Abstract
DNA polymerase (pol) ι is a Y-family polymerase involved in translesion synthesis, exhibiting higher catalytic activity with Mn2+ than Mg2+. The human germline R96G variant impairs both Mn2+-dependent and Mg2+-dependent activities of pol ι, whereas the Δ1–25 variant selectively enhances its Mg2+-dependent activity. We analyzed pre-steady-state kinetic and structural effects of these two metal ions and genetic variations on pol ι using pol ι core (residues 1–445) proteins. The presence of Mn2+ (0.15 mm) instead of Mg2+ (2 mm) caused a 770-fold increase in efficiency (kpol/Kd,dCTP) of pol ι for dCTP insertion opposite G, mainly due to a 450-fold decrease in Kd,dCTP. The R96G and Δ1–25 variants displayed a 53-fold decrease and a 3-fold increase, respectively, in kpol/Kd,dCTP for dCTP insertion opposite G with Mg2+ when compared with wild type, substantially attenuated by substitution with Mn2+. Crystal structures of pol ι ternary complexes, including the primer terminus 3′-OH and a non-hydrolyzable dCTP analogue opposite G with the active-site Mg2+ or Mn2+, revealed that Mn2+ achieves more optimal octahedral coordination geometry than Mg2+, with lower values in average coordination distance geometry in the catalytic metal A-site. Crystal structures of R96G revealed the loss of three H-bonds of residues Gly-96 and Tyr-93 with an incoming dNTP, due to the lack of an arginine, as well as a destabilized Tyr-93 side chain secondary to the loss of a cation-π interaction between both side chains. These results provide a mechanistic basis for alteration in pol ι catalytic function with coordinating metals and genetic variation.
Keywords: DNA enzyme, DNA polymerase, enzyme kinetics, metal ion-protein interaction, pre-steady-state kinetics, X-ray crystallography, genetic variation, translesion DNA synthesis, kinetics, crystal structure
Introduction
Genomic DNA is continuously attacked by a variety of endogenous and exogenous agents in cells, and the persistent unrepaired lesions can lead to genomic mutations and related diseases such as cancer. DNA polymerases (pols),2 as well as DNA repair enzymes, are key enzymes for maintaining or altering genomic integrity against DNA lesions during various DNA transactions in organisms. The DNA replicative mechanisms linked to DNA damage and repair are believed to contribute to producing various mutational signatures in human cancer genomes (1). At least 17 different human DNA polymerases have been identified to date, which differ in their functions in DNA replication, repair, recombination, and damage tolerance (2, 3).
Y-family DNA polymerases, including pols η, ι, κ, and REV1, are specialized in replicating through DNA lesions, so-called translesion DNA synthesis (TLS). These polymerases have low fidelity with undamaged DNA templates but have spacious and solvent-accessible active sites to allow the accommodation and replicative bypass of bulky and distorted DNA lesions (4). Individual Y-family polymerases play error-free or error-prone roles in TLS, depending on DNA lesion types in cells (5). At bulky carcinogen-derived N2-G DNA lesions (e.g. benzo[a]pyrene-diol epoxide-G), both pol κ and REV1 catalyze error-free TLS, but both pols η and ι catalyze error-prone TLS (6–14). By contrast, at UV-induced cyclobutane thymine dimers (T-T), only pol η (but not the other Y-family pols) can catalyze error-free TLS (15, 16). Therefore, the overall balance toward error-free TLS with all working polymerases at various DNA lesions might be crucial in preventing mutations from numerous genotoxic agents. Recently, we reported that catalytic (either hypoactive or hyperactive) alterations are found in a considerable number of human germline non-synonymous variants of Y-family pols κ, ι, and REV1 (17–19), which might potentially influence on the overall TLS capacity in the affected individuals.
pol ι is exceptionally error-prone in DNA synthesis among polymerases, particularly opposite template bases G and T, due to its uniquely restricted active-site and related non-Watson-Crick base pairing (20–22). pol ι is able to catalyze nucleotide insertion opposite a variety of DNA lesions, including N2- and O6-alkyl and aralkyl G adducts, 8-oxo-7,8-dihydroG (8-oxoG), pyrimidine dimers, and abasic sites, but mediates largely miscoding TLS (albeit occasionally accurate) with varied nucleotide selectivity depending on lesion type (13, 23, 24). Both C and T are inserted opposite template N2- and O6-alkyl and aralkyl G adducts by pol ι (13, 25). C is only slightly favored over G opposite template 8-oxoG by pol ι, A is favored opposite the 3′ T of (6-4) T-T photoproducts, and both G and T are favored opposite 5′ T of (6-4) T-T photoproducts and abasic sites (23, 24). A possible implication of pol ι in mutation and cancer has been suggested by substantial evidence from several knock-out mouse studies (26–28), as well as from multiple studies verifying frequent pol ι dysregulation in various types of human cancers (29–32). In this respect, the appropriate catalytic function of pol ι in cells might be required for preventing cancer. One of the distinctive catalytic properties of pol ι is the metal ion preference. Unlike other Y-family polymerases, pol ι is known to prefer Mn2+ over Mg2+ as the metal ion cofactor for catalysis (33, 34), but the structural mechanistic details remain speculative. In addition, substantial alterations were reported in metal-dependent DNA polymerase activity from two rare human germline pol ι variants, i.e. severe impairment of both Mg2+-dependent and Mn2+-dependent activities in the R96G variant and moderate enhancement of only the Mg2+-dependent activity in the Δ1–25 variant for matched and mismatched nucleotide incorporations opposite normal and lesion templates (17). Detailed structural and kinetic mechanisms of catalytic alterations of pol ι by these genetic variants still remain unclear. Any disease associations have not been reported yet, but the catalytically altered pol ι genetic variants might be of potential importance in that they would change the TLS capacity of pol ι and consequently modify mutation phenotypes to genotoxic agents in genetically predisposed individuals.
To elucidate both the kinetic and the structural basis for alterations in the catalytic function of pol ι by different two metal ions, Mg2+ and Mn2+, as well as by two human germline non-synonymous variants, R96G and Δ1–25, we performed pre-steady-state kinetic analysis for nucleotide insertion by pol ι and also determined x-ray crystal structures of ternary pol ι complexes in the presence of either Mg2+ or Mn2+ ions, using the recombinant human pol ι core (residues 1–445) proteins of wild type and two variants with a simple model of a correct dCTP incorporation opposite normal G. The combined pre-steady-state kinetic and structural results indicate that Mn2+ enables pol ι to adopt more ideal octahedral coordination in the active site and achieve much higher catalytic efficiency than Mg2+, whereas the R96G variant results in the loss of hydrogen bond interactions of residues Gly-96 and Tyr-93 with an incoming nucleotide as well as conferring a much greater reduction in its catalytic efficiency. Our detailed kinetic and structural results are discussed in the context of understanding the possible mechanistic and functional aspects of metal ions and genetic variations on pol ι.
Results
Pre-steady-state Kinetic Analysis of dCTP Incorporation Opposite G by pol ι(1–445), pol ι(26–445), and R96G pol ι(1–445) Enzymes in the Presence of Mg2+ or Mn2+
Pre-steady-state kinetic methods were used to quantify the alterations in catalytic efficiency and the apparent nucleotide binding affinity of pol ι by metal ions (Mg2+ or Mn2+) and known human genetic variants (Δ1–25 or R96G). Pre-steady-state kinetic parameters were determined for dCTP incorporation opposite template G into 18-mer/36-mer duplexes by pol ι(1–445), pol ι(26–445), and R96G pol ι(1–445) enzymes under single turnover conditions (where pol ι was present in 10-fold excess over DNA substrate), in the presence of either 0.15 mm MnCl2 or 2 mm MgCl2, which is in the optimal range for pol ι activity (33, 34), using a rapid quench flow instrument. Analysis of the change of the observed rate (kobs) as a function of increasing dCTP concentration yielded kpol, the maximal rate of nucleotide incorporation, and Kd,dCTP, a measure of the binding affinity of dCTP to the pol·DNA binary complex to form a ternary complex poised for catalysis (Table 1 and supplemental Fig. S1). pol ι(1–445) displayed a kpol of 0.74 ± 0.06 s−1 and a Kd,dCTP of 1.5 ± 0.4 μm in the presence of Mn2+. Thus, the catalytic efficiency (kpol/Kd,dCTP) of pol ι(1–445) with Mn2+ was estimated to be 4.9 × 105 m−1 s−1, which was 770-fold higher than that with Mg2+, mainly due to a 450-fold lower Kd,dCTP. Similar trends of kinetic results were also observed with pol ι(26–445) and the R96G variant, indicating that pol ι binds nucleotide much more tightly and catalyzes nucleotide insertion much more efficiently in the presence of Mn2+ than in the presence of Mg2+. pol ι(26–445) had a kpol/Kd,dCTP value similar to that of pol ι(1–445) in the presence of Mn2+ but displayed a 3-fold increase in kpol/Kd,dCTP in the presence of Mg2+ when compared with pol ι(1–445), indicating a slight enhancement only in the Mg2+-dependent catalytic efficiency of pol ι due to the deletion of N-terminal 25 residues. The R96G variant showed a 53-fold decrease in kpol/Kd,dCTP for dCTP insertion opposite G in the presence of Mg2+ when compared with that with wild type, while showing a 9-fold decrease of that value in the presence of Mn2+. This mitigation effect of Mn2+ on the Mg2+-dependent kpol/Kd,dCTP reduction in the R96G variant seemed to be mainly due to the full restoration of the apparent dCTP binding affinity to the level comparable with that of wild type by Mn2+.
TABLE 1.
Pre-steady-state kinetic parameters for dCTP incorporation opposite G by pol ι(1–445), pol ι(26–445), and R96G pol ι(1–445) in the presence of either Mg2+ or Mn2+
| Metal | pol | kpol | Kd,dCTP | kpol/Kd,dCTP | Relative efficiencya |
|---|---|---|---|---|---|
| s−1 | μm | m−1 s−1 | |||
| MgCl2 (2 mm) | pol ι (1–445) | 0.43 ± 0.02 | 670 ± 100 | 6.4 × 102 | 1 |
| pol ι (26–445) | 0.77 ± 0.024 | 390 ± 40 | 2.0 × 103 | 3.1 | |
| R96G pol ι (1–445) | 0.063 ± 0.007 | 5200 ± 1300 | 1.2 × 10 | 0.019 | |
| MnCl2 (150 μm) | pol ι (1–445) | 0.74 ± 0.06 | 1.5 ± 0.4 | 4.9 × 105 | 1 |
| pol ι (26–445) | 0.79 ± 0.03 | 1.8 ± 0.2 | 4.4 × 105 | 0.90 | |
| R96G pol ι (1–445) | 0.077 ± 0.002 | 1.4 ± 0.1 | 5.5 × 104 | 0.11 |
a Relative efficiency, calculated as the ratio of the kpol/Kd,dCTP of each pol ι for dCTP insertion opposite G to the kpol/Kd,dCTP of pol ι(1–445) for dCTP insertion opposite G.
Crystal Structures of Human pol ι Pre-catalytic Ternary Complexes Incorporating Nonhydrolyzable dCTP Analogue (dCMPNPP) Opposite Template G in the Presence of Mg2+ or Mn2+
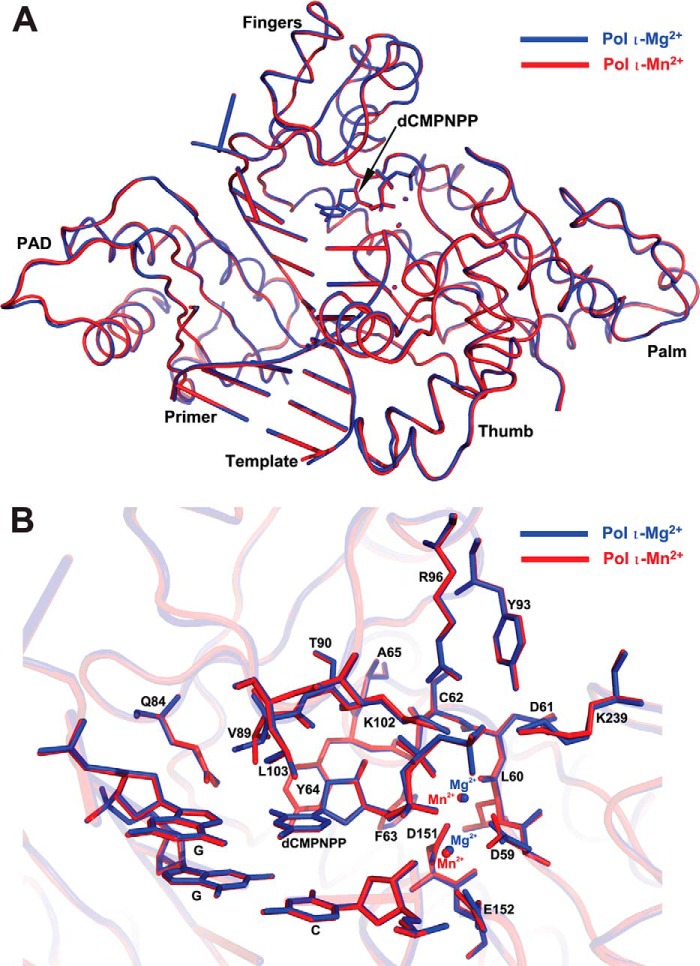
To observe the structural alterations of pol ι by metal ions (Mg2+ or Mn2+) and genetic variants (Δ1–25 or R96G), we determined six crystal structures of pre-insertion ternary complexes of pol ι(1–445), pol ι(26–445), and R96G pol ι(1–445) with DNA containing template G and dCMPNPP in the presence of either Mg2+ or Mn2+. The strategy, employing a non-hydrolyzable dCTP analogue, dCMPNPP, as well as DNA substrate having an intact 3′-OH at the primer end, was utilized to capture pre-catalytic ternary pol ι complexes that preserve metal-coordinating ligands in the active site while preventing catalysis in the presence of active-site metal ions, as successfully applied with pol β (35). Crystals of ternary complexes of pol ι(26–445), pol ι(1–445), and R96G pol ι(1–445) diffracted to about 2.5–2.6, 3.2–3.6, and 2.8 Å resolution, respectively (Table 2). All ternary complex structures of pol ι contained two metal ions with relatively lower occupancy in the A-site metal ions in the active site, except for the R96G pol ι(1–445)·Mg2+ ternary complex that missed an Mg2+ ion at the metal A-site near the primer terminus 3′-OH. To our knowledge, our structures represent the first ternary pol ι structures containing the primer end 3′-OH entity, as well as defining the position of two active-site metal ions of either Mg2+ or Mn2+, which provides geometric information in the pre-catalytic state. However, electron density was not observed for the N-terminal 25 residues in all pol ι(1–445) ternary complexes, suggesting the disordered nature of this negatively charged N-terminal region. Thus, all the refined structures of pol ι ternary complexes contained pol ι residues 51–439 as observed previously with the ternary pol ι(26–445) complex structure (36). The overall structures of six ternary pol ι complexes were almost identical, except for subtle variations near metal ions in the active site and at the amino acid substitution site (supplemental Fig. S2). Three pol ι structures with the Mn2+ ions were superimposed with root mean square deviations (RMSD) of 0.20–0.27 Å among the positions of Cα atoms (supplemental Fig. S2, A and B), whereas three pol ι structures with Mg2+ ions were superimposed with RMSD of 0.31–0.35 Å, indicating slightly more backbone variations in the presence of Mg2+ (supplemental Fig. S2, A–D). Superposition of relatively high resolution (2.5 and 2.6 Å, respectively) structures of the pol ι(26–445)·Mg2+ and pol ι(26–445)·Mn2+ ternary complexes showed almost identical overall structures between them with an RMSD of 0.20 Å over Cα atoms but displayed slight differences in positions of metal ions as well as side chains of nearby residues Asp-59 and Glu-152 in the active site (Fig. 1B), indicating possible differences between Mg2+ and Mn2+ coordination in the pol ι active site.
TABLE 2.
Crystal data, data collection parameters, and structure refinement statistics
| Complex |
||||||
|---|---|---|---|---|---|---|
| pol ι(26–445) G·dCMPNPP (Mg2+) | pol ι(26–445) G·dCMPNPP (Mn2+) | pol ι(1–445) G·dCMPNPP (Mg2+) | pol ι(1–445) G·dCMPNPP (Mn2+) | R96G pol ι(1–445) G·dCMPNPP (Mg2+) | R96G pol ι(1–445) G·dCMPNPP (Mn2+) | |
| Data collection | ||||||
| Wavelength (Å) | 0.97872 | 0.97872 | 0.97872 | 0.97872 | 0.97872 | 0.97872 |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Resolution (Å)a | 50.00–2.49 (2.53–2.49) | 50.00–2.64 (2.69–2.64) | 50.00–3.56 (3.62–3.56) | 50.00–3.15 (3.20–3.15) | 50.00–2.78 (2.83–2.78) | 50.00–2.80 (2.85–2.80) |
| Unit cell (a, b, c) (Å) | 97.61, 97.61, 203.00 | 97.83, 97.83, 202.89 | 98.27, 98.27, 201.66 | 97.74, 97.74, 202.01 | 98.12, 98.12, 202.72 | 97.54, 97.54, 202.21 |
| Unit cell (α, β, γ) (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| No. of unique reflectionsa | 20,810 (1006) | 17,727 (847) | 7564 (363) | 10,495 (487) | 15,324 (729) | 14,993 (720) |
| Completeness (%)a | 99.6 (99.4) | 100 (100) | 99.9 (100) | 99.8 (99.6) | 100 (100) | 99.9 (99.9) |
| I/σ(I)a | 36.2 (3.5) | 31.8 (4.0) | 11.6 (2.5) | 22.9 (4.5) | 30.4 (2.8) | 30.1 (3.7) |
| Wilson B-factor (Å2) | 32.2 | 33.7 | 62.1 | 40.0 | 46.7 | 41.9 |
| Rmergea,b | 0.121 (0.914) | 0.090 (0.784) | 0.222 (0.990) | 0.180 (0.811) | 0.100 (0.922) | 0.117 (0.842) |
| Redundancya | 21.3 (22.0) | 21.2 (21.8) | 14.9 (14.4) | 20.8 (21.1) | 17.7 (17.4) | 21.0 (21.3) |
| Refinement | ||||||
| Rwork | 0.220 | 0.204 | 0.204 | 0.202 | 0.217 | 0.224 |
| Rfree | 0.249 | 0.248 | 0.244 | 0.236 | 0.250 | 0.266 |
| No. of atoms | ||||||
| Protein/DNA | 2941/345 | 2934/345 | 2928/329 | 2926/345 | 2890/345 | 2887/345 |
| dNTP/Metal ions | 28/3c | 28/3c | 28/2 | 28/2 | 28/1 | 28/2 |
| Water | 85 | 65 | 0 | 0 | 22 | 10 |
| B-factor (Å2) | ||||||
| Average | 53.9 | 51.6 | 68.9 | 46.4 | 65.0 | 51.4 |
| Protein/DNA | 54.9/50.4 | 52.5/47.3 | 69.5/64.9 | 47.2/41.0 | 65.4/63.9 | 51.9/49.6 |
| dNTP/Metal ions | 34.9/36.7 | 32.4/59.0 | 54.3/44.9 | 26.3/43.9 | 47.8/40.7 | 31.6/30.6 |
| Water | 42.6 | 41.1 | − | − | 48.7 | 34.1 |
| Root mean square deviations | ||||||
| Bonds (Å) | 0.005 | 0.009 | 0.003 | 0.004 | 0.003 | 0.003 |
| Angles (°) | 0.886 | 1.024 | 0.633 | 0.676 | 0.653 | 0.629 |
| Ramachandran | ||||||
| Favored (%) | 98.7 | 97.9 | 97.1 | 97.3 | 98.4 | 97.6 |
| Allowed (%) | 1.1 | 1.9 | 2.9 | 2.7 | 1.6 | 2.4 |
| Outliers (%) | 0.3 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 |
| PDB code | 5KT2 | 5KT3 | 5KT6 | 5KT7 | 5KT4 | 5KT5 |
a Values for highest resolution shell are given in parentheses.
b Rmerge: Rlinear = SUM(ABS(I − 〈I〉))/SUM(I), where I is the integrated intensity of a given reflection.
c One non-catalytic metal ion is present in the structure.
FIGURE 1.
Structures of pol ι·DNA·nucleotide ternary complexes in the presence of Mg2+ or Mn2+. A, superposition of the overall structures of ternary complexes of pol ι(26–445) with DNA containing template G and incoming non-hydrolyzable dCMPNPP in the presence of Mg2+ (blue) or Mn2+ (red). pol ι is shown as ribbons, DNA is shown as tube and ladder, the nucleotide is shown as sticks, and metal ions are shown as spheres. B, superposition of the active sites of the pol ι(26–445)·G·dCMPNPP·Mg2+ (blue) and pol ι(26–445)·G·dCMPNPP·Mn2+ (red) complexes. Active-site residues, DNA, and nucleotide are shown as sticks, and metal ions are shown as spheres.
Coordination of Two Divalent Mg2+ or Mn2+ Ions in the Active Sites of Human pol ι(26–445) Ternary Complexes with Primer 3′-OH and dCMPNPP
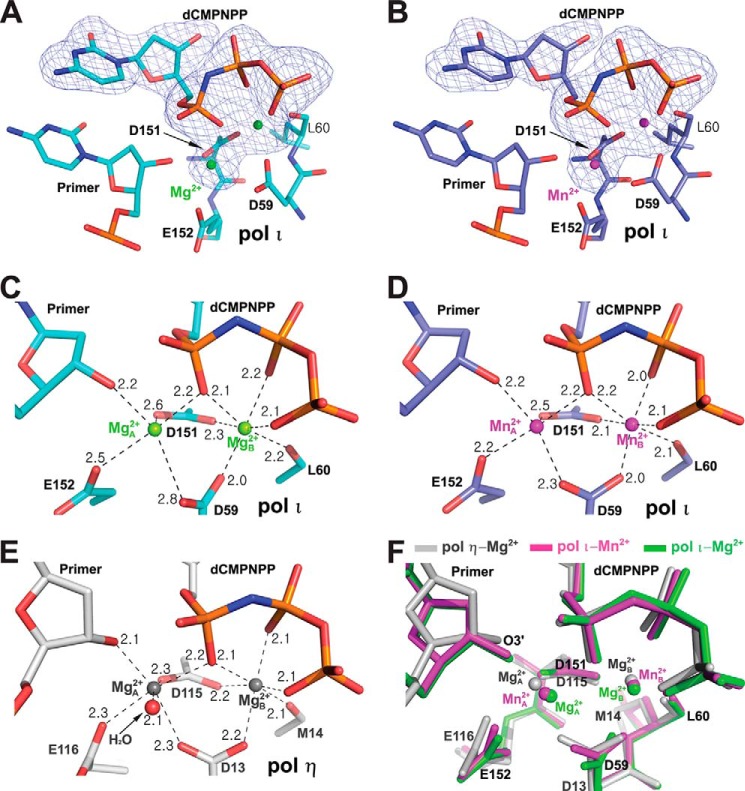
To understand the differences in the metal coordination geometry between Mg2+ and Mn2+ ions in the pol ι active site, we compared the coordination structures of Mg2+ and Mn2+ ions in the pol ι active site utilizing relatively high resolution structures of the pol ι(26–445)·Mg2+ and pol ι(26–445)·Mn2+ ternary complexes. The Fo − Fc omit maps for the incoming nucleotide and metal ions showed clear density for the incoming dCMPNPP and two Mg2+ (or Mn2+) ions bound in the pol ι active site (Fig. 2, A and B). Two Mg2+ or Mn2+ ions were present in both the catalytic metal site (A-site) and the nucleotide binding metal site (B-site) in the pol ι active site, as typically observed with polymerases (37), although having partial occupancies (63 and 67%, respectively) at the A-site. Both Mg2+ and Mn2+ at the B-site in the pol ι active site showed nearly perfect octahedral coordination geometry with six ligands, which had similar values of average coordination distance (2.14 and 2.06 Å, respectively) and distance RMSD (0.230 and 0.214 Å, respectively), albeit slightly lower with Mn2+ (Table 3 and Fig. 2, C–E), as observed with the B-site Mg2+ in the previously reported structures of normal G·dCMPNPP ternary complexes of pols β and η (38, 39). The A-site Mn2+ in the pol ι ternary complex also showed good octahedral coordination geometry involving five ligands, although missing one ligand (possibly a water molecule observed with pols β and η ternary complexes) (38, 39), yielding average coordination distance (2.29 Å) and distance RMSD (0.305 Å) values that were similar to those (2.22–2.34 and 0.187–0.293 Å, respectively) observed with the A-site Mg2+ in the normal G·dCMPNPP ternary complexes of pols β and η (38, 39) (Table 3 and Fig. 2, D and E). In strong contrast, the Mg2+ at the A-site in the pol ι active site showed a considerable deviation from the ideal octahedral coordination geometry, yielding average coordination distance (2.45 Å) and distance RMSD (0.474 Å) values that were substantially higher than those with Mn2+ (Table 3 and Fig. 2, C and D). These more optimal features with Mn2+ than Mg2+ for octahedral coordination geometry at the A-site were similarly observed with the refined pol ι(1–445) ternary complex structures (Table 3), albeit having lower resolution (3.6 and 3.2 Å). The poor coordination with Mg2+ at the A-site appeared to be related to subtle displacement of the side-chain carboxyl group of Asp-59 away from the A-site Mg2+ as well as a slight shift of the A-site Mg2+ to the B-site Mg2+, yielding a 0.2 Å shortening of inter-metal distance when compared with that with Mn2+ in the pol ι active site (Fig. 2, C, D, and F). Interestingly, the C3′-endo conformation was equally observed at the sugar moieties at the 3′ primer end (Fig. 2), as well as the nucleotide 5′ to the primer end, and three nucleotide pairs at positions n-2 to n-4 of the primer/template duplex in both pol ι ternary complex structures, unlike the previously reported structures of the pol ι binary and ternary complexes lacking the primer end 3′-OH (Protein Data Bank (PDB) IDs 2FLP and 2ALZ) (36, 40), indicating a distinctive pattern of sugar pucker changes induced in the pre-catalytic pol ι ternary complex.
FIGURE 2.
Comparison of Mg2+ and Mn2+ coordinations in the pol ι active site. A and B, Fo − Fc simulated annealing omit maps (blue) contoured at 3.0σ for the incoming dCMPNPP and Mg2+ (A) or Mn2+ (B) ions in the active sites of pol ι. The electron density is superimposed on the refined modeled dCMPNPP and metal ions. Mg2+ and Mn2+ ions are shown as green and magenta spheres, respectively. C and D, close-up view of Mg2+ (C) or Mn2+ (D) coordination in the active-site metal binding sites of pol ι·G·dCMPNPP complexes. Metal ion coordination is shown as dashed lines, and the coordination distances are indicated. E, close-up view of Mg2+ coordination in the active-site metal binding site of pol η·G·dCMPNPP complex (PDB ID 4DL3). Mg2+ ions are shown as gray spheres. F, superposition of the active-site metal binding sites of the pol ι(26–445)·G·dCMPNPP·Mg2+ (green), pol ι(26–445)·G·dCMPNPP·Mn2+ (magenta), and pol η·G·dCMPNPP·Mg2+ (PDB ID 4DL3, gray) complexes. Red, blue, and orange colors indicate oxygen, nitrogen, and phosphorus atoms, respectively. Cyan (in A and C), light blue (in B and D), and light gray (in E) colors indicate carbon atoms.
TABLE 3.
Active site metal coordination distances and angles in normal G·dCMPNPP ternary complex structures of pols ι, η, and β in the presence of divalent metal ions
| pol | Metal ion | Inter-metal distance | A-site metal coordination |
B-site metal coordination |
||||
|---|---|---|---|---|---|---|---|---|
| na | Distanceb (Å) | Distance RMSDc (Å) | n | Distance (Å) | Distance RMSD (Å) | |||
| pol ι(26–445) | Mg2+ | 3.23 | 5 | 2.45 ± 0.12 | 0.474 | 6 | 2.14 ± 0.04 | 0.230 |
| Mn2+ | 3.43 | 5 | 2.29 ± 0.06 | 0.305 | 6 | 2.06 ± 0.04 | 0.214 | |
| pol ι(1–445) | Mg2+ | 3.02 | 5 | 2.37 ± 0.14 | 0.869 | 6 | 2.13 ± 0.05 | 0.257 |
| Mn2+ | 3.33 | 5 | 2.22 ± 0.18 | 0.409 | 6 | 2.10 ± 0.07 | 0.336 | |
| pol ηd | Mg2+ | 3.55 | 6 | 2.34 ± 0.06 | 0.256 | 6 | 2.17 ± 0.04 | 0.236 |
| pol ηe | Mg2+ | 3.72 | 6 | 2.22 ± 0.04 | 0.187 | 6 | 2.13 ± 0.03 | 0.215 |
| pol βf | Mg2+ | 3.44 | 6 | 2.23 ± 0.06 | 0.293 | 6 | 2.04 ± 0.02 | 0.183 |
Active-site Structures of Human R96G pol ι(1–445) Ternary Complexes in the Presence of Mg2+ or Mn2+
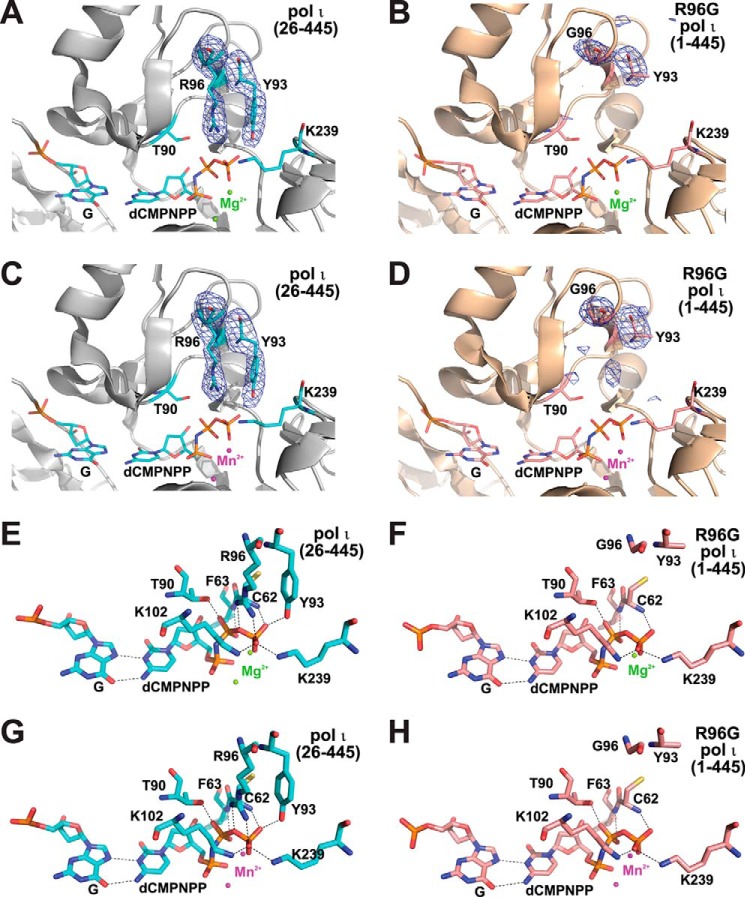
To reveal the structural mechanism of severe catalytic impairments in the R96G pol ι variant, we compared the active-site structures of R96G pol ι(1–445) ternary complexes with those of pol ι(26–445) ternary complexes of relatively high resolution. Interestingly, the R96G variant structures showed substantial alterations not only at the amino acid substitution site (Gly-96) but also at the nearby residue site (Tyr-93), when compared with pol ι(26–445) (Fig. 3). The Fo − Fc simulated annealing omit maps for two key residues (Gly-96 and Tyr-93) in R96G pol ι·Mg2+ and R96G pol ι·Mn2+ ternary complexes showed no clear electron density for two long side chains of Arg-96 and Tyr-93 (Fig. 3, B and D), which were obviously observed in those with the pol ι(26–445) complexes (Fig. 3, A and C). This collateral destabilization of the Tyr-93 side chain seems to be related to the loss of a cation-π interaction between Arg and Tyr side chains due to the loss of an Arg side chain in the R96G variant. Accordingly, two hydrogen bonds of the Arg-96 side chain with a β,γ-bridging oxygen and a γ-phosphate oxygen of dCMPNPP, as well as one hydrogen bond of the Tyr-93 side chain with a γ-phosphate oxygen, were lost in R96G variant structures when compared with those in pol ι(26–445) ternary complex structures (Fig. 3, E–H). Interestingly, only Mn2+ but not Mg2+ was observed at the A-site in the R96G pol ι active site (Fig. 3, B and D), possibly reflecting an inferior A-site binding ability of Mg2+ when compared with Mn2+, which might in part be attributed to a 6-fold greater decrease in catalytic efficiency of the R96G variant with Mg2+ than that with Mn2+ (Table 1).
FIGURE 3.
The active sites of R96G pol ι ternary complexes in the presence of Mg2+ or Mn2+. A–D, Fo − Fc simulated annealing omit maps contoured at 3.0σ for the residues Arg-96 (or Gly-96) and Tyr-93 in the active sites of pol ι(26–445)·G·dCMPNPP·Mg2+ (A); R96G pol ι(1–445)·G·dCMPNPP·Mg2+ (B); pol ι(26–445)·G·dCMPNPP·Mn2+ (C); and R96G pol ι(1–445)·G·dCMPNPP·Mn2+ (D) complexes. The electron density (blue mesh) is superimposed on the refined modeled residues Arg-96 (or Gly-96) and Tyr-93. Mg2+ and Mn2+ ions are shown as green and magenta spheres. E–H, close-up view of the active sites of pol ι(26–445)·G·dCMPNPP·Mg2+ (D); R96G pol ι(1–445)·G·dCMPNPP·Mg2+ (F); pol ι(26–445)·G·dCMPNPP·Mn2+ (G); and R96G pol ι(1–445)·G·dCMPNPP·Mn2+ (H) complexes. Hydrogen-bonding interactions are shown as dashed lines. Red, blue, orange, and yellow colors indicate oxygen, nitrogen, phosphorus, and sulfur atoms, respectively. Cyan (in A, C, E, and G) and light pink (in B, D, F, and H) colors indicate carbon atoms.
Discussion
In this study, we provide structural and pre-steady-state kinetic evidence that Mn2+ is more optimal for the two-metal ion binding site configuration and catalysis of pol ι than Mg2+. Our pol ι ternary complex structures containing intact coordinating ligands such as the primer end 3′-OH reveal that Mn2+ ions achieve more ideal octahedral coordination geometry than Mg2+ ions in the pol ι active site, specifically in the catalytic metal A-site. Moreover, our pre-steady-state kinetic data demonstrate that Mn2+ ions confer much higher (2–3 orders of magnitude) efficiency of pol ι catalysis than Mg2+ ions, mainly through augmenting nucleotide binding affinity. We also confirmed that the R96G variant, displaying a severe reduction in catalytic efficiency, loses three hydrogen bond interactions of two residues (Gly-96 and Tyr-93) with an incoming nucleotide in the pol ι active site. Interestingly, Mn2+ substantially rescued the catalytic impairment in the Gly-96 variant by restoring the apparent nucleotide binding affinity. The Δ1–25 variant displayed a small alteration (3-fold increase) only in Mg2+-dependent catalytic efficiency, although the structural effects of N-terminal 25 residues are not discernible due to the absence of their electron density.
The key differences between the pol ι(26–445)·Mg2+ and pol ι(26–445)·Mn2+ ternary complex structures were located at and near the catalytic metal A-site in the pol ι active site, particularly the configuration of the Asp-59 side chain as well as the positioning of the A-site metal (Fig. 2F), which substantially modified the A-site metal coordination. First, in the presence of Mg2+ ions as opposed to Mn2+ ions, the side-chain carboxyl group of Asp-59 is rotationally displaced to locate a ligand atom (one carboxyl oxygen of Asp-59) more distant (0.5 Å) from the A-site metal (despite no alteration in coordination distance between the other carboxyl oxygen and the B-site metal) in the pol ι active site. Second, Mg2+ (when compared with Mn2+) at the A-site was positionally shifted closer (∼0.2 Å) to the B-site metal in the pol ι active site, yielding an inter-metal distance shorter than that with Mn2+ ions (Table 3). Thus, in the pol ι active site, only Mn2+ ions but not Mg2+ ions seem to achieve an inter-metal distance similar to that with Mg2+ ions usually observed in active sites of ternary complex structures of other human pols η and β with normal G·dCMPNPP (38, 39) (Table 3). Superposition of the active-site metal binding sites of pol ι·Mg2+, pol ι·Mn2+, and pol η·Mg2+ (PDB ID 4DL3) ternary complexes appears to reflect some gradational changes in the extent of positional shift of the A-site metal (toward the B-site metal) as well as in the angle of rotational displacement of the side-chain carboxyl group of Asp-59 (or Asp-13 for pol η), in the order of pol η·Mg2+ < pol ι·Mn2+ < pol ι·Mg2+. (Fig. 2F). Consequently, Mg2+ ions led to a considerable deviation from the ideal octahedral coordination geometry at the A-site, yielding the values of average coordination distance and distance RMSD (2.45 and 0.474 Å, respectively), which were quite higher than those with Mn2+ (Table 3). These coordination parameters with Mg2+ ions seem to be slightly improved in the aspect of average coordination distance but worsened in the aspect of distance RMSD by the presence of the primer terminus 3′-OH, when compared with those (2.66 and 0.456 Å, respectively) of the A-site Mg2+ (albeit involving only four coordinating atoms) in the previously reported structure (PDB ID 2ALZ) of pol ι ternary complex lacking the primer end 3′-OH (36). This Mg2+-induced geometric alteration at the A-site in the pol ι active site contrasts with the near perfect octahedral coordination of the A-site Mg2+ observed in ternary complex structures of other pols η and β with the correct incoming non-hydrolyzable nucleotide (Table 3) (38, 39).
The Mn2+ requirement for achieving the optimal octahedral coordination geometry at the A-site in ternary complex structures with the correct incoming nucleotide seems to be unique in pol ι. Other DNA polymerases such as bacteriophage pol RB69 and human pol β achieve good octahedral coordination geometry not only with Mg2+ but also with Mn2+ at the A-site, as observed in their ternary complex structures with the correct incoming non-hydrolyzable nucleotide (PDB IDs 3SJJ, 3SPY, 2FMS, and 3C2K) (35, 41, 42). It is notable that the superior coordinating ability of Mn2+ when compared with Mg2+ at the A-site is observed in the previously reported structures of ternary pol β complexes (PDB IDs 4PGQ, 4PGX, 4PHA, and 4PHD) with an incoming incorrect non-hydrolyzable nucleotide (43). Our data and that of others suggest that Mn2+ is more tolerant of atypical pol active sites (e.g. the inherently restricted pol ι active site and the distorted pol active site due to base pair mismatch) than Mg2+) and thus able to form good octahedral coordination geometry not only at the B-site but also at the A-site in the pol active site. This effect might be attributed to a relaxed coordination requirement of Mn2+ when compared with Mg2+ (37). It is of interest to perform further studies to verify whether our results of pol ι with normal base pairs of template G and incoming dCTP are valid for other base pairs involving DNA lesions or mismatches such as G:T and T:G pairs.
The Mg2+-induced distortion of the A-site coordination geometry in the pol ι active-site structure seems to be closely related to a severe (220–770-fold) diminution in catalytic efficiency (kpol/Kd,dCTP) of pol ι in the presence of Mg2+ when compared with Mn2+ (Table 1). This severe catalytic impairment with Mg2+ was mainly due to a severe reduction in the affinity of productive nucleotide binding of pol ι in the presence of Mg2+ when compared with Mn2+, as reflected by large (220–450-fold) increases in the apparent equilibrium dissociation constant (Kd,dCTP) for incoming dCTP (Table 1). These results suggest that substitution of Mn2+ for Mg2+ might boost the catalytic efficiency of pol ι for correct nucleotide insertion, mainly through improving the binding affinity of nucleotide, by achieving optimal octahedral coordination at the A-site in the active site. Our finding is in good agreement with previous studies with other polymerases, e.g. RB69 and pol β (42, 44). It is also notable that the extent of increases (220–450-fold) in the nucleotide binding affinity by Mn2+ substitution appears to be much higher with pol ι (Table 3) than those (3–4- and 8–19-fold, respectively) observed with RB69 and pol β (42, 44), implying a more marked effect of Mn2+ on pol ι than other polymerases. Our combined structural and kinetic data suggest that the optimal octahedral coordination of two active-site metal ions is essential for proper catalytic function of pol ι, and Mn2+ is superior in this respect when compared with Mg2+, particularly at the A-site in the pol ι active site. The superiority of Mn2+ for pol ι function is also supported by the biochemical property of pol ι(1–445) to more tightly bind DNA substrates in the presence of a low level of Mn2+ than Mg2+ (17). Although DNA polymerases most likely utilize physiologically abundant Mg2+ ions for catalysis in vivo, from our and previous studies (17, 33, 34), it may be relevant to postulate that pol ι would inherently employ physiologically low levels of Mn2+ for catalysis in kinetic and structural preference to Mg2+ in vivo. Similarly, pol λ has been also suggested to use Mn2+ as the preferred activating metal ion in vivo (45). Mn2+ has also been reported to increase the activity of non-canonical DNA polymerases such as Sulfolobus solfataricus Dpo4 and human PrimPol in vitro (46, 47). We also note a very recent study suggesting the requirement of a third metal ion for pol η catalysis, with slight preference for Mg2+ (48). It would be of interest to investigate whether this is the case in pol ι catalysis.
The loss of hydrogen-bonding interactions of two structurally altered residues, the substituted Gly-96 and the nearby destabilized Tyr-93, with the incoming nucleotide in the crystal structures of the R96G pol ι ternary complexes (Fig. 3) may provide a molecular explanation for severe diminution of the catalytic efficiency in the R96G variant (Table 1). The disordered electron density of the Tyr-93 side chain in R96G pol ι crystal structures seems to be related to the destabilization of the Tyr-93 side chain due to the absent cation-π interaction with Gly-96. Both Arg-96 and Tyr-93 residues, which are conserved and important residues for nucleotide binding in all Y-family pols (20), seem to co-stabilize their side chain conformations by forming the cation-π interaction between their parallel side chains as observed in our pol ι ternary complexes (Fig. 3). In good accordance with our data, individual missense mutations of two homologous Arg-67 and Tyr-64 residues in yeast pol η severely diminish its catalytic activity (49). It can be postulated that the weakened interaction of the R96G variant with incoming nucleotide likely diminishes its nucleotide binding affinity and catalytic efficiency. Interestingly, the attenuating effect of the R96G variation on the apparent nucleotide binding affinity of pol ι was observed much more strongly in the presence of Mg2+ (Table 1). Mn2+ substitution fully restored the apparent nucleotide binding affinity of the R96G variant to a level similar to that of wild-type pol ι (Table 1), indicating a rescuing effect of Mn2+ to offset the destabilized nucleotide interaction in the R96G variant. Accordingly, the extent of reduction of catalytic efficiency in the R96G variant was considerably lessened in the presence of Mn2+ when compared with Mg2+ (Table 1).
The variant of pol ι(26–445) lacking the N-terminal 25 residues retained an Mn2+-dependent catalytic efficiency almost similar to that of the pol ι(1–445) but displayed a 3-fold increase in selectivity in Mg2+-dependent catalytic efficiency when compared with that of the pol ι(1–445) (Table 1). These results are in good agreement with our previous steady-state kinetic data (17). However, the structural effects of the N-terminal 25 residues are not clear due to their disordered nature in the x-ray crystal structure. Although it may not be obvious due to its low resolution (3.6 Å), interestingly, our refined crystal structure of pol ι(1–445) ternary complex in the presence of Mg2+ appeared to have slightly altered conformations of Arg-96 and Tyr-93 side chains when compared with the pol ι(26–445) ternary complex (supplemental Fig. S1C). This subtle structural alteration, observed only with Mg2+ ions in the pol ι(1–445) active site, may in part explain a slightly lower efficiency for Mg2+-dependent catalysis by pol ι(1–445) when compared with pol ι(26–445).
In summary, we have investigated the effects of different metal ions (Mg2+ and Mn2+) and genetic variations (R96G and Δ1–25) on both the structure and the catalytic function of pol ι. Here we report the first x-ray crystal structures acquired in the presence of either Mg2+ or Mn2+ of pol ι ternary complexes having intact coordinating metals and ligands such as the primer terminus 3′-OH. Comparisons of active-site conformations between pol ι·Mg2+ and pol ι·Mn2+ ternary complexes revealed that pol ι adopts near perfect octahedral coordination geometries for two metal ions in the active site only in the presence of Mn2+. This structural feature with Mn2+ is clearly consistent with the pre-steady-state kinetic observation that Mn2+ greatly bolsters the apparent nucleotide binding affinity and the catalytic efficiency of pol ι when compared with observations with Mg2+. Moreover, our combined structural and pre-steady-state kinetic analysis also revealed that the catalytic impairment in the R96G variant is related to the lack of hydrogen-bonding interactions of Gly-96 and Tyr-93 with incoming nucleotides in the active site. Our comparison between pol ι(1–445) and pol ι(26–445) also suggests a potential role of the disordered N-terminal 25 amino acids in selectively improving (albeit slightly) the Mn2+-dependent catalytic efficiency in the wild-type pol ι. Overall, our study provides insights into the delicate structural and kinetic features of different metal coordination and genetic variants that contribute to understanding of the molecular basis of the catalytic function of pol ι.
Experimental Procedures
Materials
T4 polynucleotide kinase and dNTPs were purchased from New England Biolabs (Ipswich, MA). [γ-32P]ATP (specific activity 3 × 103 Ci/mmol) was from PerkinElmer Life Sciences. DNA oligonucleotides were from Sigma-Genosys. Micro Bio-Spin columns were from Bio-Rad. Protease inhibitor cocktail tablets were from Roche Applied Science. FPLC columns and PreScission protease were from GE Healthcare (Uppsala, Sweden). The wild-type, Δ1–25, and R96G variant forms of recombinant human pol ι core proteins (residues 1–445) were purified using previously described protocols (17). The non-hydrolyzable dCTP analogue dCMPNPP was from Jena Bioscience (Jena, Germany). Polyethylene glycol monomethyl ether 5000 was from Hampton Research (Aliso Viejo, CA).
Pre-steady-state Reactions
The 18-mer primer (5′-AGC CAG CCG CAG ACG CAG-3′) was 5′ end-labeled using T4 polynucleotide kinase with [γ-32P]ATP and annealed with 36-mer template (3′-CGG AGC TCG GTC GGC GTC TGC GTC GCT CCT GCG GCT-5′). Rapid quench experiments were performed using a model RQF-3 KinTek Quench Flow instrument (KinTek Corp., Snow Shoe, PA). All DNA polymerase reactions were performed in 50 mm Tris-HCl (pH 7.5) buffer containing 5 mm dithiothreitol, 100 μg ml−1 BSA (w/v), 10% glycerol (v/v), and 2 mm MgCl2 (or 0.15 mm MnCl2). Reactions were initiated by rapid mixing of 32P-primer/template/polymerase mixtures (18-mer/36-mer, 100 nm; pol ι, 1 μm, in 10-fold excess to DNA substrate to ensure single turnover conditions) with the metal-dCTP mixtures (2 mm MgCl2 or 0.15 mm MnCl2; dCTP, in varying concentrations), and then quenched with 0.15 m EDTA at time intervals from 0.15 to 30–120 s (or from 2 to 240–480 s for the R96G variant). MgCl2 was supplemented by as much as the increase of dCTP to counterbalance the Mg2+-chelating effect of dCTP for the reactions at high levels of dCTP. Reaction products were mixed with formamide-dye solution (20 mm EDTA, 95% formamide (v/v), 0.5% bromphenol blue (w/v), and 0.05% xylene cyanol (w/v)) and separated using an 8 m urea-containing denaturing gel with 16% polyacrylamide (w/v), and then quantified by a Bio-Rad Personal Molecular Imager instrument and the Quantity One software. Pre-steady-state data obtained under the single turnover condition were fit to the single-exponential equation y = A(1 − exp(−kobst)), where y = concentration of product, A = reaction amplitude, kobs = observed rate of nucleotide incorporation, and t = time (50, 51), using nonlinear regression analysis in GraphPad Prism software.
Determination of kpol and Kd,dCTP
The pre-steady-state kinetic parameters kpol and Kd,dCTP were estimated by analyzing the dCTP dependence on the observed pre-steady-state rates of dCTP insertion under single turnover conditions. A graph of the observed rate (kobs) versus dCTP concentration was fit to the hyperbolic equation kobs = [kpol[dNTP]/([dNTP] + Kd)], where kpol is the maximal rate of nucleotide incorporation and Kd,dCTP is the equilibrium dissociation constant for dCTP (50, 51).
Crystallization of pol ι·dG-DNA·dCMPNPP Ternary Complexes in the presence of Mg2+ or Mn2+
Crystals were obtained under the conditions previously reported (34, 36) with slight modifications described below. The purified pol ι enzymes (0.22 mm) were incubated with annealed self-complementary 18-mer DNA (5′-TCT GGG GTC CTA GGA CCC-3′, 0.26 mm) and 20 mm dCMPNPP (or 4 mm dCMPNPP for MnCl2) in the presence of 10 mm MgCl2 (or 2 mm MnCl2) on ice. The ternary complexes were crystallized in 0.1 m MES (pH 6.5), 0.2–0.4 m (NH4)2SO4, and 10–22% polyethylene glycol 5000 (w/v) using the hanging-drop vapor diffusion method at 4 °C. Crystals were typically observed in 1–3 days. Crystals started to form with reservoir solutions containing 13–15, 18, and 18–20% polyethylene glycol monomethyl ether 5000 (w/v), respectively, for pol ι(26–445), pol ι(1–445), and R96G pol ι(1–445) ternary complexes. Crystals were mounted in nylon loops and cryoprotected in a reservoir solution containing 25% glycerol (v/v), and then flash-frozen in liquid nitrogen.
Structure Determination and Refinement
X-ray diffraction data were collected on the 21-ID-F (Life Sciences Collaborative Access Team (LS-CAT)) beam line at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Collected data were indexed, integrated, and scaled using HKL2000 (52). All crystal types belonged to space group P6522. Structures were determined by molecular replacement phasing using the program Phaser MR (53) and the pol ι structure (PDB ID 2ALZ) as a search model. Structure refinements and model building were performed using PHENIX (54) and COOT (55). Metal coordination geometry was analyzed with UCSF Chimera (56). Structural illustrations were prepared with PyMOL (Schrödinger, LLC).
Author Contributions
F. P. G. and J.-Y. C. conceived the study and designed the experiments. J.-Y. C. and M. Y. purified the enzymes. M. Y., J.-Y. C., and Q. Z. conducted the kinetic experiments. J.-Y. C. and A. P. crystallized the protein complexes. Y.-S. L., J.-Y. C., and A. P. solved the structures. J.-Y. C., F. P. G., M. E., and Y.-S. L. analyzed the data and wrote the paper.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES010375 and R01 ES010546 (to F. P. G.) and P01 CA160032 (to M. E.). This work was also supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (Grant NRF-2015R1D1A1A01057577) (to J.-Y. C.) and the DGIST R&D Program of the Ministry of Science, ICT and Technology of Korea (20160165) (to Y.-S. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1 and S2.
- pol
- polymerase
- dCMPNPP
- 2′-deoxycytidine-5′-[(α,β)-imido]triphosphate
- RMSD
- root mean square deviation(s)
- TLS
- translesion synthesis
- 8-oxoG
- 8-oxo-7,8-dihydroG.
References
- 1. Helleday T., Eshtad S., and Nik-Zainal S. (2014) Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Loeb L. A., and Monnat R. J. Jr. (2008) DNA polymerases and human disease. Nat. Rev. Genet. 9, 594–604 [DOI] [PubMed] [Google Scholar]
- 3. Lange S. S., Takata K., and Wood R. D. (2011) DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ling H., Boudsocq F., Woodgate R., and Yang W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 5. Choi J.-Y., Eoff R. E., and Guengerich F. P. (2011) Bypass DNA polymerases. in Chemical Carcinogenesis (Penning T. M., ed), pp. 345–373, Humana Press, New York, NY [Google Scholar]
- 6. Zhang Y., Wu X., Guo D., Rechkoblit O., Geacintov N. E., and Wang Z. (2002) Two-step error-prone bypass of the (+)- and (−)-trans-anti-BPDE-N2-dG adducts by human DNA polymerases η and κ. Mutat. Res. 510, 23–35 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Wu X., Rechkoblit O., Geacintov N. E., Taylor J. S., and Wang Z. (2002) Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 30, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frank E. G., Sayer J. M., Kroth H., Ohashi E., Ohmori H., Jerina D. M., and Woodgate R. (2002) Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase ι. Nucleic Acids Res. 30, 5284–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Avkin S., Goldsmith M., Velasco-Miguel S., Geacintov N., Friedberg E. C., and Livneh Z. (2004) Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase κ. J. Biol. Chem. 279, 53298–53305 [DOI] [PubMed] [Google Scholar]
- 10. Klarer A. C., Stallons L. J., Burke T. J., Skaggs R. L., and McGregor W. G. (2012) DNA polymerase η participates in the mutagenic bypass of adducts induced by benzo[a]pyrene diol epoxide in mammalian cells. PLoS ONE 7, e39596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi J.-Y., Angel K. C., and Guengerich F. P. (2006) Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 281, 21062–21072 [DOI] [PubMed] [Google Scholar]
- 12. Choi J.-Y., and Guengerich F. P. (2005) Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 352, 72–90 [DOI] [PubMed] [Google Scholar]
- 13. Choi J.-Y., and Guengerich F. P. (2006) Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase ι. J. Biol. Chem. 281, 12315–12324 [DOI] [PubMed] [Google Scholar]
- 14. Choi J.-Y., and Guengerich F. P. (2008) Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J. Biol. Chem. 283, 23645–23655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., and Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 16. Yoon J. H., Prakash L., and Prakash S. (2009) Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J., Song I., Jo A., Shin J.-H., Cho H., Eoff R. L., Guengerich F. P., and Choi J.-Y.(2014) Biochemical analysis of six genetic variants of error-prone human DNA polymerase ι involved in translesion DNA synthesis. Chem. Res. Toxicol. 27, 1837–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song I., Kim E.-J., Kim I.-H., Park E.-M., Lee K. E., Shin J.-H., Guengerich F. P., and Choi J.-Y.(2014) Biochemical characterization of eight genetic variants of human DNA polymerase κ involved in error-free bypass across bulky N2-guanyl DNA adducts. Chem. Res. Toxicol. 27, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yeom M., Kim I. H., Kim J. K., Kang K., Eoff R. L., Guengerich F. P., and Choi J.-Y. (2016) Effects of twelve germline missense variations on DNA lesion and G-quadruplex bypass activities of human DNA polymerase REV1. Chem. Res. Toxicol. 29, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nair D. T., Johnson R. E., Prakash S., Prakash L., and Aggarwal A. K. (2004) Replication by human DNA polymerase ι occurs by Hoogsteen base-pairing. Nature 430, 377–380 [DOI] [PubMed] [Google Scholar]
- 21. Choi J. Y., Lim S., Eoff R. L., and Guengerich F. P. (2009) Kinetic analysis of base-pairing preference for nucleotide incorporation opposite template pyrimidines by human DNA polymerase ι. J. Mol. Biol. 389, 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirouac K. N., and Ling H. (2009) Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase ι. EMBO J. 28, 1644–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vaisman A., Frank E. G., Iwai S., Ohashi E., Ohmori H., Hanaoka F., and Woodgate R. (2003) Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase ι. DNA Repair (Amst.) 2, 991–1006 [DOI] [PubMed] [Google Scholar]
- 24. Choi J.-Y., Lim S., Kim E. J., Jo A., and Guengerich F. P. (2010) Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 404, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi J.-Y., Chowdhury G., Zang H., Angel K. C., Vu C. C., Peterson L. A., and Guengerich F. P. (2006) Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 281, 38244–38256 [DOI] [PubMed] [Google Scholar]
- 26. Dumstorf C. A., Clark A. B., Lin Q., Kissling G. E., Yuan T., Kucherlapati R., McGregor W. G., and Kunkel T. A. (2006) Participation of mouse DNA polymerase ι in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl. Acad. Sci. U.S.A. 103, 18083–18088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iguchi M., Osanai M., Hayashi Y., Koentgen F., and Lee G. H. (2014) The error-prone DNA polymerase ι provides quantitative resistance to lung tumorigenesis and mutagenesis in mice. Oncogene 33, 3612–3617 [DOI] [PubMed] [Google Scholar]
- 28. Ohkumo T., Kondo Y., Yokoi M., Tsukamoto T., Yamada A., Sugimoto T., Kanao R., Higashi Y., Kondoh H., Tatematsu M., Masutani C., and Hanaoka F. (2006) UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol. Cell Biol. 26, 7696–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albertella M. R., Lau A., and O'Connor M. J. (2005) The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst.) 4, 583–593 [DOI] [PubMed] [Google Scholar]
- 30. Yang J., Chen Z., Liu Y., Hickey R. J., and Malkas L. H. (2004) Altered DNA polymerase ι expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 64, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 31. Yuan F., Xu Z., Yang M., Wei Q., Zhang Y., Yu J., Zhi Y., Liu Y., Chen Z., and Yang J. (2013) Overexpressed DNA polymerase ι regulated by JNK/c-Jun contributes to hypermutagenesis in bladder cancer. PLoS ONE 8, e69317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou J., Zhang S., Xie L., Liu P., Xie F., Wu J., Cao J., and Ding W. Q. (2012) Overexpression of DNA polymerase ι (pol ι) in esophageal squamous cell carcinoma. Cancer Sci. 103, 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frank E. G., and Woodgate R. (2007) Increased catalytic activity and altered fidelity of human DNA polymerase ι in the presence of manganese. J. Biol. Chem. 282, 24689–24696 [DOI] [PubMed] [Google Scholar]
- 34. Pence M. G., Blans P., Zink C. N., Hollis T., Fishbein J. C., and Perrino F. W. (2009) Lesion bypass of N2-ethylguanine by human DNA polymerase ι. J. Biol. Chem. 284, 1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., and Wilson S. H. (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 14, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2005) Human DNA polymerase ι incorporates dCTP opposite template G via a G.C+ Hoogsteen base pair. Structure 13, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 37. Yang W., Lee J. Y., and Nowotny M. (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 [DOI] [PubMed] [Google Scholar]
- 38. Zhao Y., Biertümpfel C., Gregory M. T., Hua Y. J., Hanaoka F., and Yang W. (2012) Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin. Proc. Natl. Acad. Sci. U.S.A. 109, 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koag M. C., Lai L., and Lee S. (2014) Structural basis for the inefficient nucleotide incorporation opposite cisplatin-DNA lesion by human DNA polymerase β. J. Biol. Chem. 289, 31341–31348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2006) An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure 14, 749–755 [DOI] [PubMed] [Google Scholar]
- 41. Xia S., Wang M., Blaha G., Konigsberg W. H., and Wang J. (2011) Structural insights into complete metal ion coordination from ternary complexes of B family RB69 DNA polymerase. Biochemistry 50, 9114–9124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., and Wilson S. H. (2008) Structures of DNA polymerase β with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koag M. C., Nam K., and Lee S. (2014) The spontaneous replication error and the mismatch discrimination mechanisms of human DNA polymerase β. Nucleic Acids Res. 42, 11233–11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vashishtha A. K., and Konigsberg W. H. (2016) Effect of different divalent cations on the kinetics and fidelity of RB69 DNA polymerase. Biochemistry 55, 2661–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanca G., Shevelev I., Ramadan K., Villani G., Spadari S., Hübscher U., and Maga G. (2003) Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn2+ dependence: a kinetic and thermodynamic study. Biochemistry 42, 7467–7476 [DOI] [PubMed] [Google Scholar]
- 46. Vaisman A., Ling H., Woodgate R., and Yang W. (2005) Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24, 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. García-Gómez S., Reyes A., Martínez-Jiménez M. I., Chocrón E. S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I. J., and Blanco L. (2013) PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52, 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao Y., and Yang W. (2016) Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science 352, 1334–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson R. E., Trincao J., Aggarwal A. K., Prakash S., and Prakash L. (2003) Deoxynucleotide triphosphate binding mode conserved in Y family DNA polymerases. Mol. Cell Biol. 23, 3008–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson K. A. (1995) Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 249, 38–61 [DOI] [PubMed] [Google Scholar]
- 51. Joyce C. M. (2010) Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta 1804, 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 53. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.