Abstract
Nα-terminal arginylation (Nt-arginylation) of proteins is mediated by the Ate1 arginyltransferase (R-transferase), a component of the Arg/N-end rule pathway. This proteolytic system recognizes proteins containing N-terminal degradation signals called N-degrons, polyubiquitylates these proteins, and thereby causes their degradation by the proteasome. The definitively identified (“canonical”) residues that are Nt-arginylated by R-transferase are N-terminal Asp, Glu, and (oxidized) Cys. Over the last decade, several publications have suggested (i) that Ate1 can also arginylate non-canonical N-terminal residues; (ii) that Ate1 is capable of arginylating not only α-amino groups of N-terminal residues but also γ-carboxyl groups of internal (non-N-terminal) Asp and Glu; and (iii) that some isoforms of Ate1 are specific for substrates bearing N-terminal Cys residues. In the present study, we employed arrays of immobilized 11-residue peptides and pulse-chase assays to examine the substrate specificity of mouse R-transferase. We show that amino acid sequences immediately downstream of a substrate's canonical (Nt-arginylatable) N-terminal residue, particularly a residue at position 2, can affect the rate of Nt-arginylation by R-transferase and thereby the rate of degradation of a substrate protein. We also show that the four major isoforms of mouse R-transferase have similar Nt-arginylation specificities in vitro, contrary to the claim about the specificity of some Ate1 isoforms for N-terminal Cys. In addition, we found no evidence for a significant activity of the Ate1 R-transferase toward previously invoked non-canonical N-terminal or internal amino acid residues. Together, our results raise technical concerns about earlier studies that invoked non-canonical arginylation specificities of Ate1.
Keywords: proteasome, protein degradation, ubiquitin, ubiquitin ligase, ubiquitylation (ubiquitination), Ate1, arginyltransferase, N-end rule
Introduction
The N-end rule pathway is a set of intracellular proteolytic systems whose unifying feature is the ability to recognize and polyubiquitylate proteins containing N-terminal (Nt)2 degradation signals called N-degrons, thereby causing the processive degradation of these proteins by the proteasome (Fig. 1, A and B) (1–13). Recognition components of the N-end rule pathway are called N-recognins. In eukaryotes, N-recognins are E3 ubiquitin (Ub) ligases that can target N-degrons. Some N-recognins contain several substrate-binding sites and thereby can recognize (bind to) not only N-degrons but also specific internal (non-N-terminal) degradation signals (Fig. 1A) (14–18). The main determinant of a protein's N-degron is either an unmodified or chemically modified N-terminal residue. Another determinant of an N-degron is an internal Lys residue(s). It functions as a site of protein polyubiquitylation, is often engaged stochastically (in competition with other “eligible” lysines), and tends to be located in a conformationally disordered region (2, 9, 19, 20). Bacteria also contain the N-end rule pathway, but Ub-independent versions of it (21–26).
FIGURE 1.
The mammalian N-end rule pathway and isoforms of the mouse Ate1 R-transferase. See the Introduction for references and descriptions of the pathway's mechanistic aspects and functions. Amino acid residues are denoted by single-letter abbreviations. A yellow oval denotes the rest of a protein substrate. A, the mammalian Arg/N-end rule pathway. It targets proteins for degradation through their specific unacetylated N-terminal residues. Primary, secondary, and tertiary refer to mechanistically distinct classes of destabilizing N-terminal residues. Ntan1 and Ntaq1 are N-terminal amidases (Nt-amidases) that convert the tertiary destabilizing N-terminal residues Asn and Gln to Asp and Glu, respectively. The Ate1 R-transferase (arginyltransferase or Arg-tRNA-protein transferase) conjugates Arg, a primary destabilizing residue, to N-terminal Asp, Glu, and (oxidized) Cys (the “canonical” N-terminal residues). Type 1 and type 2 refer, respectively, to two sets of primary destabilizing N-terminal residues, basic (Arg, Lys, and His) and bulky hydrophobic (Leu, Phe, Trp, Tyr, Ile, and also Met, if the latter is followed by a bulky hydrophobic residue (Φ)). These sets of N-terminal residues are recognized by distinct substrate-binding sites of N-recognins (the pathway's E3 ubiquitin ligases) Ubr1, Ubr2, Ubr4, and Ubr5. B, the mammalian Ac/N-end rule pathway. It targets proteins through their Nα-terminally acetylated (Nt-acetylated) residues. The red arrow on the left indicates the cotranslational removal of the N-terminal Met residue by Met-aminopeptidases (MetAPs). N-terminal Met is retained if a residue at position 2 is larger than Val. See the Introduction for references and additional aspects of the Ac/N-end rule pathway. C, the exons, including alternative exons (1A/1B and 7A/7B) of the mouse Ate1 gene, which encodes isoforms of R-transferase. The deduced lengths (in amino acid residues) of the encoded Ate1 exons are indicated at the top. D, mouse R-transferase isoforms that are produced through alternative splicing of Ate1 pre-mRNA. The terminology of these isoforms (Ate11A7A, Ate11A7B, Ate11B7A, Ate11B7B, Ate11A7AB, and Ate11B7AB) (9, 79) is based on the presence or absence of the alternative exons 1A/1B and 7A/7B. Ate11A7AB and Ate11B7AB are minor isoforms that result from the retention of both variants of exon 7 (79).
Regulated degradation of proteins and their natural fragments by the N-end rule pathway has been shown to mediate a strikingly broad range of biological functions, including the sensing of heme (27), nitric oxide (NO), oxygen (4, 7, 28, 29), and short peptides (14, 15, 30–35); the control, through subunit-selective degradation, of the input stoichiometries of subunits in oligomeric protein complexes (5, 36); the elimination of misfolded and otherwise abnormal proteins (6, 17, 18, 37–41); the degradation of specific proteins after their translocation to the cytosol from membrane-enclosed compartments such as mitochondria (42); the regulation of apoptosis and repression of neurodegeneration (43–46); the regulation of DNA repair, transcription, replication, and chromosome cohesion/segregation (14, 16, 47–52); the regulation of G proteins, cytoskeletal proteins, autophagy, peptide import, meiosis, immunity, circadian rhythms, fat metabolism, cell migration, cardiovascular development, spermatogenesis, and neurogenesis (4, 14, 53–59); the functioning of adult organs, including the brain, muscle, testis, and pancreas (57, 60–64); and the regulation of leaf and shoot development, leaf senescence, oxygen/NO sensing, and many other processes in plants (Fig. 1, A and B) (7–13, 22, 28, 65–69).
In eukaryotes, the N-end rule pathway consists of two branches. One branch, called the Ac/N-end rule pathway, targets proteins for degradation through their Nα-terminally acetylated (Nt-acetylated) residues (Fig. 1B) (5, 6, 13, 36, 56, 70, 71). Degradation signals and E3 Ub ligases of the Ac/N-end rule pathway are called Ac/N-degrons and Ac/N-recognins, respectively. Nt-acetylation of cellular proteins is apparently irreversible, in contrast to cycles of acetylation-deacetylation of proteins' internal Lys residues. About 90% of human proteins are cotranslationally Nt-acetylated by ribosome-associated Nt-acetylases (72). Posttranslational Nt-acetylation takes place as well. Ac/N-degrons are present in many, possibly most, Nt-acetylated proteins (Fig. 1B). Natural Ac/N-degrons are regulated through their reversible shielding in cognate protein complexes (6, 36, 56).
The pathway's other branch, called the Arg/N-end rule pathway, targets specific unacetylated N-terminal residues (Fig. 1A) (4, 6, 29, 42–44, 54, 73–75). The “primary” destabilizing N-terminal residues Arg, Lys, His, Leu, Phe, Tyr, Trp, and Ile are directly recognized by N-recognins. The unacetylated N-terminal Met, if it is followed by a bulky hydrophobic (Φ) residue, also acts as a primary destabilizing residue (Fig. 1A) (6). In contrast, the unacetylated N-terminal Asn, Gln, Asp, and Glu (as well as Cys, under some metabolic conditions) are destabilizing due to their preliminary enzymatic modifications, which include N-terminal deamidation (Nt-deamidation) of Asn and Gln and Nt-arginylation of Asp, Glu, and oxidized Cys (Fig. 1A) (7–9, 76). In the yeast Saccharomyces cerevisiae, the Arg/N-end rule pathway is mediated by the Ubr1 N-recognin, a 225-kDa RING-type E3 Ub ligase and a part of the multisubunit targeting complex comprising the Ubr1-Rad6 and Ufd4-Ubc4/5 E2-E3 holoenzymes (9, 77). In multicellular eukaryotes, several E3 Ub ligases, including Ubr1, function as N-recognins of the Arg/N-end rule pathway (8, 9) (Fig. 1A).
Nt-arginylation of proteins is mediated by the Ate1-encoded arginyltransferase (Arg-tRNA-protein transferase; R-transferase) (Fig. 1A) (3, 4, 27, 78–81). The “canonical” (definitively identified and characterized) Nt-arginylatable residues are N-terminal Asp, Glu, and Cys, the latter after its oxidation to Cys-sulfinate or Cys-sulfonate through reactions that involve NO, oxygen, and/or N-terminal Cys oxidases (4, 7, 27, 78, 79, 82, 83). Because N-terminal Cys has to undergo two consecutive modifications, an oxidation and Nt-arginylation, before the recognition of a corresponding N-degron by N-recognins, Cys is classed as a “tertiary” destabilizing residue (Fig. 1A).
In mammals such as mice, there are at least six isoforms of R-transferase, produced through alternative splicing of the Ate1 pre-mRNA (Fig. 1, C and D) (9, 78, 79). At present, about 20 mammalian proteins, including natural protein fragments, have been identified as either constitutively or conditionally short-lived physiological substrates of the Ate1 R-transferase and the post-arginylation part of the Arg/N-end rule pathway. All of these substrates have in common the presence of a canonical (Nt-arginylatable) N-terminal residue, either Asp, Glu, or Cys (Fig. 1A) (4, 29, 42–44, 49, 50, 73, 84, 85). In addition, there are hundreds of mammalian proteins, including natural protein fragments, that are predicted to be Nt-arginylated, based on the presence, in a protein or its natural fragment, of N-terminal Asp, Glu, or Cys (7, 43, 44, 49, 73, 86).
Several publications over the last decade, largely by the laboratory of A. Kashina, have reported that Ate1 R-transferase can also arginylate, to a significant extent, a variety of “non-canonical” (non-Asp, non-Glu, non-Cys) N-terminal residues in cellular proteins (87–92). The same group described evidence that some isoforms of R-transferase were specific for substrates bearing N-terminal Cys (93). Other recent publications by the same laboratory suggested, on the basis of MS analyses, that the Ate1 R-transferase is capable of arginylating not only α-amino groups of specific N-terminal residues but also γ-carboxyl groups of internal (non-N-terminal) Asp or Glu (94, 95). This suggestion followed a report, by another group, that described a modified 13-residue neurotensin hormone that appeared to be arginylated at its internal Glu-4 residue (96). Thus far, studies by other laboratories have failed to find evidence for either a significant arginylation of non-canonical N-terminal amino acid residues or a significant arginylation of internal residues (85, 97–99).
In the present work, we used purified isoforms of the mouse Ate1 R-transferase, arrays of C-terminally immobilized synthetic peptides, and pulse-chase degradation assays to address the substrate specificity of arginylation by mouse Ate1. We show here that a sequence immediately downstream of a substrate's canonical (Nt-arginylatable) N-terminal residue, and particularly a residue at position 2, can significantly affect the rate of Nt-arginylation by R-transferase and thereby the rate of degradation of a substrate protein. We also show that the four major isoforms of the mouse R-transferase have similar substrate specificities in vitro, contrary to the earlier claim that some isoforms of Ate1 are specific for substrates bearing N-terminal Cys (93). Furthermore, and also contrary to the other cited reports (87–89, 94, 100, 101), we found no evidence for a significant activity of the Ate1 R-transferase toward the previously suggested non-canonical residues.
A caveat to otherwise direct contradictions between our findings and those of the earlier studies is that our results were produced in vitro (with purified Ate1 R-transferase and peptide arrays), whereas most reports about non-canonical arginylation of cellular proteins have been about analyses of natural, in vivo-produced proteins (87–92, 94–96).
Results and Discussion
Use of CelluSpotsTM Peptide Arrays to Analyze the Substrate Specificity of R-transferase
CelluSpots arrays are produced by the synthesis of C-terminally immobilized peptides on a modified cellulose support that is dissolvable in a trifluoroacetic acid solution, thereby allowing the synthesized peptides to be spotted onto coated microscope slides. This technology makes it possible to produce a number of virtually identical copies of a peptide array from a single set of peptide-synthesizing reactions (102). CelluSpots arrays have been successfully used to examine the specificity of histone-modifying enzymes for amino acid sequences in histones; to analyze, in vitro, the binding specificity of antibodies; to characterize specific binding of SH2 (Src homology 2 domain) protein regions to phosphotyrosine-containing peptides; and to survey the specificities of phosphokinases and kinase inhibitors (103–105).
To analyze the substrate specificity of the mouse Ate1 R-transferase, we performed 14C-arginylation assays using identical CelluSpots peptide arrays, each of which comprised 96 immobilized peptides that were largely 11 residues long (two peptides were 12 residues long) (Fig. 2 and Table 1). To the best of our knowledge, CelluSpots have not been previously used to study arginylation. The unlimited sequence diversity of synthetic peptides, the availability of virtually identical copies of a given peptide array, and technically straightforward, reproducible conditions of arginylation assays make CelluSpots a particularly helpful method for analyzing the in vitro specificity of Ate1 R-transferase isoforms (3, 78–81).
FIGURE 2.
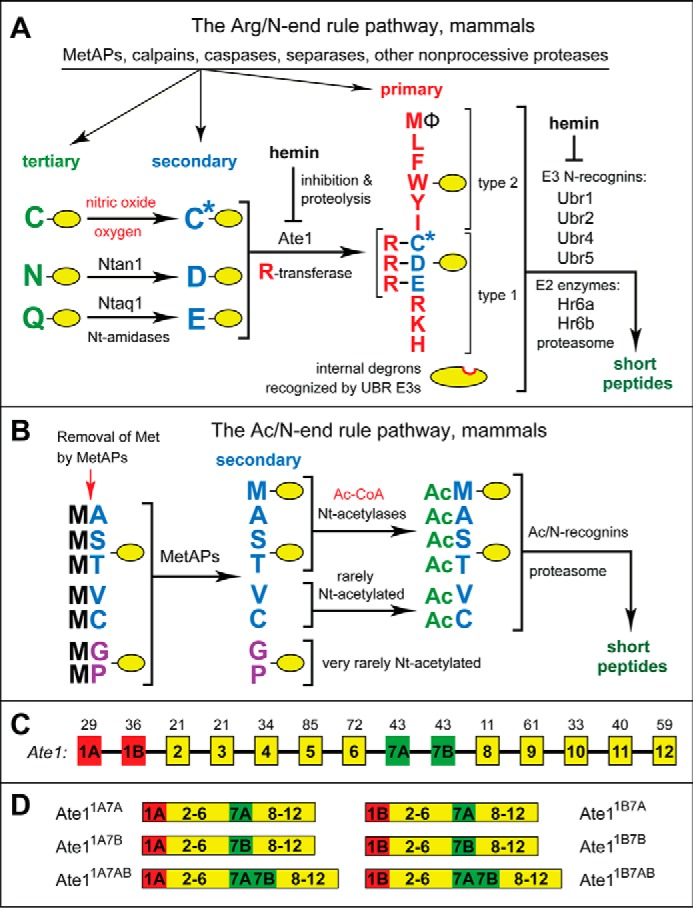
Design and validation of CelluSpots arginylation assays. A, layout of two virtually identical copies of a CelluSpots 96-spot array of C-terminally immobilized 11-residue synthetic peptides that are produced and spotted onto coated microscope slides (102). Each peptide spot is denoted by a letter (A–D) and a number (1–24). For the complete sequences of these 96 peptides, see Table 1. For the five N-terminal residues of these 11-residue peptides, see the bottom of B. Each individual CelluSpots array (left panel versus right panel) was “bounded” through the use of a hydrophobic pen, making it possible for two arrays on a slide to be used for two separate enzymatic reactions. B, rectangle on the left, an arginylation assay was carried out with a CelluSpots peptide array containing a complete reaction mix (including [14C]Arg; see “Experimental Procedures”) save for the absence of Ate1 R-transferase. Note the negligible (essentially undetectable) levels of [14C]Arg incorporation under these conditions. B, rectangle on the right, the otherwise identical assay was carried out in the presence of purified mouse Ate11B7A R-transferase. Note that “negative” peptide spots, containing 11-residue peptides bearing non-canonical (non-Asp, non-Glu, and non-Cys) N-terminal residues, exhibited negligible (undetectable) levels of [14C]Arg incorporation at this level of autoradiographic exposure. Significantly longer autoradiographic exposures made background levels of [14C]Arg incorporation detectable in the presence of Ate1 but still negligible (undetectable) in the absence of Ate1 (left). See “Results” for possible causes of very low but detectable background level [14C]Arg incorporation in spots of peptides bearing non-canonical N-terminal residues. The bottom of B recapitulates the arrangement of 96 peptide spots in a rectangle of A and, in addition, cites the first five residues of each 11-residue peptide. Spot numbers are in green. The canonical (Nt-arginylatable) N-terminal residues Asp, Glu, and Cys (the latter a precursor of Nt-arginylatable oxidized Cys) are in red. Table 1 cites complete sequences of 11-residue peptides. (Two peptides, at C1 and C5, were 12 residues long.)
TABLE 1.
Location and complete sequences of CelluSpots array peptides (Fig. 2)
| Spot | Sequence | Name | N terminus | |
|---|---|---|---|---|
| 1 | A1 | DGGSGFGPASG | DG-GSG | Amine |
| 2 | A2 | DAGSGFGPASG | DA-GSG | Amine |
| 3 | A3 | DSGSGFGPASG | DS-GSG | Amine |
| 4 | A4 | DTGSGFGPASG | DT-GSG | Amine |
| 5 | A5 | DCGSGFGPASG | DC-GSG | Amine |
| 6 | A6 | DVGSGFGPASG | DV-GSG | Amine |
| 7 | A7 | DLGSGFGPASG | DL-GSG | Amine |
| 8 | A8 | DIGSGFGPASG | DI-GSGS | Amine |
| 9 | A9 | DMGSGFGPASG | DM-GSG | Amine |
| 10 | A10 | DPGSGFGPASG | DP-GSG | Amine |
| 11 | A11 | DFGSGFGPASG | DF-GSG | Amine |
| 12 | A12 | DYGSGFGPASG | DY-GSG | Amine |
| 13 | A13 | RGGSGFGPASG | RG-GSG | Amine |
| 14 | A14 | RAGSGFGPASG | RA-GSG | Amine |
| 15 | A15 | RSGSGFGPASG | RS-GSG | Amine |
| 16 | A16 | RTGSGFGPASG | RT-GSG | Amine |
| 17 | A17 | RCGSGFGPASG | RC-GSG | Amine |
| 18 | A18 | RVGSGFGPASG | RV-GSG | Amine |
| 19 | A19 | RLGSGFGPASG | RL-GSG | Amine |
| 20 | A20 | RIGSGFGPASG | RI-GSG | Amine |
| 21 | A21 | RMGSGFGPASG | RM-GSG | Amine |
| 22 | A22 | RPGSGFGPASG | RP-GSG | Amine |
| 23 | A23 | RFGSGFGPASG | RF-GSG | Amine |
| 24 | A24 | RYGSGFGPASG | RY-GSG | Amine |
| 25 | B1 | DWGSGFGPASG | DW-GSG | Amine |
| 26 | B2 | DDGSGFGPASG | DD-GSG | Amine |
| 27 | B3 | DEGSGFGPASG | DE-GSG | Amine |
| 28 | B4 | DNGSGFGPASG | DN-GSG | Amine |
| 29 | B5 | DQGSGFGPASG | DQ-GSG | Amine |
| 30 | B6 | DHGSGFGPASG | DH-GSG | Amine |
| 31 | B7 | DKGSGFGPASG | DK-GSG | Amine |
| 32 | B8 | DRGSGFGPASG | DR-GSG | Amine |
| 33 | B9 | DGPSGFGPASG | DGP3-GSG | Amine |
| 34 | B10 | DGGPGFGPASG | DGP4-GSG | Amine |
| 35 | B11 | DGGSPFGPASG | DGP5-GSG | Amine |
| 36 | B12 | DGGSGPGPASG | DGP6-GSG | Amine |
| 37 | B13 | RWGSGFGPASG | RW-GSGS | Amine |
| 38 | B14 | RDGSGFGPASG | RD-GSGS | Amine |
| 39 | B15 | REGSGFGPASG | RE-GSGS | Amine |
| 40 | B16 | RNGSGFGPASG | RN-GSGS | Amine |
| 41 | B17 | RQGSGFGPASG | RQ-GSGS | Amine |
| 42 | B18 | RHGSGFGPASG | RH-GSGS | Amine |
| 43 | B19 | RKGSGFGPASG | RK-GSGS | Amine |
| 44 | B20 | RRGSGFGPASG | RR-GSGS | Amine |
| 45 | B21 | RGPSGFGPASG | RGP3-GSG | Amine |
| 46 | B22 | RGGPGFGPASG | RGP4-GSG | Amine |
| 47 | B23 | RGGSPFGPASG | RGP5-GSG | Amine |
| 48 | B24 | RGGSGPGPASG | RGP6-GSG | Amine |
| 49 | C1 | RCKGLAGLPASC | Rgs4 Arg | Amine |
| 50 | C2 | DKGLAGLPASC | Rgs4 C2D | Amine |
| 51 | C3 | CKGLAGLPASC | Rgs4 | Amine |
| 52 | C4 | C(sulfonate)KGLAGLPASC | Rgs4, N-terminal Cys-sulfonate. | Amine |
| 53 | C5 | RMDPLNDNIATL | Myh9, Arg | Amine |
| 54 | C6 | MDPLNDNIATL | Myh9 | Amine |
| 55 | C7 | M(sulfoxide)DPLNDNIATL | Myh9, Met(sulfoxide) | Amine |
| 56 | C8 | M(sulfone)DPLNDNIATL | Myh9, Met(sulfone) | Amine |
| 57 | C9 | ALYENKPRRPY | NT1–11, Q1A | Acetyl |
| 58 | C10 | ALYDNKPRRPY | NT1–11, Q1A,E4D | Acetyl |
| 59 | C11 | ALYNNKPRRPY | NT1–11, Q1A,E4N | Acetyl |
| 60 | C12 | ALYQNKPRRPY | NT1–11, Q1A,E4Q | Acetyl |
| 61 | C13 | DSGSGFGPASG | GSG, G1D | Amine |
| 62 | C14 | DDGSGFGPASG | GSG, G1D,A2D | Amine |
| 63 | C15 | DKGSGFGPASG | GSG, G1D,A2K | Amine |
| 64 | C16 | DPGSGFGPASG | GSG, G1D,A2P | Amine |
| 65 | C17 | CSGSGFGPASG | GSG, G1C | Amine |
| 66 | C18 | CDGSGFGPASG | GSG, G1C,A2D | Amine |
| 67 | C19 | CKGSGFGPASG | GSG, G1C,A2K | Amine |
| 68 | C20 | CPGSGFGPASG | GSG, G1C,A2P | Amine |
| 69 | C21 | ESGSGFGPASG | GSG, G1C | Amine |
| 70 | C22 | EDGSGFGPASG | GSG, G1E,A2D | Amine |
| 71 | C23 | EKGSGFGPASG | GSG, G1E,A2K | Amine |
| 72 | C24 | EPGSGFGPASG | GSG, G1E,A2P | Amine |
| 73 | D1 | DKGLAGLPASC | Rgs4, C2D | Amine |
| 74 | D2 | MCDRKAVIKNA | Dynein, cytoplasmic, light peptide | Amine |
| 75 | D3 | LEVLNFFNNQI | Ras suppressor protein 1 | Amine |
| 76 | D4 | PVLCFTQYEES | Properdin factor (complement) | Amine |
| 77 | D5 | AWGKIGGHGAE | Hemoglobin α1 chain | Amine |
| 78 | D6 | KPVYDELFYTL | Mpast2, an EH domain-containing protein | Amine |
| 79 | D7 | GDDGAEYVVES | Similar to GAPDH | Amine |
| 80 | D8 | NSALQCLSNTA | Ubiquitin-specific protease 4 | Amine |
| 81 | D9 | FYNELRVAPEE | β-Actin | Amine |
| 82 | D10 | HSQAVEELADQ | Myosin, heavy polypeptide 9, non-muscle isoform | Amine |
| 83 | D11 | VLVGDVKDRVA | Phosphoribosyl pyrophosphate synthase 1 (Prps1) | Amine |
| 84 | D12 | DKGLAGLPASC | Rgs4, C2D | Amine |
| 85 | D13 | DRGSGFGPASG | DR1-GSG | Amine |
| 86 | D14 | DRRRRFGPASG | DR4-GSG | Amine |
| 87 | D15 | RRGSGFGPASG | RR1-GSG | Amine |
| 88 | D16 | RRRRRFGPASG | RR4-GSG | Amine |
| 89 | D17 | DDGSGFGPASG | DD1-GSG | Amine |
| 90 | D18 | DDDDDFGPASG | DD4-GSG | Amine |
| 91 | D19 | RDGSGFGPASG | RD1-GSG | Amine |
| 92 | D20 | RDDDDFGPASG | RD4-GSG | Amine |
| 93 | D21 | DAGSGFGPASG | DA1-GSG | Amine |
| 94 | D22 | DAAAAFGPASG | DA4-GSG | Amine |
| 95 | D23 | RAGSGFGPASG | RA1-GSG | Amine |
| 96 | D24 | RAAAAFGPASG | RA4-GSG | Amine |
Amino acid sequences of 11-residue peptides in CelluSpots arrays were designed to examine several specificity aspects of the mouse R-transferase, including effects of a second-position residue of a substrate; effects of charge clusters immediately downstream of the (arginylatable) N-terminal residue; effects of the Pro residue (an imino acid) close to the N-terminal residue; and the influence of oxidation of N-terminal Cys on its efficacy as an acceptor of Arg in Ate1-catalyzed reactions. We also wished to address, using the conceptually independent and technically straightforward technology of peptide arrays, several earlier reports, based on MS data, that suggested the ability of Ate1 R-transferase to arginylate non-canonical amino acid residues, both N-terminal and internal ones (87, 94, 96).
As a part of initial quality controls, and also to verify that peptides on CelluSpots arrays could actually serve as efficacious substrates of purified mouse Ate1 R-transferase isoforms, the arginylation reaction was optimized on slides containing duplicate copies of a peptide array, in either the presence or the absence of a purified mouse Ate1 R-transferase isoform (Fig. 2 and see “Experimental Procedures”). The expected “positive” peptides (bearing canonical N-terminal residues; see the Introduction) and the putative “negative” peptides (bearing non-canonical N-terminal residues) were interspersed with other peptides throughout a CelluSpots array (Fig. 2). Some peptides were included in duplicate, making it easier to assess the extent of variability of these arginylation assays within each assay. Spots A1–A24 and B1–B24 (Fig. 2) were of 11-residue peptides whose sequences were variable over the first five residues and constant afterward, specifically Xaa1-Xaa2-Xaa3-Xaa4-Xaa5-Xaa6-Gly-Pro-Ala-Ser-Gly. The sequences of 11-residue peptides in other spots (C1–C24 and D1–D24) contained, in general, varying residues both before and after position 6, as described in Table 1. Two peptides, at C1 and C5, were 12 residues long (Fig. 2 and Table 1).
Under the conditions used (see “Experimental Procedures”), the otherwise complete reaction mix that included l-[14C]arginine ([14C]Arg) but lacked the added Ate1 R-transferase resulted in a uniformly negligible level of [14C]Arg incorporation (Fig. 2B, left panel versus right panel). In contrast, the same assay, with a virtually identical copy of the CelluSpots array but carried out in the presence of a purified Ate1 R-transferase isoform, such as Ate11B7A (Fig. 1C), yielded high levels of [14C]Arg conjugation in spots whose peptides contained a canonical Nt-arginylatable residue, such as N-terminal Asp or Glu (e.g. see Fig. 2B (right), spots A7, A8, A11, A12, B6–B11, C15, C23, D12, D13, D21, and D22). These and related results (Figs. 2–5) illustrate high signal-to-noise levels and a broad dynamic range of CelluSpots arginylation assays.
FIGURE 3.
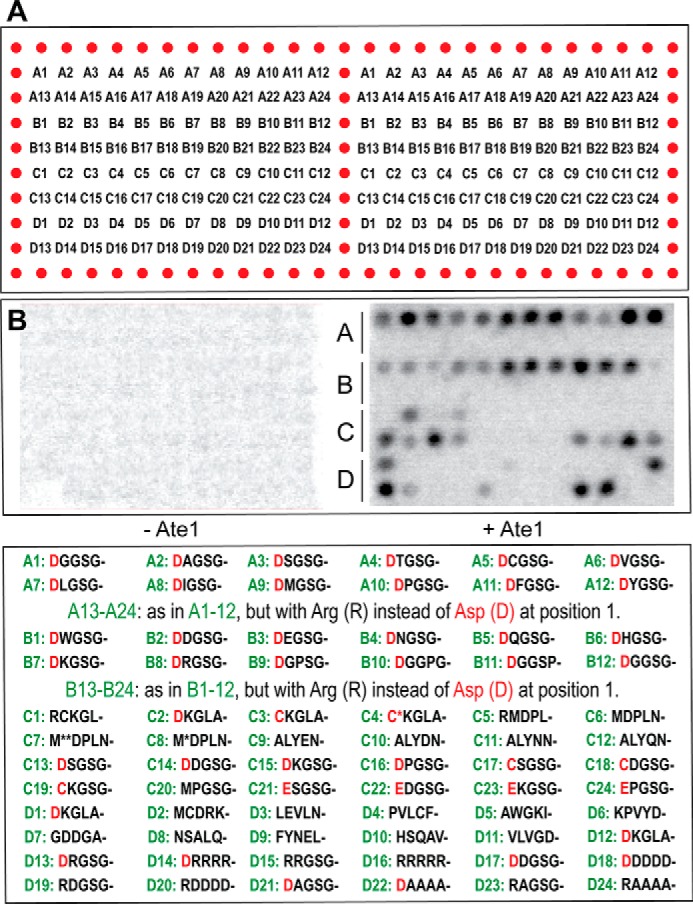
Effect of an amino acid residue at position 2 on the arginylation activity of R-transferase. A, [14C]Arg autoradiograms of CelluSpots arginylation assays with the four purified Ate1 R-transferase isoforms, Ate11A7A, Ate11A7B, Ate11B7A, and Ate11B7B, and with 11-residue peptides XZGSGFGPASG(where X represents Asp or Arg and Z is Gly, Ala, Ser, Thr, Cys, Val, Leu, Ile, Met, Pro, Tyr, Trp, Asp, Glu, Asn, Gln, His, Lys, or Arg). Identical assays were carried out, separately, with Ate11A7A, Ate11A7B, Ate11B7A, and Ate11B7B. B, quantification of results in A, using [14C]Arg autoradiography and normalizing the level of arginylation to arginylation of the N-terminal Asp-Gly (DG)-bearing 11-residue peptide. This quantification, in the present figure and in Figs. 4C and 5B, involved subtraction of the background level of [14C]Arg, a single value for a given peptide array, defined by spots with the lowest levels of [14C]Arg. This and other arginylation assays (Figs. 4 and 5 (A and B)) were carried out, independently, at least two times and yielded results within 10% of the data shown in B.
FIGURE 4.
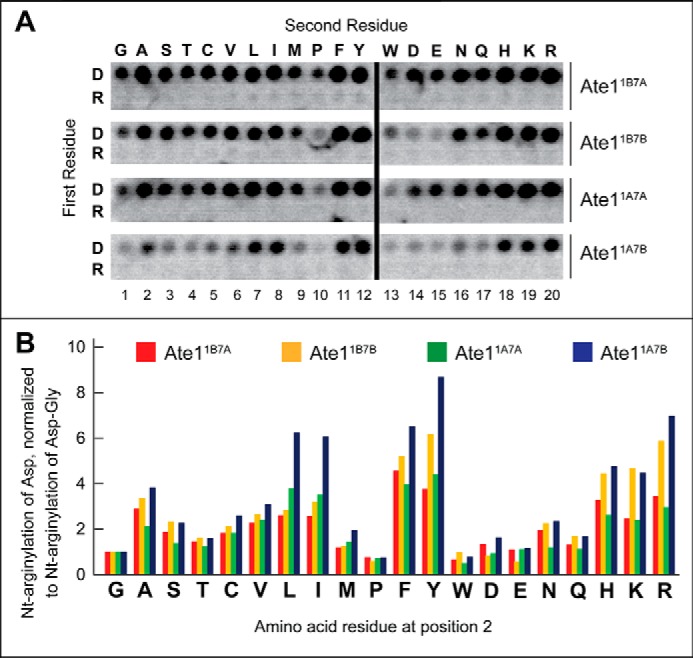
Influence of charge clusters adjoining the N-terminal residue on the rate of arginylation of CelluSpots peptides. A, complete sequences of the examined 11-residue peptides. The N-terminal Asp residue, in a subset of peptides, is shown in red. B, [14C]Arg autoradiograms of CelluSpots arginylation assays with the four purified Ate1 R-transferase isoforms, Ate11A7A, Ate11A7B, Ate11B7A, and Ate11B7B, and with the 11-residue peptides shown in A. C, quantification of results in B, using [14C]Arg autoradiography and normalizing the level of arginylation to arginylation of the Asp-Ala-bearing 11-residue peptide (DA1). These and other quantified arginylation assays (Figs. 3 and 5 (A and B)) were carried out, independently, at least two times and yielded results within 10% of the data shown in B.
FIGURE 5.
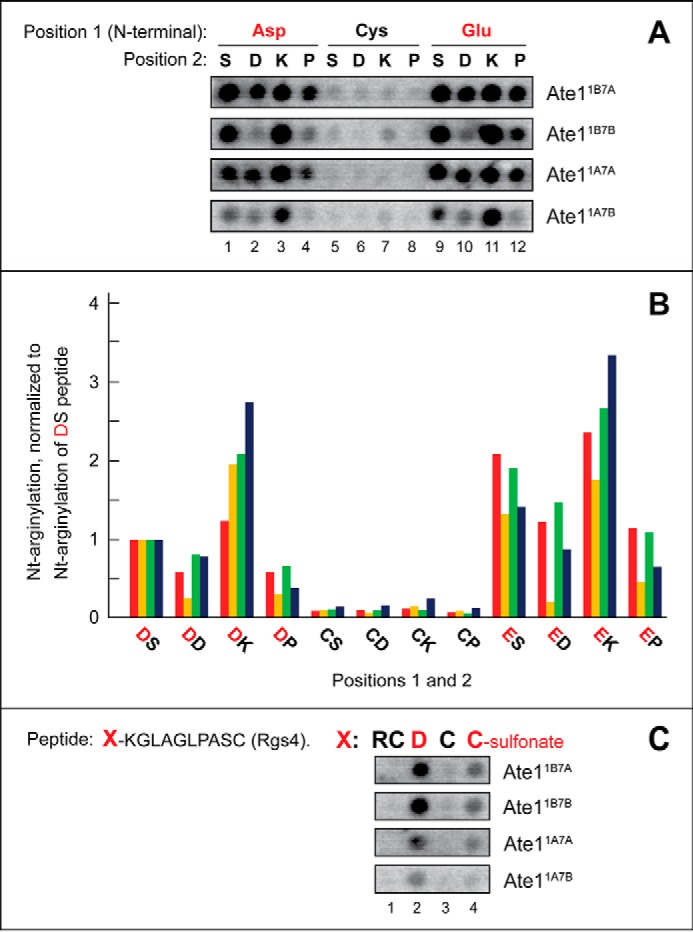
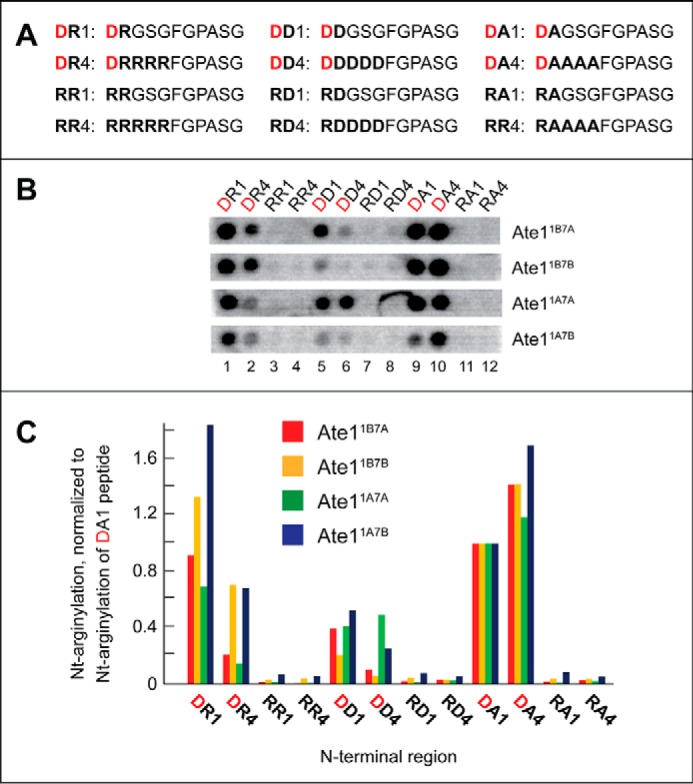
Examination of the relative efficacy of arginylation of the unmodified N-terminal Cys and a varying residue at position 2. A, [14C]Arg autoradiograms of CelluSpots arginylation assays with the four purified Ate1 R-transferase isoforms, Ate11A7A, Ate11A7B, Ate11B7A, and Ate11B7B, and with the 11-residue peptides XZGSGFGPASG (where X represents Asp, Cys, or Glu and Z is Ser, Asp, Lys, or Pro). B, quantification of results in A, using [14C]Arg autoradiography and normalizing the level of Nt-arginylation to Nt-arginylation of the Asp-Ser-bearing 11-residue peptide. These and other quantified arginylation assays (Figs. 3 and 4) were carried out, independently, at least two times and yielded results within 10% of the data shown in A. C, [14C]Arg autoradiograms of CelluSpots arginylation assays with the four purified Ate1 R-transferase isoforms, Ate11A7A, Ate11A7B, Ate11B7A, and Ate11B7B, and with the 11-residue peptides XKGLAGLPASC (where X represents Arg-Cys, Asp, Cys, or Cys-sulfonate). The sequence CKGLAGLPASC is the N-terminal sequence of mouse Rgs4 (see “Results”).
The autoradiogram in Fig. 2B (right) exhibited negligible (undetectable) levels of [14C]Arg incorporation in “negative” spots (i.e. those containing 11-residue peptides with non-canonical (non-Asp, non-Glu, non-Cys) N-terminal residues). Significantly longer autoradiographic exposures of the same or independently produced CelluSpots 14C-arginylation assays allowed the detection of (low) [14C]Arg incorporation in the “negative” peptide spots of these arrays. Examples of background level but detectable [14C]Arg incorporation that can be seen upon longer autoradiographic exposures are in Figs. 3A, 4B, and 5A.
Interestingly, a comparably long autoradiographic overexposure of the otherwise complete [14C]Arg-containing reaction mix that lacked the added Ate1 R-transferase still yielded negligible (undetectable visually) levels of [14C]Arg incorporation (Fig. 2B, left) (data not shown). These data (Figs. 2–5 and 8) suggested that background levels of [14C]Arg incorporation that could be detected in “negative” peptide spots of CelluSpots arginylation assays upon relatively long autoradiographic exposures (e.g. Figs. 3A, 4B, and 5A; compare with Fig. 2B (right)) were likely to signify a low but actual Ate1-mediated conjugation of [14C]Arg.
FIGURE 8.
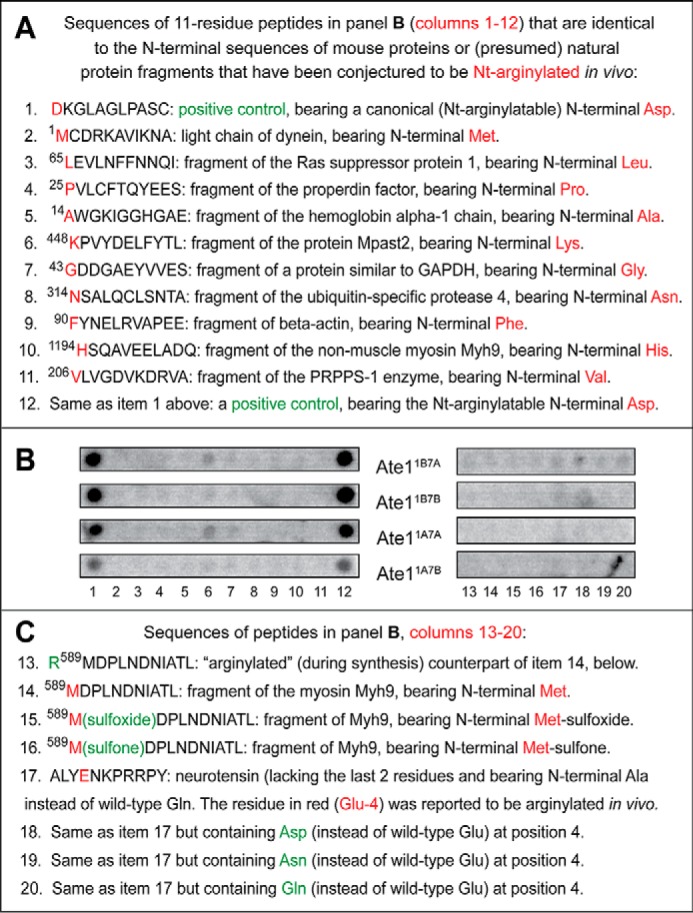
Use of CelluSpots assays to address reports about arginylation of non-canonical N-terminal residues and specific internal residues of cellular proteins. A, list of 11-residue CelluSpots peptides whose sequences were identical to the N-terminal sequences of either a full-length protein (item 2) or presumed natural fragments of other proteins (items 3–11) that have been reported to be Nt-arginylated by Ate1 at their non-canonical (non-Asp, non-Glu, non-Cys) N-terminal residues (87–92). See “Results” for a detailed list of these peptides, which contains additional information and relevant references. Items 1–12 refer to a positive-control peptide that contained N-terminal Asp, a canonical (Nt-arginylatable) residue. B, [14C]Arg autoradiograms of CelluSpots arginylation assays with the four purified Ate1 R-transferase isoforms, Ate11A7A, Ate11A7B, Ate11B7A, and Ate11B7B, and with 20 peptides shown in A (items 1–12) and C (items 13–20). Column 1, the positive-control peptide bearing the canonical (Nt-arginylated) N-terminal Asp residue (see item 1 in A). Columns 2–11, the sequences of these peptides are shown, respectively, in items 2–11 in A. Column 12, the same positive-control peptide (bearing the canonical N-terminal Asp residue) that was also present in peptide spots of column 1 (see item 1 in A). Columns 13–16, the sequences of these peptides shown in items 13–16 in C. Column 17, the peptide shown in item 17 in C. The sequence of this peptide, ALYENKPRRPY, was identical to the 13-residue sequence of the mature neurotensin hormone (QLYENKPRRPYIL) (96), save for the absence of the C-terminal Ile-Leu sequence of neurotensin and the presence of N-terminal Ala, instead of wild-type N-terminal Gln. A fraction of the QLYENKPRRPYIL neurotensin was reported to be arginylated at its internal Glu-4 residue (96); hence, the use of CelluSpots arginylation assays and the four purified isoforms of the Ate1 R-transferase with ALYENKPRRPY, a close analog of the 13-residue neurotensin, to determine whether the reported in vivo arginylation of neurotensin at the internal Glu-4 residue (96) could be observed in vitro as well. Columns 18–20, the sequences of these peptides are shown, respectively, in items 18–20 in C. C, items 13–20 are the continuation of the list of CelluSpots peptides in A that have been examined, as indicated in B, for the extent of their non-canonical arginylation in CelluSpots assays. Note the background levels of [14C]Arg incorporation with all of the examined peptides (columns 2–11 and 13–20), save for the (expected) efficacious arginylation of a positive-control peptide (see items 1 and 12 in A and columns 1 and 12 in B). See the “Results” for additional details.
If so, what might be the reason for a uniformly low but still detectable (upon longer autoradiographic exposures) incorporation of [14C]Arg in spots of peptides bearing non-canonical N-terminal residues? Two mechanistically different and non-alternative (mutually compatible) explanations of background level [14C]Arg incorporation are as follows. In the first interpretation, R-transferase might be able to conjugate Arg, at a very low, presumably physiologically irrelevant efficacy (still detectable in 14C-arginylation assays) to at least some non-canonical N-terminal residues. A different and non-alternative interpretation is that background level amounts of [14C]Arg in spots of peptides bearing non-canonical N-terminal residues reflect the presence, in the corresponding peptide spots, of non-zero (but very low) admixtures of canonical Nt-arginylatable N-terminal residues, such as Asp and/or Glu. It would be interesting to rigorously distinguish between these interpretations by carrying out analogous arginylation assays with individual (i.e. non-arrayed) short peptides that would be synthesized from particularly pure amino acid stocks and would be purified and authenticated more extensively and rigorously than is possible with arrays of immobilized peptides.
Different Ate1 Isoforms Exhibit Similar Substrate Preferences
A subset of 11-residue peptides in our CelluSpots arrays was designed to examine the effects of a second-position residue on the relative efficacy of Ate1-mediated arginylation. Peptides of that subset contained N-terminal Asp. This is a canonical (Nt-arginylatable) residue and apparently the most efficacious acceptor of Arg, among N-terminal Asp, Glu, or oxidized Cys (Fig. 1A) (9). N-terminal Asp was followed by a varying second-position amino acid residue (Xaa), by the Gly-Ser-Gly sequence at positions 3–5, and by the sequence (chosen quasi-randomly) Phe-Gly-Pro-Ala-Ser-Gly at positions 6–11 (see Fig. 3 and its legend). In addition, the array's “control” peptide spots contained 11-residue peptides of otherwise identical sequences but bearing N-terminal Arg, a non-canonical residue, instead of N-terminal Asp (Fig. 3A). The invariant sequence Gly-Ser-Gly-Phe-Gly-Pro-Ala-Ser-Gly at positions 3–11 is not significantly similar to sequences in natural mouse proteins. Arginylation assays with these peptide arrays indicated that all of the above peptides, which contained the canonical (Nt-arginylatable) N-terminal Asp followed by a varying position 2 residue (Xaa) and the sequence Gly-Ser-Gly-Phe-Gly-Pro-Ala-Ser-Gly (positions 3–11), could be arginylated by mouse R-transferase to different but significant extents, whereas none of the otherwise identical peptides bearing the non-canonical N-terminal Arg residue were arginylated above background levels (Fig. 3).
Ate11B7A, Ate11B7B, Ate11A7A, and Ate11A7B, the four major splicing-derived isoforms of the mouse Ate1 R-transferase, differ by the presence or absence of exon 1a versus the sequelogous (similar in sequence (106)) exon 1b and also exon 7a versus the sequelogous exon 7b (Fig. 1D) (3, 78, 79). Carrying out four identical arginylation assays with four Ate1 isoforms, we found that the relative activities of the purified Ate11B7A, Ate11B7B, Ate11A7A, and Ate11A7B R-transferases toward the 11-residue peptides with N-terminal Asp and a varying residue at position 2 were substantially similar (Fig. 3). Although one isoform, Ate11A7B, exhibited a significantly lower overall arginylation activity (per μg of added purified R-transferase) than the other three isoforms, the substrate preferences of Ate11A7B, vis-à-vis the examined set of peptides, were substantially similar to those of the other three isoforms (Fig. 3; see the legends to Figs. 3–5 for statistical aspects of [14C]Arg quantification in these arrays).
These arginylation assays also showed that all four Ate1 isoforms exhibited a significant preference for arginylating N-terminal Asp-bearing peptides that contained either a hydrophobic residue other than Trp (specifically Ala, Val, Leu, Ile, Phe, or Tyr) or a basic residue (His, Lys, or Arg) at position 2, in comparison with either acidic residues (Asp or Glu) or the Pro residue at this position (Fig. 3). Pro and Trp at position 2 were the least favorable residues at position 2, in the otherwise identical sequence contexts of N-terminal Asp-bearing Asp-Xaa-Gly-Ser-Gly-Phe-Gly-Pro-Ala-Ser-Gly peptides (Fig. 3). It should be noted that the isoform Ate11B7B was more sensitive to the presence of Asp or Glu at position 2 than the other three Ate1 isoforms, a rare instance of a specificity difference among Ate1 isoforms (Figs. 3 and 4).
The Overall Charge of a Region Adjoining N-terminal Residue Affects the Efficacy of Arginylation
All four Ate1 R-transferase isoforms exhibited significantly decreased efficacies of arginylation with 11-residue peptides in which N-terminal Asp was followed by four basic (Arg) residues, in comparison with a single Arg at position 2 (Fig. 4). Three of the four Ate1 isoforms were also significantly less efficacious when N-terminal Asp was followed by four acidic (Asp) residues, in comparison with a single Asp at position 2 (Fig. 4). An interesting exception was the Ate11A7A isoform, which arginylated the N-terminal sequence DDGSG comparably with the sequence DDDDD (Fig. 4).
In contrast, a difference in the efficacy of arginylation, for all four Ate1 isoforms, was much smaller between the N-terminal sequences DAGSG and DAAAA, and the run of four Ala residues after N-terminal Asp was even more favorable for arginylation than a single Ala at position 2 (Fig. 5). Notably, even a single position 2 Asp residue made the resulting peptide a significantly less efficacious arginylation substrate than the otherwise identical peptide in which N-terminal Asp was followed by either a single positive (Arg) residue or a single small uncharged (Ala) residue (Fig. 4; see the legends to Figs. 3–5 for statistical aspects of [14C]Arg quantification in these arrays).
Negligible Efficacy of Unoxidized N-terminal Cys as a Substrate for Arginylation
Earlier studies of conditional N-degrons bearing N-terminal Cys have shown that the Ate1-mediated Nt-arginylation of Cys required its preliminary oxidation to Cys-sulfinate or Cys-sulfonate (3, 4, 29). (Although it is a priori likely that R-transferase can also Nt-arginylate the (less oxidized) N-terminal Cys-sulfinate, because its side chain “resembles” the side chain of Asp more than Cys-sulfonate does, there is no rigorous evidence, so far, that bears on this conjecture, inasmuch as Cys-sulfinate is readily oxidized in vitro to Cys-sulfonate.) In agreement with those findings, our results with CelluSpots peptide arrays indicated that the unmodified N-terminal Cys residue is either a very poor arginylation substrate of the Ate1 R-transferase or not a substrate at all (Fig. 2B, right, spots C1–C4; Fig. 5, A (columns 5–8), B, and C; see the legends to Figs. 3–5 for statistical aspects of [14C]Arg quantification in these arrays). The latter possibility would obtain if the observed, essentially background levels of [14C]Arg with N-terminal Cys-bearing CelluSpots peptides (Fig. 5, A (columns 5–8), B, and C) resulted from a partial nonenzymatic oxidation of Cys in these peptides during preparation and handling of CelluSpots arrays.
A small increase over background levels of [14C]Arg incorporation could be observed with 11-residue peptides bearing unmodified N-terminal Cys when the second residue was basic (Lys) (Fig. 5, A (columns 5–8) and B). It remains to be determined whether this small effect resulted from a weak increase in the otherwise negligible activity of R-transferase toward unoxidized N-terminal Cys when the second residue was basic (Lys) or whether, non-alternatively, this effect stemmed from a (possibly) increased propensity of N-terminal Cys to be slowly and nonenzymatically oxidized to Cys-sulfinate and/or Cys-sulfonate by oxygen in the air if a residue at position 2 was basic.
Oxidation of N-terminal Cys Allows Its Arginylation
To compare Ate1 R-transferase isoforms by their ability to arginylate the unmodified versus oxidized N-terminal Cys residue, we used an 11-residue peptide whose sequence, CKGLAGLPASC, was identical to the N-terminal sequence of wild-type mouse Rgs4. We also used the otherwise identical peptides that differed at position 1 (Fig. 5C). Rgs4, a regulator of specific G proteins, has been shown to be a conditionally Nt-arginylated (and therefore conditionally short-lived) physiological substrate of the Arg/N-end rule pathway (3, 4, 29, 107). N-terminal positions of examined 11-residue peptides contained either an unmodified (i.e. not deliberately oxidized) N-terminal Cys (the same as in wild-type Rgs4) or the oxidized N-terminal Cys (Cys-sulfonate), which was substituted for unmodified Cys during the synthesis of these peptides. Two other (control) peptides contained, instead of N-terminal Cys, either N-terminal Asp (an unconditionally Nt-arginylatable (canonical) N-terminal residue; see the Introduction) or the 2-residue sequence Arg-Cys, bearing N-terminal Arg, a non-canonical residue. The latter peptide was a 12-residue one, in contrast to 11-residue peptides in the other spots, due to the addition of N-terminal Arg (Fig. 5C).
As expected (given the previous evidence about oxidation of N-terminal Cys being a prerequisite for its Nt-arginylation (3, 4, 29)), we found that the peptide bearing N-terminal Cys-sulfonate (incorporated during peptide synthesis) was a significant arginylation substrate. In contrast, the arginylation, with four examined Ate1 isoforms, of the otherwise identical peptide bearing the unoxidized (more accurately, not deliberately oxidized) N-terminal Cys residue was at background levels (Fig. 5C, column 4 versus column 3). Also, as expected, the otherwise identical peptide bearing the canonical N-terminal Asp residue (instead of Cys) was arginylated much more efficaciously than even the oxidized N-terminal Cys residue, whereas the otherwise identical 12-residue peptide bearing the 2-residue N-terminal sequence Arg-Cys (N-terminal Arg is a non-canonical residue) was not arginylated above background levels (Fig. 5C; see also Fig. 2B (right), spots C1–C4).
We found that all four examined isoforms of the mouse Ate1 R-transferase exhibited similar relative efficacies of arginylation of peptides in this N-terminal Cys-based setting (Fig. 5). These and other results of this section disagreed with data of an earlier report indicating that some isoforms of Ate1 are nearly specific for N-terminal Cys (93). In particular, none of the four examined Ate1 isoforms in CelluSpots assays could arginylate the unoxidized N-terminal Cys residue above background levels of [14C]Arg incorporation (Fig. 5). In addition, these Ate1 isoforms were similar in their relative ability to arginylate the oxidized N-terminal Cys (Cys-sulfonate) residue (Fig. 5C).
The Rate of Ate1-dependent Protein Degradation Is Influenced by the Overall Charge of a Region Adjoining the N Terminus
We asked whether the observed influence of second-position residues and adjoining clusters of charged residues on the efficacy of Nt-arginylation in CelluSpots assays (Fig. 4) was also relevant to the rate of Ate1-dependent protein degradation. Pulse-chase assays were carried out in rabbit reticulocyte extract, a previously characterized transcription-translation-degradation system that contains the Arg/N-end rule pathway (44, 49, 73). The Arg/N-end rule reporter substrates were derivatives of Rgs4, a regulator of G proteins that has been identified as an Ate1-dependent physiological substrate of the Arg/N-end rule pathway (4, 29, 107). Wild-type mammalian Rgs4 proteins bear N-terminal Cys, after the cotranslational removal of their N-terminal Met by Met-aminopeptidases. The N-terminal Cys residue of Rgs4 has been shown to be oxidized, by NO and oxygen, either in vivo or in reticulocyte extract to (ultimately) Cys-sulfonate, which can be Nt-arginylated by the Ate1 R-transferase. The resulting Nt-arginylated Rgs4 is processively destroyed to short peptides by the post-arginylation part of the Arg/N-end rule pathway (Fig. 1A) (3, 4).
Degradation assays in reticulocyte extract employed the Ub reference technique (URT), derived from the Ub fusion technique (Fig. 6A) (1, 108). In this method, the cotranslational cleavage of a URT-based protein fusion by deubiquitylases in reticulocyte extract produces, at the initially equimolar ratio, a test protein with a desired N-terminal residue and a “reference” protein, in the present case fDHFR-UbK48R, a flag-tagged derivative of the mouse dihydrofolate reductase (Fig. 6A). In URT-based assays, a pulse-labeled test protein is quantified by measuring its level relative to that of a stable reference protein, fDHFR-UbK48R, at each time point of a chase. The URT-based, “built-in” reference protein increases the accuracy of degradation assays (Fig. 6A) (108–110).
FIGURE 6.
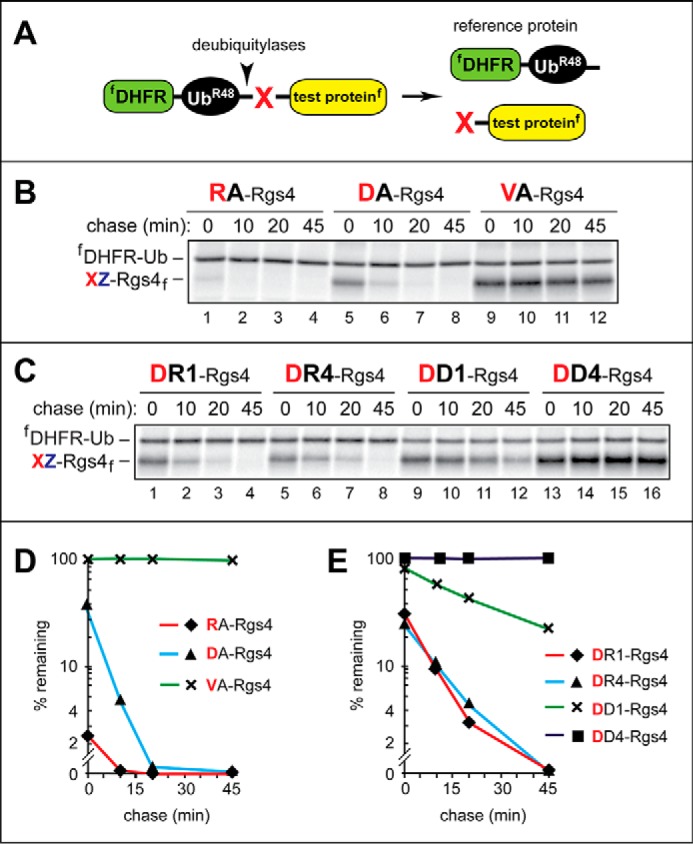
Pulse-chase degradation assays using Rgs4-based Arg/N-end rule substrates and the ubiquitin reference technique. A, URT. For its description and references, see “Results.” B, 35S-pulse-chase analyses, in rabbit reticulocyte extract, of XZGSG-Rgs4 (where X represents Arg, Asp, or Val and Z is Ala), a set of mouse Rgs4-based test proteins. Rgs4 is a regulator of G proteins and a conditionally short-lived physiological substrate of the Arg/N-end rule pathway (see “Results” for references and additional information). In these test proteins, the sequence Gly-Ser-Gly (GSG) was inserted at positions 3–5 to make possible its replacements, in test proteins of C, with the sequences RRR or DDD (see below). C-terminally flag-tagged XZGSG-Rgs4f proteins were coexpressed, in reticulocyte extract, with the N-terminally flag-tagged fDHFR-Ub reference protein, initially as a single Ub fusion (see A), and were labeled for 10 min at 30 °C with [35S]Met/Cys, followed by a chase for 45 min, immunoprecipitation with anti-flag antibody, SDS-PAGE of immunoprecipitated proteins, and autoradiography. The bands of 35S-labeled XZ-Rgs4f and fDHFR-Ub are shown on the left. Lanes 1–4, RAGSG-Rgs4f. Lanes 5–8, DAGSG-Rgs4f. Lanes 9–12, VAGSG-Rgs4f. C, same as in B, but with XZ-Rgs4f (where X represents Asp or Arg and Z is Arg, Arg4, Asp, or Asp4) test proteins, in some of which the sequences RRR or DDD replaced the sequence GSG at positions 3–5 (see above; see “Results”). Lanes 1–4, DRGSG-Rgs4f. Lanes 5–8, DRRRR-Rgs4f (denoted as DR4-Rgs4f). Lanes 9–12, DDGSG-Rgs4f. Lanes 13–16, DDDDD-Rgs4f (denoted as DD4-Rgs4f). D, quantification of data in B. E, quantification of data in C. These and other quantified pulse-chases (Fig. 7) were carried out, independently, at least three times and yielded results within 10% of the data values shown in D and E.
To simplify the mechanics of initial targeting (i.e. to bypass the N-terminal Cys oxidation step) (it is required for degradation of wild-type Rgs4 (3, 4, 29)), the natural N-terminal Cys of Rgs4 was replaced by Asp in the context of URT fusion (Fig. 6A). In addition, to place an uncharged residue at position 2 and to allow changes of residues 2–5 without altering the length of the reporter, the (charged) Lys residue at position 2 of wild-type mouse Rgs4 was replaced by Ala, and the Gly-Ser-Gly sequence was inserted at positions 3–5. As described below, the set of constructed and examined Rgs4 derivatives (Figs. 6 and 7) involved alterations of residues at positions 1–5. The C-terminally flag-tagged DAGSG-Rgs4f (specifically fDHFR-UbK48R-Asp-Ala-Rgs4f, before the cotranslational cleavage, by deubiquitylases, at the UbK48R-Asp junction; Fig. 6A) (108–110)) was Nt-arginylated by Ate1 in reticulocyte extract (Fig. 1A). The sequence of the DAGSG-Rgs4f test protein was identical to that of wild-type Rgs4, save for the replacement of wild-type Cys-Lys by Asp-Ala and the insertion of Gly-Ser-Gly.
FIGURE 7.
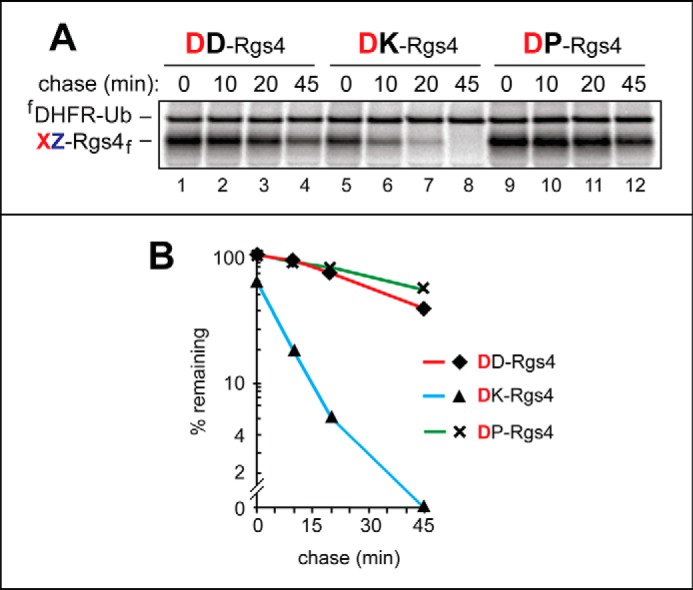
Efficacy of N-terminal arginylation and its influence on the degradation rate of Arg/N-end rule substrates. A, URT (see Fig. 6A and “Results”) and 35S-pulse-chases in reticulocyte extract were used to examine the arginylation-dependent degradation of the DZ-Rgs4f test proteins (where Z represents Asp, Lys, or Pro) (in the presence of the fDHFR-Ub reference protein) by the Arg/N-end rule pathway. The bands of 35S-labeled DZ-Rgs4f and fDHFR-Ub are shown on the left. Lanes 1–4, DD-Rgs4f. Lanes 5–8, DK-Rgs4f. Lanes 9–12, DP-Rgs4f. As shown in Figs. 3 and 5 (A and B), the relative efficacy of Nt-arginylation of N-terminal Asp is a function of the identity of a residue at position 2 (also see “Results”). B, quantification of data in A. These and other quantified pulse-chases (Fig. 6) were carried out, independently, at least three times, and yielded results within 10% of the data values shown in A.
In reticulocyte extract, DAGSG-Rgs4f was rapidly degraded (t½ < 10 min; Fig. 6, B (lanes 5–8) and D). The otherwise identical RAGSG-Rgs4f, with the N-terminal sequence Arg-Ala, was degraded even faster (t½ < 1 min; Fig. 6, B (lanes 1–4) and D), most likely because in this case the preliminary step of Nt-arginylation was neither possible nor necessary prior to the recognition of RAGSG-Rgs4f by a mammalian N-recognin such as Ubr1 (Fig. 1A). In contrast, the otherwise identical VAGSG-Rgs4f (bearing the N-terminal sequence Val-Ala, which is not recognized by the Arg/N-end rule pathway) was stable during the chase (t½ ≫ 45 min; Fig. 6, B (lanes 9–12) and D). In addition, the initial, time 0 (pre-chase) levels of DAGSG-Rgs4f and (particularly) RAGSG-Rgs4f were much lower than the relative level of VAGSG-Rgs4f, indicating a substantial early (pre-chase) degradation of DAGSG-Rgs4f and RAGSG-Rgs4f but not of VAGSG-Rgs4f (Fig. 6, B and D). In sum, this set of otherwise identical Rgs4-based test proteins, which differ solely at their N terminus-proximal positions, is a particularly suitable setting for examining effects of N-terminal changes on the rate of proteolysis by the Arg/N-end rule pathway (Fig. 6, B and D; see the legends to Figs. 6 and 7 for statistical aspects of quantifying protein degradation in these assays).
We also compared degradation rate effects of a second-position basic residue (Lys) versus either an acidic residue (Asp) or an imino residue (Pro) in DXGSG-Rgs4f test proteins (where X represents Asp, Pro, Lys). In agreement with a significantly less efficient arginylation of N-terminal Asp-bearing CelluSpots peptides containing either an acidic (Asp) or Pro residue at position 2 (in comparison with a basic residue at that position), the degradation of DKGSG-Rgs4f, with Lys at position 2, was much faster than the degradation of either DDGSG-Rgs4f or DPGSG-Rgs4f (Fig. 7; see the legends to Figs. 6 and 7 for statistical aspects of quantifying protein degradation in these assays). In sum, the identity of a position 2 residue, in an N-degron that requires Nt-arginylation, can influence the degradation rate of a corresponding protein, at least in part by altering the rate of Nt-arginylation (Fig. 1A).
We also constructed and examined derivatives of DAGSG-Rgs4f that contained either one or four Asp residues at positions 2–5 or, alternatively, either one or four Arg residues at the same positions. (These changes of Rgs4 were done through substitutions of its wild-type residue 2 and of the previously inserted Gly-Ser-Gly at positions 3–5 (see above) by either four Asp or four Arg residues.) Interestingly, DDDDD-Rgs4f, in which a stretch of four acidic (Asp) residues followed N-terminal Asp, was found to be long-lived in reticulocyte extract (t½ ≫ 45 min; Fig. 6, C (lanes 13–15) and E), despite the fact that N-terminal Asp, when it is present in a less disfavorable downstream sequence context (e.g. in DAGSG-Rgs4f), is a short-lived substrate of the Arg/N-end rule pathway (Fig. 6, B (lanes 1–4) and D; see the legends to Figs. 6 and 7 for statistical aspects of quantifying protein degradation in these assays). Predictably, the N-terminal sequence, Asp-Ala, was a good substrate for Nt-arginylation in CelluSpots peptide arrays (e.g. see Fig. 3A). The absence of significant degradation of DDDDD-Rgs4f was in qualitative agreement with the relative extents of Nt-arginylation of N-terminal Asp in the context of CelluSpots peptide arrays, in which the N-terminal DDDDD … sequence was arginylated much less efficaciously than the N-terminal DDGSG … sequence (Fig. 4).
When four basic (Arg) residues followed N-terminal Asp, the rate of degradation of the resulting DRRRR-Rgs4f was nearly identical to that of DR-Rgs4f, which contained a single Arg residue after N-terminal Asp (followed by the wild-type GLA sequence of Rgs4 at positions 3–5) (Fig. 6, C (lanes 5–8 versus lanes 1–4) and E). These degradation assay results were in qualitative agreement with the extents of Nt-arginylation of N-terminal Asp in the context of CelluSpots assays, in which both the DRGSG … and DRRRR … peptides were arginylated, with the DRGSG … peptide being a more efficacious Ate1 substrate than the DRRRR … peptide (Fig. 4).
Together, the results of arginylation assays on peptide arrays and protein degradation assays in reticulocyte extract (Figs. 2–7) indicated that multiple acidic residues immediately downstream of a canonical (arginylatable) N-terminal residue such as Asp could significantly attenuate its Ate1-mediated Nt-arginylation and, consequently, the rate of degradation of a corresponding N-terminal Asp-bearing test protein by the Arg/N-end rule pathway. Although multiple basic residues (Arg) downstream of N-terminal Asp also decreased the efficacy of Nt-arginylation, this decrease was less severe than the one of several acidic (Asp) residues and did not strongly influence the rate of degradation of a corresponding N-end rule substrate (Figs. 4–6).
Use of CelluSpots Assays to Address Reports about Arginylation of Non-canonical N-terminal Residues and Specific Internal Residues of Cellular Proteins
As mentioned in the Abstract and Introduction, several publications, over the last decade, have suggested, largely on the basis of MS-based analyses of isolated cellular proteins (digested by trypsin or other proteases), that the arginylation of specific proteins or their natural fragments can involve, to a significant extent, non-canonical (non-Asp, non-Glu, non-Cys) N-terminal residues, including N-terminal Met, Leu, Phe, Val, Ala, Thr, Gly, Asn, Lys, and Pro (87–92). In addition, a 2005 study (96) described a modified 13-residue neurotensin hormone that appeared to be arginylated, in vivo, at its internal Glu-4 residue. Neurotensin is produced through the processing of a larger precursor protein in the endoplasmic reticulum and Golgi. These compartments are distinct from the currently known locations of the cytosolic/nuclear Ate1 R-transferase (96). A subsequent publication suggested that Ate1 is capable of arginylating not only the α-amino groups of N-terminal residues but also γ-carboxyl groups of internal (non-N-terminal) Asp and Glu in natural proteins and described several proteins, including actin, dystrophin, myosin, titin, and tubulin, that appeared to contain, according to MS-based data, internal Asp or Glu residues conjugated to Arg (94).
To the best of our knowledge, none of the MS-based evidence about significant levels of protein arginylation either at non-canonical N-terminal residues or at specific internal residues (87–92, 94–96) has been examined, thus far, using independent, non-MS methods. We addressed the possibility of non-canonical arginylation by asking, using C-terminally immobilized 11-residue CelluSpots peptides and [14C]arginylation assays, whether specific amino acid sequences that encompass the previously reported non-canonical arginylation sites (87–92, 94–96) are capable of being significantly arginylated by the purified Ate1 R-transferase.
One set of [14C]arginylation assays was carried out with the peptide ALYENKPRRPY and its derivatives at position 4. The 11-residue sequence ALYENKPRRPY is identical to the 13-residue sequence of mature neurotensin (QLYENKPRRPYIL) (96), save for the absence of the C-terminal Ile-Leu sequence of neurotensin and the presence of N-terminal Ala, instead of wild-type N-terminal Gln, in the 11-residue CelluSpots peptide. The Gln → Ala N-terminal replacement was necessary due to the possibility of a nonenzymatic deamidation of wild-type N-terminal Gln (76), because it would yield N-terminal Glu, a canonical substrate of Ate1 R-transferase (Fig. 1A). The CelluSpots arrays contained not only the above 11-residue ALYENKPRRPY peptide but also the otherwise identical peptides that differed by bearing, at position 4, either Asp, Asn, or Gln, instead of wild-type Glu (Fig. 8, B (columns 17–20) and C). The earlier MS-based evidence (96) for the in vivo arginylation of the neurotensin's internal Glu-4 residue was summarized above.
We found that none of the neurotensin-based peptides (including the one containing the wild-type Glu-4 residue) generated a [14C]arginylation signal above background levels of CelluSpots assays with four individually examined purified Ate1 R-transferase isoforms, indicating the inability of any one among these peptides to serve as Ate1 substrates at least in vitro (Fig. 8, B (columns 17–20) and C (items 17–20)). Given this robustly negative and reproducible result, we did not use CelluSpots assays to examine analogous claims, by another laboratory, about arginylation of internal Asp or Glu in actin, myosin, and other cellular proteins (94, 95).
To address a related but different set of earlier reports about non-canonical specificities of Ate1, we carried out CelluSpots arginylation assays with 11 C-terminally immobilized 11-residue peptides (Table 1, entries 53–56 and 74–83) whose sequences were identical to the N-terminal sequences of some among nearly 50 proteins that have been described previously as in vivo Ate1 substrates that were Nt-arginylated at non-canonical (non-Asp, non-Glu, non-Cys) N-terminal residues (87). In most cases, Nt-arginylation of this kind was reported to involve non-canonical N-terminal residues of presumed (i.e. neither isolated nor otherwise characterized) natural protein fragments, generated in vivo by unknown proteases (87).
The putative Nt-arginylation substrates listed below were selected from a much larger set of claimed non-canonical Ate1 substrates (87) to encompass a broad range of reported non-canonical N-terminal residues, specifically Met, Leu, Pro, Ala, Lys, Gly, Asn, Phe, His, and Val. The 11 resulting 11-residue peptides were identical to the N-terminal sequences of either a full-length protein or (presumed) protein fragments that have been reported as substrates of non-canonical Nt-arginylation (87). The CelluSpots arginylation assays with these peptides also contained, as items 1 and 12, a positive (N-terminal Asp-bearing) control.
DKGLAGLPASC: the Rgs4-derived, 11-residue peptide, a positive control. It contains N-terminal Asp, a canonical (Nt-arginylatable) residue, and was present in the array at two positions, as described in Fig. 8, A (items 1 and 12) and B (columns 1 and 12).
MCDRKAVIKNA: the light chain of cytoplasmic dynein (Dynnl1); NP_062656.2. The reported Nt-arginylated residue is the initial Met-1 of Dynnl1 (87). We note that the N-terminal Met residue, in the sequence Met-Cys, is expected to be cotranslationally cleaved off, from a nascent protein, by Met-aminopeptidases (9). The authors of Ref. 87, which reported Nt-arginylation of the Met-1 residue of the full-length Dynnl1, did not mention this issue.
LEVLNFFNNQI: a presumed fragment of the Ras suppressor protein 1; NP_033131.2. The Nt-arginylated residue is Leu-65 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Leu-65).
PVLCFTQYEES: a presumed fragment of the properdin factor (complement); NP_032849.1. The Nt-arginylated residue is Pro-25 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Pro-25).
AWGKIGGHGAE: a presumed fragment of the hemoglobin α-1 chain; NP_032244.1. The Nt-arginylated residue is Ala-14 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Ala-14).
KPVYDELFYTL: a presumed fragment of the EH domain containing protein Mpast2; AAH12272.1. The arginylated residue is Lys-448 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Lys-448).
GDDGAEYVVES: a presumed fragment of the protein similar to GAPDH; XP_979290.1. The Nt-arginylated residue is Gly-43 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Gly-43).
NSALQCLSNTA: a presumed fragment of the ubiquitin-specific protease 4; NP_035808.1. The Nt-arginylated residue is Asn-314 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Asn-314).
FYNELRVAPEE: a presumed fragment of β-actin; NP_031419.1. The Nt-arginylated residue is Phe-90 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Phe-90).
HSQAVEELADQ: a presumed fragment of the non-muscle myosin Myh9, heavy polypeptide 9, isoform 1; NP_071855.1. The Nt-arginylated residue is His-1194 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal His-1194).
VLVGDVKDRVA: a presumed fragment of the phosphoribosyl pyrophosphate synthase-1 (Prps1); NP_067438.1. The Nt-arginylated residue is Val-206 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Val-206).
MDPLNDNIATL: the same protein as in item 10 above, but a (presumed) different fragment of the non-muscle myosin Myh9 (NP_071855.1). The Nt-arginylated residue of this fragment is Met-589 (i.e. the authors (87) reported that their protein preparation contained a (presumed) fragment of this protein that was Nt-arginylated at its N-terminal Met-589).
We found that 10 of these 11 CelluSpots peptides, corresponding to N-terminal sequences of some among the previously reported non-canonical substrates of the Ate1 R-transferase (87), generated background levels of [14C]Arg incorporation with four individually examined purified Ate1 isoforms, indicating the inability of these peptides to serve as significant Ate1 substrates at least in vitro (Fig. 8, A (items 2–11 and 14) and B (columns 2–11 and 14)).
Importantly, the CelluSpots arrays containing these 11-residue peptides were flanked by spots of the positive-control peptide DKGLAGLPASC (item 1 above), which contained N-terminal Asp, a canonical (Nt-arginylatable) residue. As expected, DKGLAGLPASC was efficaciously Nt-arginylated by Ate1 isoforms, in striking contrast to the absence of significant arginylation of the other 11-residue peptides (Fig. 8, A (items 1 and 12) and B (columns 1 and 12); compare with Fig. 8, A (items 2–11 and 14) and B (columns 2–11 and 14)). Possible causes of very low but detectable background level arginylation in CelluSpots peptide arrays, as distinguished from negligible (undetectable) levels of [14C]Arg incorporation, are discussed above.
The single and weak potential exception, in which a slightly above-background level of [14C]Arg incorporation was observed with two of the four examined Ate1 isoforms, was the peptide KPVYDELFYTL, corresponding to the (presumed) internal fragment of the Mpast2 protein (see item 6 above) (Fig. 8, A (item 6) and B (column 6)). The near-background levels of [14C]Arg incorporation with the peptide KPVYDELFYTL (Fig. 8B, column 6; compare with columns 1 and 12) were significantly lower, in absolute amounts of incorporated [14C]Arg, than the levels of [14C]Arg vis-à-vis 11-residue peptides bearing the unmodified N-terminal Cys residue (Fig. 5, A and C). As described above, these N-terminal Cys-bearing peptides were classed as non-arginylatable, in agreement with earlier data (4, 7, 29).
We also tried to determine whether the report about non-canonical Nt-arginylation of, for example, the N-terminal Met-bearing sequence MDPLNDNIATL (87) (see item 12 above) could have stemmed from in vivo oxidation of N-terminal Met to either Met-sulfoxide or Met-sulfone, a change that might have converted the non-arginylatable N-terminal Met into a possible substrate of Ate1. However, we found that the 11-residue peptides M(sulfoxide)DPLNDNIATL and M(sulfone)DPLNDNIATL were just as incapable of arginylation by the four individually examined purified Ate1 isoforms as was the otherwise identical unmodified MDPLNDNIATL peptide, thereby precluding the above possibility at least in vitro (Fig. 2B, right, spots C6–C8; Fig. 8, B (columns 14–16) and C (items 14–16)). In addition and as expected, the 12-residue negative-control peptide RMDPLNDNIATL, which contained the non-canonical N-terminal Arg residue (added during peptide synthesis to the 11-residue MDPLNDNIATL), also yielded background levels of [14C]Arg incorporation in these assays (Fig. 2B, right, spot C5; Fig. 8, B (column 13) and C (item 13)).
It was previously suggested that the proposed ability of the Ate1 R-transferase to arginylate a variety of non-canonical N-terminal as well as internal residues in cellular proteins may be conferred by (unknown) Ate1 cofactors that can be active both in vivo and in cell extracts (87, 91). If correct, this hypothesis may at least partly reconcile the direct and extensive discrepancy between the cited earlier reports (87–92) and our arginylation data with peptide arrays and purified Ate1 (Figs. 2–5 and 8). Therefore, we modified the CelluSpots arginylation assays by adding an extract from mouse cells (the N2a cell line) to reaction mixes, either in the absence or the presence of the exogenous purified Ate11B7A isoform. This modification of arginylation assays did not alter any of our findings.
In particular, CelluSpots 14C-arginylation assays with extract alone, containing only the endogenous Ate1, produced arginylation patterns similar to those generated in the presence of both the extract and the added purified Ate11B7A isoform (data not shown). Because the absolute levels of 14C-arginylation were significantly lower in N2a extract-only assays (in the absence of added exogenous Ate1), we cannot rule out the possibility that procedures for preparing the extract either damaged or diluted the postulated (unknown) Ate1-modifying factors. With that caveat, although explanations of the above discrepancy based on the presence, in vivo, of (unknown) factors that may alter the specificity of Ate1 remain formally possible, they were not supported by the above assays.
Conclusions
In the present study, CelluSpots arrays of C-terminally immobilized synthetic 11-residue peptides, were used, in conjunction with protein degradation assays, to analyze the specificity of arginylation by purified isoforms of the mouse Ate1 R-transferase, a component of the Arg/N-end rule pathway (Figs. 1 and 2). We showed, among other things, that amino acid sequences immediately downstream of a substrate's canonical (Nt-arginylatable) N-terminal residue (Asp, Glu, or oxidized Cys), and particularly a residue at position 2, can affect the rate of Nt-arginylation by R-transferase and thereby the rate of degradation of a substrate protein (Figs. 3–7).
Other CelluSpots arginylation assays of the present study (Fig. 8) were designed to examine specific interpretations of MS-based data that have been published over the last decade, largely by the Kashina laboratory, about the ability of the Ate1 R-transferase to arginylate a variety of non-canonical (non-Asp, non-Glu, non-Cys) N-terminal residues and also specific internal (non-N-terminal) Asp or Glu residues in many cellular proteins (87–92, 94–96). The chief reason for concerns about the validity of the cited reports is a technical one, inasmuch as the evidence for non-canonical arginylation specificities of Ate1 remained almost entirely MS-based (87–92, 94–96), without a significant attempt, until the present study with CelluSpots arrays, to examine interpretations of MS data by other, independent methods.
We addressed the 2005 report by the Sillard laboratory about arginylation of the hormone neurotensin at its internal Glu-4 residue (96) by carrying out CelluSpots arginylation assays using four purified Ate1 isoforms and 11-residue analogs of the 13-residue neurotensin. The robustly negative results of these arginylation assays (Fig. 8, B (columns 17–20) and C (items 17–20)) did not contradict the above findings (96) directly, given, in particular, the in vitro nature of the CelluSpots approach. Nevertheless, our results (Fig. 8) do raise concerns about the validity of MS-based interpretations that suggested the in vivo arginylation of internal (as distinguished from N-terminal) Asp or Glu residues in a number of cellular proteins (94–96).
In addition and independently, our analyses, using CelluSpots arginylation assays and four purified Ate1 isoforms, of Nt-arginylation of non-canonical N-terminal residues encompassed 11 different 11-residue peptides. These peptides were identical to the N-terminal sequences of either full-length proteins or (presumed) protein fragments that have been reported to undergo Nt-arginylation at non-canonical N-terminal residues (87). Positive controls, a part of the CelluSpots arginylation assays, were 11-residue peptides bearing a canonical (Nt-arginylatable) N-terminal residue, such as Asp (Figs. 2, 3, and 8, A (items 1 and 12) and B (columns 1 and 12)). As described above, the results of these arginylation assays were reproducibly and robustly negative. Specifically, we observed background levels of 14C-arginylation of the 11 examined 11-residue peptides (Fig. 8). Their sequences were identical to the N-terminal sequences of 11 among nearly 50 proteins that have been previously described as substrates of Ate1 that were Nt-arginylated at non-canonical (non-Asp, non-Glu, non-Cys) N-terminal residues (87).
A caveat to the otherwise direct contradictions between our findings and those in the cited earlier studies is that our results were produced in vitro (with purified Ate1 R-transferase and peptide arrays), whereas most reports about non-canonical arginylation of cellular proteins were about analyses of natural, in vivo-produced proteins (87–92, 94–96). Nevertheless, given the clear absence of in vitro support for MS-based interpretations that suggested significant levels of non-canonical arginylation of both N-terminal and internal residues in cellular proteins, it would be particularly important to verify the above interpretations definitively, using methods that do not rely on MS alone.
Experimental Procedures
Plasmids, cDNAs, and Primers
The NEB Turbo Escherichia coli strain (New England Biolabs, Ipswich, MA) was used for cloning and maintaining plasmids. Phusion High-Fidelity DNA polymerase (New England Biolabs) was used for PCR. All constructed plasmids (Table 2) were verified by DNA sequencing. pKP496, the parental plasmid used for constructing URT-based plasmids, has been described previously (44).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or Reference |
|---|---|---|
| pcDNA3 | AmpR; NeoR; Expression vector for cloning the gene of interest | Invitrogen |
| pH10UE | AmpR , pET15b-based vector with His10-Ub fusion cassette | Ref. 111 |
| pKP496 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha Ub-MCS-flag under the control of CMV promoter. MCS has SacII, EcoRI, XhoI, ClaI, and EcoRV unique cloning sites | Ref. 43 |
| pBW359 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DRGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW360 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DRRRR-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW361 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DDGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW362 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DDDDD-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW363 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DAGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW364 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DAAAA-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW365 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-RAGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW363 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-VAGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW367 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DSGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW368 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DKGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pBW369 | AmpR; NeoR; pcDNA3.0-based plasmid encoding flag-DHFR-ha-Ub-DPGSG-Gly3mRgs4-flag under the control of CMV promoter | This study |
| pCB407 | AmpR; pH10UE-based vector used for the bacterial expression of a fusion between His10-ubiquitin and Ate11B7A | Ref. 81 |
| pCB408 | AmpR; pH10UE-based vector used for the bacterial expression of a fusion between His10-ubiquitin and Ate11B7B | Ref. 81 |
| pCB409 | AmpR; pH10UE-based vector used for the bacterial expression of a fusion between His10-ubiquitin and Ate11A7A | Ref. 81 |
| pCB410 | AmpR; pH10UE-based vector used for the bacterial expression of a fusion between His10-ubiquitin and Ate11A7B | Ref. 81 |
Purification of Mouse Ate1 R-transferase from E. coli
The plasmids pCB407 (Ate11B7A), pCB408 (Ate11B7B), pCB409 (Ate11A7A), and pCB410 (Ate11A7B) (Table 2) expressed isoform-specific (Fig. 1C) mouse His10-Ub Ate1 fusions that could be purified and deubiquitylated in vitro using a technique described previously (111).
1-liter cultures of BL21 (DE2) E. coli transformed with one of the above plasmids were grown to an A600 of ∼0.6 at 37 °C in LB containing 50 μg/ml ampicillin. Cultures were cooled on ice for 45 min and then induced by the addition of isopropyl-β-d-thiogalactoside to a final concentration of 0.25 mm for 6 h at room temperature. Cells were harvested by centrifugation at 2,000 × g for 10 min at 4 °C. Pelleted cells were resuspended in 15 ml of PBS containing 30% (v/v) glycerol, 0.3 m NaCl, 12 mm imidazole, 20 mm β-mercaptoethanol, and 1 mg/ml lysozyme, followed by freezing in liquid nitrogen. Samples were then thawed slowly on ice and centrifuged at 27,000 × g for 30 min at 4 °C. The N-terminal His10-Ub fusions of Ate1 isoforms were then purified by nickel-nitrilotriacetic acid chromatography, with elution by 0.3 m imidazole. Eluted proteins were dialyzed overnight against 5% (v/v) glycerol, 0.3 m NaCl, 2 mm β-mercaptoethanol, and 50 mm Na2HPO4-NaH2PO4 (pH 8.0), containing 1× Complete EDTA-free protease inhibitor mixture (Roche Applied Science). The N-terminal His10-Ub moiety was cleaved off by incubating dialyzed samples for 1 h at 37 °C with purified Usp2cc deubiquitylase (111). The latter was added at the 1:10 molar ratio. The resulting samples were dialyzed against 5% (v/v) glycerol, 0.15 m NaCl, 10 mm β-mercaptoethanol, 50 mm Tris (pH 7.4), followed by further purification of (deubiquitylated) Ate1 using Mono-S chromatography (Pharmacia Biotech catalog no. 9723125), with an elution gradient between 0.15 and 1 m NaCl in the above dialysis buffer. Peak Ate1 fractions were pooled; dialyzed overnight at 4 °C against 30% (v/v) glycerol, 0.15 m NaCl, 10 mm β-mercaptoethanol, and 50 mm Tris-HCl (pH 7.4); and stored at −80 °C.
CelluSpots-based Arginylation Assays
CelluSpots peptide arrays spotted on glass slides were synthesized by Intavis (Köln, Germany). Each slide contained two side-by-side identical copies of the array (Fig. 2, A and B). Mouse Ate1-dependent arginylation assays were performed as described previously (79), with slight modifications. Individual (rectangular) peptide arrays were “outlined” with an ImmEdge hydrophobic barrier pen (Vector Laboratories, Burlingame, CA) to minimize the required reaction volume. 90-μl reaction samples containing 5.8 μm [14C]Arg (PerkinElmer Life Sciences, NEC267E050UC; 346 mCi/mmol), purified total E. coli tRNA (0.6 mg/ml), and E. coli aminoacyl-tRNA synthetases (800 units/ml; Sigma-Aldrich) in 5 mm ATP, 0.15 m KCl, 10 mm MgCl2, 1 mm DTT, 50 mm Tris-HCl (pH 8.0) were preincubated at 37 °C for 15 min to allow the formation of Arg-tRNA. Thereafter, a specific purified Ate1 isoform (2 μg) was added, and the resulting mixtures (final volumes of 0.1 ml) were added to each array. Arginylation was allowed to proceed for 1 h at 37 °C in a humidified chamber with gentle rocking. Reactions were quenched by rinsing the arrays twice with a large volume excess (10 ml) of Tris-buffered saline (TBS) (1.15 m NaCl, 50 mm Tris-HCl, pH 7.5) containing, in addition, 1% Tween 20 and RNase A (10 μg/μl). Arrays were then washed three times for 20 min each in the same buffer (including RNase A) at 37 °C, followed by an overnight (∼16-h) wash, at room temperature, in TBS containing 1% Tween 20 but no RNase A. The resulting slides were washed once with 10 mm Tris, pH 7.5, and were allowed to air-dry, followed by autoradiography for varying times (usually ∼2 weeks), using a PhosphorImager-type screen. The exposed screens were scanned using a Storm-860 scanner (Molecular Dynamics, Caesarea, Israel). Quantification of 14C spot intensities was performed using the array analysis module of ImageQuant TL software (GE Healthcare, Little Chalfont, UK).
In Vitro Transcription-Translation-Degradation Assays
The TNT T7 coupled transcription/translation system (Promega, Madison, WI) was used to carry out transcription-translation-degradation assays, largely as described previously (44, 50, 73) (see also the Tansey Laboratory Protocol, “Ultimate Mammalian Denaturing IP,” available on the World Wide Web). Nascent proteins in reticulocyte extract were pulse-labeled with l-[35S]methionine (0.55 mCi/ml, 1,000 Ci/mmol; MP Biomedicals, Santa Ana, CA) for 10 min at 30 °C in the total volume of 30 μl. The labeling was quenched by the addition of cycloheximide and unlabeled methionine to final concentrations of 0.1 mg/ml and 5 mm, respectively, bringing the total volume to 40 μl. Samples of 10 μl were removed at the indicated time points. The reaction was immediately terminated by the addition of 80 μl of TSD buffer (1% SDS, 5 mm DTT, 50 mm Tris-HCl, pH 7.4). A sample was snap-frozen in liquid nitrogen and was stored at −80 °C until further processing, which comprised heating the samples at 95 °C for 10 min, followed by dilution with 1 ml of TNN buffer (0.5% Nonidet P-40, 0.25 m NaCl, 5 mm Na-EDTA, 50 mm Tris-HCl, pH 7.4) containing 1× Complete protease inhibitor mixture (Roche Diagnostics). Samples were thereafter immunoprecipitated using 10 μl of anti-flag M2 magnetic beads (Sigma-Aldrich), with rocking at 4 °C for 4 h, followed by four washes in TNN buffer, resuspension in 20 μl of SDS-sample buffer, and heating at 95 °C for 10 min. Immunoprecipitated proteins were fractionated by SDS-4–15% PAGE, followed by autoradiography. Quantification of autoradiograms was carried out using a Storm 860 scanner (Molecular Dynamics).
Author Contributions
B. W., K. I. P., C. S. B., and A. V. designed the experiments. B. W. performed the experiments with participation by K. I. P., and C. B., B. W., and A. V. wrote the paper. All authors discussed the results and commented on the manuscript.
Acknowledgments
We thank the present and former members of the Varshavsky laboratory for helpful discussions during this study.
This study was supported by National Institutes of Health Grants R01-DK039520 and R01-GM031530 (to A. V.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Nt
- N-terminal
- R-transferase
- arginyltransferase or Arg-tRNA-protein transferase
- N-degron
- an N-terminal degradation signal recognized by the N-end rule pathway
- N-recognin
- an E3 ubiquitin ligase that can recognize at least some N-degrons
- Ub
- ubiquitin
- URT
- Ub reference technique.
References
- 1. Bachmair A., Finley D., and Varshavsky A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 2. Bachmair A., and Varshavsky A. (1989) The degradation signal in a short-lived protein. Cell 56, 1019–1032 [DOI] [PubMed] [Google Scholar]
- 3. Kwon Y. T., Kashina A. S., Davydov I. V., Hu R.-G., An J. Y., Seo J. W., Du F., and Varshavsky A. (2002) An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 [DOI] [PubMed] [Google Scholar]
- 4. Hu R.-G., Sheng J., Qi X., Xu Z., Takahashi T. T., and Varshavsky A. (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 [DOI] [PubMed] [Google Scholar]
- 5. Hwang C. S., Shemorry A., and Varshavsky A. (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim H. K., Kim R. R., Oh J. H., Cho H., Varshavsky A., and Hwang C. S. (2014) The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gibbs D. J., Bacardit J., Bachmair A., and Holdsworth M. J. (2014) The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24, 603–611 [DOI] [PubMed] [Google Scholar]
- 8. Tasaki T., Sriram S. M., Park K. S., and Kwon Y. T. (2012) The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eldeeb M., and Fahlman R. (2016) The N-End rule: the beginning determines the end. Protein Pept. Lett. 23, 343–348 [DOI] [PubMed] [Google Scholar]
- 11. Dougan D. A., Micevski D., and Truscott K. N. (2012) The N-end rule pathway: from recognition by N-recognins to destruction by AAA+ proteases. Biochim. Biophys. Acta 1823, 83–91 [DOI] [PubMed] [Google Scholar]
- 12. Varshavsky A. (2008) Discovery of cellular regulation by protein degradation. J. Biol. Chem. 283, 34469–34489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee K. E., Heo J. E., Kim J. M., and Hwang C. S. (2016) N-terminal acetylation-targeted N-end rule proteolytic system: the Ac/N-end rule pathway. Mol. Cells 39, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turner G. C., Du F., and Varshavsky A. (2000) Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405, 579–583 [DOI] [PubMed] [Google Scholar]
- 15. Du F., Navarro-Garcia F., Xia Z., Tasaki T., and Varshavsky A. (2002) Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. U.S.A. 99, 14110–14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hwang C. S., Shemorry A., and Varshavsky A. (2009) Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc. Natl. Acad. Sci. U.S.A. 106, 2142–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heck J. W., Cheung S. K., and Hampton R. Y. (2010) Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U.S.A. 107, 1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eisele F., and Wolf D. H. (2008) Degradation of misfolded proteins in the cytoplasm by the ubiquitin ligase Ubr1. FEBS Lett. 582, 4143–4146 [DOI] [PubMed] [Google Scholar]
- 19. Suzuki T., and Varshavsky A. (1999) Degradation signals in the lysine-asparagine sequence space. EMBO J. 18, 6017–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Inobe T., and Matouschek A. (2014) Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 24, 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tobias J. W., Shrader T. E., Rocap G., and Varshavsky A. (1991) The N-end rule in bacteria. Science 254, 1374–1377 [DOI] [PubMed] [Google Scholar]
- 22. Mogk A., Schmidt R., and Bukau B. (2007) The N-end rule pathway of regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 17, 165–172 [DOI] [PubMed] [Google Scholar]
- 23. Rivera-Rivera I., Román-Hernández G., Sauer R. T., and Baker T. A. (2014) Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates. Proc. Natl. Acad. Sci. U.S.A. 111, E3853–E3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Piatkov K. I., Vu T. T., Hwang C.-S., and Varshavsky A. (2015) Formyl-methionine as a degradation signal at the N-termini of bacterial proteins. Microbial Cell 2, 376–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Humbard M. A., Surkov S., De Donatis G. M., Jenkins L. M., and Maurizi M. R. (2013) The N-degradome of Escherichia coli: limited proteolysis in vivo generates a large pool of proteins bearing N-degrons. J. Biol. Chem. 288, 28913–28924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Graciet E., Hu R. G., Piatkov K., Rhee J. H., Schwarz E. M., and Varshavsky A. (2006) Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc. Natl. Acad. Sci. U.S.A. 103, 3078–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu R.-G., Wang H., Xia Z., and Varshavsky A. (2008) The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. U.S.A. 105, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Dongen J. T., and Licausi F. (2015) Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66, 345–367 [DOI] [PubMed] [Google Scholar]
- 29. Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., and Kwon Y. T. (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alagramam K., Naider F., and Becker J. M. (1995) A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 15, 225–234 [DOI] [PubMed] [Google Scholar]
- 31. Byrd C., Turner G. C., and Varshavsky A. (1998) The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hwang C. S., and Varshavsky A. (2008) Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 19188–19193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kitamura K., and Fujiwara H. (2013) The type-2 N-end rule peptide recognition activity of Ubr11 ubiquitin ligase is required for the expression of peptide transporters. FEBS Lett. 587, 214–219 [DOI] [PubMed] [Google Scholar]
- 34. Xia Z., Webster A., Du F., Piatkov K., Ghislain M., and Varshavsky A. (2008) Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283, 24011–24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xia Z., Turner G. C., Hwang C.-S., Byrd C., and Varshavsky A. (2008) Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter. J. Biol. Chem. 283, 28958–28968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shemorry A., Hwang C. S., and Varshavsky A. (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Theodoraki M. A., Nillegoda N. B., Saini J., and Caplan A. J. (2012) A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J. Biol. Chem. 287, 23911–23922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khosrow-Khavar F., Fang N. N., Ng A. H. M., Winget J. M., Comyn S. A., and Mayor T. (2012) The yeast Ubr1 ubiquitin ligase participates in a prominent pathway that targets cytosolic thermosensitive mutants for degradation. G3 Genes 2, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prasad R., Kawaguchi S., and Ng D. T. (2012) Biosynthetic mode can determine the mechanism of protein quality control. Biochem. Biophys. Res. Commun. 425, 689–695 [DOI] [PubMed] [Google Scholar]
- 40. Sultana R., Theodoraki M. A., and Caplan A. J. (2012) UBR1 promotes protein kinase quality control and sensitizes cells to Hsp90 inhibition. Exp. Cell Res. 18, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nillegoda N. B., Theodoraki M. A., Mandal A. K., Mayo K. J., Ren H. Y., Sultana R., Wu K., Johnson J., Cyr D. M., and Caplan A. J. (2010) Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell 21, 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamano K., and Youle R. J. (2013) PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Piatkov K. I., Brower C. S., and Varshavsky A. (2012) The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci. U.S.A. 109, E1839–E1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brower C. S., Piatkov K. I., and Varshavsky A. (2013) Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol. Cell 50, 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Z., Payoe R., and Fahlman R. P. (2012) The C-terminal proteolytic fragment of the breast cancer susceptibility type 1 protein (BRCA1) is degraded by the N-end rule pathway. J. Biol. Chem. 287, 7495–7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eldeeb M. A., and Fahlman R. P. (2014) The anti-apoptotic form of tyrosine kinase Lyn that is generated by proteolysis is degraded by the N-end rule pathway. Oncotarget 5, 2714–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jentsch S., McGrath J. P., and Varshavsky A. (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329, 131–134 [DOI] [PubMed] [Google Scholar]
- 48. Rao H., Uhlmann F., Nasmyth K., and Varshavsky A. (2001) Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955–959 [DOI] [PubMed] [Google Scholar]
- 49. Piatkov K. I., Colnaghi L., Békés M., Varshavsky A., and Huang T. T. (2012) The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol. Cell 48, 926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y. J., Liu C., Chang Z., Wadas B., Brower C. S., Song Z. H., Xu Z. L., Shang Y. L., Liu W. X., Wang L. N., Dong W., Varshavsky A., Hu R. G., and Li W. (2016) Degradation of the separase-cleaved Rec8, a meiotic cohesin subunit, by the N-end rule pathway. J. Biol. Chem. 291, 7426–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ouyang Y., Kwon Y. T., An J. Y., Eller D., Tsai S.-C., Diaz-Perez S., Troke J. J., Teitell M. A., and Marahrens Y. (2006) Loss of Ubr2, an E3 ubiquitin ligase, leads to chromosome fragility and impaired homologous recombinational repair. Mutat. Res. 596, 64–75 [DOI] [PubMed] [Google Scholar]
- 52. An J. Y., Kim E., Zakrzewska A., Yoo Y. D., Jang J. M., Han D. H., Lee M. J., Seo J. W., Lee Y. J., Kim T. Y., de Rooij D. G., Kim B. Y., and Kwon Y. T. (2012) UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLoS One 7, e37414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim S. T., Tasaki T., Zakrzewska A., Yoo Y. D., Sa Sung K., Kim S. H., Cha-Molstad H., Hwang J., Kim K. A., Kim B. Y., and Kwon Y. T. (2013) The N-end rule proteolytic system in autophagy. Autophagy 9, 1100–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cha-Molstad H., Sung K. S., Hwang J., Kim K. A., Yu J. E., Yoo Y. D., Jang J. M., Han D. H., Molstad M., Kim J. G., Lee Y. J., Zakrzewska A., Kim S. H., Kim S. T., Kim S. Y., Lee H. G., Soung N. K., Ahn J. S., Ciechanover A., Kim B. Y., and Kwon Y. T. (2015) Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 17, 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tasaki T., Kim S. T., Zakrzewska A., Lee B. E., Kang M. J., Yoo Y. D., Cha-Molstad H. J., Hwang J., Soung N. K., Sung K. S., Kim S. H., Nguyen M. D., Sun M., Yi E. C., Kim B. Y., and Kwon Y. T. (2013) UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc. Natl. Acad. Sci. U.S.A. 110, 3800–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Park S. E., Kim J. M., Seok O. H., Cho H., Wadas B., Kim S. Y., Varshavsky A., and Hwang C. S. (2015) Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 347, 1249–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. An J. Y., Kim E.-A., Jiang Y., Zakrzewska A., Kim D. E., Lee M. J., Mook-Jung I., Zhang Y., and Kwon Y. T. (2010) UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 107, 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kurosaka S., Leu N. A., Zhang F., Bunte R., Saha S., Wang J., Guo C., He W., and Kashina A. (2010) Arginylation-dependent neural crest cell migration is essential for mouse development. PLoS Genet. 6, e1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wadas B., Borjigin J., Huang Z., Oh J.-H., Hwang C.-S., and Varshavsky A. (2016) Degradation of serotonin N-acetyltransferase, a circadian regulator, by the N-end rule pathway. J. Biol. Chem. 291, 17178–17196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zenker M., Mayerle J., Lerch M. M., Tagariello A., Zerres K., Durie P. R., Beier M., Hülskamp G., Guzman C., Rehder H., Beemer F. A., Hamel B., Vanlieferinghen P., Gershoni-Baruch R., Vieira M. W., et al. (2005) Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat. Genet. 37, 1345–1350 [DOI] [PubMed] [Google Scholar]
- 61. Hwang C. S., Sukalo M., Batygin O., Addor M. C., Brunner H., Aytes A. P., Mayerle J., Song H. K., Varshavsky A., and Zenker M. (2011) Ubiquitin ligases of the N-end rule pathway: assessment of mutations in UBR1 that cause the Johanson-Blizzard syndrome. PLoS One 6, e24925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang F., Cheng Y., An J. Y., Kwon Y. T., Eckardt S., Leu N. A., McLaughlin K. J., and Wang P. J. (2010) The ubiquitin ligase Ubr2, a recognition E3 component of the N-end rule pathway, stabilizes Tex.19.1 during spermatogenesis. PLoS One 5, e14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kwon Y. T., Xia Z., An J. Y., Tasaki T., Davydov I. V., Seo J. W., Sheng J., Xie Y., and Varshavsky A. (2003) Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 23, 8255–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. An J. Y., Seo J. W., Tasaki T., Lee M. J., Varshavsky A., and Kwon Y. T. (2006) Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 6212–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Graciet E., and Wellmer F. (2010) The plant N-end rule pathway: structure and functions. Trends Plant Sci. 15, 447–453 [DOI] [PubMed] [Google Scholar]
- 66. Licausi F., Kosmacz M., Weits D. A., Giuntoli B., Giorgi F. M., Voesenek L. A. C. J., Perata P., and van Dongen J. T. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 [DOI] [PubMed] [Google Scholar]
- 67. Holman T. J., Jones P. D., Russell L., Medhurst A., Ubeda Tomás S., Talloji P., Marquez J., Schmuths H., Tung S. A., Taylor I., Footitt S., Bachmair A., Theodoulou F. L., and Holdsworth M. J. (2009) The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4549–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sasidharan R., and Mustroph A. (2011) Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell 23, 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang G., Lin R. K., Kwon Y. T., and Li Y. P. (2013) Signaling mechanism of tumor cell-induced up-regulation of E3 ubiquitin ligase UBR2. FASEB J. 27, 2893–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Aksnes H., Hole K., and Arnesen T. (2015) Molecular, cellular, and physiological significance of N-terminal acetylation. Int. Rev. Cell. Mol. Biol. 316, 267–305 [DOI] [PubMed] [Google Scholar]
- 71. Dörfel M. J., and Lyon G. J. (2015) The biological functions of Naa10: from amino-terminal acetylation to human disease. Gene 567, 103–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Starheim K. K., Gevaert K., and Arnesen T. (2012) Protein N-terminal acettyltransferases: when the start matters. Trends Biochem. Sci. 37, 152–161 [DOI] [PubMed] [Google Scholar]
- 73. Piatkov K. I., Oh J.-H., Liu Y., and Varshavsky A. (2014) Calpain-generated natural protein pragments as short-lived substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 111, E817–E826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Choi W. S., Jeong B.-C., Joo Y. J., Lee M.-R., Kim J., Eck M. J., and Song H. K. (2010) Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 17, 1175–1181 [DOI] [PubMed] [Google Scholar]
- 75. Matta-Camacho E., Kozlov G., Li F. F., and Gehring K. (2010) Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 17, 1182–1187 [DOI] [PubMed] [Google Scholar]
- 76. Wang H., Piatkov K. I., Brower C. S., and Varshavsky A. (2009) Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol. Cell 34, 686–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hwang C. S., Shemorry A., Auerbach D., and Varshavsky A. (2010) The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol. 12, 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kwon Y. T., Kashina A. S., and Varshavsky A. (1999) Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol. Cell. Biol. 19, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hu R.-G., Brower C. S., Wang H., Davydov I. V., Sheng J., Zhou J., Kwon Y. T., and Varshavsky A. (2006) Arginyl-transferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J. Biol. Chem. 281, 32559–32573 [DOI] [PubMed] [Google Scholar]
- 80. Brower C. S., and Varshavsky A. (2009) Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brower C. S., Rosen C. E., Jones R. H., Wadas B. C., Piatkov K. I., and Varshavsky A. (2014) Liat1, an arginyltransferase-binding protein whose evolution among primates involved changes in the numbers of its 10-residue repeats. Proc. Natl. Acad. Sci. U.S.A. 111, E4936–E4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Weits D. A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H. M., Riegler H., Hoefgen R., Perata P., van Dongen J. T., and Licausi F. (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gibbs D. J., Md Isa N., Movahedi M., Lozano-Juste J., Mendiondo G. M., Berckhan S., Marín-de la Rosa N., Vicente Conde J., Sousa Correia C., Pearce S. P., Bassel G. W., Hamali B., Talloji P., Tomé D. F., Coego A., et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53, 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang F., Saha S., Shabalina S. A., and Kashina A. (2010) Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329, 1534–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hoernstein S. N., Mueller S. J., Fiedler K., Schuelke M., Vanselow J. T., Schuessele C., Lang D., Nitschke R., Igloi G. L., Schlosser A., and Reski R. (2016) Identification of targets and interaction partners of arginyl-tRNA protein transferase in the moss Physcomitrella patens. Mol. Cell. Proteomics 15, 1808–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Varshavsky A. (2012) Augmented generation of protein fragments during wakefulness as the molecular cause of sleep: a hypothesis. Protein Sci. 21, 1634–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wong C. C. L., Xu T., Rai R., Bailey A. O., Yates J. R. 3rd, Wolf Y. I., Zebroski H., and Kashina A. (2007) Global analysis of posttranslational protein arginylation. PLos Biol. 5, e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rai R., Wong C. C., Xu T., Leu N. A., Dong D. W., Guo C., McLaughlin K. J., Yates J. R. 3rd, and Kashina A. (2008) Arginyltransferase regulates α cardiac actin function, myofibril formation and contractility during heart development. Development 135, 3881–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Xu T., Wong C. C., Kashina A., and Yates J. R. 3rd (2009) Identification of N-terminally arginylated proteins and peptides by mass spectrometry. Nat. Protoc. 4, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Saha S., Wong C. C., Xu T., Namgoong S., Zebroski H., Yates J. R. 3rd, and Kashina A. (2011) Arginylation and methylation double up to regulate nuclear proteins and nuclear architecture in vivo. Chem. Biol. 18, 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang J., Han X., Saha S., Xu T., Rai R., Zhang F., Wolf Y. I., Wolfson A., Yates J. R. 3rd, and Kashina A. (2011) Arginyltransferase is an ATP-independent self-regulating enzyme that forms distinct functional complexes in vivo. Chem. Biol. 18, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kurosaka S., Leu N. A., Pavlov I., Han X., Ribeiro P. A., Xu T., Bunte R., Saha S., Wang J., Cornachione A., Mai W., Yates J. R. 3rd, Rassier D. E., and Kashina A. (2012) Arginylation regulates myofibrils to maintain heart function and prevent dilated cardiomyopathy. J. Mol. Cell. Cardiol. 53, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rai R., and Kashina A. (2005) Identification of mammalian arginyltransferases that modify a specific subset of protein substrates. Proc. Natl. Acad. Sci. U.S.A. 102, 10123–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang J., Han X., Wong C. C., Cheng H., Aslanian A., Xu T., Leavis P., Roder H., Hedstrom L., Yates J. R. 3rd, and Kashina A. (2014) Arginyltransferase ATE1 catalyzes midchain arginylation of proteins at side chain carboxylates in vivo. Chem. Biol. 21, 331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cornachione A. S., Leite F. S., Wang J., Leu N. A., Kalganov A., Volgin D., Han X., Xu T., Cheng Y. S., Yates J. R. 3rd, Rassier D. E., and Kashina A. (2014) Arginylation of myosin heavy chain regulates skeletal muscle strength. Cell Rep. 8, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Eriste E., Norberg Å., Nepomuceno D., Kuei C., Kamme F., Tran D.-T., Strupat K., Jörnvall H., Liu C., Lovenberg T. W., and Sillard R. (2005) A novel form of neurotensin post-translationally modified by arginylation. J. Biol. Chem. 280, 35089–35097 [DOI] [PubMed] [Google Scholar]
- 97. Zhang H., Deery M. J., Gannon L., Powers S. J., Lilley K. S., and Theodoulou F. L. (2015) Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots. Proteomics 15, 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Majovsky P., Naumann C., Lee C.-W., Lassowskat I., Trujillo M., Dissmeyer N., and Hoehenwarter W. (2014) Targeted proteomics analysis of protein degradation in plant signaling on an LTQ-Orbitrap mass spectrometer. J. Proteome Res. 13, 4246–4258 [DOI] [PubMed] [Google Scholar]
- 99. Ebhardt H. A., Nan J., Chaulk S. G., Fahlman R. P., and Aebersold R. (2014) Enzymatic generation of peptides flanked by basic amino acids to obtain MS/MS spectra with 2× sequence coverage. Rapid Commun. Mass Spectrom. 28, 2735–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang F., Saha S., and Kashina A. (2012) Arginylation-dependent regulation of a proteolytic product of talin is essential for cell-cell adhesion. J. Cell Biol. 197, 819–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang F., Patel D. M., Colavita K., Rodionova I., Buckley B., Scott D. A., Kumar A., Shabalina S. A., Saha S., Chernov M., Osterman A. L., and Kashina A. (2015) Arginylation regulates purine nucleotide biosynthesis by enhancing the activity of phosphoribosyl pyrophosphate synthase. Nat. Commun. 6, 7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wu C., and Li S. S. (2009) CelluSpots: a reproducible means of making peptide arrays for the determination of SH2 domain binding specificity. Methods Mol. Biol. 570, 197–202 [DOI] [PubMed] [Google Scholar]
- 103. Bock I., Kudithipudi S., Tamas R., Kungulovski G., Dhayalan A., and Jeltsch A. (2011) Application of CelluSpots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails. BMC Biochem. 12, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bock I., Dhayalan A., Kudithipudi S., Brandt O., Rathert P., and Jeltsch A. (2011) Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 6, 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., and Jeltsch A. (2010) Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38, 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Varshavsky A. (2004) Spalog and sequelog: neutral terms for spatial and sequence similarity. Curr. Biol. 14, R181–R183 [DOI] [PubMed] [Google Scholar]
- 107. Davydov I. V., and Varshavsky A. (2000) RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem. 275, 22931–22941 [DOI] [PubMed] [Google Scholar]
- 108. Varshavsky A. (2005) Ubiquitin fusion technique and related methods. Methods Enzymol. 399, 777–799 [DOI] [PubMed] [Google Scholar]
- 109. Varshavsky A. (2000) Ubiquitin fusion technique and its descendants. Methods Enzymol. 327, 578–593 [DOI] [PubMed] [Google Scholar]
- 110. Lévy F., Johnsson N., Rümenapf T., and Varshavsky A. (1996) Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. U.S.A. 93, 4907–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Catanzariti A.-M., Soboleva T. A., Jans D. A., Board P. G., and Baker R. T. (2004) An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 13, 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]


