Abstract
N-terminal tails of histones H3 and H4 are known to bind several different Importins to import the histones into the cell nucleus. However, it is not known what binding elements in the histone tails are recognized by the individual Importins. Biochemical studies of H3 and H4 tails binding to seven Importins, Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα, show the H3 tail binding more tightly than the H4 tail. The H3 tail binds Kapβ2 and Imp5 with KD values of 77 and 57 nm, respectively, and binds the other five Importins more weakly. Mutagenic analysis shows H3 tail residues 11–27 to be the sole binding segment for Impβ, Kapβ2, and Imp4. However, Imp5, Imp7, Imp9, and Impα bind two separate elements in the H3 tail: the segment at residues 11–27 and an isoleucine-lysine nuclear localization signal (IK-NLS) motif at residues 35–40. The H4 tail also uses either one or two basic segments to bind the same set of Importins with a similar trend of relative affinities as the H3 tail, albeit at least 10-fold weaker. Of the many lysine residues in the H3 and H4 tails, only acetylation of the H3 Lys14 substantially decreased binding to several Importins. Lastly, we show that, in addition to the N-terminal tails, the histone fold domains of H3 and H4 and/or the histone chaperone Asf1b are important for Importin-histone recognition.
Keywords: chromatin regulation, histone, histone acetylation, nuclear transport, protein import, importin, karyopherin, nuclear import, nuclear localization signal
Introduction
New nucleosomes are assembled in the nucleus during S phase as new core histones H2A, H2B, H3, and H4 are synthesized, assembled into H2A/H2B and H3/H4 dimers in the cytoplasm, and then imported into the nucleus for deposition onto replicating chromatin (1–8). Little is known about cytoplasmic assembly/processing of H2A and H2B, but assembly/processing of H3 and H4 are better understood. In the cytoplasm, H3 and H4 are passed from one histone chaperone to another for folding, assembly into H3/H4 dimers, and acetylation. Acetylated H3/H4 is finally transferred to histone chaperone antisilencing function protein 1 (Asf1)2 and bound by Importins for transport into the nucleus (9–18). In the nucleus, two H3/H4 dimers are deposited onto DNA followed by two H2A/H2B dimers to form the nucleosome particle (4–6).
Importins bind and transport H3 and H4 into the nucleus (20–25). There are a total of at least 10 different Importins in human cells (26, 27). Co-immunoprecipitation, in vitro binding with recombinant proteins, and nuclear localization studies in yeast and permeabilized HeLa cells showed that several Importins can bind and import H3 and H4. These Importins are yeast Kap95, Kap104, Kap123, and Kap121; their human homologs Impβ, Kapβ2, Imp4, and Imp5; and three additional human Importins, Imp7, Imp9, and the Importin adaptor Impα (12–17, 28, 29). Although multiple Importins can bind and import H3 and H4, Imp4 is consistently the most abundant Importin that co-purifies with the histones, suggesting that it is the major/primary nuclear importer of H3/H4 in human cells (17, 28, 30). Similarly, the homolog of Imp4 in yeast, Kap123, is also the most abundant Importin that co-purifies with H3 and H4 from yeast cytosol (16).
Histones H3 and H4 each consist of an N-terminal disordered tail region followed by a globular histone fold domain (1, 2) (Fig. 1A). Previous studies showed that N-terminal tails of histones H3 and H4 are necessary and sufficient for nuclear import, consistent with the presence of NLS-like sequences in the tails. Removal of either the H3 or H4 tail does not prevent nuclear import, but simultaneous removal of both tails produced non-viable S. cerevisiae and caused loss of nuclear H3-H4 in Physarum polycephalum (12, 31). Of the Importins that bind and import H3 and H4, only classes of NLSs for Impα/Impβ, Kapβ2, and Imp5 are known. Impα binds directly to the classical NLS (c-NLS), Kapβ2 binds the entirely distinct PY-NLS, and Imp5 recognizes a short lysine-rich NLS named the IK-NLS (also distinct from c-NLS) (24, 26, 27). Classical NLSs contain either one or two clusters of basic residues (consensus sequences K(K/R)X(K/R) or (K/R)(K/R)X10–12(K/R)3/5 where X is any amino acid and (K/R)3/5 is three lysines or arginines in five consecutive residues) (32–34). The PY-NLS is defined by loose sequence motifs (N-terminal hydrophobic or basic motifs and a C-terminal (R/K/H)X2–5PY motif), structural disorder, and an overall basic charge (35). The IK-NLS is defined by the consensus motif K(V/I)XKX1–2(K/H/R) (36). Examination of sequences in the H3 and H4 tails revealed no recognizable c-NLS or PY-NLS (35, 37). A previous report suggested that residues 35–40 of the H3 tail resemble an Imp5-specific IK-NLS (36). Classes of NLS that bind Imp4, Imp7, and Imp9 have not yet been defined.
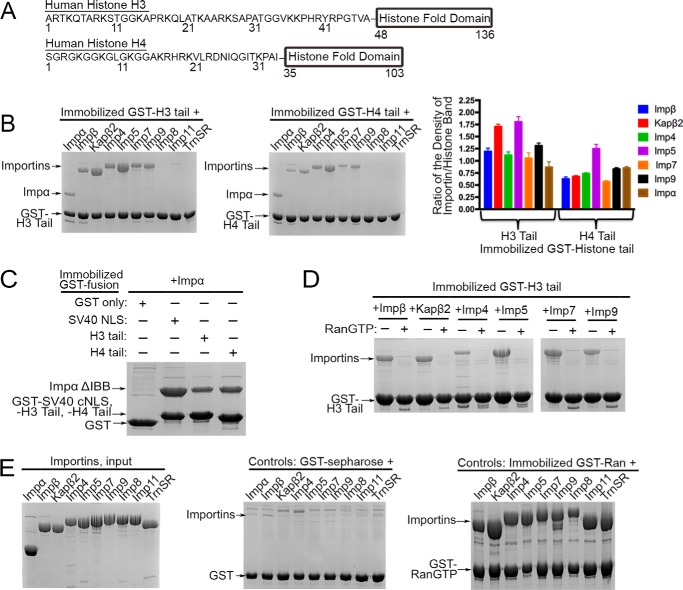
FIGURE 1.
Seven different human Importins bind the H3 and H4 tails. A, domain organization of human histones H3 and H4. B, pulldown binding assays of immobilized GST-H3 tail and GST-H4 tail (each 20 nmol; 250 nm) with a 500 nm concentration of each Importin (SDS-PAGE/Coomassie Blue). Relative densities of the gel bands from three separate experiments are plotted in histograms. One-way ANOVA tests were performed (H3 tail, p < 0.0001; H4 tail, p < 0.0001). Error bars represent S.D. C, Impα binding to immobilized GST-SV40 c-NLS, GST-H3 tail, and GST-H4 tail. Impα binds more tightly to the c-NLS than to the histone tails. D, binding assays of immobilized GST-H3 tail with Impβ, Kapβ2, Imp4, Imp5, Imp7, and Imp9 in the presence and absence of RanGTP (SDS-PAGE/Coomassie Blue). E, control experiments. Left panel, input samples of purified recombinant Importins (5 μl; 8 μm concentration of each protein; ∼10% of proteins used in B and in Figs. 2; 3C; 4, B–D; 5; and 6). Middle panel, Importin (∼4 μm) binding to GST protein (0.8 nmol; ∼2 μm) immobilized on glutathione-Sepharose. Right panel, Importin binding to immobilized GST-RanGTP. All six β-Importins bind RanGTP tightly.
20–30% of cytoplasmic H3 histones are acetylated at Lys14 and/or Lys18, and all cytoplasmic H4 are acetylated at both Lys5 and Lys12 (30). However, the effect of H3 and H4 tail acetylation on histone import is controversial. Mutations of H3 and H4 tail lysines to glutamines (acetylation mimics) in yeast abolished nuclear accumulation, slowed growth, or caused loss of viability, suggesting that acetylation impairs nuclear import (12). In contrast, H3 and H4 acetylation in P. polycephalum led to increased nuclear accumulation, and acetylated H4 tail peptides were reported to bind Imp4 better than unacetylated peptides, suggesting that acetylation may promote nuclear import (17, 31).
Here, we biochemically map binding determinants in the H3 and H4 tails for Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα. Structural analysis revealed that Kapβ2 binds residues 11–27 of the H3 tail, which resemble a basic PY-NLS that is missing its PY epitope (19). This basic segment of H3 is also important for binding Impβ, Imp4, Imp5, Imp7, Imp9, and Impα. In addition, an IK-NLS-like motif at H3 residues 35–40 is used to bind Imp5, Imp7, Imp9, and Impα. The first 20 residues of the H4 tail, enriched in basic and glycine residues, interact with Impβ, Kapβ2, Imp4, Imp7, and Imp9. The H4 tail also uses an IK-NLS-like motif to bind Imp5, Imp7, Imp9, and Impα. As we uncovered the Importin binding determinants, we also examined effects of histone tail acetylation on Importin interactions. Finally, we assembled a complex of the histone chaperone Asf1b bound to the full-length H3/H4 dimer and compared Importin interactions of this three-protein complex with those of the histone tails alone.
Results
Seven Different Human Importins Bind the H3 and H4 Tails
H3 and H4 tails were shown to mediate nuclear import of the histones by binding to several different Importins (12, 16–18, 29). First, pulldown binding assays were performed using immobilized GST-H3 tail (residues 1–47) or immobilized GST-H4 tail (residues 1–34) and nine different recombinant human Importins (Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, Imp11, Imp8, and Impα) (Fig. 1B). Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα bind strongly to both GST-H3 tail and GST-H4 tail (Coomassie-stained SDS-PAGE). Imp11, Imp8, and TrnSR do not bind either the H3 or H4 tail. Activities of the eight β-Importins (Impβ, Kapβ2, Imp4, Imp5, Imp7, and Imp9) were verified by RanGTP binding, and activity of Impα was verified by classical NLS binding (Fig. 1, C and E). Furthermore, Importins do not bind empty glutathione-Sepharose beads or GST protein immobilized on glutathione-Sepharose beads (Fig. 1E and supplemental Fig. 1). Importin-histone tail interactions are sensitive to RanGTP, indicative of specific Importin-cargo-like interactions (Fig. 1D).
Dissociation constants (KD values) of Kapβ2-H3 tail interactions were previously measured by isothermal titration calorimetry (ITC) (19). H3 tail binds Kapβ2 tightly with a KD of 77.1 nm, which is comparable with affinities of known Kapβ2-PY-NLS interactions (35, 38–42). Here, we measured the KD values of MBP-H3 tail and MBP-H4 tail binding to Imp5 and Kapβ2 by ITC. MBP-H3 tail binds Imp5 with a KD of 57.2 (43.9, 89.2) (Table 1; numbers in parentheses represent the 68.3% confidence intervals on KD as calculated using F-statistics and error-surface projection method). The H4 tail binds Kapβ2 and Imp5 ∼10-fold more weakly (H4 tail-Kapβ2, KD = 871 nm (737.3, 924.5); H4 tail-Imp5, KD = 619 nm (506.4, 692.3)). To verify that the N-terminal MBP tag has no effect on Importin binding, we first showed by ITC that MBP alone does not bind Imp5 (supplemental Fig. 6). ITC analysis of Imp5 and the H3 tail peptide (residues 1–47 with no MBP tag) produces a KD of 60.4 nm, which is very similar to the KD of 57 nm for Imp5 binding to MBP-H3 tail.
TABLE 1.
Binding affinities of Imp5 with H3 tails by ITC
ND, not determined.
| MBP-H3 tail(1–47) | KD determined by ITC |
|---|---|
| nm | |
| MBP alone | ND |
| Wild type | 57.2 (43.9, 89.2)a |
| K14A/R17A/K18A/K23A/R26A/K27A | 861.6 (661.8, 942.2)a |
| K14A/R17A/K18A | 26.1 (15.3, 52.3)a |
| K23A/R26A/K27A | 119.1 (74.0, 165.6)a |
| V35A/K36A/K37A/P38A/H39A/R40A | 198.1 (150.1, 241.5)a |
| K14/R17/K18/K23/R26/K27/35VKKPHR40 to Ala | ND |
| H3 tail(1–28) | 811.5 (664.4, 984.50)a |
| H3 tail (no MBP tag) | 60.4 (27.7, 92.4)a |
| GST-H3 tail | 47.1 (28.6, 76.9)a |
a Numbers in parentheses represent the 68.3% confidence intervals on KD as calculated using F-statistics and error-surface projection method.
We were not able to measure KD values of histone tail binding to Impβ, Imp4, Imp7, Imp9, or Impα by either ITC, microscale thermophoresis, or fluorescence anisotropy. The Importins aggregate in conditions for these biophysical experiments. For example, the Importins aggregated as a result of stirring in the ITC cell due to heating during microscale thermophoresis experiments and at higher titration concentrations necessary for the fluorescence anisotropy experiments. Therefore, band densities for bound Importins and histone tails in pulldown binding assays were measured, and their ratios were compared in a histogram to estimate relative strengths of H3 tail and H4 tail binding to Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα (Fig. 1B). Neither immobilization to glutathione-Sepharose or dimerization of the GST tag in GST-H3 tail has any effect on Importin binding as the KD values obtained by titrating Imp5 onto immobilized GST-H3 tail via pulldown binding assays (apparent KD = 66 nm; supplemental Fig. 3) and by ITC of Imp5 with GST-H3 tail (KD = 47.1 nm; Table 1 and supplemental Fig. 6) are similar to the KD obtained by ITC for the monomeric MBP-H3 tail (Table 1).
Comparison of bound Importin bands suggests that Importin-H3 tail interactions can be roughly divided into two groups: 1) Kapβ2 and Imp5, known from ITC to bind tightly (KD values <100 nm), and 2) Impβ, Imp4, Imp7, Imp9, and Impα, which all seem to bind more weakly. Apparent KD values estimated by fitting pulldown titration data for Impα, Impβ, Imp4, Imp7, and Imp9 (∼150–500 nm) are consistent with the Importin-H3 tail affinity trend shown in Fig. 1B (supplemental Fig. 3). Impα binds the histone tails more weakly than the SV40 c-NLS (Fig. 1C). The Importin-H4 tail affinity trend is similar, but H4 tail binds at least 10-fold more weakly than H3 tail as we compare KD values for Kapβ2 and Imp5. H4 tail binds most strongly to Imp5 (KD = 619 nm (506.4, 692.3)) and more weakly to the other six Importins. We validated the Importin-histone tail affinity trends by performing pulldown assays similar to those in Fig. 1B but using the amount of proteins (gel stained with SYPRO Ruby protein stain; supplemental Fig. 4).
Interactions of H3 Tail Residues 11–27 with Importins
A crystal structure of Kapβ2 bound to the entire H3 tail showed only H3 residues 11–27 bound to the PY-NLS binding site of the Importin (19). H3 residues 11TGGKAPRKN19 bind in extended conformation and contribute a significant portion of the total binding energy (Lys14 is a hot spot for binding Kapβ2), whereas residues 20LATKAARK27 form an α-helix (19). We used a series of H3 tail mutants, ITC, and qualitative pulldown binding assays to examine the contributions of H3 residues 11–27 to the binding of different Importins. The large majority of H3 tail side chains that contact Kapβ2 are from basic residues (19). Interestingly, interactions of the H3 tail with the other six Importins also appear to be dominated by electrostatic interactions as observed by the salt dependence of binding strengths (supplemental Fig. 5). Therefore, we mutated individual basic residues and all basic residues within H3 residues 11–27 and examined effects of the mutations on Importin binding by ITC (Imp5 (Table 1) and Kapβ2 (19)) and by pulldown binding assays (Fig. 2A). Significance of changes in Importin binding due to H3 tail mutations were assessed by performing one-way ANOVA tests and t tests on the raw Importin/histone ratios (Fig. 2A). Single site mutant H3 tail(K14A) shows large decreases in binding to Impβ, Kapβ2, Imp4, Imp7, and Imp9 but does not affect Impα and Imp5 binding. Single mutations of Arg17 and Lys23 moderately decrease binding to Impβ, Kapβ2 (R17A only), Imp4 (R17A only), Imp7, and Imp9 but do not affect Impα and Imp5 binding. In contrast, single mutations of Lys18, Arg26, and of Lys27 show little to no effect in binding any of the Importins.
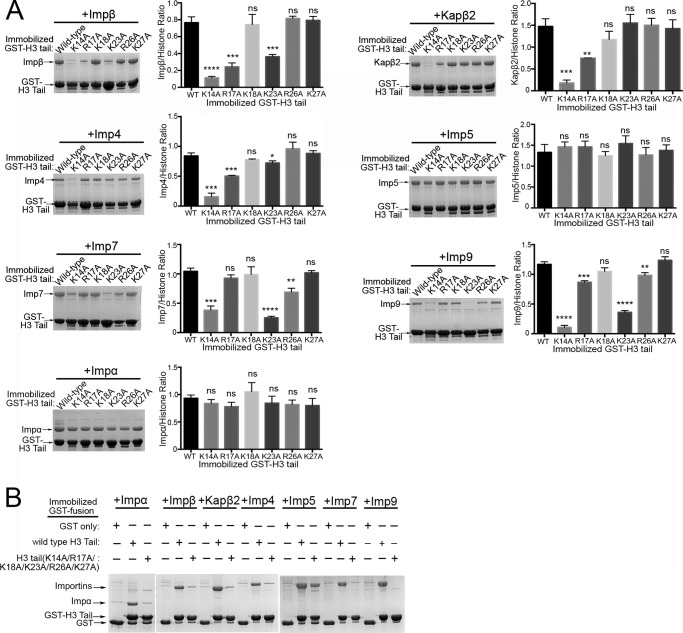
FIGURE 2.
The basic segment at residues 11–27 of the H3 tail is important for binding Importins. A, pulldown binding assays of immobilized GST-H3 tail proteins (wild type and single site alanine mutants of Lys14, Arg17, Lys18, Lys23, Arg26, and Lys27) with Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα (SDS-PAGE/Coomassie). Densities of the gel bands from experiments performed in triplicate are plotted in histograms. t tests were performed to compare each mutant with the wild-type protein (ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001). Significance of mutations was also validated by one-way ANOVA tests (Impα, p > 0.05; Impβ, p ≤ 0.0001; Kapβ2, p ≤ 0.0001; Imp4, p ≤ 0.0001; Imp5, p > 0.05; Imp7, p ≤ 0.0001; and Imp9, p ≤ 0.0001). Error bars represent S.D. B, pulldown binding assay of immobilized GST-H3 tail proteins (wild type and the H3 tail(K14A/R17A/K18A/K23A/R26A/K27A) mutant) with Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα (SDS-PAGE/Coomassie).
ITC data show that mutation of all basic residues within H3 residues 11–27 (mutant MBP-H3 tail(K14A/K17A/K18A/K23A/R26A/K27A) decreases Imp5 affinity ∼15-fold (KD of 862 nm for the mutant versus 57 nm for wild-type H3 tail; Table 1). Consistently, pulldown of this mutant (GST-H3 tail(K14A/K17A/K18A/K23A/R26A/K27A)) still shows Imp5 binding, suggesting additional binding element(s) beyond H3 residues 11–27 (Fig. 2B). Mutation of all six basic residues abolishes binding to Impβ, Kapβ2, Imp4, Imp7, Imp9, and Impα, suggesting that the same region of H3 that binds Kapβ2 (residues 11–27) is also critical for binding these other Importins (Fig. 2B). Interestingly, although mutation of all six basic residues disrupts Impα binding (Fig. 2B), no single mutation affects Impα binding (Fig. 2A), suggesting that the energy for binding Impα is distributed across the entire H3 segment possibly through involvement of both side chain and main chain atoms reminiscent of c-NLS interactions.
In summary, the basic segment that spans H3 residues 11–27 is important for binding Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα. Here, residue Lys14 is a hot spot for binding all the Importins except for Imp5 and Impα. This basic segment is likely not the sole binding element for Imp5.
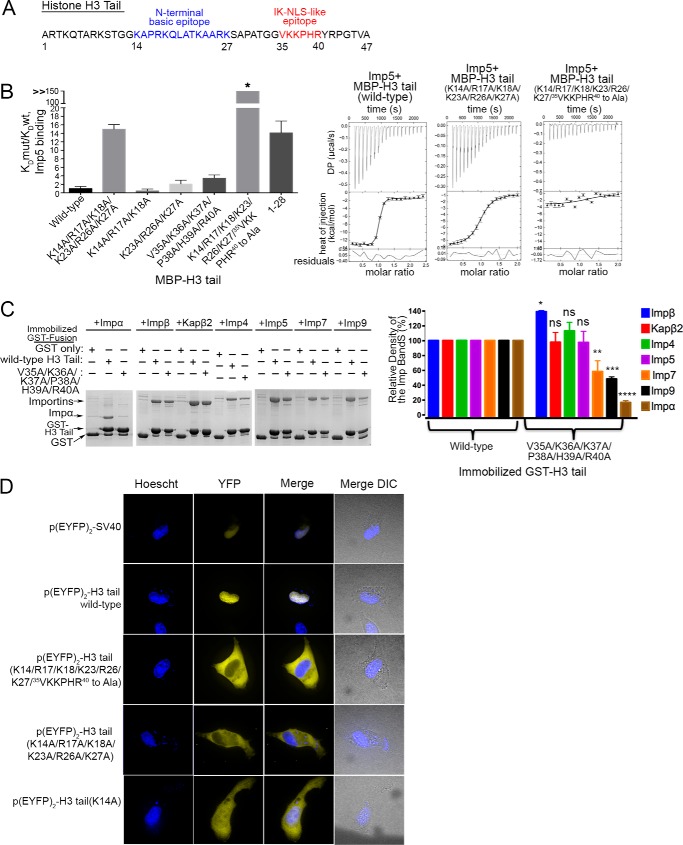
An IK-NLS-like Epitope in the H3 Tail
Matsuura and co-worker (36) had reported previously that 35VKKPHR40 in the H3 tail resembles the consensus sequence K(V/I)XKX1–2(K/H/R) for the Kap121- or Imp5-specific IK-NLS motif (Fig. 3A). To determine whether the IK-NLS-like motif is indeed important for Imp5 binding, we mutated 35VKKPHR40 in the H3 tail to alanines (H3 tail(V35A/K36A/K37A/P38A/H39A/R40A) and analyzed binding to Imp5 by ITC and pulldown assays (Table 1 and Fig. 3, B and C). MBP-H3 tail(V35A/K36A/K37A/P38A/H39A/R40A) binds Imp5 ∼4-fold more weakly than wild-type H3 tail, suggesting that the IK-NLS motif does indeed contribute to Imp5 binding (Table 1). Interestingly, the energetic contribution of the IK-NLS motif to Imp5 binding is less than that of all basic residues within residues 11–27 as the MBP-H3 tail(K14A/K17A/K18A/K23A/R26A/K27A) mutant decreases Imp5 affinity by 15-fold (Table 1). Mutation of both the basic segment spanning residues 11–27 and the IK-NLS epitope (MBP-H3 tail(K14/K17/K18/K23/R26/K27/35VKKPHR40 to Ala) mutant) completely abolishes Imp5 binding (Table 1 and Fig. 3B).
FIGURE 3.
35VKKPHR40 of the H3 tail contributes to binding Imp5, Imp7, Imp9, and Impα. A, a proposed Imp5-specific IK-NLS motif in H3 tail. B, ITC analysis of Imp5 and H3 tail showing that 35VKKPHR40 is also important for Imp5 binding. KD(mutant (mut))/KD(WT) for H3 tail mutants (KD values from triplicate ITC experiments) are shown in a histogram. C, pulldown binding assays of immobilized GST-H3 tail proteins (wild type and the H3 tail(V35A/K36A/K37A/P38A/H39A/R40A) mutant) with Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα (SDS-PAGE/Coomassie). Relative densities of the gel bands from experiments performed in triplicate are plotted in histograms. t tests were performed to assess significance of changes in Importin binding for the 36VKKPHR40 to Ala mutation (ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001). Error bars represent S.D. D, nuclear localization of EYFP2-H3 tail proteins in HT1080 cells. YFP (pseudocolored in yellow), Hoechst (pseudocolored in blue), and merged images were captured using spinning disk confocal microscopy of live HT1080 cells (100×). The image shown is a representative of four images.
A truncated H3 tail with only residues 1–28, which lacks the IK-NLS motif and the intervening 29APATGG34, binds Imp5 with lower affinity than the H3 tail(V35A/K36A/K37A/P38A/H39A/R40A) mutant (KD = 811 nm for MBP-H3 tail(1–28) versus KD = 198 nm for MBP-H3 tail(V35A/K36A/K37A/P38A/H39A/R40A); Table 1 and Fig. 3B), suggesting either contributions from the intervening 29APATGG34 segment and/or cooperativity between residues 11–27 and the IK-NLS epitope. Truncation of the H3 tail to residues 1–28 has no effect on Kapβ2 binding, consistent with structural observations that binding elements for this Importin reside within residues 11–27 of the H3 tail (19). Pulldown binding assays of the H3 tail(V35A/K36A/K37A/P38A/H39A/R40A) mutant show no effect on Impβ, Kapβ2, and Imp4 binding, but Impα binding is significantly decreased, and Imp7 and Imp9 binding are moderately decreased (Fig. 3C). In summary, Impβ, Kapβ2, and Imp4 appear to mostly bind the basic region in residues 11–27 of H3. Imp5, Imp7, Imp9, and Impα bind at least two regions of the H3 tail: 1) the basic region from residues 11 to 27 and 2) the IK-NLS epitope.
Mutations of the H3 Tail and Nuclear Localization in Cells
Nuclear localization of EYFP2-H3 tail proteins (wild type and mutants) was examined in live HT1080 cells using a spinning disk confocal microscope (Fig. 3D). Like the SV40 NLS, wild-type H3 tail localizes exclusively to the nucleus. However, the H3 tail(K14A/K17A/K18A/K23A/R26A/K27A) and H3 tail(K14/K17/K18/K23/R26/K27/35VKKPHR40 to Ala) mutants localize to the cytoplasm. The single site H3(K14A) mutant shows decreased nuclear accumulation, probably due to decreased binding to most but not all Importins as the mutant protein is still able to bind Impα and Imp5.
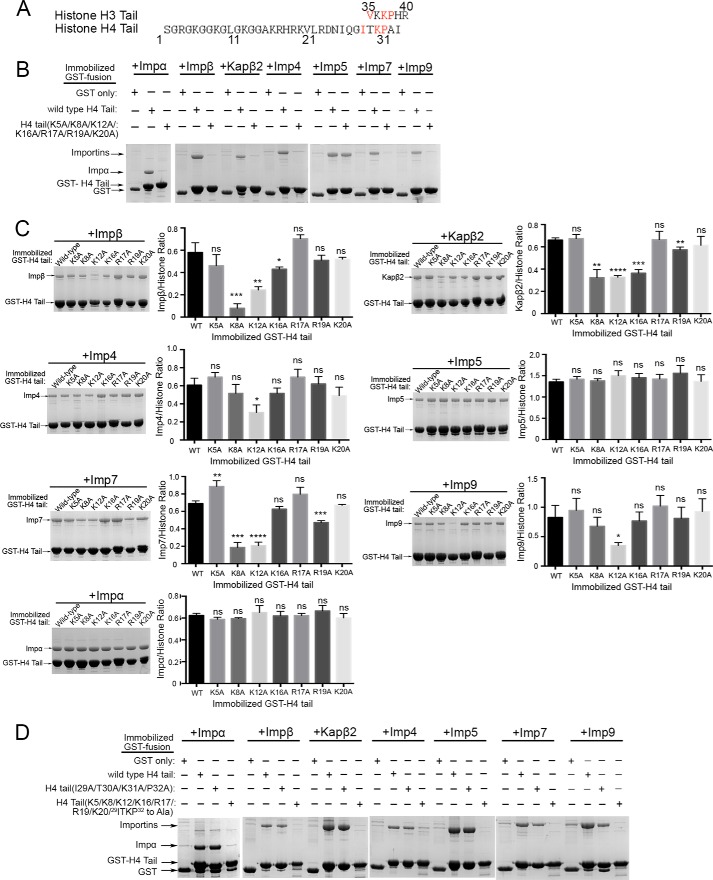
Basic Epitope(s) in the H4 Tail That Bind Importins
The same Importins that bind the H3 tail also bind the H4 tail but with much lower affinities (Fig. 1B). Binding determinants in the H4 tail for Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα were biochemically mapped. The first 20 residues of the H4 tail are rich in glycine and basic residues (Fig. 4A). When all basic residues in this segment are mutated to alanines, the resulting GST-H4 tail(K5A/K8A/K12A/K16A/R17A/R19A/K20A) mutant no longer binds Impβ, Kapβ2, Imp4, Imp7, Imp9, and Impα (Fig. 4B). Single site mutants of the individual basic residues (Lys5, Lys8, Lys12, Lys16, Arg17, Arg19, and Lys20) were analyzed by pulldown assays (Fig. 4C). As for the H3 tail, the significance of the changes in Importin binding due to H4 tail mutations was assessed with one-way ANOVA tests and t tests (Fig. 4C). The K5A, R17A, and K20A mutations have no effect on any of the Importins. The K8A and K12A mutations decrease binding to Impβ, Kapβ2, Imp4, Imp7, and Imp9 (Fig. 4C). The K16A and R19A mutations moderately affect Kapβ2 binding. The R19A mutation also affects Imp7 binding. These results suggest that the H4 tail uses basic side chains within residues 5–20 to bind Impβ, Kapβ2, Imp4, Imp7, Imp9, and Impα.
FIGURE 4.
Two basic segments within residues 5–20 and residues 29–34 of the histone H4 tail are used to bind Importins. A, alignment of the H3 IK-NLS motif with the H4 tail sequence. The H4 tail has an IK-NLS-like motif at residues 29–34. B, pulldown binding assays of immobilized GST-H4 tail proteins (wild type and the H4 tail(K5A/K8A/K12A/K16A/R17A/R19A/K20A) mutant) with Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα (SDS-PAGE/Coomassie). C, pulldown binding assays of immobilized GST-H4 tail proteins (wild type and single site alanine mutants of Lys5, Lys8, Lys16, Arg17, Arg19, and Lys20) with Importins (SDS-PAGE/Coomassie). Densities of the gel bands from experiments performed in triplicate are plotted in histograms. t tests were performed to assess effects of mutations (ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001). Significance of mutations was also validated by one-way ANOVA tests (Impα, p > 0.05; Impβ, p ≤ 0.0001; Kapβ2, p ≤ 0.0001; Imp4, p ≤ 0.0001; Imp5, p > 0.05; Imp7, p ≤ 0.0001; Imp9, p ≤ 0.01). Error bars represent S.D. D, pulldown binding assays of immobilized GST-H4 tail proteins (wild type, H4 tail(I29A/T30A/K31A/P32A) mutant, and H4 tail(K5/K8/K12/K16/R17/R19/K20/29ITKP32 to Ala) mutant) with Importins (SDS-PAGE/Coomassie).
Binding to Imp5 is not affected by mutations of any or all of the basic residues within the first 20 amino acids of the H4 tail (Fig. 4, B and C). No IK-NLS was evident in the H4 tail, but we aligned the IK-NLS of the H3 tail (35VKKPHR40) to the H4 tail sequence and found residues 29ITKP32 that match part of the IK-NLS consensus sequence of K(V/I)XKX1–2(K/H/R) (Fig. 4A). H4 residues 29ITKP32 were mutated to alanines (H4 tail(I29A/T30A/K31A/P32A)) to determine whether the IK-NLS-like motif contributes to Imp5 binding. The GST-H4 tail(I29A/T30A/K31A/P32A) mutant does not affect Imp5 binding (Fig. 4D). However, when combined with mutations of basic residues in residues 5–20, the mutant GST-H4 tail(K5/K8/K12/K16/R17/R19/K20/29ITKP32 to Ala) no longer binds Imp5 (Fig. 4D). As expected from results in Fig. 4B, the GST-H4 tail(K5/K8/K12/K16/R17/R19/K20/29ITKP32 to Ala) mutant does not bind Impβ, Kapβ2, Imp4, Imp7, and Imp9.
In summary, a basic region spanning residues 5–20 of the H4 tail is important for binding Impβ, Kapβ2, Imp4, Imp7, Imp9, and Impα. Here, Lys12 is the binding hot spot for most of the Importins. The H4 tail uses at least two basic segments to bind Imp5: 1) the basic epitope within residues 5–20 and 2) an IK-NLS-like motif at residues 29ITKP32.
Acetylation of H3 and H4 Tails and Importin Binding
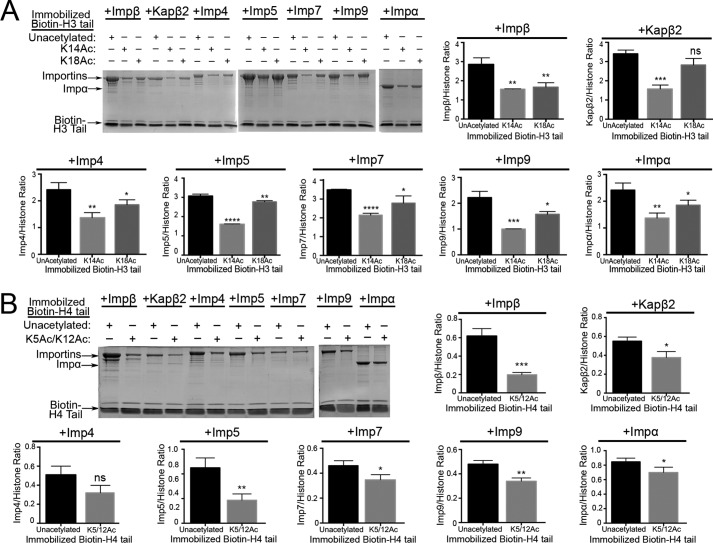
Previous mass spectrometry studies showed that 20–30% of cytoplasmic H3 is acetylated at Lys14 and/or Lys18, whereas all cytoplasmic H4 is acetylated at Lys5 and Lys12 (30). H3 Lys14 is important for H3 tail binding to Impβ, Kapβ2, Imp4, Imp7, and Imp9 (Fig. 2A). Similarly, H4 Lys12 is important for binding to Impβ, Kapβ2, Imp4, Imp7, and Imp9 (Fig. 4C). Pulldown binding assays were performed using immobilized synthetic acetylated H3 and acetylated H4 tail peptides (biotin-H3 tail(K14Ac), biotin-H3 tail(K18Ac) and biotin-H4 tail(K5Ac/K12Ac)) (Fig. 5, A and B). H3 tail(K14Ac) shows decreased binding to all Importins compared with unacetylated H3 tail (Fig. 5A). H3 tail(K18Ac) shows mild decreases in binding Impα, Impβ, and Imp5 but no effect on the other Importins (Fig. 5A). H4 tail(K5Ac/K12Ac) shows moderate to mild decreases in binding Impβ, Imp5 and Imp9 but shows no confident effect on the other Importins (Fig. 5B). These results suggest that Lys14 acetylation decreases H3 tail binding to Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα. In contrast, H4 tail diacetylation on Lys5 and Lys12, marks of newly synthesized histones, has little effect on binding to most Importins.
FIGURE 5.
Interactions of acetylated H3 and H4 tails with Importins. Pulldown assays of Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα binding to immobilized biotin-tagged unacetylated and acetylated histone H3 tail and H4 tail peptides (SDS-PAGE/Coomassie) were performed. Densities of the gel bands from experiments performed in triplicate are plotted in histograms. t tests were performed to assess effects of acetylation (ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001). A, pulldown binding assays of biotin-H3 tail (unacetylated) versus biotin-H3 tails acetylated at either Lys14 or Lys18. B, pulldown binding assays of biotin-H4 tail (unacetylated) versus biotin-H4 tail acetylated at both Lys5 and Lys12. Error bars represent S.D.
Importins Binding to the Asf1-H3/H4 Complex
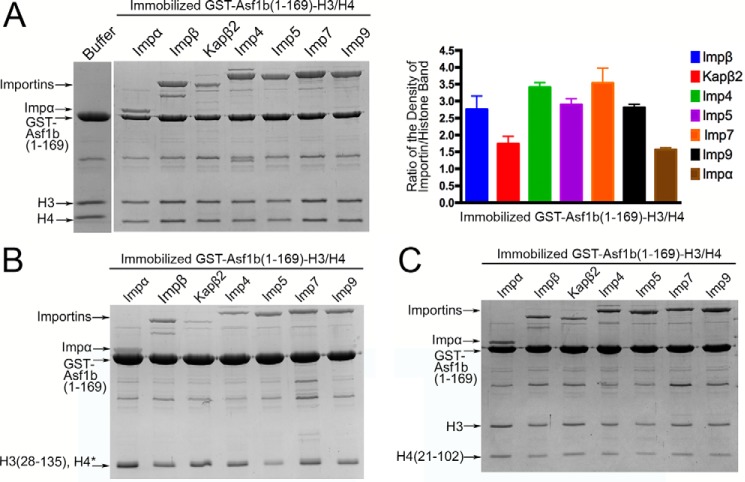
Previous co-immunoprecipitation studies showed that H3 and H4 are imported as a dimer in complex with the histone chaperone Asf1 (9–18). However, no information is available about how Importins interact with the Asf1-bound H3/H4 dimer. It is not known whether Importins also make contact with the histone fold domains of H3/H4 and/or with Asf1. Full-length human Asf1b is very sensitive to proteolytic degradation. Therefore, we performed pulldown binding assays with immobilized GST-Asf1b(1–169), which covers only the conserved N-terminal core domain of Asf1 (the highly divergent C-terminal tail of Asf1 is removed), and with immobilized GST-Asf1b(1–169)-H3/H4 (Fig. 6A and supplemental Fig. 8). None of the Importins bind GST-Asf1b(1–169) (supplemental Fig. 8). Analysis of the GST-Asf1b(1–169)-H3/H4 pulldown data shows that Importins may be qualitatively divided into three groups depending on their affinities for the histone-chaperone complex (from high to low affinity): Imp4, Imp7 > Impβ, Imp5, Imp9 > Impα, Kapβ2 (Fig. 6A). The Importin-Asf1b(1–169)-H3/H4 binding trend is quite different from those of the N-terminal H3 and H4 tails alone (Kapβ2, Imp5 > Impβ, Imp4, Imp7, Imp9, Impα; Fig. 1B), suggesting that histone fold domains of the dimer and/or Asf1b (only in the context of the complex) are also important for Importin-histone binding. Gel filtration chromatography of the Imp4-Asf1b-H3/H4 complex shows a stable high affinity assembly (supplemental Fig. 9).
FIGURE 6.
Importin binding to the Asf1b-H3/H4 complex. A, pulldown binding assays of Importin binding to immobilized GST-Asf1b(1–169)-H3/H4. Band densities from three separate experiments are plotted (one-way ANOVA: p < 0.0001). Error bars represent S.D. B and C, pulldown binding assays of Importin binding to immobilized GST-Asf1b(1–169)-H3(28–135)/H4 (*, bands for H3(28–135) and H4 co-migrate on SDS-polyacrylamide gel) (B) and GST-Asf1b(1–169)-H3/H4(21–102) (C).
Similar pulldown assays were performed with H3(28–135) and H4 tail(21–102), which lack tail basic epitopes that bind Importins (Fig. 6, B and C). Removal of the basic epitopes decreased binding to all seven Importins, but the overall binding affinity trend remains similar to that for the Asf1b(1–169)-bound full-length H3/H4 dimer. In summary, along with the basic epitopes in the N-terminal tails, the histone fold domains of H3 and H4 and/or the histone chaperone Asf1b is important for Importin-histone recognition and Importin specificity.
Discussion
We have mapped sequence elements in H3 and H4 tails that bind Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα. The results inform on both the NLS organization in the histone tails and general sequence elements that bind different Importins.
Importin-Binding Epitopes in H3 and H4 Tails
The basic segment in H3 residues 11–27 contributes most of the binding energy for interactions with Impβ, Kapβ2, and Imp4, but interactions with Imp5, Imp7, Imp9, and Impα involve an additional downstream IK-NLS-like motif. The H4 tail also uses either one or two basic regions to bind multiple Importins. Impβ, Kapβ2, Imp4, Imp7, Imp9, and Impα bind the basic segment between H4 residues 5 and 20, whereas Imp5 binds an additional IK-NLS-life motif.
Nuclear import cargoes usually have specific NLSs that bind one of the 10 Importins (26). However, two classes of highly abundant proteins, histones and ribosomal proteins, are exceptions as they can bind multiple Importins (12–16, 28, 29, 43). For example, the ribosomal protein L23A (rpL23A) binds Impα/β, Kapβ2, Imp5, and Imp7 through a β-like import receptor binding (BIB) sequence (43). Ribosomal proteins like rpS7 and rpL5 also have BIB-like sequences and bind multiple Importins (43). BIBs were suggested to have originated from ancestral nuclear import signals prior to the divergence of Importins to gain specialized/distinct NLS binding sites (43). Although Importins have evolved to bind distinct signals, many of them may retain binding to these ancestral non-specialized BIB sequences. 59KYPRKSAPRRNK70 in the rpL23A BIB aligns with 14KAPRKQLATKAAR26 in the H3 tail and 8KGLGKGGAKRHR20 in the H4 tail. Sequence similarity and the ability to bind multiple Importins suggest that the N-terminal basic segments of H3 and H4 tails may be ancestral NLSs like BIBs.
NLSs in the Histone Tails That bind Kapβ2, Impβ, and Imp4
Kapβ2, Impβ, and Imp4 bind solely to the N-terminal basic segments of the H3 and H4 tails. Impβ is known to bind NLSs of very different lengths, sequences, and structural elements, but electrostatic contacts are common features in all these Impβ-NLS interactions (24, 26, 27, 44–47). Electrostatics are also important for H3 and H4 tails binding to Impβ, but in the absence of Impβ-histone tail structures, we cannot predict the NLS conformations or the locations of their binding sites on Impβ. A Kapβ2-H3 tail structure shows that H3 residues 11–27 form a PY-NLS variant that is missing the canonical PY motif (19). No structural information is available at this time for how Imp4 binds cargoes/NLSs, but binding to only the N-terminal basic segments of the H3 and H4 tails suggests that NLSs for Imp4 may be generally compact and monopartite.
NLSs in the Histone Tails That Bind Impα, Imp5, Imp7, and Imp9
Interactions of the H3 tail with Imp5, Imp7, Imp9, and Impα are bipartite, involving the basic segment at residues 11–27 and an IK-NLS motif at 35VKKPHR40. The H4 tail also uses two analogous elements to bind Imp5. Bipartite interaction with Impα is not unexpected as the import adaptor binds bipartite c-NLSs (48). The H3 tail sequence that binds Impα, 11TGGKAPRKQLATKAARK27. . . . .35VKKPHR40 (possible bipartite c-NLS consensus residues are underlined), does not match the bipartite c-NLS consensus of (K/R)(K/R)X10–12(K/R)3/5) because linkers between basic segments are either too short or too long. If the bipartite NLS is 17RKQLATKAARK27 or 26RK. . . . .35VKKPHR40, the seven-residue linkers are too short to match the c-NLS consensus. If the NLS is 17RKQLA. . . . .35VKKPHR40, the 16-residue linker is too long. Impα can recognize bipartite c-NLSs with artificial linkers as short as eight residues and linkers up to 40 residues (32, 48–51). Therefore, the range of linker length in the current bipartite c-NLS consensus is probably too restrictive, and the H3 tail does indeed contain a bipartite c-NLS.
The second epitope of the H3 bipartite c-NLS is in the IK-NLS motif (35VKKPHR40) that is also used to bind Imp5. Structures of Kap121 (yeast Imp5) suggested that the IK-NLS is the most important binding element in Kap121 and Imp5 cargoes (36). Interestingly, our results suggest that IK-NLS motifs in H3 and H4 tails are only a portion of their NLSs for Imp5 with relatively minor contribution to binding energy. Furthermore, unlike IK-NLSs in Kap121 cargoes Pho4p, Spo12p, and binding partner Nup53p, which are specific for Kap121 (36), IK-NLS motifs in H3 and H4 are also part of bipartite c-NLSs that bind Impα. Finally, structures of Imp7 and Imp9 are not yet available, but our results suggest that both Importins can bind long sprawling NLSs with multiple basic epitopes.
Importin Interactions beyond the Histone Tails
Several groups reported that H3 and H4 tails are key for histones import (12, 16–18, 29). However, H3 and H4 import is likely coordinated with upstream and downstream histone processing/nucleosome assembly events and thus may involve other protein players (28, 30, 31, 52–56). Furthermore, H3 and H4 dimerize and bind histone chaperone b before binding Importins in cells (17, 28). Asf1 binds the histone folds of the H3/H4 dimer, thus leaving H3 and H4 tails free to bind Importins (9, 10). Furthermore, although many different Importins can bind and import H3 and H4 tails, Imp4 and its yeast homolog Kap123 are consistently the most abundant Importins that co-purify with H3 and H4 in lysates (16, 17, 28, 30). Our H3 and H4 tail binding data show no specificity for Imp4 over the other Importins. Imp4 is in fact one of the weakest binding Importins. The specificity of H3/H4 for Imp4 in cells must therefore lie outside of the histone tails, perhaps in the histone folds, and/or involves Asf1. Binding studies of Asf1b-H3/H4 showed that Imp4 is one of the strongest binders for the histone-chaperone complex, suggesting that the H3/H4 dimer histone fold and/or Asf1b must be important for Imp4 specificity. Kap123 is the most abundant Importin in budding yeast. Its abundance may be key in accomplishing a nuclear import rate that is 5–12-fold more rapid than other Importins (57). If Imp4 is similarly abundant in human cells, its high cellular concentration may be an advantage for Imp4-mediated H3/H4 import.
Acetylation of H3 Lys14 Impairs Importin Binding
The Importin-binding segments in H3 and H4 tails contain several lysine residues that are acetylated to different degrees in the cytoplasm (H3 Lys14, H3 Lys18, H4 Lys5, and H4 Lys12) (28, 30, 31, 52–54). The role of histone acetylation in nuclear import was controversial with reports of both promoting and inhibiting histone import (12, 17, 31).
When Lys14 (binding hot spot for Impβ, Kapβ2, Imp4, Imp7, and Imp9) in the H3 tail is acetylated, binding to Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, and Impα is decreased. Therefore, nuclear import of the small pool of cytoplasmic H3/H4 with acetylated H3 tail may be affected. In contrast, all new H4 is persistently acetylated prior to nuclear import. We showed that diacetylation of H4 tail Lys5 and Lys12 has little effect on binding to most Importins. It is important to note that unacetylated H4 tail binds Importins weakly (KD values >600 nm) and may contribute little toward nuclear import of the H3/H4 dimer. It is currently difficult to predict how acetylation of the full-length H3/H4 dimer or the Asf1b-H3/H4 complex affects Importin binding. Studies of Importin-histone complexes beyond histone tails will be important to resolve current controversies regarding histone acetylation and nuclear import.
Experimental Procedures
Plasmids
GST fusion constructs were generated by inserting PCR fragments corresponding to the regions of the genes of interest into the pGEX-TEV plasmid, which is a pGEX4T3 vector (GE Healthcare) that encodes an N-terminal GST and is modified to include a TEV protease cleavage site (58). GST-Importin constructs were generated for full-length human Impα(ΔIBB), Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, Imp11, Imp8, and TrnSR (42). GST-histone tail constructs of H3 (residues 1–47) and the H4 tail (residues 1–34) were kindly provided by B. Li (University of Texas Southwestern). MBP-histone tail constructs of H3 (residues 1–47) and H4 (residues 1–34) and shorter fragments of the histone tails were subcloned into the pMALTEV vector (pMAL (New England BioLabs, Ipswich, MA); N-terminal MBP followed by a TEV cleavage site) (59). H3 tail and H4 tail mutations were made by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and all constructs were sequenced. Acetylated H3 tail (K14Ac and K18Ac) peptides and the acetylated H4 tail (K5Ac/K12Ac) peptide were purchased from Biomatik.
Expression and Purification of Histones and Importins
Human Impα(ΔIBB), Impβ, Kapβ2, Imp4, Imp5, Imp7, Imp9, Imp11, Imp8, and TrnSR were expressed as GST fusion proteins. Human H3 tail (residues 1–47) and H4 tail (residues 1–34) were also expressed as GST fusion and MBP fusion proteins. All recombinant proteins were expressed separately in BL21(DE3) Escherichia coli cells (induced with 0.4 mm isopropyl β-d-1-thiogalactoside for 12 h at 25 °C). Bacteria were lysed with the EmulsiFlex-C5 cell homogenizer (Avestin, Ottawa, Canada) in buffer containing 50 mm Tris, pH 7.5, 200 mm NaCl, 20% glycerol, 2 mm DTT, 1 mm EDTA, and protease inhibitors.
GST-Importin proteins were purified by affinity chromatography on GSH-Sepharose (GE Healthcare), eluting with buffer containing 50 mm Tris, pH 7.5, 75 mm NaCl, 20% glycerol, 2 mm DTT, and 20 mm l-glutathione. GST-Importins were cleaved with TEV protease, and the Importin proteins were further purified using ion exchange and gel filtration chromatography in TB buffer (20 mm HEPES, pH 7.3, 150 mm sodium chloride, 2 mm DTT, 2 mm magnesium acetate, 10% glycerol, and 1 mm EGTA).
For pulldown binding assays, bacteria expressing the GST-histone tail proteins were lysed by sonication and centrifuged. The supernatants were incubated with GSH-Sepharose followed by extensive washes with TB buffer containing 10% glycerol. Immobilized GST-histone tail proteins were stored in TB buffer containing 40% glycerol at −20 °C.
To purify MBP-histone tail proteins, bacterial lysates were incubated with amylose beads (New England BioLabs), and the fusion proteins were eluted with buffer containing 20 mm Tris, pH 7.5, 50 mm NaCl, 2 mm EDTA, 2 mm DTT, 10% glycerol, and 10 mm maltose. Eluted proteins were further purified by ion exchange chromatography.
Recombinant Xenopus laevis H3 and H4 were be expressed in E. coli BL21(DE3) pLysS as His6-histone fusion proteins using the pET3a vector. H3 and H4 proteins were purified from inclusion bodies and refolded to obtain H3/H4 tetramers as described previously (60). The N-terminal core domain of human Asf1b (residues 1–169) was expressed in E. coli BL21(DE3) as GST-Asf1b(1–169) from a pGEX-TEV vector. GST-Asf1b(1–169) was purified using affinity and gel filtration chromatography. GST-Asf1b(1–169)-H3/H4 complexes were assembled by mixing purified GST-Asf1b and H3/H4 in a 1:5 molar ratio followed gel filtration chromatography as described previously (9). Concentrations of purified proteins were determined by measuring absorbance at 280 nm using a Thermo Fisher Genesys 10S UV-visible spectrometer and by Bradford assays.
Pulldown Binding Assays
Purity of Importins for pulldown assays was verified by SDS-PAGE and Coomassie Blue staining (Fig. 1E). Normalization of Coomassie Blue staining and confirmation of linearity of staining were performed for GST-H3 tail (supplemental Fig. 2). Qualitative comparisons of Importin-histone tail binding in Fig. 1B were performed by incubating immobilized GST-H3 tail or GST-H4 tail on glutathione-Sepharose beads (20 nmol of GST-histone tail in each binding assay) with a 500 nm concentration of each purified Importin in TB buffer in a total volume of 800 μl for 30 min at 4 °C followed by extensive washing with the same buffer. Bound proteins were visualized using SDS-PAGE/Coomassie Blue. Gels were subject to densitometry analysis using ImageJ. The density of the Importin band was divided by the density of the GST-histone tail band in the same gel lane. Importin inputs were visualized by SDS-PAGE/Coomassie Blue to ensure that similar concentrations of Importins were used for all binding assays, and excess unbound Importins in the flow-through were also monitored.
All other pulldown binding assays were performed by incubating of 0.8 nmol immobilized GST-H3 tail or GST-H4 tail proteins (∼2 μm) with an ∼4 μm concentration of each purified Importin in TB buffer in a total volume of 100 μl for 30 min at 4 °C followed by extensive washing with the same buffer. For RanGTP dissociations assays, ∼10 μm purified RanGTP was added to immobilized GST-H3 tail proteins that are bound to Importins followed by extensive washing. Activities of Impβ, Kapβ2, Imp4, Imp5, Imp7, and Imp9 were verified by their binding to RanGTP, and the activity of Impα was verified by binding to the classical NLS of the SV40 T antigen. Bound proteins were visualized using SDS-PAGE/Coomassie Blue. Gels were subjected to densitometry analysis using ImageJ. Density of the Importin band was divided by the density of the GST-histone tail band in the same gel lane. The density ratios were then normalized to the ratio of the Importin band over the wild-type GST-H3 tail band or wild-type GST-H4 tail band. Relative band densities of experiments performed in triplicate are plotted with standard errors in histograms generated with GraphPad Prism. One-way ANOVA and t tests were performed on all pulldown binding assay data.
KD values were also estimated from pulldown assays. 20 nm to 1 μm concentrations of each Importin in total volumes of 0.4–15 ml (to ensure a molar excess of Importins to the H3 tail) were titrated onto 0.2 nmol of GST-H3 tail immobilized on glutathione-Sepharose beads in a series of pulldown binding assays. Relative densities of the gel bands from three separate experiments were measured using ImageJ. The data were fitted to a simple bimolecular equilibrium relationship in GraphPad Prism to obtain the KD values.
Measuring Dissociation Constants with Isothermal Titration Calorimetry
Binding affinities of MBP-H3 tail proteins to Kapβ2 and Imp5 were measured using ITC. ITC experiments were performed with a Malvern ITC200 calorimeter (Malvern Instruments, Worcestershire, UK). Proteins were dialyzed against buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, and 2 mm β-mercaptoethanol. 200–400 μm MBP-H3 tail proteins were titrated into a sample cell containing 20–40 μm recombinant Kapβ2 or Imp5. ITC experiments were performed at 20 °C with 19 rounds of 4-μl injections. Data were plotted and analyzed using NITPIC and Sedphat, and the data were visualized using GUSSI. For error reporting, we used F-statistics and error-surface projection method to calculate the 68.3% confidence intervals of the fitted data (61). Histograms to compare KD values were generated by GraphPad Prism.
Nuclear-Cytoplasmic Localization of H3 Tail Proteins in Cells
Cellular localization of EYFP2-H3 tail fusion proteins overexpressed in HT1080 cells were observed as described previously (62). EYFP2-H3 tail expression constructs were cloned into the pEYFP2 vector. Live cell images were collected using a spinning disk confocal microscope system (Nikon-Andor, Nikon, NY) and MetaMorph software. Image analysis was performed similarly with ImageJ. Experiments were performed in duplicates or triplicates with a total of >150 transfected cells.
Author Contributions
M. S. and T. C. conducted the experiments. M. S. and Y. M. C. designed the experiments and wrote the paper.
Supplementary Material
Acknowledgments
We thank Bing Li for histone constructs, Chad Brautigam and Thomas Scheuermann from the Macromolecular Biophysics Resource at University of Texas Southwestern for assistance in ITC experiments, Diana Tomchick and James Chen from Structural Biology Laboratory at University of Texas Southwestern for assistance in crystallographic data collection, Ho Yee Joyce Fung and Szu-Chin Fu for comments, and Xian-Jin Xie for help with statistical analysis. The use of SBC 19ID beamline at Advanced Photon Source is supported by United States Department of Energy Contract DE-AC02-06CH11357.
This work is supported by National Institutes of Health Grants R01 GM069909 (to Y. M. C.) and U01 GM98256-01 (to Y. M. C.), Welch Foundation Grant I-1532 (to Y. M. C.), a Leukemia and Lymphoma Society scholar award (to Y. M. C.), and the University of Texas Southwestern Endowed Scholars Program (to Y. M. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. 1–9.
- Asf1
- antisilencing function protein 1
- NLS
- nuclear localization signal
- c-NLS
- classical NLS
- ITC
- isothermal titration calorimetry
- MBP
- maltose-binding protein
- ANOVA
- analysis of variance
- EYFP
- enhanced YFP
- rp
- ribosomal protein
- BIB
- β-like import receptor binding
- TEV
- tobacco etch virus
- TrnSR
- transportin-SR.
References
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. White C. L., Suto R. K., and Luger K. (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eickbush T. H., and Moudrianakis E. N. (1978) The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry 17, 4955–4964 [DOI] [PubMed] [Google Scholar]
- 4. Annunziato A. T. (2013) Assembling chromatin: the long and winding road. Biochim. Biophys. Acta 1819, 196–210 [DOI] [PubMed] [Google Scholar]
- 5. Burgess R. J., and Zhang Z. (2013) Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keck K. M., and Pemberton L. F. (2012) Histone chaperones link histone nuclear import and chromatin assembly. Biochim. Biophys. Acta 1819, 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakagawa T., Bulger M., Muramatsu M., and Ito T. (2001) Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J. Biol. Chem. 276, 27384–27391 [DOI] [PubMed] [Google Scholar]
- 8. Smith S., and Stillman B. (1991) Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., and Senda T. (2007) Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 10. English C. M., Adkins M. W., Carson J. J., Churchill M. E., and Tyler J. K. (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mousson F., Lautrette A., Thuret J. Y., Agez M., Courbeyrette R., Amigues B., Becker E., Neumann J. M., Guerois R., Mann C., and Ochsenbein F. (2005) Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. U.S.A. 102, 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blackwell J. S. Jr, Wilkinson S. T., Mosammaparast N., and Pemberton L. F. (2007) Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J. Biol. Chem. 282, 20142–20150 [DOI] [PubMed] [Google Scholar]
- 13. Baake M., Bäuerle M., Doenecke D., and Albig W. (2001) Core histones and linker histones are imported into the nucleus by different pathways. Eur. J. Cell Biol. 80, 669–677 [DOI] [PubMed] [Google Scholar]
- 14. Mühlhäusser P., Müller E. C., Otto A., and Kutay U. (2001) Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2, 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greiner M., Caesar S., and Schlenstedt G. (2004) The histones H2A/H2B and H3/H4 are imported into the yeast nucleus by different mechanisms. Eur. J. Cell Biol. 83, 511–520 [DOI] [PubMed] [Google Scholar]
- 16. Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., and Pemberton L. F. (2002) Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 277, 862–868 [DOI] [PubMed] [Google Scholar]
- 17. Alvarez F., Muñoz F., Schilcher P., Imhof A., Almouzni G., and Loyola A. (2011) Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286, 17714–17721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baake M., Doenecke D., and Albig W. (2001) Characterisation of nuclear localisation signals of the four human core histones. J. Cell. Biochem. 81, 333–346 [PubMed] [Google Scholar]
- 19. Soniat M., and Chook Y. M. (2016) Karyopherin-β2 recognition of a PY-NLS variant that lacks the proline-tyrosine motif. Structure, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cook A., Bono F., Jinek M., and Conti E. (2007) Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 21. Görlich D., and Kutay U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 [DOI] [PubMed] [Google Scholar]
- 22. Chook Y. M., and Blobel G. (2001) Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 [DOI] [PubMed] [Google Scholar]
- 23. Weis K. (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441–451 [DOI] [PubMed] [Google Scholar]
- 24. Xu D., Farmer A., and Chook Y. M. (2010) Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr. Opin. Struct. Biol. 20, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mosammaparast N., and Pemberton L. F. (2004) Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 26. Soniat M., and Chook Y. M. (2015) Nuclear localization signals for four distinct karyopherin-β nuclear import systems. Biochem. J. 468, 353–362 [DOI] [PubMed] [Google Scholar]
- 27. Chook Y. M., and Süel K. E. (2011) Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campos E. I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W. H., Seepany H., Gao Z., Day L. A., Greenblatt J. F., and Reinberg D. (2010) The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson-Saliba M., Siddon N. A., Clarkson M. J., Tremethick D. J., and Jans D. A. (2000) Distinct importin recognition properties of histones and chromatin assembly factors. FEBS Lett. 467, 169–174 [DOI] [PubMed] [Google Scholar]
- 30. Jasencakova Z., Scharf A. N., Ask K., Corpet A., Imhof A., Almouzni G., and Groth A. (2010) Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 [DOI] [PubMed] [Google Scholar]
- 31. Ejlassi-Lassallette A., Mocquard E., Arnaud M. C., and Thiriet C. (2011) H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol. Biol. Cell 22, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dingwall C., Sharnick S. V., and Laskey R. A. (1982) A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 30, 449–458 [DOI] [PubMed] [Google Scholar]
- 33. Kalderon D., Richardson W. D., Markham A. F., and Smith A. E. (1984) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 [DOI] [PubMed] [Google Scholar]
- 34. Lanford R. E., and Butel J. S. (1984) Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37, 801–813 [DOI] [PubMed] [Google Scholar]
- 35. Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., and Chook Y. M. (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kobayashi J., and Matsuura Y. (2013) Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J. Mol. Biol. 425, 1852–1868 [DOI] [PubMed] [Google Scholar]
- 37. Kosugi S., Hasebe M., Tomita M., and Yanagawa H. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z. C., and Chook Y. M. (2012) Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc. Natl. Acad. Sci. U.S.A. 109, 12017–12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Süel K. E., and Chook Y. M. (2009) Kap104p imports the PY-NLS-containing transcription factor Tfg2p into the nucleus. J. Biol. Chem. 284, 15416–15424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Süel K. E., Gu H., and Chook Y. M. (2008) Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 6, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cansizoglu A. E., Lee B. J., Zhang Z. C., Fontoura B. M., and Chook Y. M. (2007) Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 14, 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Z. C., Satterly N., Fontoura B. M., and Chook Y. M. (2011) Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1. Mol. Biol. Cell 22, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jäkel S., and Görlich D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cingolani G., Bednenko J., Gillespie M. T., and Gerace L. (2002) Molecular basis for the recognition of a nonclassical nuclear localization signal by importin β. Mol. Cell 10, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 45. Lott K., and Cingolani G. (2011) The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1578–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cingolani G., Petosa C., Weis K., and Müller C. W. (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 [DOI] [PubMed] [Google Scholar]
- 47. Lam M. H., Briggs L. J., Hu W., Martin T. J., Gillespie M. T., and Jans D. A. (1999) Importin β recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin α. J. Biol. Chem. 274, 7391–7398 [DOI] [PubMed] [Google Scholar]
- 48. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., and Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 49. Lange A., McLane L. M., Mills R. E., Devine S. E., and Corbett A. H. (2010) Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11, 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giesecke A., and Stewart M. (2010) Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J. Biol. Chem. 285, 17628–17635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McLane L. M., Pulliam K. F., Devine S. E., and Corbett A. H. (2008) The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res. 36, 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y., Zhang L., Liu T., Chai C., Fang Q., Wu H., Agudelo Garcia P. A., Han Z., Zong S., Yu Y., Zhang X., Parthun M. R., Chai J., Xu R. M., and Yang M. (2014) Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 28, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., and Laue E. D. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Verreault A., Kaufman P. D., Kobayashi R., and Stillman B. (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8, 96–108 [DOI] [PubMed] [Google Scholar]
- 55. Alekseev O. M., Widgren E. E., Richardson R. T., and O'Rand M. G. (2005) Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J. Biol. Chem. 280, 2904–2911 [DOI] [PubMed] [Google Scholar]
- 56. Hartl F. U., and Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 57. Timney B. L., Tetenbaum-Novatt J., Agate D. S., Williams R., Zhang W., Chait B. T., and Rout M. P. (2006) Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175, 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chook Y. M., and Blobel G. (1999) Structure of the nuclear transport complex karyopherin-β2-Ran x GppNHp. Nature 399, 230–237 [DOI] [PubMed] [Google Scholar]
- 59. Chook Y. M., Jung A., Rosen M. K., and Blobel G. (2002) Uncoupling Kapβ2 substrate dissociation and Ran binding. Biochemistry 41, 6955–6966 [DOI] [PubMed] [Google Scholar]
- 60. Luger K., Rechsteiner T. J., and Richmond T. J. (1999) Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 [DOI] [PubMed] [Google Scholar]
- 61. Bevington P. R., and Robinson D. K. (1992) Data Reduction and Error Analysis for the Physical Sciences, McGraw-Hill, New York [Google Scholar]
- 62. Xu D., Marquis K., Pei J., Fu S. C., Cağatay T., Grishin N. V., and Chook Y. M. (2015) LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 31, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.