Abstract
MicroRNA-7 (miR-7)has been characterized as an anti-oncogenic microRNA (miRNA) in several cancers, including hepatocellular carcinoma (HCC). However, the mechanism for the regulation of miR-7 production in tumors remains unclear. Here, we identified nuclear factor 90 (NF90) and NF45 complex (NF90-NF45) as negative regulators of miR-7 processing in HCC. Expression of NF90 and NF45 was significantly elevated in primary HCC tissues compared with adjacent non-tumor tissues. To examine which miRNAs are controlled by NF90-NF45, we performed an miRNA microarray and quantitative RT-PCR analyses of HCC cell lines. Depletion of NF90 resulted in elevated levels of mature miR-7, whereas the expression of primary miR-7-1 (pri-miR-7-1) was decreased in cells following knockdown of NF90. Conversely, the levels of mature miR-7 were reduced in cells overexpressing NF90 and NF45, although pri-miR-7-1 was accumulated in the same cells. Furthermore, NF90-NF45 was found to bind pri-miR-7-1 in vitro. These results suggest that NF90-NF45 inhibits the pri-miR-7-1 processing step through the binding of NF90-NF45 to pri-miR-7-1. We also found that levels of the EGF receptor, an oncogenic factor that is a direct target of miR-7, and phosphorylation of AKT were significantly decreased in HCC cell lines depleted of NF90 or NF45. Of note, knockdown of NF90 or NF45 caused a reduction in the proliferation rate of HCC cells. Taken together, NF90-NF45 stimulates an elevation of EGF receptor levels via the suppression of miR-7 biogenesis, resulting in the promotion of cell proliferation in HCC.
Keywords: epidermal growth factor receptor (EGFR), hepatocellular carcinoma, microRNA (miRNA), microRNA biogenesis, RNA-binding protein
Introduction
MicroRNAs (miRNAs)2 are non-coding small RNAs that function as repressors for gene expression by binding to the 3′-untranslated regions (3′UTRs) of target mRNAs (1). This binding causes degradation and/or translational inhibition of target mRNAs, depending upon the degree of sequence complementarity between miRNAs and the 3′UTRs of target mRNAs. miRNAs regulate the expression of many genes that are involved in cell proliferation, apoptosis, development, and differentiation, as well as tumorigenesis (2–6). In mammals, miRNA genes are initially transcribed by RNA polymerase II as long primary miRNAs (pri-miRNAs). The pri-miRNAs are first processed into precursor miRNAs (pre-miRNAs) by a microprocessor complex in the nucleus. The microprocessor complex contains the RNase III enzyme Drosha and a double-stranded RNA-binding protein DGCR8 (7). The pre-miRNAs are exported from the nucleus to the cytoplasm by exportin-5 and RanGTP transporter. In the cytoplasm, pre-miRNAs are then processed to miRNA duplex forms by a cytoplasmic RNase III enzyme Dicer and a double-stranded RNA-binding protein TRBP (8). Following this processing, one strand of the miRNA duplex is incorporated into the RNA-induced silencing complex. The single strand of miRNA (mature miRNA) guides the RNA-induced silencing complex to the target mRNA with sequence complementarity, which leads to either mRNA degradation or to translational repression (9). In the first processing step, the function of the microprocessor complex is negatively regulated by many RNA-binding proteins, including Lin28B, Hu antigen R, and nuclear factor 90 (NF90, also referred to as ILF3, NFAR1, and DRBP76) and nuclear factor 45 (NF45) complex (10–12).
NF90 is a double-stranded (ds) RNA-binding protein that forms a heterodimeric complex with NF45 (13). NF90 protein contains two dsRNA-binding motifs, a nuclear localization signal, and a single nucleic acid-binding RGG motif. NF90 is known to participate in transcription, mRNA stability, translational inhibition, and RNA export (14–18). In addition, we previously demonstrated that the NF90-NF45 complex negatively regulates the pri-miRNA processing step, resulting in a reduction of mature miRNA production (12, 19).
In recent studies, a prevailing feature observed in cancers is the global decrease in miRNA expression compared with adjacent normal tissues (20–22). It is believed that miRNAs that are down-regulated in cancers function as tumor suppressors. These miRNAs are referred to as anti-oncogenic miRNAs (anti-oncomirs). Therefore, inhibition of anti-oncomir expression has potential as a therapeutic target in cancers. However, the detailed molecular mechanism of anti-oncomir biogenesis remains unclear. Until now, it has been reported that NF90 expression is elevated in breast, non-small lung, epithelial ovarian cancer, and also hepatocellular carcinoma (HCC) (23–27). As mentioned above, NF90-NF45 is known to function as a negative regulator in miRNA biogenesis (12). Therefore, we hypothesized that the NF90-NF45 complex plays a role in the promotion of tumorigenesis via suppression of anti-oncomir expression.
Here, we found that the processing pathway of an anti-oncomir, miR-7, was repressed by high expression of NF90-NF45. Epidermal growth factor receptor (EGFR), an oncogenic factor, is known to be a direct target of miR-7 (28, 29). Furthermore, downstream of EGFR signaling is the AKT signaling pathway, which is involved in cell proliferation and apoptosis resistance (30, 31). We observed that EGFR expression was significantly decreased in cells depleted of NF90 or NF45, resulting in down-regulation of AKT signaling. In addition, knockdown of NF90 or NF45 resulted in significant proliferation retardation in HCC cell lines. These findings support the idea that a complex of NF90-NF45 facilitates tumorigenesis through the repression of anti-oncomir biogenesis.
Results
Expression of NF90 and NF45 Is Elevated in the Majority of HCC Tissues
To address our working hypothesis that the NF90-NF45 complex plays a critical role in the promotion of tumorigenesis via suppression of anti-oncomir biogenesis, we first measured the expression levels of NF90 and NF45 in 18 pairs of primary HCC tumor tissues versus adjacent non-tumor tissues by immunoblotting. NF90 and NF45 expression was elevated by more than 2-fold in 72.2% (13/18) of the HCC tumor tissues, respectively, compared with adjacent matched non-tumor tissues (Fig. 1A). The expression levels of NF90 and NF45 in the tumor tissues were obviously higher than that in the paired non-tumor tissues in the majority the cases (Fig. 1B). These results were consistent with recent findings (27) and support the hypothesis that overexpression of NF90-NF45 enhances tumorigenesis through repression of anti-oncomiR biogenesis.
FIGURE 1.
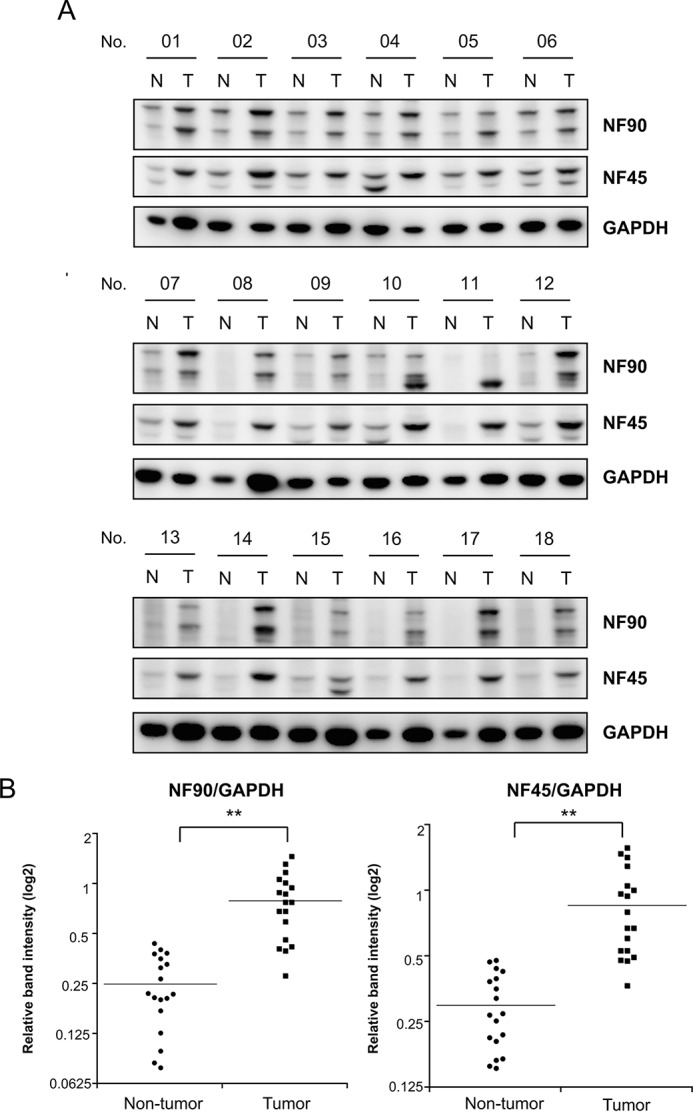
HCC exhibits high levels of NF90 and NF45 expression. A, expression levels of NF90 and NF45 were examined in 18 HCC tissues or adjacent non-tumors by immunoblotting. GAPDH was used as an internal control. T, tumor tissues in HCC; N, adjacent non-tumor tissues in HCC. B, intensities of immunoreactive bands were measured by densitometry and are presented as dot plots. GAPDH was used as an internal control and for normalization of data. **, p < 0.005 relative to adjacent non-tumor tissues by a two-tailed Student's t test.
NF90-NF45 Complex Functions as a Negative Regulator in miR-7 Biogenesis
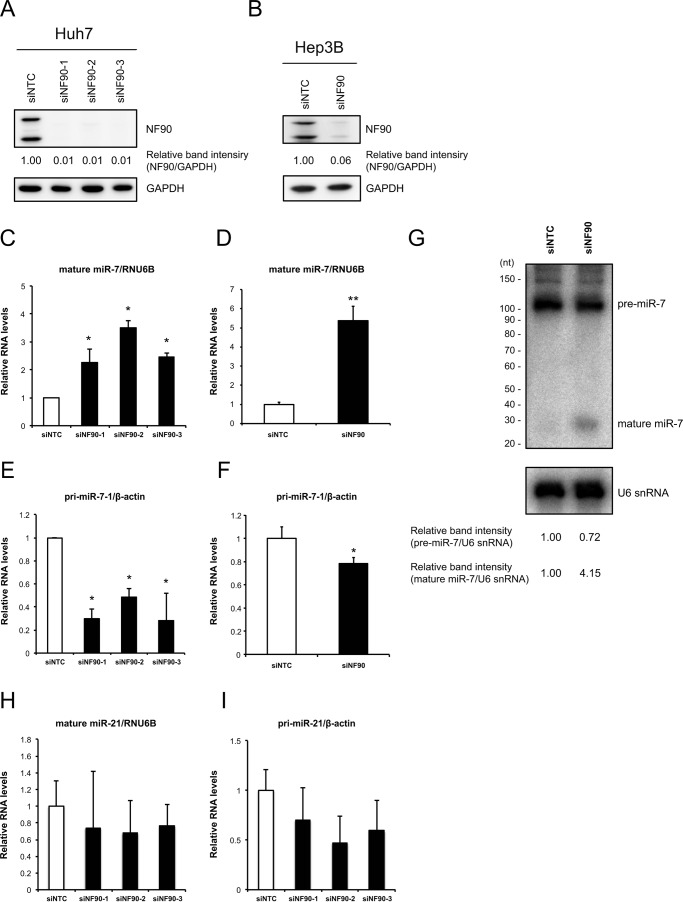
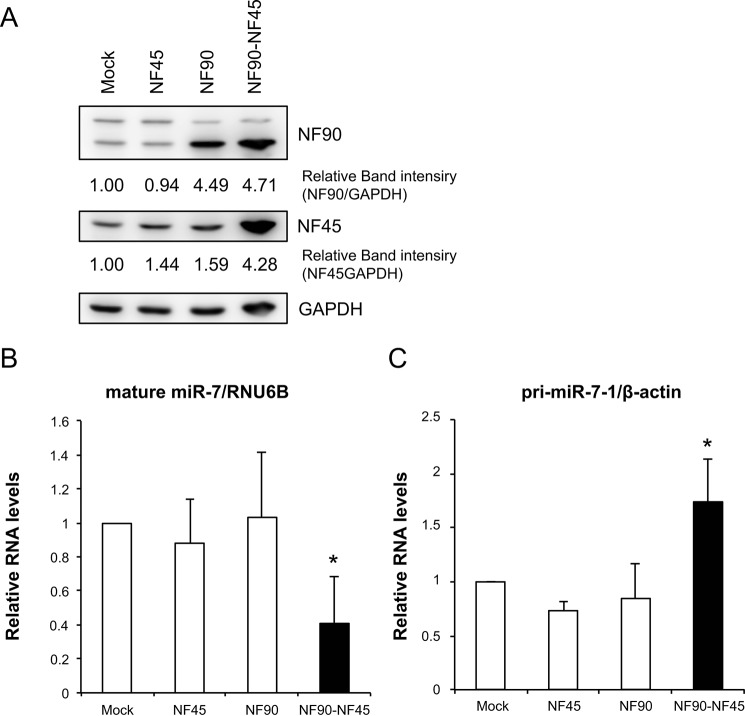
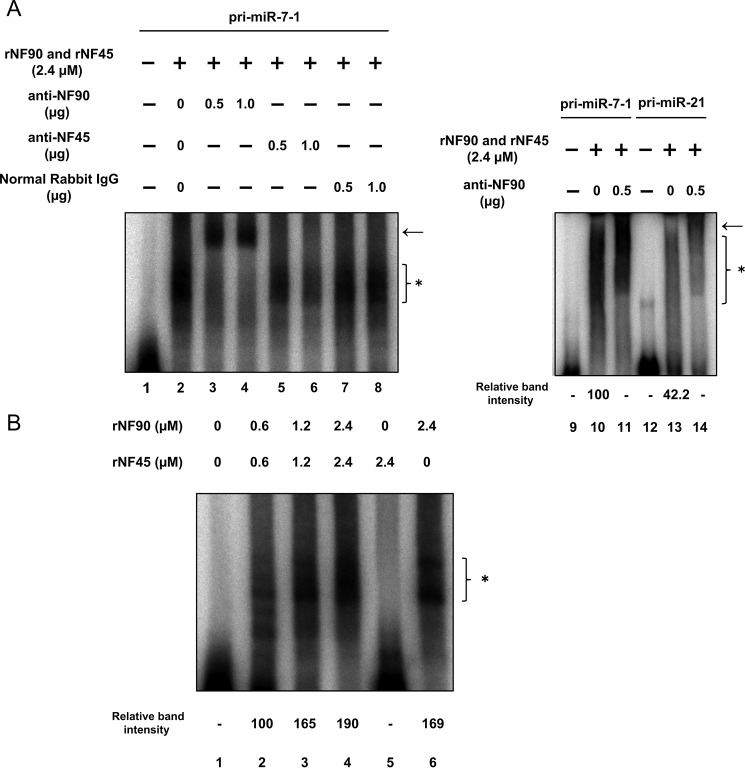
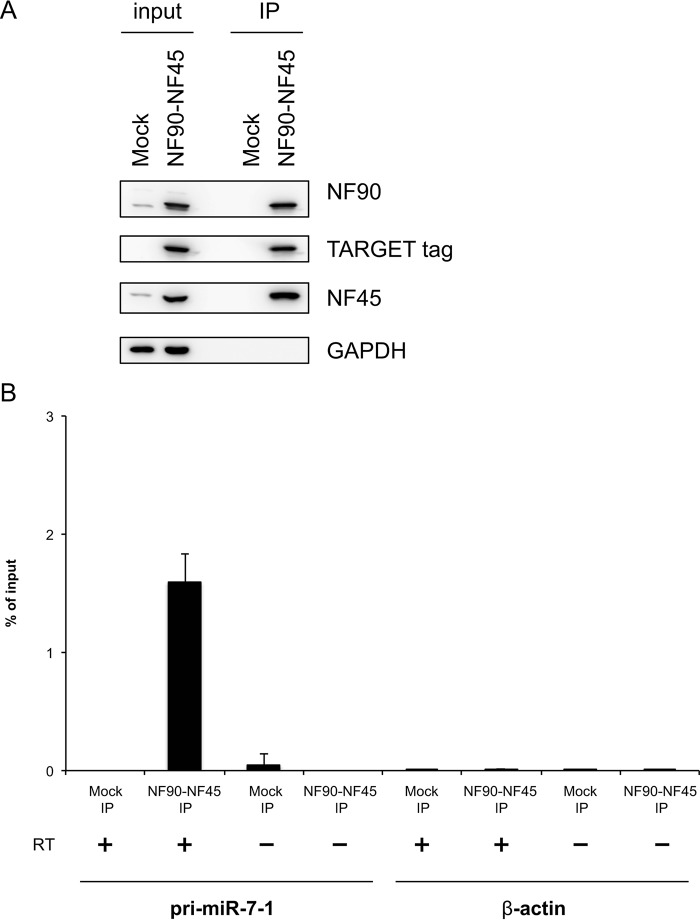
Our previous study demonstrated that the NF90-NF45 complex inhibits miRNA biogenesis through negative regulation of the pri-miRNA processing step (12, 19). To examine miRNA biogenesis suppression by NF90-NF45 in HCC, we performed an miRNA microarray analysis using Huh7 HCC cells that had been depleted of NF90. When siRNAs targeting NF90 were transfected into Huh7 cells, NF90 expression was markedly decreased (Fig. 2A). GAPDH was used as an internal control (Fig. 2A). In the miRNA microarray analysis, we noted miRNAs that had adequate signaling levels. Consequently, seven miRNAs levels were elevated by 2-fold in NF90-knockdown cells compared with control cells (Table 1 and GEO accession number GSE67411). Among the up-regulated miRNAs in NF90-knockdown cells, we focused on miR-7, which is known as an anti-oncomir targeting EGFR, IGF-1R, and mTOR (28, 32, 33). To confirm reliability of the microarray analysis, we measured levels of mature miR-7 and pri-miR-7-1 in Huh7 and Hep3B cells depleted of NF90. Knockdown of NF90 exhibited significant increments in mature miR-7, although the amount of pri-miR-7-1 was decreased in cells in which NF90 had been knocked down (Fig. 2, C–F). In addition, we measured the levels of mature miR-21 and pri-miR-21 in the NF90-knockdown cells as a negative control to confirm the specific effect on miR-7 biogenesis by NF90-NF45 because there was no significant difference in the level of miR-21 between control cells and the NF90-knockdown cells (supplemental Table S1, sheet 2). As shown in Fig. 2, H and I, knockdown of NF90 did not significantly influence the levels of both mature miR-21 and pri-miR-21. These results together with our previous findings (12, 19) suggest that the regulation of pri-miRNA processing by NF90-NF45 would be coupled with the production of mature miRNA. To verify the inhibitory effect on pri-miR-7 processing by NF90-NF45, we measured the level of pre-miR-7 in the NF90-knockdown cells using Northern blotting analysis. As shown in Fig. 2G, knockdown of NF90 induced the reduction of pre-miR-7 level, whereas the level of mature miR-7 was elevated in the NF90-knockdown cells compared with the control cells. These results suggest that NF90-NF45 would inhibit the processing of pri-miR-7 to pre-miR-7 in HCC cell lines. To further confirm the negative regulation of miR-7 biogenesis by NF90-NF45, we generated Huh7 cell lines stably expressing NF90 and/or NF45. Overexpression of NF90 and/or NF45 in the stable cell lines was confirmed by immunoblotting (Fig. 3A). qRT-PCR analysis demonstrated that mature miR-7 levels were significantly reduced in both NF90- and NF45-overexpressing cells compared with control cells and cells transfected with NF90 or NF45 plasmids (Fig. 3B). Moreover, pri-miR-7-1 levels were significantly elevated in cells overexpressing both NF90 and NF45 compared with control cells and NF90 or NF45 alone overexpressing cells (Fig. 3C). Taken together, these findings suggest that the generation of mature miR-7 is generally repressed by NF90-NF45 through inhibition of the pri-miR-7 processing step in HCC cell lines. Subsequently, to examine whether or not NF90-NF45 directly binds pri-miR-7-1, we performed an electrophoresis mobility shift assay (EMSA) with pri-miR-7-1 probe using recombinant human NF90 (rNF90) and recombinant human NF45 (rNF45). We found a shifted band in the presence of rNF90 and rNF45 (see asterisk in Fig. 4A, lane 2). To investigate whether the shifted band was a result of the association of rNF90-rNF45 with pri-miR-7-1, we performed a supershift assay using anti-NF90 and anti-NF45 antibodies. Addition of NF90 antibody resulted in the formation of a supershifted complex (see arrow in Fig. 4A, left panel, lanes 3 and 4), whereas the band of NF90-NF45 binding pri-miR-7-1 was diminished in the presence of NF45 antibody (see asterisk in Fig. 4A, left panel, lanes 5 and 6). These results indicated that the shifted band shown in Fig. 4A (left panel, lane 2) resulted from the association of rNF90-rNF45 with pri-miR-7-1. In addition, we performed EMSA probed with pri-miR-21 as a negative control, because there was no significant difference in the level of mature miR-21 between the siNTC-transfected Huh7 cells and the NF90-knockdown cells on our miRNA microarray analysis (supplemental Table S1, sheet 2). As shown in Fig. 4A, the rNF90-rNF45 complex exhibited much lower binding activity for pri-miR-21 (less than 50%) than pri-miR-7 (right panel, asterisk, compare lane 11 to lane 13). A supershift assay using anti-NF90 antibody also revealed that the major band indicated by an asterisk was supershifted by the anti-NF90 antibody (Fig. 4A, right panel, an arrow), showing that the major band is the complex of NF90-NF45 and pri-miRNAs. Furthermore, the binding activity of rNF90-rNF45 to pri-miR-7-1 was found to be higher than that of rNF90 alone (see asterisk in Fig. 4B, compare lanes 4 and 6). NF45 alone did not bind to pri-miR-7-1 (Fig. 4B, lane 5). To confirm the data shown in Fig. 4 in vitro, we carried out RNA immunoprecipitation assay using WCEs from Huh7 cells transfected with either mock vectors or both the mammalian expression vectors, including TARGET-tagged NF90 gene or NF45 gene. As shown in Fig. 5, pri-miR-7 was detected by immunoprecipitation of TARGET-tagged NF90-NF45, but not by mock-immunoprecipitation, whereas β-actin mRNA was not identified in the immunoprecipitates of both mock and TARGET-tagged NF90-NF45, indicating that NF90-NF45 associates with endogenous pri-miR-7-1 in vivo. These findings, together with Figs. 2 and 3, suggest that the robust association of NF90-NF45 complex with pri-miR-7-1 promoted the reduction of mature miR-7 production in HCC cells.
FIGURE 2.
Knockdown of NF90 induces elevation of mature miR-7 and repression of primary miR-7 in HCC cells. A and B, Huh7 and Hep3B cells were transfected with the indicated siRNAs. Expression levels of NF90 and NF45 were detected by immunoblotting. GAPDH was used as an internal control. C–F, RNA levels of mature miR-7 (C and D) and pri-miR-7-1 (E and F) in Huh7 and Hep3B cells, respectively, transfected with the indicated siRNAs were analyzed by qRT-PCR. RNAU6B and β-actin were used as internal controls and for normalization of the data. siNTC, non-targeting control siRNA, siNF90–1, -2, and -3, siRNAs targeted to NF90; siNF90, mixed three different siRNAs targeted to NF90. Data are expressed as means ± S.D. (n = 3 per group). *, p < 0.05, and ** p < 0.01, relative to control by a two-tailed Student's t test. G, expression levels of pre-miR-7 and mature miR-7 in Huh7 cells transfected with the indicated siRNAs were detected by Northern blotting analysis. U6 snRNA was used as an internal control. H and I, RNA levels of mature miR-21 (H) and pri-miR-21 (I) in Huh7 cells transfected with the indicated siRNAs were analyzed by qRT-PCR. RNAU6B and β-actin were used as internal controls and for normalization of the data.
TABLE 1.
Up-regulated miRNAs (>2-fold change) in NF90-knockdown Huh7 cells compared with cells transfected with non-targeting control siRNA (siNTC)
| miRNA | -Fold change (siNF90–2/siNTC) | -Fold change (siNF90–3/siNTC) | Target gene | Ref. |
|---|---|---|---|---|
| has-mir-7 | 4.16 | 4.68 | EGFR | 28, 29, 37–39 |
| RAF1 | 28. 39 | |||
| IGF1R | 32. 36 | |||
| FAK | 35. 48 | |||
| IRS-1 | 29 | |||
| IRS-2 | 29 | |||
| BCL-2 | 3 | |||
| PIK3CD | 33 | |||
| mTOR | 33 | |||
| p70S6K | 33 | |||
| ACK1 | 39 | |||
| KLF4 | 40 | |||
| PAK1 | 49 | |||
| has-mir-513a-5p | 3.00 | 5.58 | XIAP | 50 |
| B7-H1 | 51 | |||
| has-mir-629* | 2.83 | 6.40 | Unclear | |
| has-mir-671–5p | 2.31 | 4.62 | SMRCB1 | 52 |
| has-mir-768–5p | 2.19 | 3.95 | Unclear | |
| has-mir-135a* | 2.16 | 3.89 | Unclear | |
| has-mir-513b | 2.11 | 3.48 | Unclear |
FIGURE 3.
Overexpression of NF90-NF45 results in accumulation of pri-miR-7-1 and suppression of mature miR-7 in HCC cells. A, Huh7 cells were transfected with stable NF90 and/or NF45 expression plasmids. Expression levels of NF90 and NF45 were detected by immunoblotting. GAPDH was used as an internal control. B and C, RNA levels of mature miR-7 (B) and pri-miR-7-1 (C) in cells transfected with the indicated expression plasmids were analyzed by qRT-PCR. RNAU6B and β-actin were used as internal controls and for normalization of data. Data are expressed as means ± S.D. (n = 3 per group). *, p < 0.05 relative to control by a two-tailed Student's t test.
FIGURE 4.
NF90-NF45 complex is bound to pri-miR-7-1 in vitro. A, RNA-EMSA performed with pri-miR-7-1 or pri-miR-21 probes and rNF90-rNF45 protein. Anti-NF90 antibody (lanes 3, 4, and 11), anti-NF45 antibody (lanes 5 and 6), or normal rabbit IgG (lanes 7 and 8) were incubated with the recombinant proteins in binding buffer prior to the addition of probe. The position of protein-RNA complex and supershifted complex was indicated by an asterisk and an arrow, respectively. B, RNA-EMSA performed with pri-miR-7-1 probe and rNF90 alone, rNF45 alone, or both rNF90 and rNF45 proteins at the indicated concentrations.
FIGURE 5.
NF90-NF45 complex is bound to endogenous pri-miR-7-1. WCEs from Huh7 cells transfected with indicated TARGET-tagged NF90 and NF45 were immunoprecipitated with anti-TARGET tag antibody. A, target-tagged NF90 and NF45 in the immunoprecipitates (IP) were detected by immunoblotting probed with anti-NF90, anti-NF45, anti-TARGET tag, and anti-GAPDH antibodies. 20% of WCEs was used as input. B, RNAs were isolated from precipitates and analyzed by qRT-PCR with specific primers for pri-miR-7-1 or β-actin. 4% of WCEs was used as input. RT− was negative control of reverse transcription. Data are expressed as the mean ± S.D. of triplicate PCRs with ranges and are representative of three independent experiments.
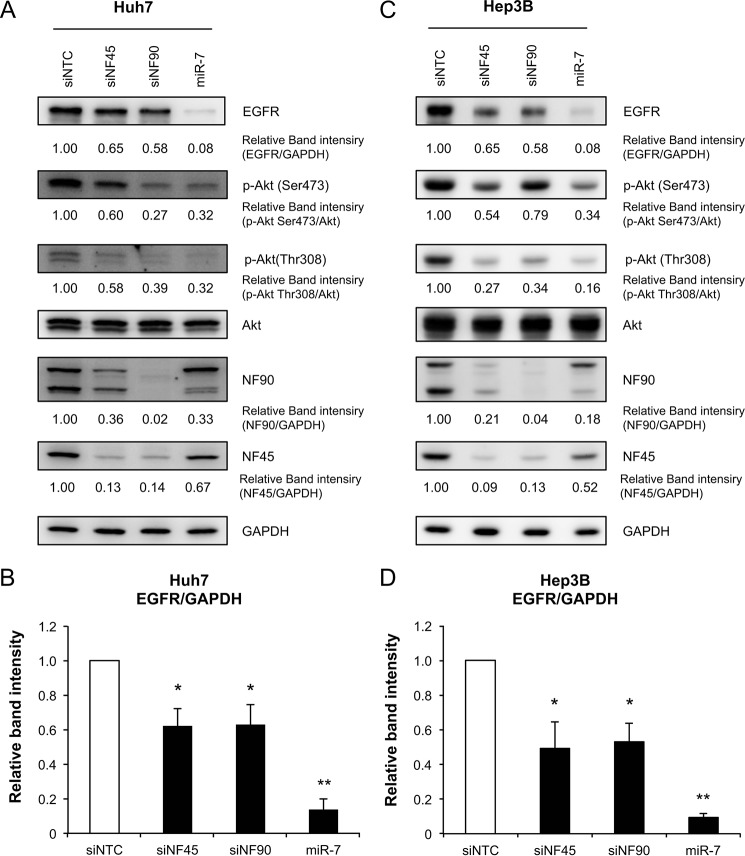
NF90 and NF45 Regulate EGFR-AKT Signaling in HCC Cell Lines
Several reports have indicated that miR-7 inhibits EGFR expression and suppresses AKT signaling, which is a downstream signal of EFGR (28, 33). To examine whether NF90-NF45 regulates EGFR-AKT signaling through the control of miR-7 biogenesis, we measured EGFR expression levels and activation of AKT in two HCC cell lines (Huh7 and Hep3B) depleted of NF90 or NF45. Immunoblot analysis revealed that EGFR expression was significantly reduced in NF90 or NF45 knockdown cells compared with control cells (Fig. 6). Moreover, phosphorylation of AKT at Ser-473 and Thr-308, which results in the active form of AKT, was decreased in cells with knockdown of NF90 or NF45 (Fig. 6, A and C). Transfection of miR-7 mimic also reduced EGFR expression and AKT phosphorylation (Fig. 6). As shown in Figs. 2 and 3, NF90-NF45 functions as a negative regulator in the miR-7 biogenesis. Therefore, these findings suggest that overexpression of NF90-NF45 would promote increased EGFR expression via suppression of miR-7 biogenesis, followed by activation of AKT signaling.
FIGURE 6.
NF90-NF45 regulates EGFR expression and AKT signaling. A, Huh7 cells were transfected with siNTC, siNF45, siNF90, or miR-7 mimic. EGFR, AKT, and phosphorylated AKT in cells transfected with the indicated siRNAs or mimic RNA were detected by immunoblotting. B, intensities of the specific bands shown in A were measured by densitometry and are presented as a graph. GAPDH was used as an internal control and for normalization of data. Data are expressed as means ± S.D. (n = 3 per group). *, p < 0.05; **, p < 0.005 relative to siNTC by a two-tailed Student's t test. C, Hep3B cells were transfected with siNTC, siNF45, siNF90, or miR-7 mimic. EGFR, AKT, and phosphorylated AKT in cells transfected with the indicated siRNA or miRNA mimic were detected by immunoblotting. D, intensities of the specific bands shown in C were measured by densitometry and are presented as a graph. GAPDH was used as an internal control and for normalization of data. Data are expressed as means ± S.D. (n = 3 per group). *, p < 0.05; **, p < 0.005 relative to siNTC by a two-tailed Student's t test.
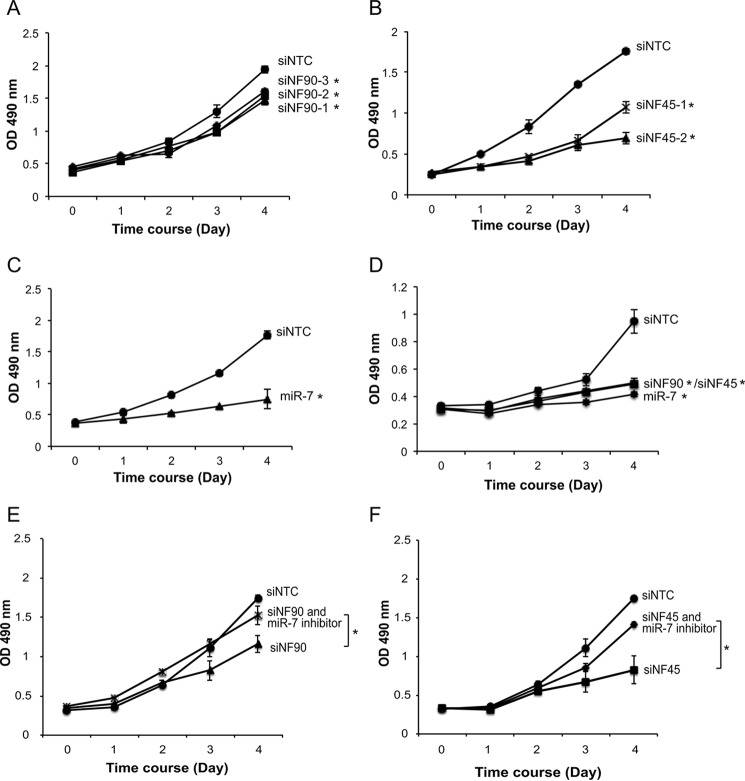
NF90 and NF45 Are Involved in Proliferation of HCC Cells
We next examined whether NF90-NF45 has a role in the proliferation of HCC cell lines through regulation of the EGFR-AKT pathway. MTS assays demonstrated that knockdown of NF90 or NF45 resulted in significant inhibition of Huh7 and Hep3B cell proliferation (Fig. 7, A, B, and D). Proliferation rates were also reduced in Huh7 and Hep3B cells transfected with miR-7 mimic (Fig. 7, C and D). Moreover, an addition of miR-7 inhibitor recovered the growth retardation by the knockdown of NF90 or NF45 (Fig. 7, E and F). These results indicate that NF90-NF45 would control the proliferation of HCC cell lines through the regulation of mature miR-7 production by these proteins.
FIGURE 7.
Knockdown of NF90 or NF45 promoted reduced HCC cell proliferation. A–C, proliferation of Huh7 cells transfected with siRNAs targeted to NF90 (A) or NF45 (B) and miRNA mimic (C) was analyzed by MTS assay. D, proliferation of Hep3B cells transfected with siRNAs targeted to NF90 or NF45 and miRNA mimic was analyzed by MTS assay. E and F, proliferation of Huh7 cells transfected with siRNAs targeted to NF90 (E) or NF45 (F) with or without miR-7 inhibitor was analyzed by MTS assay. After the indicated days of culture, the assay was performed according to the manufacturer's instructions, and absorbance was measured at 490 nm. Data are expressed as means ± S.D. (n = 3 per group). *, p < 0.05 relative to control by a two-tailed Student's t test.
Discussion
miR-7 has been characterized as an anti-oncomir in several cancers and suppresses cancer cell proliferation, survival, migration, invasion, and tumor metastasis (34). Indeed, miR-7 levels are significantly decreased in breast (35) and gastric cancers (36). Fang et al. (33) also demonstrated that 7 of 10 clinical HCC samples exhibited a reduction in miR-7. miR-7 inhibits expression of oncogenic regulatory factors, including EGFR (28, 29, 37–39), RAF-1 (28, 39), IGF1R (32, 36), BCL-2 (3), KLF-4 (40), PI3KCD, mTOR, and p70S6K (33) through its binding to the 3′UTRs of these mRNAs (Table 1). Thus, reduction of miR-7 induces enhancement of oncogene expression, resulting in tumorigenesis. However, the molecular mechanism for miR-7 down-regulation in tumors remains unclear. Previous studies show that quaking isoforms and the Hu antigen R (Hu antigen R)-Musashi homolog 2 (MSI2) complex regulate neuronal cell proliferation and differentiation, respectively, by repressing the processing of pri-miR-7 (41, 42). However, these factors have not been reported to be involved in tumorigenesis.
In this study, we demonstrated that knockdown of NF90 led to an elevation in mature miR-7 levels and down-regulation of pri-miR-7-1 levels in HCC cells (Fig. 2), although overexpression of NF90-NF45 caused a reduction in mature miR-7 levels and an accumulation of pri-miR-7-1 (Fig. 3). Furthermore, RNA-EMSA showed that rNF90-rNF45 directly bound to pri-miR-7-1 (Fig. 4). In addition, it has been apparent that NF90-NF45 is associated with endogenous pri-miR-7-1 (Fig. 5). We have previously reported that the binding of NF90-NF45 to pri-miRNAs would impair access of the microprocessor complex, which plays a pivotal role in processing of pri-miRNAs to pre-miRNAs, resulting in the inhibition of mature miRNA production (12). These findings, together with our previous findings (12), suggest that NF90-NF45 is a novel negative regulator of miR-7 biogenesis through inhibition of the pri-miR-7-1 processing step in HCC.
EGFR is a member of the HER/ErbB family of receptor tyrosine kinases. The EGF family, which belongs to a group of peptide growth factors, binds EGFR, inducing dimerization of EGFR. Dimerized EGFR causes activation of intracellular signal transduction, including the PI3K-AKT and RAS-RAF-MEK-MAPK pathways. These pathways are involved in cellular processes, including cell proliferation, survival, differentiation, and tumorigenesis (43, 44). Of note, numerous human solid tumors exhibit a high level of EGFR, which is related to poor prognosis (45). Intriguingly, miR-7 is known to inhibit EGFR expression, resulting in repressive activity of the downstream EGFR pathway in several cancer cell lines such as head and neck (39), lung, breast, and prostate (28). In this study, we show that knockdown of NF90 or NF45 leads to reduced EGFR expression, downstream AKT phosphorylation, and cell proliferation, as well as elevation of miR-7 in HCC cell lines (Figs. 2, 6, and 7). These findings imply that the NF90-NF45 complex regulates cell proliferation by alteration of EGFR expression through the control of miR-7 biogenesis.
We demonstrated that the expression of NF90 and NF45 was markedly increased in HCC tumor tissues compared with adjacent non-tumor tissues (Fig. 1). Furthermore, deletion of NF90 or NF45 caused a significant reduction in the proliferation rate of HCC cell lines, Huh7 and Hep3B (Fig. 7). Other groups have reported that the expression levels of NF90 are elevated in non-small lung, epithelial ovarian, breast cancers, and HCC (23–27). In addition, proliferation of cervical and breast cancer cell lines was depressed by knockdown of NF90 (12, 13, 23). Recently, NF90 was found to enhance hypoxia-mediated metastasis via regulation of NF90 protein stability by long non-coding RNA low expression in tumor and cell proliferation through regulation of cyclin E1 mRNA stability in HCC (26, 27). These observations, together with our findings in this study, suggest that NF90-NF45 or NF90 function as oncogenic factors in various tumors and raise the possibility that inhibitors of these proteins are novel therapeutic agents for several cancers.
In this report, we reveal that NF90-NF45 enhances the proliferation rates of tumor cells through repression of miR-7 biogenesis. Besides cell proliferation, the malignancy of tumor cells is tightly associated with cell migration and invasion. We performed whole-genome expression microarray analysis of the NF90-knockdown Huh7 cells (GEO accession no. GSE83589). In consequence, we found that the expression of 41 genes was decreased by less than ½-fold in the NF90-knockdown cells compared with the siNTC-treated cells. Among the 41 genes, we attempted to explore predicted targets of miR-513a-5p, miR-629–3p, miR-671–5p, miR-135a-3p, and miR-513b-5p, whose expressions were significantly increased by more than 2-fold in the NF90-knockdown cells compared with the siNTC-transfected cells (Table 1), using TargetScan Release 7.1, a computational tool for miRNA target prediction. Finally, we found several predicted targets of miR-513a-5p, miR-629-3p, miR-671-5p, miR-135a-3p, and miR-513b-5p. The predicted targets of each miRNAs were listed as follows: predicted miR-513a-5p targets: OLR1, VPS13C, ARL6IP1, ZSWIM6, AGXT, RHOBTB1, LMLN, STMN3, UGT2B10/11, and ILF3; predicted miR-629-3p targets: EYA3, ALDH1L2, LGSN, C7, VPS13C, ARL6IP1, ZSWIM6, RHOBTB1, SLCO1B3, and ILF3; predicted miR-671-5p targets: EYA3, ARL6IP1, ZSWIM6, RHOBTB1, and LMLN; predicted miR-135a-3p targets: PIGL, ALDH1L2, LGSN, C7, ZNF563, ARL6IP1, and ZSWIM6; predicted miR-513b-5p targets: PIGL, LGSN, C7, VPS13C, ARL6IP1, ZSWIM6, CXCL5, RHOBTB1, LMLN, and ILF3.
Subsequently, we performed gene ontology analysis of the above-mentioned targets. This analysis indicated that the possibility that OLR1, ZSWIN6, and CXCL5 may be involved in cell migration (data not shown). Elucidation of involvements of these factors in the development of HCC will be of interest to examine whether NF90-NF45 is involved in the cell migration in HCC, but it will require extensive work to address this issue in the future.
Experimental Procedures
Human Tissues Samples
Tumor and surrounding tissues from 18 cases of HCC were surgically resected at Kyoto University Hospital. Informed consent was obtained from all patients. Clinical information on the patients is listed in Table 2. The resected tissue samples for protein extraction were immediately frozen in liquid nitrogen and stored at −80 °C until use.
TABLE 2.
Clinical features of HCC patients with hepatocellular carcinoma enrolled in this study
| Sample no. | Age | Sex | Stage | Differentiationa |
|---|---|---|---|---|
| 01 | 74 | Male | IV | P |
| 02 | 66 | Male | III | W |
| 03 | 57 | Male | III | P |
| 04 | 76 | Male | III | W |
| 05 | 70 | Male | III | W |
| 06 | 56 | Male | II | W |
| 07 | 67 | Male | III | W |
| 08 | 64 | Male | II | P |
| 09 | 59 | Male | III | P |
| 10 | 77 | Female | II | P |
| 11 | 76 | Male | IV | P |
| 12 | 67 | Male | I | W |
| 13 | 74 | Female | II | P |
| 14 | 73 | Male | III | P |
| 15 | 73 | Male | III | W |
| 16 | 60 | Male | III | P |
| 17 | 58 | Male | III | P |
| 18 | 77 | Female | I | P |
a W and P indicate well and poorly differentiated tumors, respectively.
Immunoblot Analysis
Immunoblot analysis was performed as described previously (46). Anti-NF90 rabbit polyclonal antibody was prepared as described previously (12). Anti-NF45 antibody was produced by immunizing New Zealand White rabbits with a full-length His-NF45 recombinant protein as described previously (47). The following primary antibodies were obtained: anti-EGFR, anti-GAPDH, anti-AKT, anti-phospho-AKT Ser-473, and Thr-308 (catalog nos. 4267, 5174, 4691, 4060, and 2965, respectively; Cell Signaling Technology). Images were captured, and the intensities of specific bands were measured using an Las-4000 mini imaging system (Fuji Film).
Cell Culture
Human hepatocellular carcinoma cell lines Huh7 and Hep3B were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 4500 mg/liter glucose. To generate NF90 and/or NF45 stable cell lines, Huh7 cells were transfected with pEBMulti-Neo-NF90b and pEBMulti-Hyg-NF45. Three days after transfection, cells were exposed to 2000 μg/ml neomycin (Nacalai Tesque) and 1500 μg/ml hygromycin B (Wako) for 11 days.
Plasmids
To construct an expression plasmid of NF45, full-length NF45 cDNA was obtained by PCR of cDNA from Huh7 cells and subcloned into KpnI and XhoI sites of pEBMulti-Hyg vector (Wako Biochemicals). Full-length NF90b was obtained from pcDNA3.1-NF90b plasmid (provided by M. B. Mathews, University of Medical and Dentistry of New Jersey, Newark, NJ), and was inserted into KpnI and BamHI sites of the pEBMulti-Neo vector and pEBMulti-Neo TARGET tag vector (Wako Biochemicals).
qRT-PCR
Total RNA was isolated from Huh7 cells using TRIzol (Invitrogen), and contaminating genomic DNA was removed using DNA-free (Ambion). cDNA was synthesized using SuperScript III first-strand synthesis system (Invitrogen) and oligo(dT) primer according to the manufacturer's instructions (Invitrogen). For qRT-PCR, PCR samples composed of dilution cDNA (1:10), SYBR Green PCR master mix (Applied Biosystems), and 0.5 μm each of forward and reverse primers in a total volume of 10 μl were prepared. β-actin was used as an internal control. qRT-PCR for the detection of mature miRNA was carried out using a TaqMan miRNA assay kit (Applied Biosystems) and TaqMan gene expression master mix (Invitrogen) according to the manufacturer's protocol. A human RNU6B small nuclear RNA (RNU6B) was used as an internal control to normalize RNA input.
Transfection
Huh7 cells were transfected with NF90 and NF45 plasmids using X-tremeGENE HP DNA transfection reagent according to the manufacturer's instructions (Roche Applied Science). Huh7 cells were transfected with miR-7 mimic and miR-7 inhibitor at a final concentration of 6.7 nm using Lipofectamine RNAi Max according to the manufacturer's instructions (Invitrogen). An hsa-miR-7 mimic oligonucleotide and an hsa-miR-7-5p LNA inhibitor were obtained from Thermo Scientific (catalog no. C-300546-07-0005) and Exiqon (catalog no. 4100814-000), respectively. Stealth RNAi negative control (Invitrogen) was used as a negative control.
Northern Blotting
5 μg of total RNA was resolved on a 12% acrylamide, 7 m urea gel and blotted onto Hybond N+ (Amersham Biosciences). The blotted membrane was hybridized with a miRCURY LNA miR-7 detection probe (Exiqon 38485-00) and DNA oligonucleotide of U6 snRNA detection probe, 5′-CAC GAA TTT GCG TGT CAT CCT T-3′, as a control. These probe were end-labeled with [γ-32P]ATP by T4 polynucleotide kinase (New England Biolabs). The probes were washed once for 5 min at 47.8 °C in 2× SSC, 0.1% SDS, followed by washing four times for 5 min at 47.8 °C in 1.5× SSC, 0.1% SDS. the pre-miR-7, mature miR-7, and U6 snRNA were visualized by autoradiography.
RNA Interference
RNA interference analysis was performed as described previously (12). Stealth RNAi duplexes for NF90 and NF45 were obtained from Invitrogen. Stealth RNAi negative control (Invitrogen) was used as a negative control.
miRNA Microarray
Total RNA was isolated from Huh7 cells depleted of NF90 and labeled with Cy3 using an miRNA complete Labeling Reagent and Hyb kit (Agilent Technologies) according to the manufacturer's instructions. The labeled RNA was hybridized on Human miRNA Microarray Version 2 (Agilent Technologies). The array was scanned using a microarray scanner (Agilent Technologies) to measure the intensities of the microarray spots. The intensities of the spots were normalized by GeneSpring GX. All microarray data have been deposited in the Gene Expression Omnibus (GEO) database, accession no. GSE67411.
RNA-EMSA
RNA-EMSA analysis was performed as described previously (12). The pri-miR-7-1 was amplified from HEK293T cDNA by PCR with specific primers. The PCR products were subcloned into the pGEM-T-easy vector (Promega). These plasmids were linearized with PstI restriction enzyme and used for in vitro transcription to generate 32P-labeled probes.
RNA Immunoprecipitation Assay
WCEs from the Huh7 cells, transiently transfected with either mock vectors or both pEBMulti-Neo TARGET tag-NF90b vector and pEBMulti-Hyg-NF45 vector, were prepared with lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1 mm PMSF, proteinase inhibitor mixture (Nakalai Tesque)). The WCEs were immunoprecipitated with anti-TARGET tag antibody-conjugated beads (Wako Biochemicals). After the immunoprecipitates were digested by proteinase K, the coprecipitated RNAs were extracted with phenol/chloroform and ethanol precipitation. The precipitated RNA was annealed with oligo(dT) primer and reverse-transcribed with the SuperScript III first-strand synthesis system (Invitrogen). cDNA was amplified by qRT-PCR using specific primers for pri-miR-7-1 and β-actin mRNA.
MTS Assay
At 3 days post-transfection, siRNA- or miRNA mimic-transfected Huh7 and Hep3B cells were seeded at a density of 1,000 cells/well in 96-well plates. Cell viability following 0–4 days of culture was assayed using a CellTiter 96 Aqueous One Solution cell proliferation assay kit (Promega), and the absorbance was measured at 490 nm.
Sequences of all oligonucleotides are available upon request.
Author Contributions
T. H., S. S., Y. S., and T. T. designed the work. T. H., H. T., Y. S., M. O., N. T., E. H., Y. T., K. H., T. M., S. L., K. M., M. T., and S. S. acquired, analyzed, or interpreted the data. S. S., T. H., and Y. S. prepared the manuscript.
Supplementary Material
This work was supported by Japan Society for the Promotion of Science Grant-in-aid for JSPS Fellows 12J10660 and Grants-in-aid for Scientific Research (C) 25460371 and 16K08590. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- miRNA
- microRNA
- HCC
- hepatocellular carcinoma
- NF90
- nuclear factor 90
- NF45
- nuclear factor 45
- NF90-NF45
- NF90 and NF45 complex
- pri-miR-7-1
- primary miR-7–1
- pre-miRNA
- precursor miRNA
- mature miRNA
- single strand of miRNA
- anti-oncomirs
- anti-oncogenic miRNAs
- EGFR
- epidermal growth factor receptor
- qRT-PCR
- quantitative reverse transcription-PCR
- RNU6B
- RNU6B small nuclear RNA
- MSI2
- musashi homolog 2
- siNTC
- non-targeting control siRNA
- lncRNA-LET
- long non-coding RNA low expression in tumor
- WCE
- whole cell extract
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- r
- recombinant human.
References
- 1. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 2. Brennecke J., Hipfner D. R., Stark A., Russell R. B., and Cohen S. M. (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 [DOI] [PubMed] [Google Scholar]
- 3. Xiong S., Zheng Y., Jiang P., Liu R., Liu X., and Chu Y. (2011) MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int. J. Biol. Sci. 7, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R., and Ruvkun G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 [DOI] [PubMed] [Google Scholar]
- 5. Chen C. Z., Li L., Lodish H. F., and Bartel D. P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 6. Zhang B., Pan X., Cobb G. P., and Anderson T. A. (2007) microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 7. Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., and Shiekhattar R. (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 [DOI] [PubMed] [Google Scholar]
- 8. Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., and Shiekhattar R. (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krol J., Loedige I., and Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 [DOI] [PubMed] [Google Scholar]
- 10. Piskounova E., Polytarchou C., Thornton J. E., LaPierre R. J., Pothoulakis C., Hagan J. P., Iliopoulos D., and Gregory R. I. (2011) Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srikantan S., Tominaga K., and Gorospe M. (2012) Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 13, 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sakamoto S., Aoki K., Higuchi T., Todaka H., Morisawa K., Tamaki N., Hatano E., Fukushima A., Taniguchi T., and Agata Y. (2009) The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 29, 3754–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guan D., Altan-Bonnet N., Parrott A. M., Arrigo C. J., Li Q., Khaleduzzaman M., Li H., Lee C. G., Pe'ery T., and Mathews M. B. (2008) Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell. Biol. 28, 4629–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reichman T. W., Muñiz L. C., and Mathews M. B. (2002) The RNA-binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol. Cell. Biol. 22, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shim J., Lim H., R Yates J., and Karin M. (2002) Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10, 1331–1344 [DOI] [PubMed] [Google Scholar]
- 16. Harashima A., Guettouche T., and Barber G. N. (2010) Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 24, 2640–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tominaga-Yamanaka K., Abdelmohsen K., Martindale J. L., Yang X., Taub D. D., and Gorospe M. (2012) NF90 coordinately represses the senescence-associated secretory phenotype. Aging 4, 695–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gwizdek C., Ossareh-Nazari B., Brownawell A. M., Evers S., Macara I. G., and Dargemont C. (2004) Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J. Biol. Chem. 279, 884–891 [DOI] [PubMed] [Google Scholar]
- 19. Todaka H., Higuchi T., Yagyu K., Sugiyama Y., Yamaguchi F., Morisawa K., Ono M., Fukushima A., Tsuda M., Taniguchi T., and Sakamoto S. (2015) Overexpression of NF90-NF45 represses myogenic microRNA biogenesis, resulting in development of skeletal muscle atrophy and centronuclear muscle fibers. Mol. Cell. Biol. 35, 2295–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., and Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 21. Ozen M., Creighton C. J., Ozdemir M., and Ittmann M. (2008) Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 27, 1788–1793 [DOI] [PubMed] [Google Scholar]
- 22. Thomson J. M., Newman M., Parker J. S., Morin-Kensicki E. M., Wright T., and Hammond S. M. (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 20, 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu Q., Lu Y. Y., Noh H., Hong S., Dong Z., Ding H. F., Su S. B., and Huang S. (2013) Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene 32, 3933–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo N. L., Wan Y. W., Tosun K., Lin H., Msiska Z., Flynn D. C., Remick S. C., Vallyathan V., Dowlati A., Shi X., Castranova V., Beer D. G., and Qian Y. (2008) Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin. Cancer Res. 14, 8213–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo Y., Fu P., Zhu H., Reed E., Remick S. C., Petros W., Mueller M. D., and Yu J. J. (2012) Correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol. Rep. 27, 286–292 [DOI] [PubMed] [Google Scholar]
- 26. Yang F., Huo X. S., Yuan S. X., Zhang L., Zhou W. P., Wang F., and Sun S. H. (2013) Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 49, 1083–1096 [DOI] [PubMed] [Google Scholar]
- 27. Jiang W., Huang H., Ding L., Zhu P., Saiyin H., Ji G., Zuo J., Han D., Pan Y., Ding D., Ma X., Zhang Y., Wu J., Yi Q., Liu J. O., Huang H., Dang Y., and Yu L. (2015) Regulation of cell cycle of hepatocellular carcinoma by NF90 through modulation of cyclin E1 mRNA stability. Oncogene 34, 4460–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Webster R. J., Giles K. M., Price K. J., Zhang P. M., Mattick J. S., and Leedman P. J. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J. Biol. Chem. 284, 5731–5741 [DOI] [PubMed] [Google Scholar]
- 29. Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., Lee J., Fine H., Chiocca E. A., Lawler S., and Purow B. (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 68, 3566–3572 [DOI] [PubMed] [Google Scholar]
- 30. Engelman J. A. (2009) Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer 9, 550–562 [DOI] [PubMed] [Google Scholar]
- 31. Shaw R. J., and Cantley L. C. (2006) Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441, 424–430 [DOI] [PubMed] [Google Scholar]
- 32. Jiang L., Liu X., Chen Z., Jin Y., Heidbreder C. E., Kolokythas A., Wang A., Dai Y., and Zhou X. (2010) MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem. J. 432, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fang Y., Xue J. L., Shen Q., Chen J., and Tian L. (2012) MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 55, 1852–1862, [DOI] [PubMed] [Google Scholar]
- 34. Kalinowski F. C., Brown R. A., Ganda C., Giles K. M., Epis M. R., Horsham J., and Leedman P. J. (2014) MicroRNA-7: a tumor suppressor miRNA with therapeutic potential. Int. J. Biochem. Cell Biol. 54, 312–317 [DOI] [PubMed] [Google Scholar]
- 35. Kong X., Li G., Yuan Y., He Y., Wu X., Zhang W., Wu Z., Chen T., Wu W., Lobie P. E., and Zhu T. (2012) MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS One 7, e41523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao X., Dou W., He L., Liang S., Tie J., Liu C., Li T., Lu Y., Mo P., Shi Y., Wu K., Nie Y., and Fan D. (2013) MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 32, 1363–1372 [DOI] [PubMed] [Google Scholar]
- 37. Rai K., Takigawa N., Ito S., Kashihara H., Ichihara E., Yasuda T., Shimizu K., Tanimoto M., and Kiura K. (2011) Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol. Cancer Ther. 10, 1720–1727 [DOI] [PubMed] [Google Scholar]
- 38. Saydam O., Senol O., Würdinger T., Mizrak A., Ozdener G. B., Stemmer-Rachamimov A. O., Yi M., Stephens R. M., Krichevsky A. M., Saydam N., Brenner G. J., and Breakefield X. O. (2011) miRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 71, 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kalinowski F. C., Giles K. M., Candy P. A., Ali A., Ganda C., Epis M. R., Webster R. J., and Leedman P. J. (2012) Regulation of epidermal growth factor receptor signaling and erlotinib sensitivity in head and neck cancer cells by miR-7. PLoS One 7, e47067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okuda H., Xing F., Pandey P. R., Sharma S., Watabe M., Pai S. K., Mo Y. Y., Iiizumi-Gairani M., Hirota S., Liu Y., Wu K., Pochampally R., and Watabe K. (2013) miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 73, 1434–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y., Vogel G., Yu Z., and Richard S. (2013) The QKI-5 and QKI-6 RNA-binding proteins regulate the expression of microRNA 7 in glial cells. Mol. Cell. Biol. 33, 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choudhury N. R., de Lima Alves F., de Andrés-Aguayo L., Graf T., Cáceres J. F., Rappsilber J., and Michlewski G. (2013) Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 27, 24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Normanno N., Maiello M. R., and De Luca A. (2003) Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs): simple drugs with a complex mechanism of action? J. Cell. Physiol. 194, 13–19 [DOI] [PubMed] [Google Scholar]
- 44. Ono M., and Kuwano M. (2006) Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin. Cancer Res. 12, 7242–7251 [DOI] [PubMed] [Google Scholar]
- 45. Salomon D. S., Brandt R., Ciardiello F., and Normanno N. (1995) Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. Hematol. 19, 183–232 [DOI] [PubMed] [Google Scholar]
- 46. Sakamoto S., and Taniguchi T. (2001) Identification of a phorbol ester-responsive element in the interferon-γ receptor 1 chain gene. J. Biol. Chem. 276, 37237–37241 [DOI] [PubMed] [Google Scholar]
- 47. Song D., Sakamoto S., and Taniguchi T. (2002) Inhibition of poly(ADP-ribose) polymerase activity by Bcl-2 in association with the ribosomal protein S3a. Biochemistry 41, 929–934 [DOI] [PubMed] [Google Scholar]
- 48. Wu D. G., Wang Y. Y., Fan L. G., Luo H., Han B., Sun L. H., Wang X. F., Zhang J. X., Cao L., Wang X. R., You Y. P., and Liu N. (2011) MicroRNA-7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chin. Med. J. 124, 2616–2621 [PubMed] [Google Scholar]
- 49. Reddy S. D., Ohshiro K., Rayala S. K., and Kumar R. (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 68, 8195–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shin S., Moon K. C., Park K. U., and Ha E. (2012) MicroRNA-513a-5p mediates TNF-α and LPS induced apoptosis via downregulation of X-linked inhibitor of apoptotic protein in endothelial cells. Biochimie 94, 1431–1436 [DOI] [PubMed] [Google Scholar]
- 51. Wu L., Chen Z., Zhang J., and Xing Y. (2012) Effect of miR-513a-5p on etoposide-stimulating B7-H1 expression in retinoblastoma cells. J. Huazhong Univ. Sci. Technolog. Med. Sci. 32, 601–606 [DOI] [PubMed] [Google Scholar]
- 52. Papp G., Krausz T., Stricker T. P., Szendrői M., and Sápi Z. (2014) SMARCB1 expression in epithelioid sarcoma is regulated by miR-206, miR-381, and miR-671–5p on Both mRNA and protein levels. Genes Chromosomes Cancer 53, 168–176 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.