Abstract
Divalent metal ions are essential components of DNA polymerases both for catalysis of the nucleotidyl transfer reaction and for base excision. They occupy two sites, A and B, for DNA synthesis. Recently, a third metal ion was shown to be essential for phosphoryl transfer reaction. The metal ion in the A site is coordinated by the carboxylate of two highly conserved acidic residues, water molecules, and the 3′-hydroxyl group of the primer so that the A metal is in an octahedral complex. Its catalytic function is to lower the pKa of the hydroxyl group, making it a highly effective nucleophile that can attack the α phosphorous atom of the incoming dNTP. The metal ion in the B site is coordinated by the same two carboxylates that are affixed to the A metal ion as well as the non-bridging oxygen atoms of the incoming dNTP. The carboxyl oxygen of an adjacent peptide bond serves as the sixth ligand that completes the octahedral coordination geometry of the B metal ion. Similarly, two metal ions are required for proofreading; one helps to lower the pKa of the attacking water molecule, and the other helps to stabilize the transition state for nucleotide excision. The role of different divalent cations are discussed in relation to these two activities as well as their influence on base selectivity and misincorporation by DNA polymerases. Some, but not all, of the effects of these different metal ions can be rationalized based on their intrinsic properties, which are tabulated in this review.
Keywords: calcium, magnesium, manganese, nickel, protein complex
Introduction
All DNA polymerases (DNA pols)2 require Mg2+ or Mn2+ for primer extension and for excision of incorrectly incorporated dNTPs via intrinsic 3′→5′ exonuclease activity (1–5). A two-metal-ion mechanism is used by all DNA pols to catalyze nucleotide addition to a growing primer strand (6). Although DNA polymerases employ the physiologically relevant Mg2+, other divalent metal ions can substitute for Mg2+, although they tend to reduce the fidelity of DNA replication (7–10). The effect of metal ion cofactors on the fidelity of DNA replication has been studied for various DNA pols including E. coli DNA pol I (11), AMV DNA pol (12), Klenow fragment of E. coli DNA pol I (13), T4 pol (7), T7 pol (7), human pol α (7), pol β (7), and Dpo4 (8). Some metal ions have been shown to be mutagens and carcinogens probably because they reduce the base selectivity of DNA pols (7, 8, 11–15). Different divalent cations influence fidelity check points in the minimal kinetic scheme for the nucleotidyl transfer reaction (Scheme 1). Cations that can substitute for Mg2+ affect DNA pols by: 1) altering the ground-state binding affinity of incoming dNTPs to pol·DNA binary complexes (16); 2) decreasing base selectivity by promoting misincorporation during primer extension (8); 3) decreasing the rate of base excision (17); 4) altering primer extension past a mismatch at the primer-template (P/T) terminus (17). This review will address the way various metal ions increase misincorporation based on their physical properties. Our emphasis will be on the effect of different cations on the behavior of RB69 pol, which we have studied extensively in our lab (18). There are other reviews that deal with pol β, another DNA pol that has been thoroughly studied with respect to the influence of different metal ions on its structure and function (19–21).
SCHEME 1.
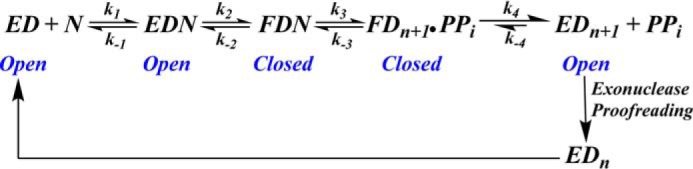
Minimal kinetic scheme for DNA polymerases depicting various fidelity checkpoints along the reaction pathway. EDN represents the open conformation of the ternary collision complex, whereas FDN represents the closed conformation. Additional details are provided in the text.
The Two-metal-ion Mechanism for Nucleotidyl Transfer Reaction
All DNA pols require two divalent metal ions for primer extension (6). One metal ion occupies the “A site” and helps to lower the pKa of the terminal 3′-OH group on the primer and coordinates both the α-phosphate of the incoming dNTP and the 3′-OH of the primer strand, which facilitates its nucleophilic attack on the α-phosphorous atom of the incoming dNTP (6). The other metal ion, occupying the “B site,” coordinates the α-, β-, and γ- non-bridging phosphate oxygens of the incoming dNTP, helping to neutralize the developing negative charge as the ternary complex approaches the transition state in the nucleotidyl transfer reaction, and assists in the departure of the PPi product. Yang et al. (22) have shown that for pol β, the dNTP·metal ion complex in the “B site” alone is unable to induce closing of the fingers, a necessary step for the phosphoryl transfer reaction to proceed. Both A and B metal ions are thus required to prepare the active site for nucleotidyl transfer (22). Tsai and co-workers (23) have used Rh+3 as an exchange-inert cation complexed with an incoming dNTP to selectively fill the “B site” in pol β so that the effect of binding a metal ion in the “A site” could be studied independently of the cation in the B site. Their results showed that the closing of the fingers could occur before occupancy of the A site but that Mg2+ could diffuse into the A site, although pol β was in the closed form (23). In contrast, studies with RB69 pol by Wang and co-workers (16) showed that although the fingers can close in the absence of an “A” metal ion, the fingers have to reopen for a divalent cation to bind in the “A site.” Recent studies by Yang and co-workers (24) observed the transient presence of a third metal ion of pol η after the nucleotidyl transfer reaction was initiated, but before release of the products, using time-resolved x-ray crystallography (24). The third transient metal ion was coordinated to four water molecules in addition to an oxygen atom (which acts as a bridge between the α- and β-phosphorous atoms) and to the non-bridging oxygen of the α-phosphate. Yang and co-workers (24) proposed that in addition to the A and B metal ions, the third metal ion (Mg2+) participates in neutralizing the negative charges built up in the transition state and is likely involved in facilitating the protonation of the pyrophosphate leaving group. Recent studies by Gao and Yang (25) have shown that the third metal ion is indeed required for catalysis by pol η.
The Nature of Metal Ion Coordination Complexes with Various DNA pols
Several groups have solved the crystal structures of DNA pols with metal ions and an incoming dNTP bound in the polymerase active site (6, 9, 26–29). RB69 pol has been one of the most extensively studied DNA pols in the B family and is the only DNA pol where all combinations of mispaired bases were captured in ternary complexes (26). For this purpose, four mutations were required adjacent to the nucleotide binding pocket. Among these the triple mutant (tm) was used to capture a ternary complex in the presence of Mn2+ and dUpNpp, a non-hydrolyzable dNTP analogue (Fig. 1) (6). In this structure, Mn2+, bound in the “B site,” has nearly ideal octahedral geometry and is coordinated by the triphosphate tail of the incoming dUpNpp along with the carboxylate side chains of Asp411 and Asp623, and the backbone carbonyl oxygen of Leu412 (Fig. 1b). Mn2+ bound in the “A site,” however, has highly distorted octahedral geometry because in this structure, it is coordinated with the 3′-OH group of the primer, the two carboxylate side chains of Asp411 and Asp623, and the oxygen of the α-phosphate of the incoming dNTP. However, the distance between the 3′-OH group and Pα of the incoming dUpNpp is too great to support phosphodiester bond formation (6). Xia et al. (6) proposed that as the reaction approaches the transition state, metal ion A helps to reduce the 3′-OH-Pα distance, facilitating covalent bond formation (Fig. 1c). Similar results have been observed when Mg2+ occupies both the A sites and B sites, suggesting that Mg2+ and Mn2+ share similar coordination geometries in RB69 pol ternary complexes (6).
FIGURE 1.
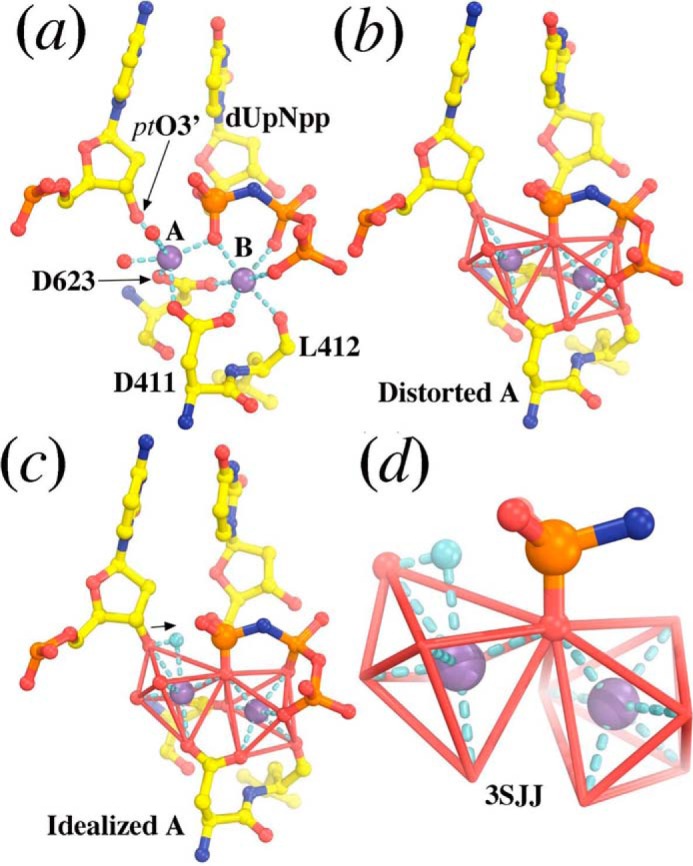
The structure of the tm RB69 pol ternary complex (Protein Data Bank (PDB) accession number 3SJJ). a, ternary complex showing dUpNpp bound in the active site with metal ions A and B. b, close-up of the tm RB69 pol active site showing the B metal ion in perfect octahedral geometry and the A metal ion in a distorted octahedral geometry. c, predicted position of 3′-OH group in an idealized octahedron during the transition state. d, a close-up of the coordination complex showing the two metal ions, the 3′-OH group, and the α phosphate of the incoming dNTP.
In addition to RB69 pol (6, 30), T7 pol (31, 32), Klenow fragment (33–35), and Dpo4 (8, 10, 36), pol β has also been extensively studied (9, 19–21, 23). Although the topology of the Palm domain of pol β, which contains the two conserved catalytic carboxylates, differs between RB69 pol and pol β, the metal ion coordination geometries are nearly identical. In fact, crystal structures of all DNA pols have the same coordination geometry of the A and B metal ions (for examples, see Fig. 2).
FIGURE 2.
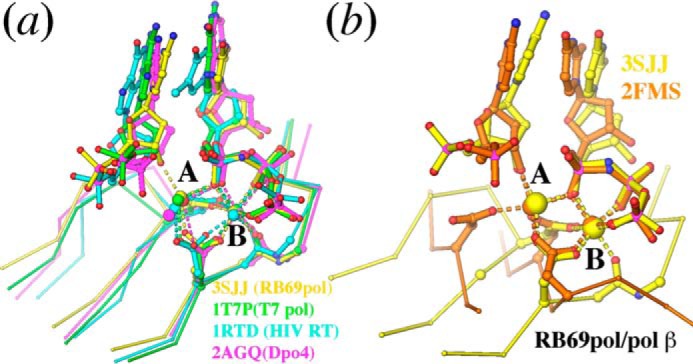
Comparison of metal ion bound structures of the tm RB69 pol with other DNA polymerases. a, superposition of the tm RB69 pol with T7 DNA pol (PDB accession number: 1T7P, green), HIV reverse transcriptase (HIV RT) (PDB accession number: 1RTD, cyan), and Dpo4 (PDB accession number: 2AGQ, magenta). b, superposition of the tm RB69 pol with pol β (PDB accession number: 2FMS, golden).
DNA Polymerases Can Use Different Metal Ions for Catalyzing Phosphoryl Transfer
Early studies by Sirover and Loeb (15, 37) measured perturbations in the fidelity of DNA synthesis using AMV DNA pol. They identified several metal ions as being mutagenic or carcinogenic including Ag+, Be2+, Cd2+, Co2+, Cr3+, Mn2+, Ni2+, and Pb2+ (15). Subsequent studies with E. coli DNA pol I (11, 38) showed that Co2+ and Mn2+ could effectively replace Mg2+ but caused an increase in misincorporation. Similar results were reported with Mn2+ and Co2+ for human pol α and pol β (39). Later studies by Snow et al. (7) showed that Ni2+, albeit active, was an inefficient activator for a variety of DNA pols including AMV pol, human pol α, T4 pol, Klenow fragment, and T7 pol.
Pelletier et al. (9) have reported structures of pol β ternary complexes in the presence of several different metal ions and showed that apart from Mg2+ and Mn2+, only Cd2+ and Zn2+ catalyzed primer extension with a blunt-end DNA substrate. Egli and co-workers (36) carried out kinetic studies using Dpo4 pol with a variety of divalent cations and found that only Mg2+, Mn2+, and Ca2+ could support Dpo4-catalyzed polymerization but that Sr2+, Ba2+, Zn2+, Cu2+, Ni2+, and Co2+ were inactive. Recent work by Vashishtha and Konigsberg (30) on RB69 pol showed that, apart from Mg2+ and Mn2+, Co2+, and to a lesser extent Ni2+, were the only divalent cations that could support both pol and exo activities. The metal ion preferences for different DNA pols are summarized in Table 1. Thus, DNA pols from different families can utilize different metal ions as cofactors, but they do not follow a pattern that can be predicted from their physical properties.
TABLE 1.
Summary of metal ion preferences of different DNA polymerases
Metal ions that activate the respective DNA polymerase are shown with +, and those that are not able to support the polymerase activity are shown with −. Bst, B. stearothermophilus.
| DNA polymerase | Metal ion |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mn2+ | Co2+ | Fe2+ | Ni2+ | Zn2+ | Cd2+ | Sr2+ | Ba2+ | Cu2+ | Cr3+ | Ca2+ | |
| DNA pol I | + | + | − | + | − | − | − | − | − | − | − |
| Human pol α | + | + | − | − | − | − | − | − | − | − | − |
| pol β | + | +a | − | − | + | + | − | − | − | − | − |
| AMV DNA pol | + | + | − | + | − | − | − | − | − | − | − |
| T4 pol | + | + | − | + | − | − | − | − | − | − | − |
| T7 pol | + | + | − | + | − | − | − | − | − | − | − |
| RB69pol | + | + | − | + | − | − | − | − | − | − | − |
| Bst pol | + | +b | − | +b | +b | +b | − | − | +b | − | − |
| Dpo4 pol | + | +a | − | − | − | − | − | − | − | − | + |
a Studies by Pelletier et al. (9) and Egli and co-workers (36) previously claimed that human pol β and Dpo4 were not able to utilize Co2+ as cofactor. However, recent studies by Vashishtha and Konigsberg (30) showed that both pol β and Dpo4 can catalyze primer extension in the presence of Co2+ as explained in the text.
b See Footnote 3.
Metal Ions Affect Various Fidelity Checkpoints during DNA Replication
Divalent metal ions can alter the fidelity of DNA replication at various points along the reaction pathway by: 1) affecting the ground-state binding affinity of correct and incorrect dNTPs to DNA pol·P/T binary complexes (16); 2) promoting misincorporation during primer extension (8); and 3) influencing exonuclease activity (17). Scheme 1 shows the various fidelity check points employed by DNA pols that minimize dNMP misincorporation. The effect of divalent cations on each of these checkpoints will be addressed in the following sections.
The Effect of Metal Ions on Ground-state Binding Affinity of Incoming dNTPs for pol·P/T Binary Complexes
Different divalent cations can affect the ground-state equilibrium dissociation constant (Kd,g) for incoming dNTPs (Scheme 2). Zhang et al. (40) used a dideoxy P/T containing 2AP opposite an incoming dNTP (the templating position) and measured the Kd,g for dTTP binding, which was 9 μm in the presence of Mg2+. This equilibrium binding assay was based on the observation that, upon formation of the pol·P/T binary complex, 2AP becomes unstacked and exists in a high fluorescence state (41, 42). The addition of dTTP results in 2AP stacking (2AP is translocated from n position to n − 1 position) and quenching of the 2AP fluorescence (41). In the same study (40), there was no change in 2AP fluorescence when it was present at a location one residue downstream from the templating position when dCTP was added opposite dG as the templating base; therefore the Kd,g for binding of the incoming dNTPs could not be determined. In a similar study carried out by Wang and co-workers (16) in the presence of Ca2+, the Kd,g values were determined to be 53 nm and 53 μm for dTTP (correct) and dCTP (incorrect) binding, respectively, opposite 2AP (when 2AP was present in the templating position), suggesting that the identity of the divalent cation cofactor has a profound effect on the ground-state equilibrium dissociation constant. Hariharan et al. (41) also reported a Kd,g value of 31 μm for dTTP binding opposite 2AP with T4 pol in the presence of Mg2+, similar to the value reported for RB69 pol (16). A more comprehensive study on the effect of different divalent cations on ground-state binding affinity for incoming dNTPs was carried out by Vashishtha and Konigsberg (30), where the Kd,g values were measured in the presence of Mg2+, Mn2+, Co2+, and Ca2+ with RB69 pol. Their results showed that the dissociation constants for dTTP binding opposite 2AP in RB69 pol ternary complexes decreased from Mg2+, Co2+, Mn2+, and Ca2+ in that order. A similar pattern of dissociation constants was observed for dCTP binding opposite 2AP. In general, the Kd,g values were substantially lower with Mn2+ as compared with Mg2+ (30). This may be due to the ability of Mn2+, in contrast to Mg2+, to accommodate base pairs other than the regular Watson-Crick base pairs in the nucleotide binding pocket. The possible reasons for this are discussed in the following sections.
SCHEME 2.
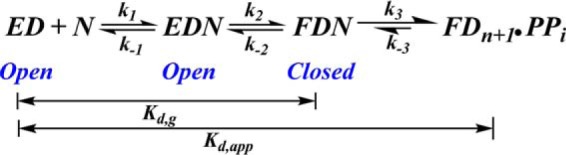
Minimal Kinetic scheme for DNA polymerases depicting the ground-state binding affinity (Kd,g) and apparent binding affinity (Kd,app) for an incoming dNTP. EDN represents the open conformation of the ternary collision complex, whereas FDN represents the closed conformation.
Effect of Metal Ions on Base Selectivity
Numerous studies have shown that various divalent cations affect the base selectivity of pols to different extents (13, 14, 43–46). Substitution of Mg2+ by Mn2+ generally results in a decrease in the fidelity of DNA pols including T4 pol (14, 47), T7 pol (31), E. coli DNA pol I (13, 48), AMV DNA pol (45), and pol β (9). Beckman et al. (48) showed that at very low [Mn2+] (<1 μm), the fidelity of DNA replication is similar to that observed with Mg2+ and that the decrease in fidelity is only observed at elevated [Mn2+] (<100 μm). The possible reasons for this behavior include the binding of Mn2+ to the DNA template at elevated [Mn2+]. Other divalent cations including Co2+ and Ni2+ have been reported to have similar effects on base selectivity as a function of the metal ion concentration (12). Pre-steady-state kinetic studies by Vashishtha and Konigsberg (30) showed that Mn2+ and Co2+ are cofactors for RB69 pol-catalyzed reactions. Surprisingly, the incorporation efficiency for correct incoming dNTPs was higher with Co2+ than with Mg2+ or Mn2+. In contrast, base selectivity was decreased with Co2+ versus Mg2+ but not nearly as much as when Mn2+ replaced Mg2+ (30). Of all the divalent cations tested, Mn2+ is the most highly mutagenic as Mn2+ promotes misincorporation by increasing the rate of incorporation (kpol or Vmax) as well as by decreasing the Kd,app or Km values for incorrect incoming dNTPs (14, 30) (where kpol is the maximum rate of dNMP incorporation, and Kd,app is the apparent equilibrium dissociation constant for [dNTP] that supports the half-maximal rate of dNMP incorporation).
Rare Tautomer Hypothesis for Mutagenesis When Mn2+ Is Present
Occasionally, replicative DNA pols incorporate mismatched nucleotides (49) via the formation of high-energy tautomers (50–52). In fact, Beese and co-workers (53) provided structural evidence for the presence of these rare tautomers using a D598A/F710Y double mutant of a catalytically competent fragment of Bacillus stearothermophilus pol. Beese and co-workers (53) showed that, in the presence of Mn2+, the C/A mismatch adopts a tautomeric cognate base pair shape that is virtually indistinguishable from the canonical, Watson-crick base pair in double-stranded DNA at the insertion site. With Mn2+, the triphosphate tail was properly aligned for catalysis, and the polymerase was in a closed conformation, facilitating the misincorporation. In contrast, in the presence of Mg2+, the C/A mismatch forms a non-cognate wobble base pair, and the polymerase adopts an “ajar” or partially closed conformation, which prevented mismatch incorporation. In addition, the triphosphate tail of the incoming incorrect dNTP was not properly aligned for catalysis, which helped to prevent misincorporation. These results provide a structural rationale for the mutagenic behavior of Mn2+ in this situation.
Effect of Metal Ions on the Exonuclease Activity
Similar to the polymerase active site, the exonuclease site also requires divalent cations to catalyze the 3′-5′-exonuclease activity associated with several DNA pols (5, 30, 32, 35, 54, 55). Divalent cations are required but have a varying effect on the exonuclease activity. Results with RB69 pol (30) showed that, as compared with Mn2+ and Co2+, Mg2+ was most effective in promoting base excision but the exo rates varied only slightly among these three metal ions. Ni2+ on the other hand caused a dramatic decrease in exo activity (33-fold with Ni2+ versus Mg2+). Similarly, the rates of base excision were reported to be nearly identical for E. coli DNA pol I with Mg2+, Mn2+, and Co2+ (11).
All DNA pols Can Utilize Co2+ as Cofactor
In addition to Mg2+, most other DNA pols can also use Mn2+and Co2+, albeit with reduced fidelity (11–13, 15, 30). Pelletier et al. (9) and Egli and co-workers (36) have shown that pol β and Dpo4 are the two known exceptions that cannot be activated by Co2+. In contrast, Vashishtha and Konigsberg (30) showed that these DNA pols can actually catalyze primer extension in the presence of Co2+. The apparent conflicting results can be rationalized based on the fact that different assay conditions were used by each of these groups. For example, Pelletier et al. (9) used blunt-ended DNA with pol β, whereas Vashishtha and Konigsberg (30) used a P/T with a four-base overhang 5′ to the templating base. In the Egli and co-workers (36) experiments, 2 mm DTT was included in their assay with Dpo4, which reduced Co2+ to Co1+ (E0 = −0.33 V for DTT versus −0.28 V for Co2+). DTT was omitted by Vashishtha and Konigsberg (30) in their assays of Dpo4, so cobalt remained as Co2+. Based on these results, it appears that Co2+ can support catalysis for all DNA pols that have been studied to date.
Properties of Divalent Cations That Can Activate DNA Polymerases
Magnesium is in the second row of the periodic table and has 2s electrons that are typically lost when it becomes Mg2+ (56). Together with two 3d orbitals, Mg2+ forms unoccupied stable sp3d2 hybrid orbitals for six coordination ligands. Because of the involvement of 3d orbitals, the coordination bonds are very strong. The average covalent length is 2.09 Å when all high-resolution Mg2+-containing protein structures are compared (6, 29, 57). When carboxylate groups are ligands, the coordination bond lengths are typically reduced slightly relative to non-carboxylate ligands. Manganese is in the third row of the periodic table and has two electrons in 3d orbitals, which are more stable than its 3s electrons. When it loses two 3s electrons, it becomes Mn2+, and leaves two 3d electrons in parallel configurations with two separate 3d orbitals in a high-spin state (56). Upon hybridization in sp3d2 orbitals, the coordination geometry is also octahedral. Due to the involvement of 3s orbitals, the coordination bond lengths increase to 2.22 Å when all the Mn2+-containing high-resolution protein structures are compared (6, 9, 57). The increased distance between adjacent oxygen ligands of Mn2+ (2.22 Å relative to 2.09 Å for Mg2+) permits the Mn2+ coordination octahedron to access ligands that have larger deviations of coordination bond lengths than Mg2+. This is also true for ligands from the triphosphate moieties of mismatched dNTPs.
Proceeding from Mg2+, to Mn2+, to Ca2+, the coordination bond lengths continue to increase from 2.09, 2.22, and 2.40 Å for corresponding metal ion-containing high-resolution protein structures (6) However, increased coordination bond lengths also pose problems for reducing the apical ptO3′-Pα distance in the transition state. In fact, RB69 pol is completely inactive with Ca2+, because the shortest estimated ptO3′-Pα distance estimated would be about 3.3 Å, which is too great for nucleophilic attack of ptO3′ on the Pα center (6). The reason why Mn2+ is catalytically active is that certain Mn2+ coordination bond lengths can be reduced to those of Mg2+ at the expense of increasing the bond lengths of its adjacent ligands, i.e. distortion of the octahedrons (6). Although Mn2+ remains in a high-spin state in most protein structures, it can be converted to low spin in the transition state where two of its 3d electrons occupy the same orbital and the coordination bond lengths are reduced to those of Mg2+ (57). This conversion can accelerate incorporation of any dNMP, correct or incorrect, once they are stabilized in a closed ternary complex. When going from Mn2+ to Co2+, two more electrons are added to the 3d orbital, which results in a low-spin state. As a consequence, the Co2+ coordination bond lengths are reduced to 2.09 Å, almost identical to those of Mg2+, which could account for activation of various DNA pols by Co2+. From Co2+ to Zn2+, the preferred coordination geometry becomes tetrahedral instead of octahedral, notably in Zn2+-binding motifs that involve Cys and His residues. Thus, neither Zn2+ nor Cd2+ can be used by most DNA pols as catalytic metal ions. pol β (9) and B. stearothermophilus DNA pol I large fragment3 are the only two DNA pols that can be activated by Zn2+ and Cd2+, but the reason for this is not known.
The ability to reduce the pKa of bound water is very similar for Mg2+ and Mn2+ but is considerably higher for Co2+, Ni2+, Zn2+, and Cd2+ (Table 2). Based on this property alone, Co2+, Ni2+, Zn2+, and Cd2+ should be more effective as cofactors than Mg2+ and Mn2+, but this does not agree with the experimental data obtained with nearly all DNA pols (11–13, 15, 30, 36–38, 58), so other issues must be involved. The ionic radii of metal ion A play a crucial role in determining the proximal distance between the 3′-hydroxyl group and α-phosphorous atom of the incoming dNTP as the transition state is approached. The ionic radii of Mn2+, Co2+, Ni2+, and Zn2+ are very close to that of Mg2+ (Table 2), enabling all these metal ions to potentially bring the 3′-hydroxyl group and α-phosphate atom of the incoming dNTP close enough for reaction as opposed to Cd2+ and Ca2+, whose ionic radii are larger than that Mg2+ (56). Despite its ability to lower the pKa of bound water and despite having similar ionic radii to Mg2+, Zn2+ is not able to activate most DNA pols. Based on its properties (Table 2), Ni2+ would be expected to substitute for Mg2+, but it is not clear why Ni2+ is such a poor cofactor for DNA pols despite having similar physical properties as Mg2+.
TABLE 2.
Ionic radii, coordination geometry, and pKa of water molecules coordinated to Mg2+, Mn2+, Co2+, Ni2+, Zn2+, Cd2+, and Ca2+
Td, tetrahedral; Sq, square planar; TBP, trigonal bipyramidal; Oct, octahedral; PBP, pentagonal bipyramid; HBP, hexagonal bipyramidal.
| Metal ion |
|||||||
|---|---|---|---|---|---|---|---|
| Mg2+ | Mn2+ | Co2+ | Ni2+ | Zn2+ | Cd2+ | Ca2+ | |
| Ionic radius (Å) | 0.86 | 0.81 | 0.89 | 0.83 | 0.88 | 0.95 | 1.1 |
| Coordination | Oct | Oct | Oct | Oct | Oct | Oct | Oct |
| Geometry | Td | Td | Td | Td | Tda | Td | PBP |
| Sq | Sq | Sq | TBP | TBP | HBP | ||
| TBP | TBP | ||||||
| pKa of the water molecule | 11.4 | 11.5 | 10.0 | 10.6 | 7.0 | 9.0 | 12.8 |
a Although Zn2+ can form octahedral complexes, the majority of Zn2+ complexes are tetrahedral.
Ca2+ does not support primer extension with DNA pols with the exception of Dpo4 (6). One possible reason for this is the inability of Ca2+ to lower the pKa of the 3′-hydroxyl group of the primer as compared with Mg2+ (12.8 versus 11.4). The ionic radius of Ca2+ is also significantly larger than that of Mg2+ (1.1 Å versus 0.86 Å), which renders Ca2+ ineffective in polarizing the hydroxyl group for nucleophilic attack. Dpo4 is the only DNA polymerase that can be activated by Ca2+, although its ability to act as a cofactor is much reduced as compared with Mg2+ (36).
Mn2+ has been reported to be highly mutagenic, and this behavior can be rationalized based on the fact that Mn2+ is a softer metal ion than Mg2+, suggesting that Mn2+ is more polarizable than Mg2+ (44). In terms of a hexahydrated complex of Mn[H2O]62+and Mg[H2O]62+, there is a greater energy penalty with Mg2+ as compared with Mn2+ when the inner sphere coordination number is changed from 6→5→4, indicating more rigid coordination requirements for Mg2+ complexes, which allows less freedom for mismatched dNTPs to be accessible to the nucleotide binding pocket (44). Transition metal ions such as Mn2+ bind more tightly to carboxylate groups and the triphosphate moiety of dNTPs as compared with Mg2+ (9), which could explain its ability to reduce base selectivity.
To summarize, DNA pols from different families are able to utilize different divalent cations as cofactors to catalyze primer extension. Crystal soaking experiments with pol β have shown that Zn2+ and Cd2+ were active (9), whereas these metal ions were not able to activate RB69 pol for catalysis (30). Ni2+ can support primer extension with all DNA polymerases, albeit with greatly reduced activity except for Dpo4 (36), human pol α (43), and pol β (9). Moreover, Dpo4 is the only polymerase that can utilize Ca2+, although Ca2+ is much less effective than Mg2+ (36). Thus, it seems that the abilities of different metal ions to lower the pKa of the primer's 3′-OH group, the respective coordination geometries, and size are the main but not the only determinants of metal ion activation of DNA pols for catalysis of nucleotidyl transfer and base excision.
This work was supported by the Frank and Suzanne Konigsberg Research Fund. This is the fourth article in the Thematic Minireview series “Metals in Biology 2016: Molecular Basis of Selection of Metals by Enzymes.” The authors declare that they have no conflicts of interest with the contents of this article.
A. K. Vashishtha, J. Wang, and W. H. Konigsberg, unpublished data.
- pol
- polymerase
- AMV
- avian myeloblastosis virus
- 2AP
- 2-aminopurine
- Dpo4
- DNA polymerase IV from Sulfolobus solfataricus
- exo
- exonuclease
- ptO3′
- terminal 3′-OH group on the primer
- RB69 pol
- bacteriophage RB69 DNA polymerase
- tm
- triple mutant of RB69 pol containing L561A, S565G, and Y567A mutations in the active site
- P/T
- primer-template
- dUpNpp
- 2′-deoxyuridine 5′-(α,β-imido)triphosphate.
References
- 1. Drake J. W. (1969) Comparative rates of spontaneous mutation. Nature 221, 1132. [DOI] [PubMed] [Google Scholar]
- 2. Goodman M. F., and Tippin B. (2000) The expanding polymerase universe. Nat. Rev. Mol. Cell Biol. 1, 101–109 [DOI] [PubMed] [Google Scholar]
- 3. Filée J., Forterre P., Sen-Lin T., and Laurent J. (2002) Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54, 763–773 [DOI] [PubMed] [Google Scholar]
- 4. Vashishtha A. K., and Kuchta R. D. (2016) Effects of acyclovir, Foscarnet, and ribonucleotides on herpes simplex virus-1 DNA polymerase: mechanistic insights and a novel mechanism for preventing stable incorporation of ribonucleotides into DNA. Biochemistry 55, 1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vashishtha A. K., and Kuchta R. D. (2015) Polymerase and exonuclease activities in herpes simplex virus type 1 DNA polymerase are not highly coordinated. Biochemistry 54, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xia S., Wang M., Blaha G., Konigsberg W. H., and Wang J. (2011) Structural insights into complete metal ion coordination from ternary complexes of B family RB69 DNA polymerase. Biochemistry 50, 9114–9124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Snow E. T., Xu L. S., and Kinney P. L. (1993) Effects of nickel ions on polymerase activity and fidelity during DNA replication in vitro. Chem. Biol. Interact 88, 155–173 [DOI] [PubMed] [Google Scholar]
- 8. Vaisman A., Ling H., Woodgate R., and Yang W. (2005) Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24, 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pelletier H., Sawaya M. R., Wolfle W., Wilson S. H., and Kraut J. (1996) A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry 35, 12762–12777 [DOI] [PubMed] [Google Scholar]
- 10. Irimia A., Loukachevitch L. V., Eoff R. L., Guengerich F. P., and Egli M. (2010) Metal-ion dependence of the active-site conformation of the translesion DNA polymerase Dpo4 from Sulfolobus solfataricus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sirover M. A., Dube D. K., and Loeb L. A. (1979) On the fidelity of DNA replication: metal activation of Escherichia coli DNA polymerase I. J. Biol. Chem. 254, 107–111 [PubMed] [Google Scholar]
- 12. Sirover M. A., and Loeb L. A. (1977) On the fidelity of DNA replication. Effect of metal activators during synthesis with avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 252, 3605–3610 [PubMed] [Google Scholar]
- 13. Miyaki M., Murata I., Osabe M., and Ono T. (1977) Effect of metal cations on misincorporation by E. coli DNA polymerases. Biochem. Biophys. Res. Commun. 77, 854–860 [DOI] [PubMed] [Google Scholar]
- 14. Goodman M. F., Keener S., Guidotti S., and Branscomb E. W. (1983) On the enzymatic basis for mutagenesis by manganese. J. Biol. Chem. 258, 3469–3475 [PubMed] [Google Scholar]
- 15. Sirover M. A., and Loeb L. A. (1976) Infidelity of DNA synthesis in vitro: screening for potential metal mutagens or carcinogens. Science 194, 1434–1436 [DOI] [PubMed] [Google Scholar]
- 16. Lee H. R., Wang M., and Konigsberg W. (2009) The reopening rate of the fingers domain is a determinant of base selectivity for RB69 DNA polymerase. Biochemistry 48, 2087–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villani G., Tanguy Le Gac N., Wasungu L., Burnouf D., Fuchs R. P., and Boehmer P. E. (2002) Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase. Nucleic Acids Res. 30, 3323–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia S., and Konigsberg W. H. (2014) RB69 DNA polymerase structure, kinetics, and fidelity. Biochemistry 53, 2752–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beard W. A., and Wilson S. H. (2014) Structure and mechanism of DNA polymerase β. Biochemistry 53, 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sobol R. W., and Wilson S. H. (2001) Mammalian DNA β-polymerase in base excision repair of alkylation damage. Prog. Nucleic Acid Res. Mol. Biol. 68, 57–74 [DOI] [PubMed] [Google Scholar]
- 21. Beard W. A., and Wilson S. H. (2000) Structural design of a eukaryotic DNA repair polymerase: DNA polymerase β. Mutat Res. 460, 231–244 [DOI] [PubMed] [Google Scholar]
- 22. Yang L., Arora K., Beard W. A., Wilson S. H., and Schlick T. (2004) Critical role of magnesium ions in DNA polymerase β's closing and active site assembly. J. Am. Chem. Soc. 126, 8441–8453 [DOI] [PubMed] [Google Scholar]
- 23. Bakhtina M., Lee S., Wang Y., Dunlap C., Lamarche B., and Tsai M. D. (2005) Use of viscogens, dNTPαS, and rhodium(III) as probes in stopped-flow experiments to obtain new evidence for the mechanism of catalysis by DNA polymerase β. Biochemistry 44, 5177–5187 [DOI] [PubMed] [Google Scholar]
- 24. Nakamura T., Zhao Y., Yamagata Y., Hua Y. J., and Yang W. (2012) Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao Y., and Yang W. (2016) Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science 352, 1334–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia S., Wang J., and Konigsberg W. H. (2013) DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J. Am. Chem. Soc. 135, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doublié S., Tabor S., Long A. M., Richardson C. C., and Ellenberger T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 28. Johnson S. J., Taylor J. S., and Beese L. S. (2003) Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 100, 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jain R., Rajashankar K. R., Buku A., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2014) Crystal structure of yeast DNA polymerase ϵ catalytic domain. PLoS ONE 9, e94835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vashishtha A. K., and Konigsberg W. H. (2016) Effect of different divalent cations on the kinetics and fidelity of RB69 DNA polymerase. Biochemistry 55, 2661–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tabor S., and Richardson C. C. (1989) Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 86, 4076–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hori K., Mark D. F., and Richardson C. C. (1979) Deoxyribonucleic acid polymerase of bacteriophage T7: characterization of the exonuclease activities of the gene 5 protein and the reconstituted polymerase. J. Biol. Chem. 254, 11598–11604 [PubMed] [Google Scholar]
- 33. Kuchta R. D., Mizrahi V., Benkovic P. A., Johnson K. A., and Benkovic S. J. (1987) Kinetic mechanism of DNA polymerase I (Klenow). Biochemistry 26, 8410–8417 [DOI] [PubMed] [Google Scholar]
- 34. Venkitaraman A. R. (1989) Use of modified T7 DNA polymerase (sequenase version 2.0) for oligonucleotide site-directed mutagenesis. Nucleic Acids Res. 17, 3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joyce C. M. (1989) How DNA travels between the separate polymerase and 3′-5′-exonuclease sites of DNA polymerase I (Klenow fragment). J. Biol. Chem. 264, 10858–10866 [PubMed] [Google Scholar]
- 36. Irimia A., Zang H., Loukachevitch L. V., Eoff R. L., Guengerich F. P., and Egli M. (2006) Calcium is a cofactor of polymerization but inhibits pyrophosphorolysis by the Sulfolobus solfataricus DNA polymerase Dpo4. Biochemistry 45, 5949–5956 [DOI] [PubMed] [Google Scholar]
- 37. Sirover M. A., and Loeb L. A. (1976) Metal-induced infidelity during DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 73, 2331–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sirover M. A., and Loeb L. A. (1976) Metal activation of DNA synthesis. Biochem. Biophys. Res. Commun. 70, 812–817 [DOI] [PubMed] [Google Scholar]
- 39. Seal G., Shearman C. W., and Loeb L. A. (1979) On the fidelity of DNA replication: studies with human placenta DNA polymerases. J. Biol. Chem. 254, 5229–5237 [PubMed] [Google Scholar]
- 40. Zhang H., Cao W., Zakharova E., Konigsberg W., and De La Cruz E. M. (2007) Fluorescence of 2-aminopurine reveals rapid conformational changes in the RB69 DNA polymerase-primer/template complexes upon binding and incorporation of matched deoxynucleoside triphosphates. Nucleic Acids Res. 35, 6052–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hariharan C., Bloom L. B., Helquist S. A., Kool E. T., and Reha-Krantz L. J. (2006) Dynamics of nucleotide incorporation: snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry 45, 2836–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frey M. W., Sowers L. C., Millar D. P., and Benkovic S. J. (1995) The nucleotide analog 2-aminopurine as a spectroscopic probe of nucleotide incorporation by the Klenow fragment of Escherichia coli polymerase I and bacteriophage T4 DNA polymerase. Biochemistry 34, 9185–9192 [DOI] [PubMed] [Google Scholar]
- 43. Chin Y. E., Snow E. T., Cohen M. D., and Christie N. T. (1994) The effect of divalent nickel (Ni2+) on in vitro DNA replication by DNA polymerase α. Cancer Res. 54, 2337–2341 [PubMed] [Google Scholar]
- 44. Bock C. W., Katz A. K., Markham G. D., and Glusker J. P. (1999) Manganese as a replacement for magnesium and zinc: functional comparison of the divalent ions. J. Am. Chem. Soc. 121, 7360–7372 [Google Scholar]
- 45. Dube D. K., and Loeb L. A. (1975) Manganese as a mutagenic agent during in vitro DNA synthesis. Biochem. Biophys. Res. Commun. 67, 1041–1046 [DOI] [PubMed] [Google Scholar]
- 46. Jin Y. H., Clark A. B., Slebos R. J., Al-Refai H., Taylor J. A., Kunkel T. A., Resnick M. A., and Gordenin D. A. (2003) Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34, 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hays H., and Berdis A. J. (2002) Manganese substantially alters the dynamics of translesion DNA synthesis. Biochemistry 41, 4771–4778 [DOI] [PubMed] [Google Scholar]
- 48. Beckman R. A., Mildvan A. S., and Loeb L. A. (1985) On the fidelity of DNA replication: manganese mutagenesis in vitro. Biochemistry 24, 5810–5817 [DOI] [PubMed] [Google Scholar]
- 49. Kunkel T. A., and Bebenek K. (2000) DNA replication fidelity. Annu. Rev. Biochem. 69, 497–529 [DOI] [PubMed] [Google Scholar]
- 50. Harris V. H., Smith C. L., Cummins W. J., Hamilton A. L., Adams H., Dickman M., Hornby D. P., and Williams D. M. (2003) The effect of tautomeric constant on the specificity of nucleotide incorporation during DNA replication: support for the rare tautomer hypothesis of substitution mutagenesis. J. Mol. Biol. 326, 1389–1401 [DOI] [PubMed] [Google Scholar]
- 51. Topal M. D., and Fresco J. R. (1976) Complementary base pairing and the origin of substitution mutations. Nature 263, 285–289 [DOI] [PubMed] [Google Scholar]
- 52. Bebenek K., Pedersen L. C., and Kunkel T. A. (2011) Replication infidelity via a mismatch with Watson-Crick geometry. Proc. Natl. Acad. Sci. U.S.A. 108, 1862–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W., Hellinga H. W., and Beese L. S. (2011) Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 17644–17648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Doetsch P. W., Chan G. L., and Haseltine W. A. (1985) T4 DNA polymerase (3′-5′) exonuclease, an enzyme for the detection and quantitation of stable DNA lesions: the ultraviolet light example. Nucleic Acids Res. 13, 3285–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kunkel T. A., and Soni A. (1988) Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-γ. J. Biol. Chem. 263, 4450–4459 [PubMed] [Google Scholar]
- 56. Cotton F. A., Wilkinson G., Murillo C. A., and Bochmann M. (1999) Advanced Inorganic Chemistry, 6th Ed., pp. 692–854, John Wiley & Sons, New York [Google Scholar]
- 57. Harding M. M. (2006) Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr. D Biol. Crystallogr. 62, 678–682 [DOI] [PubMed] [Google Scholar]
- 58. Mildvan A. S., and Loeb L. A. (1979) The role of metal ions in the mechanisms of DNA and RNA polymerases. CRC Crit. Rev. Biochem. 6, 219–244 [DOI] [PubMed] [Google Scholar]


