how viruses target these regulatory mechanisms for immune evasion.
Current Opinion in Virology 2015, 12:7–14
This review comes from a themed issue on Antiviral strategies
Edited by Michael Gale Jr. and Curt M Horvath
For a complete overview see the Issue and the Editorial
Available online 30th January 2015
http://dx.doi.org/10.1016/j.coviro.2015.01.004
1879-6257/© 2015 Elsevier B.V. All rights reserved.
Introduction
Mammalian cells possess pattern-recognition receptors (PRRs) that recognize foreign molecules, such as viral replication products or structural components of the virion, commonly known as pathogen-associated molecular patterns (PAMPs). Upon non-self recognition, the infected host cell rapidly mounts an innate immune response characterized by the induction of type I and III interferons (IFNs), interferon-stimulated genes (ISGs) and proinflammatory cytokines, to restrict viral replication and direct adaptive immune responses [1].
The RLR family, comprised of RIG-I, MDA5 and LGP2, detects viral RNA in the cytosol of most cell types. The RLRs possess a central DExD/H-box helicase domain and a C-terminal domain (CTD), which are important for binding viral RNA. RIG-I and MDA5 further harbor two N-terminal caspase activation and recruitment domains (CARDs), which are responsible for downstream signaling. LGP2 lacks the CARDs and is generally thought to play a regulatory role in RLR signaling (reviewed in [2]).
RIG-I and MDA5 recognize structurally-distinct viral RNA species. RIG-I senses RNAs possessing both panhandle structures and a 5′triphosphate moiety, while MDA5 is thought to recognize long dsRNA or web-like RNA aggregates (reviewed in [3]). In addition, a recent study indicated that a 5′diphosphate moiety in the viral RNA can also be recognized by RIG-I [4]. Extensive functional studies have demonstrated the importance of RIG-I and MDA5 in sensing RNA virus infections, with RIG-I playing a critical role in the detection of orthomyxoviruses, rhabdoviruses and arenaviruses, and MDA5 preferentially detecting picornaviruses. Moreover, many viruses (flaviviruses, paramyxoviruses, reoviruses) are sensed by both RIG-I and MDA5 [5]. Upon RNA ligand binding, RIG-I and MDA5 bind to their common adaptor MAVS/Cardif/IPS-1/VISA through CARD–CARD interactions. This leads, via TRAF3 or TRAF6, to the activation of several well-studied kinases of the IKK family, namely IKKɛ and TBK1 as well as IKKα/β/γ. Through phosphorylation steps, these kinases ultimately activate the transcription factors IFN-regulatory factor 3 and 7 (IRF3/7), NF-κB, and ATF2/c-Jun, which then induce the transcription of IFNs and proinflammatory cytokines (reviewed in [6]). Subsequently, secreted IFNs bind to their respective receptors and induce the expression of hundreds of ISGs, leading to an antiviral state.
While antiviral and proinflammatory cytokines are key to controlling viral infection, they can also lead to inflammation and tissue damage and hence must be tightly controlled. In the following sections we review the molecular and cellular processes that regulate RLR signaling, with emphasis given to recently published work.
RLR regulation by posttranslational modifications (PTMs)
In the past several years, it has become evident that the activation of RIG-I and MDA5 is a multi-step process consisting of viral RNA binding, conformational changes, and a series of PTMs. Furthermore, regulatory PTMs ensure that aberrant RLR signal transduction does not occur in the absence of a viral infection (Figure 1 ). In uninfected cells, RIG-I is kept in an auto-repressed state due to the masking of its CARDs by the helicase domain [7]. To prevent aberrant downstream signaling, RIG-I and MDA5 also undergo phosphorylation at multiple residues: S8 and T170 as well as T770 and S854/S855 in the RIG-I CARDs and CTD, respectively; and S88 in the MDA5 CARDs [8]. RIG-I is kept phosphorylated at these CARD and CTD sites by protein kinase C α/β (PKCα/β) and casein kinase II (CKII), respectively, while the kinase(s) for MDA5 phosphorylation is still unknown [9, 10]. An RNAi screen against the human phosphatome recently revealed that two highly homologous isoenzymes of phosphoprotein phosphatase 1 (PP1α and PP1γ) are responsible for RIG-I and MDA5 dephosphorylation, thereby triggering their activation [11•]. In response to viral RNA binding, PP1α/γ binds and dephosphorylates both RIG-I (S8 and T170) and MDA5 (S88) in the CARDs, allowing for MAVS binding, likely through a rearrangement of the tandem CARD after dephosphorylation. The phosphatase(s) for the removal of the phosphorylation marks in the RIG-I CTD is currently unknown. As PP1α and PP1γ dephosphorylate numerous substrates in the cell, current studies are focused on elucidating the mechanism of PP1's substrate specificity towards RLRs in infected cells.
Figure 1.
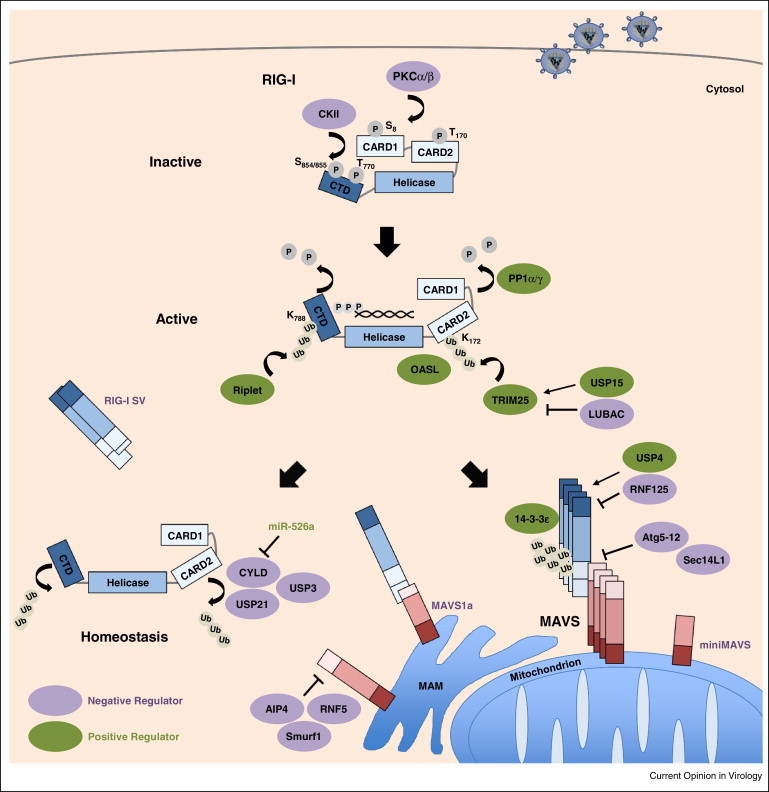
Regulation of RLR signaling, as exemplified by RIG-I. RIG-I is kept in an inactive phosphorylated state in resting cells by PKCα/β and CKII. Upon engagement of viral RNA, RIG-I undergoes a conformational change and is dephosphorylated by PP1α/γ. Subsequently, activation of RIG-I is mediated by K63-linked ubiquitination of the CTD and CARD domains by Riplet and TRIM25, respectively, promoting RIG-I tetramerization. OASL can mimic K63-linked ubiquitination to promote RIG-I activation. The adaptor protein 14-3-3ɛ mediates translocation of the active RIG-I-TRIM25 complex to mitochondrion/MAM-localized MAVS, leading to downstream signal transduction that results in type I IFN gene expression (not illustrated). The deubiquitinating enzymes CYLD, USP21 and USP3 remove K63-linked polyubiquitin chains from RIG-I as a form of homeostatic regulation to prevent aberrant IFN induction. The expression of CYLD is suppressed by miR-526a. TRIM25, RIG-I and MAVS are further regulated by degradative K48-linked ubiquitination mediated by LUBAC, RNF125, and AIP4, Smurf1, and RNF5, respectively. Conversely, USP15 and USP4 deubiquitinate TRIM25 and RIG-I, respectively, to stabilize the proteins. The Atg5–Atg12 conjugate and Sec14L1 block RIG-I–MAVS interaction to prevent antiviral signaling. A RIG-I splice variant (RIG-I SV), MAVS splice variant (MAVS1a) and miniMAVS also contribute to prevent excessive signaling.
Recent data demonstrated that, in the case of RIG-I, there is crosstalk between phosphorylation and K63-linked ubiquitination, a polyubiquitin linkage that does not trigger proteasomal degradation but facilities signal transduction events. Biochemical studies demonstrated that in uninfected cells, RIG-I is robustly phosphorylated but minimally ubiquitinated in its CARDs and CTD. However, upon stimulation of RIG-I by viral RNA binding, dephosphorylation occurs, and this triggers robust K63-ubiquitination of RIG-I by two critical ubiquitin E3 ligases, TRIM25 and Riplet. Mechanistically, Riplet first induces K63-linked ubiquitination of K788 in the CTD of RIG-I [12]. This appears to trigger a conformational change that exposes the CARDs, enabling TRIM25 to bind and to attach K63-linked ubiquitin chains to K172 in RIG-I CARD2, ultimately leading to RIG-I oligomerization and MAVS binding [13]. K63-linked ubiquitination of CARD2 by TRIM25 is critical for RIG-I activation, as loss of TRIM25 severely hampers RIG-I signaling. Furthermore, K63-linked ubiquitin chains have also been shown to bind to the CARDs non-covalently to promote RIG-I oligomerization and activation [14]. Thus, it has been unclear for quite some time how both covalent and non-covalent K63-polyubiquitin mediate RIG-I activation. This question was recently addressed by structural analysis of the RIG-I CARDs, which showed that the CARDs form a helical tetramer adopting a ‘lock-washer configuration’, in which three K63-diubiquitins are wrapped around the outer rim of the CARD tetramer [15••]. This study further showed that K172 is within the covalent linkage distance to ubiquitin (<20 Å), strongly indicating that this residue is indeed covalently ubiquitinated. Biochemical studies comparing the activation capacity of covalent versus non-covalent K63-diubiquitin showed that, while both induced RIG-I tetramerization and MAVS activation, covalent K63-diubiquitin had a stronger RIG-I activation capacity than unanchored K63-diubiquitin [15••]. It has been recently shown that RIG-I can also be activated in a ubiquitin-independent manner. Specifically, the IFN-inducible oligoadenylate synthetase-like (OASL) protein, which contains two ubiquitin-like domains, binds to RIG-I and mimics K63-linked polyubiquitin, thereby enhancing RIG-I activation [16•]. This study proposed a model in which TRIM25 and Riplet-mediated K63-linked ubiquitination is essential for RIG-I activation early during infection, while OASL activates RIG-I at later time points.
The importance of K63-linked ubiquitination for RIG-I activation was further strengthened by the identification of several deubiquitinating (DUB) enzymes that remove this ubiquitin mark from RIG-I to inhibit its signaling. CYLD (cylindromatosis) deubiquitinates RIG-I and several downstream molecules to prevent premature RIG-I activation in uninfected cells [17], while USP3 deubiquitinates RIG-I specifically after viral infection, likely serving as a negative feedback regulator [18]. In contrast, USP21 has been shown to bind and deubiquitinate RIG-I independent of viral infection [19]. Together, these studies establish K63-linked ubiquitination as a crucial activation mark for RIG-I. In contrast, the role of K63-linked ubiquitin polymers in MDA5 activation is still a subject of debate. In general, our knowledge of PTMs that regulate the signaling activity of MDA5 lags significantly. As described above, dephosphorylation by PP1α/γ has been shown to be critical for MDA5 activation [11•]. In addition, SUMOylation of the MDA5 CTD by the E3 ligase PIAS2β facilitates MDA5-mediated antiviral signaling; however, the precise mechanism of this activation mode remains unknown [20].
In contrast to K63-linked ubiquitination, which serves as an activation mark, K48-linked ubiquitination triggers proteasomal degradation to regulate the turnover of cellular proteins, including key molecules in the RLR pathway. The RING-finger protein 125 (RNF125) induces K48-linked ubiquitination and proteasomal degradation of RIG-I, MDA5 and MAVS, thereby preventing excessive RLR signaling [21]. Conversely, USP4 stabilizes RIG-I by removing K48-linked ubiquitination [22]. The stability of TRIM25 is tightly regulated by K48-linked ubiquitination mediated by the linear ubiquitin assembly complex (LUBAC), consisting of the two E3 ligases HOIL-1L and HOIP [23]. Conversely, USP15 has been recently identified as a DUB enzyme that stabilizes TRIM25, thereby ensuring effective viral clearance through sustained IFN-β production [24]. The abundance of MAVS is also delicately controlled by degradative K48-linked ubiquitination mediated by the E3 ligases AIP4 (also called ITCH), Smurf1 (SMAD ubiquitin regulatory factor 1), and RNF5 [25, 26, 27]. However, it is currently unknown how MAVS stability is dynamically regulated by these three E3 ligases, and whether they act in a temporal or cell type-specific manner.
RLR regulation by posttranscriptional mechanisms
Several posttranscriptional mechanisms modulating RLR signaling have been identified, including alternative splicing and translation, as well as regulation by microRNAs (miRNAs), small noncoding RNAs that lead to the degradation or translational repression of target mRNAs by binding to complementary sequences in their 3′ untranslated region (UTR). In most cases, posttranscriptional mechanisms are part of a negative feedback loop to dampen RLR signaling, thereby preventing excessive or sustained production of antiviral and inflammation-inducing proteins (Figure 1).
Alternative splicing has been shown to play an important role in modulating the activities of RIG-I and MAVS. A splice variant of RIG-I (RIG-I SV) is specifically induced upon viral infection or IFN stimulation [28]. RIG-I SV carries a short deletion (amino acids 36–80) in CARD1 and is therefore unable to bind TRIM25 for downstream activation. RIG-I SV suppresses antiviral signaling in a dominant-negative manner by hetero-oligomerizing with full-length RIG-I, which prevents MAVS binding. Similarly, a splice variant of MAVS (MAVS1a) strongly binds to RIG-I and inhibits its interaction with full-length MAVS for signal transduction [29]. Furthermore, it has been recently reported that the MAVS mRNA is bicistronic, and that alternative translation gives rise to a smaller MAVS protein termed ‘miniMAVS’ [30••]. MiniMAVS dampens IFN induction, but its ability to promote cell death is comparable to that of full-length MAVS. The precise mechanism of how miniMAVS acts, however, is unknown.
Innate immune signaling triggered by virus infection also leads to the upregulation of several miRNAs, which in turn modulate RIG-I activity and IFN induction. For example, miR-526a is induced in monocytes upon vesicular stomatitis virus (VSV) infection and directly suppresses the expression of CYLD, thereby enhancing K63-linked ubiquitination of RIG-I and its activation [31]. VSV infection also induces the expression of miR-146a in macrophages in a RIG-I-dependent manner. MiR-146a then acts as a negative-feedback regulator of the RLR pathway by targeting several important downstream signaling molecules, including TRAF6 [32]. Furthermore, miR-466l directly binds to the 3′UTR of IFN-α mRNAs and reduces their expression during VSV infection [33].
Regulation of RLR–MAVS signal transduction by subcellular localization and autophagy
Apart from molecular regulatory mechanisms, cellular processes control and shape the signaling activities of RLRs and MAVS. RLRs are traditionally thought to be ‘free floating’ cytosolic molecules, though recent studies indicated that RLRs are localized to cytoplasmic bodies induced by protein kinase R (PKR) and DHX36, known as antiviral stress granules (avSGs) [34]. It has been proposed that avSGs provide a platform for RLRs, other antiviral proteins (PKR, RNAseL, and OAS1), and viral RNA to interact, thereby augmenting RLR signaling. Further studies are required to determine the contribution of soluble versus avSG-associated RLRs to antiviral immunity, and whether other subcellular compartments are used by RLRs for initiating signal transduction.
In contrast to that of RLRs, the regulation of MAVS by subcellular localization is better characterized due to its membrane-bound nature. MAVS resides in multiple subcellular regions, including the outer mitochondrial membrane, mitochondrial-associated membranes (MAMs, a specialized subdomain of the ER located adjacent to mitochondria), and peroxisomes [35, 36, 37]. Functionally, while cytosolic MAVS is unable to signal, mitochondrial and MAM-associated MAVS are responsible for type I IFN induction. Furthermore, recent work has indicated that peroxisomal MAVS preferentially induces type III IFNs [38].
Upon PAMP recognition, cytosolic RLRs must interact with membrane-bound MAVS. How this translocation event occurs, however, was not known until recently. It has been shown that the activated RIG-I-TRIM25 complex requires binding to the adaptor protein 14-3-3ɛ to translocate to mitochondria/MAMs for MAVS interaction and downstream activation [39•]. It is unclear if MDA5 also requires 14-3-3ɛ for mitochondrial translocation, and if other adaptor proteins control RLR translocation to other MAVS locations such as the peroxisomes.
Autophagy, a degradation process well-known for its role in the removal of protein aggregates and organelles, also plays a critical role in innate immunity by either degrading intracellular pathogens or by homeostatically regulating innate immune signaling (reviewed in [40]). It has been reported that cells deficient in Atg5, a key regulatory protein of autophagy, are defective in autophagosome formation and accumulate damaged mitochondria, leading to increased RLR stimulation, likely due to increased release of reactive oxygen species (ROS) [41]. Another line of evidence also supports the negative regulation of RLR signaling by Atg5; however, this study suggested a different mechanism, specifically that Atg5–Atg12 conjugate suppresses IFN induction by directly interacting with the CARDs of RIG-I and MAVS [42]. Similarly, Sec14L1, a protein that is not implicated in autophagy, inhibits the CARD–CARD interaction of RIG-I and MAVS [43].
Pathogenic viruses target RLR regulation for immune evasion
Viruses and their hosts are in an active ‘arms race’ that drives continuous co-evolution. Given the importance of RLRs for an effective innate immune response, viral pathogens have evolved means to manipulate various RLR regulatory mechanisms for immune evasion (Figure 2 ). Many viruses have been shown to dysregulate the PTMs of RLRs. For example, measles and Nipah viruses, both members of the paramyxovirus family, antagonize the phosphatases PP1α/γ to prevent RLR dephosphorylation and hence activation. Mechanistically, their V protein, a well-known IFN antagonist, interacts with PP1α/γ and sequesters these phosphatases away from MDA5, thereby keeping it in the CARD-phosphorylated, inactive state [44]. Furthermore, in dendritic cells, measles virus targets PP1α/γ through a V-independent mechanism by inducing DC-SIGN signaling and formation of a negative-regulatory PP1 complex, inhibiting both RIG-I and MDA5 [45]. Previously, multiple studies have demonstrated that the V proteins of several paramyxoviruses, including parainfluenza virus 5 (PIV5), antagonize MDA5 through a direct interaction with its helicase domain, thereby blocking its ATPase activity [46, 47•]. VP35 of Ebola virus (EboV) and NS1 of influenza A virus (IAV) specifically inhibit the ATPase activity of RIG-I that is stimulated by PACT (PKR activator) [48, 49].
Figure 2.
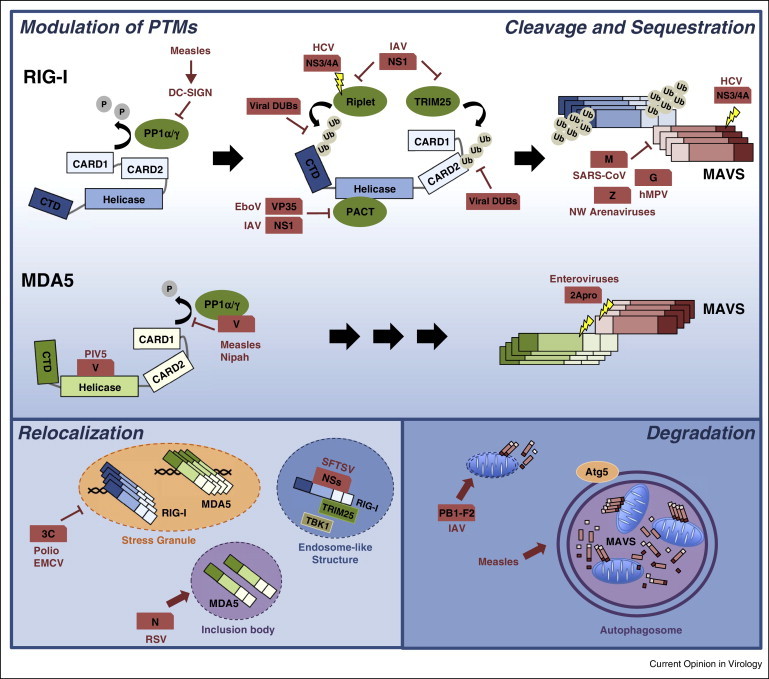
Pathogenic viruses target RLR regulation for immune evasion. There are five general strategies used by viruses to target RLR–MAVS signaling: (1) modulation of PTMs of RLRs, (2) cleavage of RLR pathway components, (3) sequestration of RLRs, (4) modulation of RLR localization, and (5) degradation of MAVS and other RLR downstream signaling molecules. The details of the viral antagonistic mechanisms are described in the text.
With regards to K63-linked ubiquitination of RIG-I, the NS1 protein of IAV binds to the ubiquitin E3 ligase TRIM25 to block ubiquitination of the RIG-I CARDs [50]. Furthermore, the NS1 proteins of some IAV strains were shown to also bind human Riplet to inhibit ubiquitination of the RIG-I CTD [51]. A recent study indicated that the NS3/4A protease complex of Hepatitis C virus (HCV) cleaves not only MAVS but also Riplet to prevent RIG-I activation [12, 37, 52]. Viruses also act directly on RIG-I ubiquitination by encoding enzymes that deubiquitinate RIG-I and hence inactivate it. Orf64, a viral DUB of Kaposi's sarcoma-associated herpesvirus (KSHV), removes K63-ubiquitin chains from the RIG-I CARDs [53]. The papain-like protease (PLP) of severe acute respiratory syndrome coronavirus (SARS-CoV) and the leader proteinase (Lpro) of foot-and-mouth disease virus (FMDV) also deubiquitinate RIG-I and other innate immune signaling molecules [54, 55]. Finally, arteriviruses and nairoviruses encode proteins with ovarian tumor (OTU)-type DUB enzymatic activities to remove K63-polyubiquitin from RIG-I [56].
Another viral strategy to escape the RLR response is modulating the expression of specific miRNAs that target critical regulatory proteins in the RLR pathway. For example, the 3C protein of Enterovirus 71 blocks the upregulation of miR-526a in infected cells, which leads to increased expression of CYLD and hence RIG-I inhibition via deubiquitination [31].
Viruses are also equipped with proteins that modulate the subcellular localization of RLRs or actively degrade components of the RLR pathway. For example, poliovirus and encephalomyocarditis virus (EMCV) use their 3C proteases to prevent the formation of RLR-containing avSGs through cleavage of the Ras-Gap SH3 domain binding protein 1 (G3BP1) [57, 58]. Other proteins sequester RLRs from MAVS. The M protein of SARS-CoV, the Z protein of New World (NW) arenaviruses, and the glycoprotein G of human metapneumovirus (hMPV) bind to RIG-I to sequester it from MAVS. Furthermore, the N protein of respiratory syncytial virus (RSV) binds specifically to MDA5, relocalizing it to large inclusion bodies [59, 60, 61, 62]. In addition, the NSs protein of severe fever with thrombocytopenia syndrome virus (SFTSV) has been recently shown to interact with RIG-I, TRIM25 and TBK1 and to relocalize them into cytoplasmic endosome-like structures for sequestration [63]. Moreover, PB1-F2 of IAV reduces the inner membrane potential of mitochondria, leading to fragmentation of these organelles and inhibition of innate immune signaling [64, 65•].
Another important evasion strategy employed by several viruses is cleavage of RIG-I, MDA5 and/or MAVS. For example, enteroviruses cleave both MDA5 and MAVS using their protease 2Apro, thereby blunting IFN-β induction [66, 67]. Measles virus infection triggers selective autophagy to degrade mitochondria (a process termed ‘mitophagy’), resulting in decreased MAVS abundance and disruption of RLR signaling [68]. Finally, the NS1 and NS2 proteins of RSV have been recently shown to trigger the degradation of RIG-I, IRF3 and many other molecules in the IFN induction pathway by assembling a large degradative complex on the mitochondria [69].
Conclusions
The past 10 years have provided new fundamental insights into the molecular mechanisms that stimulate RLR signaling, such as K63-ubiquitin-mediated assembly of CARD signaling platforms, and the diversification of RLR function due to specific subcellular localizations of MAVS. However, while many regulatory mechanisms have been unveiled for RIG-I, significantly less is known about the molecular details of how the activities of MDA5 and LGP2 are controlled during infection. Likewise, whereas the role of miRNAs in other cellular processes is well-characterized, the regulation of RLR signaling by miRNAs has just begun to be elucidated.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We apologize to all colleagues whose important contributions could not be cited due to space constraints. Current research in the Gack laboratory is supported by National Institutes of Health Grants (AI087846, AI097699, and AI104415), the Alexander and Margaret Stewart Trust Foundation, and a John and Virginia Kaneb Fellowship.
References
- 1.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez K.R., Bruns A.M., Horvath C.M. MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction. J Virol. 2014;88:8194–8200. doi: 10.1128/JVI.00640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlee M. Master sensors of pathogenic RNA — RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goubau D., Deddouche S., Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang J.J., Davis M.E., Gack M.U. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev. 2014;25:491–505. doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Gack M.U., Nistal-Villan E., Inn K.S., Garcia-Sastre A., Jung J.U. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 2010;84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maharaj N.P., Wies E., Stoll A., Gack M.U. Conventional protein kinase C-alpha (PKC-alpha) and PKC-beta negatively regulate RIG-I antiviral signal transduction. J Virol. 2012;86:1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z., Ren H., Liu Y., Teeling J.L., Gu J. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol. 2011;85:1036–1047. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Wies E., Wang M.K., Maharaj N.P., Chen K., Zhou S., Finberg R.W., Gack M.U. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uncovers PP1α and PP1γ as activators of both RIG-I and MDA5.
- 12.Oshiumi H., Miyashita M., Matsumoto M., Seya T. A distinct role of riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gack M.U., Shin Y.C., Joo C.H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 14.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides structural insights into RIG-I activation by K63-polyubiquitin chains.
- 16•.Zhu J., Zhang Y., Ghosh A., Cuevas R.A., Forero A., Dhar J., Ibsen M.S., Schmid-Burgk J.L., Schmidt T., Ganapathiraju M.K. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reveals a novel role for OASL in mimicking ubiquitin chains for RIG-I activation.
- 17.Friedman C.S., O’Donnell M.A., Legarda-Addison D., Ng A., Cardenas W.B., Yount J.S., Moran T.M., Basler C.F., Komuro A., Horvath C.M. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J., Song Y., Li Y., Zhu Q., Tan P., Qin Y., Wang H.Y., Wang R.F. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24:400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y., Mao R., Yu Y., Liu S., Shi Z., Cheng J., Zhang H., An L., Zhao Y., Xu X. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211:313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu J., Xiong Y., Xu Y., Cheng G., Tang H. MDA5 is SUMOylated by PIAS2beta in the upregulation of type I interferon signaling. Mol Immunol. 2011;48:415–422. doi: 10.1016/j.molimm.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Zhao W., Zhang M., Wang P., Zhao K., Zhao X., Yang S., Gao C. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J Virol. 2013;87:4507–4515. doi: 10.1128/JVI.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inn K.S., Gack M.U., Tokunaga F., Shi M., Wong L.Y., Iwai K., Jung J.U. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauli E.K., Chan Y.K., Davis M.E., Gableske S., Wang M.K., Feister K.F., Gack M.U. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You F., Sun H., Zhou X., Sun W., Liang S., Zhai Z., Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Tong X., Ye X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol. 2012;189:5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 27.Zhong B., Zhang Y., Tan B., Liu T.T., Wang Y.Y., Shu H.B. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol. 2010;184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 28.Gack M.U., Kirchhofer A., Shin Y.C., Inn K.S., Liang C., Cui S., Myong S., Ha T., Hopfner K.P., Jung J.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lad S.P., Yang G., Scott D.A., Chao T.H., Correia J. da S., de la Torre J.C., Li E. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. Mol Immunol. 2008;45:2277–2287. doi: 10.1016/j.molimm.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 30••.Brubaker S.W., Gauthier A.E., Mills E.W., Ingolia N.T., Kagan J.C. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell. 2014;156:800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that the MAVS transcript is bicistronic and further identify other antiviral truncated variants using genome-wide ribosomal profiling.
- 31.Xu C., He X., Zheng Z., Zhang Z., Wei C., Guan K., Hou L., Zhang B., Zhu L., Cao Y. Downregulation of microRNA miR-526a by enterovirus inhibits RIG-I-dependent innate immune response. J Virol. 2014;88:11356–11368. doi: 10.1128/JVI.01400-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6 IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Fan X., He X., Sun H., Zou Z., Yuan H., Xu H., Wang C., Shi X. MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cell Mol Immunol. 2012;9:497–502. doi: 10.1038/cmi.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo J.S., Takahasi K., Ng C.S., Ouda R., Onomoto K., Yoneyama M., Lai J.C., Lattmann S., Nagamine Y., Matsui T. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog. 2014;10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., Kagan J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Liu H.M., Loo Y.M., Horner S.M., Zornetzer G.A., Katze M.G., Gale M., Jr. The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that 14-3-3ɛ regulates the translocation of RIG-I to mitochondria/MAMs for MAVS interaction and signal transduction.
- 40.Deretic V., Saitoh T., Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M.T., Di W., Xu H., Yang Y.K., Chen H.W., Zhang F.X., Zhai Z.H., Chen D.Y. Negative regulation of RIG-I-mediated innate antiviral signaling by SEC14L1. J Virol. 2013;87:10037–10046. doi: 10.1128/JVI.01073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis M.E., Wang M.K., Rennick L.J., Full F., Gableske S., Mesman A.W., Gringhuis S.I., Geijtenbeek T.B., Duprex W.P., Gack M.U. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe. 2014;16:19–30. doi: 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mesman A.W., Zijlstra-Willems E.M., Kaptein T.M., de Swart R.L., Davis M.E., Ludlow M., Duprex W.P., Gack M.U., Gringhuis S.I., Geijtenbeek T.B. Measles virus suppresses RIG-I-like receptor activation in dendritic cells via DC-SIGN-mediated inhibition of PP1 phosphatases. Cell Host Microbe. 2014;16:31–42. doi: 10.1016/j.chom.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez K.R., Horvath C.M. Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: a single arginine distinguishes MDA5 from RIG-I. J Virol. 2013;87:2974–2978. doi: 10.1128/JVI.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Motz C., Schuhmann K.M., Kirchhofer A., Moldt M., Witte G., Conzelmann K.K., Hopfner K.P. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]; This study solves the crystal structure of the MDA5 ATP-hydrolysis domain in complex with the V protein of PIV5, revealing that the interaction induces mutual structural unfolding.
- 48.Luthra P., Ramanan P., Mire C.E., Weisend C., Tsuda Y., Yen B., Liu G., Leung D.W., Geisbert T.W., Ebihara H. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe. 2013;14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tawaratsumida K., Phan V., Hrincius E.R., High A.A., Webby R., Redecke V., Hacker H. Quantitative proteomic analysis of the influenza A virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor. J Virol. 2014;88:9038–9048. doi: 10.1128/JVI.00830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E., Farzan M., Inoue S., Jung J.U., Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajsbaum R., Albrecht R.A., Wang M.K., Maharaj N.P., Versteeg G.A., Nistal-Villan E., Garcia-Sastre A., Gack M.U. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inn K.S., Lee S.H., Rathbun J.Y., Wong L.Y., Toth Z., Machida K., Ou J.H., Jung J.U. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol. 2011;85:10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q., Luo R., Liu X., Li K., Chen H. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol. 2011;85:3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci U S A. 2013;110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng C.S., Jogi M., Yoo J.S., Onomoto K., Koike S., Iwasaki T., Yoneyama M., Kato H., Fujita T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Siu K.L., Kok K.H., Ng M.H., Poon V.K., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan L., Briese T., Lipkin W.I. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J Virol. 2010;84:1785–1791. doi: 10.1128/JVI.01362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bao X., Kolli D., Ren J., Liu T., Garofalo R.P., Casola A. Human metapneumovirus glycoprotein G disrupts mitochondrial signaling in airway epithelial cells. PLoS One. 2013;8:e62568. doi: 10.1371/journal.pone.0062568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bao X., Liu T., Shan Y., Li K., Garofalo R.P., Casola A. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 2008;4:e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santiago F.W., Covaleda L.M., Sanchez-Aparicio M.T., Silvas J.A., Diaz-Vizarreta A.C., Patel J.R., Popov V., Yu X.J., Garcia-Sastre A., Aguilar P.V. Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J Virol. 2014;88:4572–4585. doi: 10.1128/JVI.03021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Varga Z.T., Grant A., Manicassamy B., Palese P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol. 2012;86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Yoshizumi T., Ichinohe T., Sasaki O., Otera H., Kawabata S., Mihara K., Koshiba T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun. 2014;5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]; This study shows that PB1-F2 translocates into the mitochondrial inner membrane space and accelerates mitochondria fragmentation, thereby impairing RLR–MAVS signaling.
- 66.Wang B., Xi X., Lei X., Zhang X., Cui S., Wang J., Jin Q., Zhao Z. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog. 2013;9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng Q., Langereis M.A., Lork M., Nguyen M., Hato S.V., Lanke K., Emdad L., Bhoopathi P., Fisher P.B., Lloyd R.E. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia M., Gonzalez P., Li C., Meng G., Jiang A., Wang H., Gao Q., Debatin K.M., Beltinger C., Wei J. Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. J Virol. 2014;88:5152–5164. doi: 10.1128/JVI.03851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goswami R., Majumdar T., Dhar J., Chattopadhyay S., Bandyopadhyay S.K., Verbovetskaya V., Sen G.C., Barik S. Viral degradasome hijacks mitochondria to suppress innate immunity. Cell Res. 2013;23:1025–1042. doi: 10.1038/cr.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]


